Abstract
The standardization and broad-scale integration of dynamic susceptibility contrast (DSC)-magnetic resonance imaging (MRI) have been confounded by a lack of consensus on DSC-MRI methodology for preventing potential relative cerebral blood volume inaccuracies, including the choice of acquisition protocols and postprocessing algorithms. Therefore, we developed a digital reference object (DRO), using physiological and kinetic parameters derived from in vivo data, unique voxel-wise 3-dimensional tissue structures, and a validated MRI signal computational approach, aimed at validating image acquisition and analysis methods for accurately measuring relative cerebral blood volume in glioblastomas. To achieve DSC-MRI signals representative of the temporal characteristics, magnitude, and distribution of contrast agent-induced T1 and T2* changes observed across multiple glioblastomas, the DRO's input parameters were trained using DSC-MRI data from 23 glioblastomas (>40 000 voxels). The DRO's ability to produce reliable signals for combinations of pulse sequence parameters and contrast agent dosing schemes unlike those in the training data set was validated by comparison with in vivo dual-echo DSC-MRI data acquired in a separate cohort of patients with glioblastomas. Representative applications of the DRO are presented, including the selection of DSC-MRI acquisition and postprocessing methods that optimize CBV accuracy, determination of the impact of DSC-MRI methodology choices on sample size requirements, and the assessment of treatment response in clinical glioblastoma trials.
Keywords: dynamic susceptibility contrast MRI, digital reference object, brain tumor perfusion
Introduction
Dynamic susceptibility contrast (DSC)-magnetic resonance imaging (MRI) noninvasively measures brain tumor cerebral blood flow (CBF) and cerebral blood volume (CBV), and it has found increasing clinical applications for patient management (1–18). To facilitate multi-institutional comparability and consistency, national initiatives, including National Cancer Institute's Quantitative Imaging Network, Radiological Society of North America's Quantitative Imaging Biomarkers Alliance, and the National Brain Tumor Society's Jumpstarting Brain Tumor Drug Development Coalition, are underway to standardize acquisition and analysis protocols for DSC-MRI (19, 20). A challenge to such efforts is the relative paucity of studies systematically evaluating the influence of DSC-MRI methodology on CBV accuracy. In practice, such validation studies are difficult to perform in patients because of the need for multiple contrast agent (CA) injections and lack of a noninvasive gold standard CBV measure for reference. As an alternative to in vivo validation, in silico digital reference objects (DROs) provide a means for computing synthetic MRI signals and derived kinetic parameters for a range of clinically relevant input conditions. Such a DRO was recently developed for dynamic contrast-enhanced MRI to investigate the biases and variances of algorithms used for image analysis (21).
The goal of this report is to describe the development of a DSC-MRI DRO that recapitulates the heterogeneous signal characteristics measured in glioblastomas. In general, there are two underlying strategies that can be pursued for DROs emulating MRI data. When the primary objective is to establish multisite analysis consistency, synthetic signals can be computed using simple heuristic models approximating the underlying biophysics of signal formation, as the endpoint is to assess the agreement between software estimates of a parameter such as CBV that is explicitly defined by the “ground truth” time course. However, if the intention is to optimize acquisition protocols and CA dosing schemes, such as those used in DSC-MRI, or if the accuracy of the analysis is dependent upon certain physical or physiological assumptions, the synthetic signals should accurately reflect the biophysics of the MRI signal. For the DSC-MRI DRO, we pursued the latter strategy because it enables a more accurate and comprehensive investigation into the DSC-MRI methodology.
In brain tumor DSC-MRI, the acquired signals reflect a complex combination of T1, T2, and T2* changes that depend upon numerous features including CA kinetic parameters (CBF, permeability, intra- and extravascular volume fractions), precontrast T1 and T2*, vascular architecture, cellular microstructure (size, shape, spatial distribution), transvascular and transcellular water exchange, and CA T1 and T2* relaxivity. The sensitivity of the DSC-MRI signal to relaxation time variations is influenced by the acquisition parameters (repetition time [TR], echo time [TE], flip angle [FA], pulse sequence type) and CA dosing scheme (preload and bolus dose and timing). Accordingly, for the DRO to yield realistic signals, its design must reasonably approximate the magnitude and heterogeneity of these physical and physiological parameters in vivo. To that end, we developed a DSC-MRI DRO that is driven by a validated computational strategy to compute MRI signals for realistic 3-dimensional (3D) tissue structures (22); partially constrained by parameter inputs defined from in vivo data; and, for unknown parameters, trained using a public database of DSC-MRI data in glioblastomas.
Methodology
The computational approach used herein, termed the finite perturber finite difference method (FPFDM) (22, 23), models the effects of water protons diffusing in heterogeneous magnetic field medium based on a 3D tissue structure. The FPFDM computes magnetic field perturbations induced by susceptibility variations between the simulated tissue compartments, and it determines the resulting gradient echo transverse relaxation rates. In addition to a 3D matrix that defines the tissue structure (eg, blood vessels and cells), requisite FPFDM inputs include the static magnetic field strength, the CA concentration in each compartment for determining intercompartment susceptibility differences, the water proton diffusion coefficient, and the DSC-MRI pulse sequence parameters. To ensure clinical relevancy, the DRO derived from these input parameters should replicate the magnitude and heterogeneity of CA-induced T1 and T2* changes during bolus passage through vessels and into the extravascular extracellular space (EES).
CA Kinetics
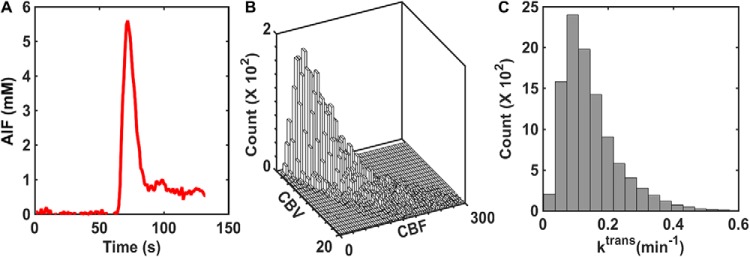
The 2-compartment pharmacokinetic model described by Brix et al. (24) was used to simulate concentration–time profiles in plasma (Cp) and the EES (Ce). Inputs to the Brix model include vascular volume fraction, blood flow, CA transfer coefficient (Ktrans), and volume fraction of the EES (ve). Rather than use previously reported mean CBF and CBV values in glioblastoma, our simulated kinetic curves better represented clinical data if the DRO voxels matched the paired, voxel-wise distribution of these parameters across patients (as compared with randomly distributed unpaired parameters). Accordingly, we extracted DSC-MRI data from 23 patients with glioblastoma (>40 000 voxels) in The Cancer Imaging Archive (TCIA) database for characterizing the distribution of paired CBF and CBV values. For this patient cohort, DSC-MRI was acquired at 3T, consisting of General Electric (General Electric Healthcare, Waukesha, WI, USA) (n = 14) and Siemens (Siemens Medical Systems, Erlangen, Germany) (n = 9) scanners using single-echo gradient echo-planar imaging (TR = 1–1.25 seconds, TE = 30 milliseconds, FA = 70–80°, field of view = 240 × 240 mm2, section thickness = 4–5 mm, matrix = 962 or 1282) before, during, and after administration of 0.1 mmol/kg gadopentetate dimeglumine (Gd-DTPA) infusion at 4 ml/s followed by a saline flush. Five minutes before bolus injection, a 0.05 mmol/kg Gd-DTPA preload was administered to minimize T1 leakage effects. Residual leakage effects were corrected using the Boxerman–Schmainda–Weisskoff approach (25). Voxel-wise relative CBV and CBF maps were calculated from the leakage-corrected DSC-MRI data and an automated measure of the arterial input function (AIF), using circular singular value decomposition-based deconvolution (26–29). The voxel-wise distributions of Ktrans and ve were characterized using a retrospective analysis of dynamic contrast-enhanced MRI signals extracted from a dual-echo DSC-MRI data set in 11 glioblastomas (30). Because DSC-MRI data yield relative tumor CBV and CBF measures, their values were scaled using data obtained from dynamic computed tomography perfusion imaging (31). In addition, the AIF used as input for the DRO's kinetic modeling was computed as the average AIF among all patients in the TCIA data. Figure 1, A, B, and C shows the average AIF values, CBV and CBF paired distribution, and Ktrans distribution, respectively.
Figure 1.
Summary of input parameters for the pharmacokinetic model used in the development of the digital reference object (DRO). The population-based arterial input function (AIF) was computed from 23 glioblastomas (A). The paired distribution of cerebral blood volume (CBV) and cerebral blood flow (CBF) was derived from the same database and scaled by dynamic computed tomography (CT) perfusion data (B). The distribution of Ktrans was derived from previously published dynamic CT perfusion data (C).
To define a computationally manageable number of tissue models in the DRO that still accurately reflected the in vivo voxel-wise heterogeneity, the 2-dimensional paired distribution of CBF and CBV was first binned into intervals of 5 ml/100 g/min and 1 ml/100 g, respectively. The resulting distribution was then scaled to yield 100 combinations of CBF and CBV pairs, which were then used to define the number and vascular properties of the tissue structures.
Tissue Structures
Although the component of DSC-MRI signal associated with CA-induced T1 changes is easily calculated by assuming fast water exchange (32–34), the CA-induced T2* changes depend on vascular and cellular microstructural geometry, precluding use of a simple analytical model. To reflect this complexity, we modeled tissue structures using ellipsoids (cells) (22, 23, 35) packed around randomly oriented cylinders (vessels) (36–45). Previously, we showed that modeling cells as ellipsoids rather than spheres provides a more accurate estimate of the magnitude of T2* changes observed in clinical DSC-MRI studies (22, 23, 35), whereas modeling the vasculature structure as randomly oriented cylinders has been shown to accurately estimate the T2* effects that occur when CA is distributed within blood vessels (36–45). The cylindrical vascular volume fraction was fixed using the in vivo extracted CBV values, and vessel sizes varied from 5 to 30 μm (46). Tumor cell volume fractions were allowed to vary within a physiologically relevant range (45%–65%) (47), and the mean cellular axis radii for a given voxel varied between 4 and 15 μm (46). Figure 2 shows a representative 3D volume rendering of 2 tissue structures, one with homogeneous ellipsoids with a constant aspect ratio (Figure 2A) and one showing ellipsoids with heterogeneous shapes (Figure 2B).
Figure 2.
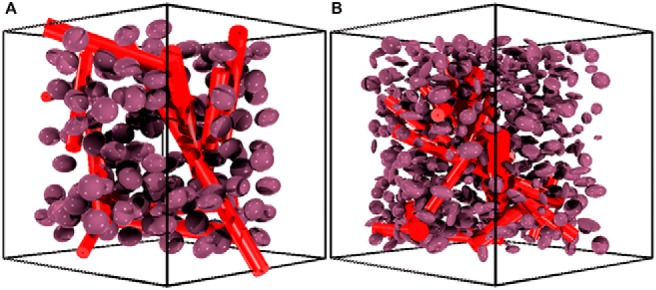
Sample 3-dimensional (3D) tissue structures used in the development of the DRO. Tissue structure composed of homogenously shaped ellipsoids packed around randomly oriented cylindrical vessels (A). Tissue structure containing heterogeneously shaped ellipsoids (B). To aid in structure visualization, these images depict a smaller number of ellipsoids compared with what was actually used for the simulations.
Computation of DSC-MRI Signal
The susceptibility differences between the vascular and extravascular compartments were computed using Δχ = χm·[CA], where [CA] is the compartmental CA concentration (Cp and Ce) and χm is the CA molar susceptibility (0.027 × 106 mM−1) (48). In addition to all the aforementioned input parameters, the FPFDM calculates the DSC-MRI signal as described previously (22) using a water proton diffusion rate (D) of 1.3 × 10−3 mm2/s (49), relevant pulse parameters (TE, B0, FA, TR), and precontrast T10 values ranging from 1 to 2.2 seconds. Figure 3 shows representative simulated Cp and Ce time curves (Figure 3A), and the corresponding gradient echo DSC-MRI signal ratio (S/S0) time curves (Figure 3B) for the 2 tissue voxels are shown in Figure 2.
Figure 3.
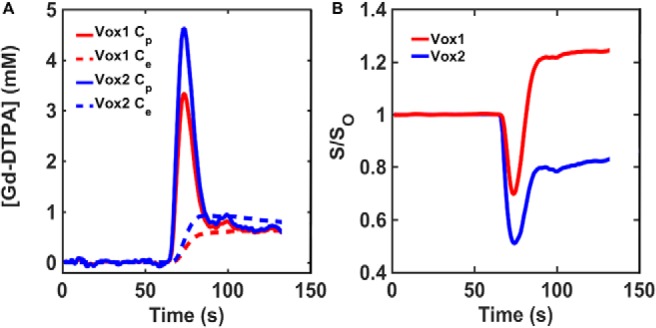
Representative concentration–time and signal–time curves found in the DRO. Simulated Cp and Ce curves derived using the 2-compartment model for the tissue structures with homogeneous (red) and heterogeneous (blue) ellipsoids illustrated in Figure 2 (A). Corresponding gradient echo signal ratio (S/S0) computed using the finite perturber finite difference method (FPFDM) (B). The tissues consisting of homogeneous and heterogeneous ellipsoids yielded signals exhibiting predominantly T1 and T2* leakage effects, respectively.
DRO Training
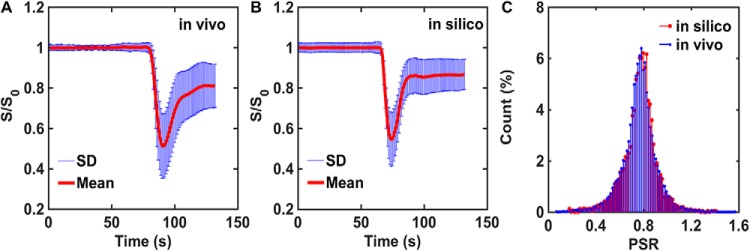
Given the large number of input parameters and a wide range of potential permutations, it is critical to ensure that the DRO's simulated DSC-MRI signals accurately represent the temporal characteristics, magnitude, and distribution of CA-induced T1 and T2* changes observed across typical glioblastomas. To achieve this, we used the voxel-wise TCIA data described above (>40 000 voxels) for identifying the appropriate combination of input parameters. In particular, all computed signals, for an equivalent preload dosing scheme and pulse sequence parameters to those in the TCIA data set, underwent a selection criteria process based on their percent signal recovery (PSR) and the mean and standard deviation of the signals across the DRO. The PSR is a useful metric for comparison because it reflects the magnitude of the signal drop during bolus passage and the postbolus signal recovery. The DRO's input tissue structure (eg, cell size, shape), kinetic parameters (eg, CBF, Ktrans), and physical parameters (precontrast T1) were systematically permutated until the distribution of PSR values and the mean and standard deviation of signals across the DRO agreed with those found in the voxel-wise TCIA data. The PSR agreement was evaluated using a 2-sample Kolmogorov–Smirnov test. In addition, a 95% agreement between the FWHM and the maximum signal drop was used to determine the agreement between the mean signals. To achieve this level of agreement, the iterative process required a DRO consisting of ∼10 000 unique voxels. The data training based on this selection criterion ensured the removal of computed signals from the DRO, because of an unrealistic combination of tissue parameters. Figure 4A–B shows the agreement between the in vivo and in silico mean and standard deviation of the signal. The distribution of PSR values obtained from the training data set and the DRO is shown in Figure 4C. The 2-sample Kolmogorov–Smirnov test yielded a P-value of .69, indicating agreement between the 2 distributions. Table 1 summarizes the final tissue parameter values that were identified through the DRO training.
Figure 4.
A database of DSC-MRI performed in 23 glioblastomas was used to train input parameter permutations for the DRO. After training, the mean and standard deviation values of the in vivo (>40 000 voxels) and in silico (10 000 voxels) DSC-MRI signals are in strong agreement (A–B). The training phase ensured concordance of the percent signal recovery (PSR) distributions for in vivo and in silico data, supported by 2-sample Kolmogorov–Smirnov test (C).
Table 1.
Input Parameters for Tumor and Normal Tissue
| CBV (%) | CBF (ml/100 g/min) | Ktrans (min−1) | T10 (s) | ve (%) | Cell Radii (μm) | Vessel Radii (μm) | |
|---|---|---|---|---|---|---|---|
| WM | 3.5 ± 0.6 | 38.4 ± 8.3 | 0.0 ± 0.0 | 1.8 ± 0.2 | 25.4 ± 0.3 | 8.4 ± 2.7 | 9.9 ± 1.0 |
| (2.5–4.6) | (26.6–55.2) | (0.0–0.0) | (1.3–2.5) | (24.8–26.0) | (4.4–12.6) | (6.0–13.6) | |
| Tumor | 6.1 ± 2.8 | 150.3 ± 55.6 | 0.19 ± 0.08 | 1.8 ± 0.2 | 24.3 ± 1.5 | 8.4 ± 2.7 | 9.9 ± 2.5 |
| (1.5–13.8) | (43.9–268.2) | (0.03–0.47) | (1.0–2.6) | (22.3–26.5) | (4.4–12.6) | (1.2–19.7) |
Values are expressed as mean ± SD and (minimum–maximum).
DRO Validation
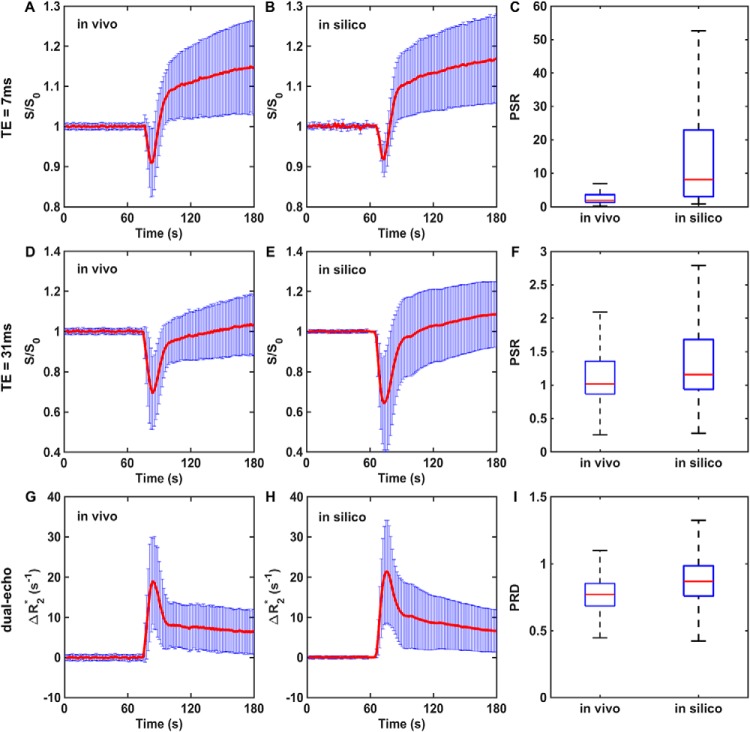
To validate the DRO's ability to produce reliable signals for pulse sequences and that the CA dosing schemes are different from those in the training data set, we compared simulated dual-echo signals with those found in an in vivo dual-echo DSC-MRI “validation” data set. The validation data set was acquired in patients with glioblastoma (n = 3) at 3T using a dual gradient echo-planar imaging protocol with the following parameters: TR = 1.5 seconds, TE1/TE2 = 7.0/31.0 milliseconds, field of view = 240 × 240 mm2, section thickness = 5 mm, matrix = 962. Measurements were taken before, during, and after administration of Gd-DTPA (0.1 mmol/kg Gd-DTPA, 4 ml/s infusion rate followed by 20 ml of saline flush). In the simulation, the structural and kinetic inputs derived during the training phase remained the same, but the acquisition parameters and dosing scheme were chosen to match those used in the patient data. The goal of this validation study was to determine whether the DRO fully captures the heterogeneity (eg, magnitude and temporal characteristics such as PSR) of the DSC-MRI signals acquired in this separate (and smaller) cohort of patients. To identify this subset of voxels within the DRO, a correlation analysis was performed between the signals in the in vivo and DRO data. The range of PSR values found in the in vivo and DRO data was compared to ensure that the DRO captured the signal heterogeneity measured in the validation set for both TEs. A parameter termed percent relaxation drop (PRD) was also formulated in a similar fashion as PSR using the derived dual-echo ΔR2* time courses and compared between the in vivo and DRO data.
All simulations were performed using Matlab (MathWorks, Natick, MA) running on a high-performance 32-core system with 2.3 GHz processors and 128 GB of RAM.
Results
Validation
Figure 5 compares simulated and in vivo dual-echo DSC-MRI data. The DRO could accurately recapitulate the TE = 7 milliseconds and TE = 31 milliseconds signals and the derived dual-echo ΔR2* time courses, which remove T1 leakage effects but retain T2* leakage effects. The PSR and PRD heterogeneity of the in vivo data was also fully reflected in the DRO. This indicates that the trained DRO can accurately model the underlying CA-induced T1 and T2* effects and the associated DSC-MRI signals for different sets of pulse sequence parameters and CA dosing schemes.
Figure 5.
Validation of the DRO. A correlation analysis identified DRO voxels accurately reflecting the mean and standard deviation of voxel-wise dual-echo dynamic susceptibility contrast-magnetic resonance imaging (DSC-MRI) signals, and the computed dual-echo ΔR2*, in the validation data set. Agreement is seen between the in vivo and simulated signals and PSR values at echo time (TE) = 7 milliseconds (A–C) and TE = 31 milliseconds (D–F), and between the in vivo and simulated dual-echo ΔR2* and the computed PRD values (G–I). The PSR and PRD distributions across all voxels in the in vivo validation data set are a subset of those found in the DRO. These results reflect the broader PSR and PRD heterogeneity exhibited across the much larger number of tumors used to train the DRO compared with that used in the validation data set.
Application 1: Influence of Acquisition and Postprocessing Methods on CBV Accuracy
It is well established that T1 and T2* CA leakage effects confound the reliable measurement of CBV (25, 50). DSC-MRI acquisition strategies have been proposed to reduce T1 leakage effects, including the use of preload CA administration, low FAs, long TEs and TRs, and dual-echo pulse sequences. In addition, postprocessing methods have been developed that eliminate residual T1 and/or T2* leakage effects (25, 51–59). However, validation of these acquisition and postprocessing strategies in vivo has been limited because of the lack of a reliable gold standard reference. A potential application of the population-based DRO is the systematic investigation of the acquisition and postprocessing methods that influence the reliability of CBV measurements.
To this end, we computed the percentage difference between tumor CBV simulated with and without (Ktrans = 0) CA leakage effects for a single-dose bolus injection protocol (no preload), FA = 30° and 90°, TE = 30 milliseconds, and TR = 1 and 2 seconds. We also compared CBV accuracy with and without the application of postprocessing leakage correction using the Boxerman–Schmainda–Weisskoff approach. Results of this analysis are shown in Figure 6. For TR = 1 second, FA = 30° yielded more accurate CBV values than FA = 90°, with and without postprocessing leakage correction (Figure 6A). As expected, the uncorrected 90° FA data yielded substantially underestimated CBV across the DRO voxels, reflecting the strong sensitivity to T1 leakage effects. For TR = 2 seconds, a greater fraction of voxels overestimate CBV, indicating a shift toward T2*-dominated leakage effects due to reduced T1 sensitivity (Figure 6B). Leakage correction improved CBV accuracy across all acquisition parameters. A similar approach could be used to systematically investigate the influence of a range of acquisition and postprocessing methods on CBV accuracy.
Figure 6.
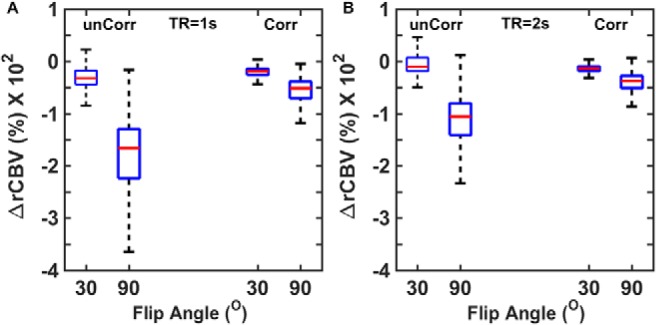
Use of the DRO to investigate the influence of acquisition and postprocessing strategy on CBV accuracy. CA leakage-corrected (Corr) and -uncorrected (unCorr) CBV percentage error for DSC-MRI simulated at 2 flip angles (FA) and repetition time (TR) values (A–B). For all parameter combinations, CA leakage correction increases CBV accuracy. Leakage-corrected CBV estimates acquired with lower FAs and longer TRs provided more accurate CBV estimates.
Application 2: In Silico Optimization of DSC-MRI for Use in Clinical Trials
The population-based DRO can also be used to optimize DSC-MRI for assessment of treatment response in clinical trials. For example, the influence of acquisition and postprocessing methods on the sensitivity of DSC-MRI to a given CBV change can be used to determine protocols that minimize the sample size needed to power a clinical trial. In this context, the DRO serves as an atlas of possible tumor DSC-MRI signals. By using the correlation analysis discussed in the validation section, a virtual patient DSC-MRI data set can be generated by replacing voxel-wise in vivo tumor signals with an atlas-matched version. This analysis can be propagated across an existing clinical trial database to compute in silico pre- and post-treatment DSC-MRI data. Because the simulated signals for a given voxel originate from a unique set of input conditions, the DSC-MRI signals can be recomputed for any combination of acquisition parameters, such as a new FA or CA dosing scheme. This permits systematic investigation of how acquisition and postprocessing methods influence the inter- and intrasubject CBV heterogeneity, pre- and post-therapy. Alternatively, an assumed effect size distribution (eg, 20% ± 5% decrease in a tumor's CBV) could be applied to the untreated cohort of virtual patients and can be used to identify, within the DRO, the “treated” DSC-MRI signals for each voxel.
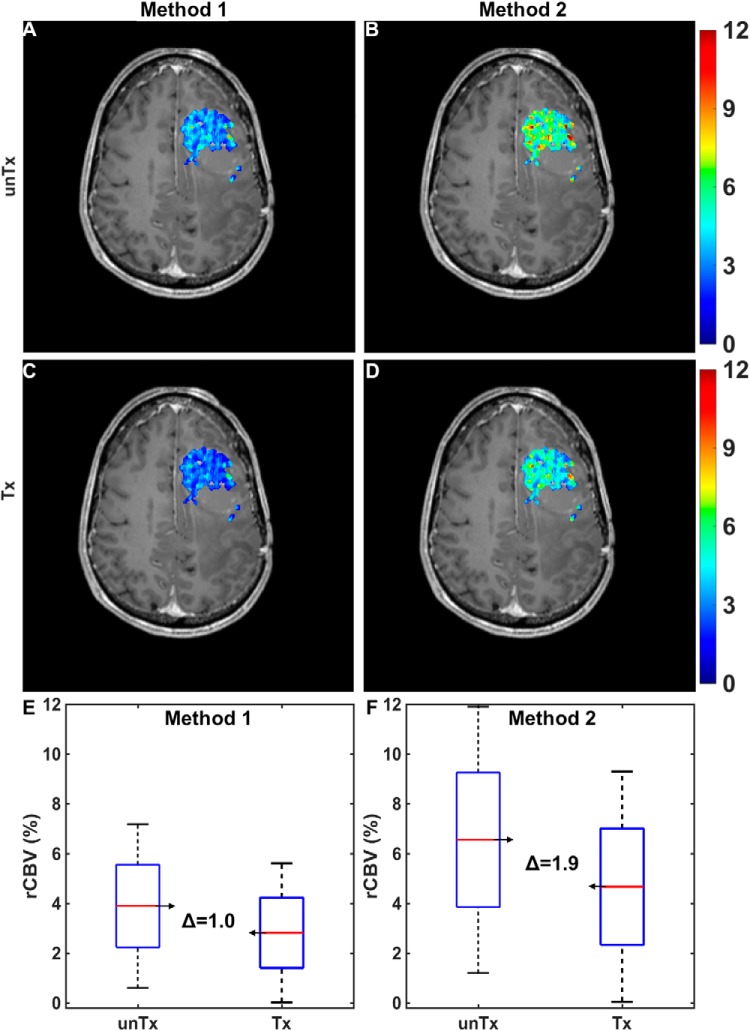
Figure 7A–B illustrates a simulated pretreatment CBV map for a virtual patient computed using 2 different CA dosing schemes: a single-bolus dose with no preload (method 1) and a single-dose preload preceding a single-bolus dose (method 2). The corresponding treated CBV maps (modeled as a 20% mean reduction in tumor CBV) for both methods are shown in Figure 7C–D. The pre- and post-treatment CBV distributions across the entire tumor region of interest for both acquisition methods are shown in Figure 7E–F. In this example, CBV estimates derived from method 2 were more sensitive to treatment response compared with those derived from method 1, as indicated by the change in CBV. Similar analyses could be extended to cohorts of virtual patients to identify the most robust and sensitive DSC-MRI acquisition and postprocessing strategies for use in clinical trials.
Figure 7.
Application of the DRO to optimize DSC-MRI for assessment of treatment response in clinical trials. Simulated pre- (A, B) and post-treatment (C, D) CBV maps in a virtual patient computed using 2 contrast agent (CA) dosing schemes: a single-bolus dose with no preload (method 1; A, C) and a single-dose preload preceding a single-bolus dose (method 2; B, D). Pre- and post-treatment CBV changes within tumor for the 2 methods (E and F). Method 1 yielded a narrower intratumoral range of pre- and post-treatment CBV values but a smaller mean difference compared with method 2.
Discussion
We have described the development of a DRO that recapitulates the DSC-MRI signal characteristics observed in human glioblastomas. The DRO enables signals to be computed for ranges of physiological, physical, and acquisition parameters. Clinical relevance is ensured through the use of a training data set. Furthermore, we validated the DRO's ability to produce reliable signals for different CA dosing schemes and acquisition parameters. Although in silico models may be limited by the accuracy of the biophysical model used, they provide a feasible and robust alternative to in vivo studies, which, in the case of DSC-MRI, may require multiple contrast injections and MRI scans and often lack a reliable “ground truth” for establishing accuracy.
Two key features of the proposed DRO are instrumental to its ability to provide signals that emulate clinical data. First, the DSC-MRI signals are derived using a validated computational approach that enables the incorporation of realistic tissue structures. Unlike heuristic models of DSC-MRI (34), this approach does not make assumptions regarding the voxel-wise CA T2* relaxivity, a parameter that is highly dependent upon vascular and cellular microstructure. In the proposed DRO, the voxel-wise microstructure determines the compartmental volume fractions and the associated CA relaxivity. Second, the training phase ensures that the range of simulated signals reflects the heterogeneity observed in vivo. Without training, there is the potential to introduce bias into the optimization of acquisition and postprocessing methods, as such methods may have not have uniform accuracy across the range of parameters.
Although we have presented two potential applications for the proposed DRO, there exist numerous opportunities for its use. Studies seeking to characterize and explore the biophysical basis of DSC-MRI data in brain tumors have yielded new biomarkers sensitive to the underlying tumor microstructure (eg, morphological features of vessels and cells) (23, 60–62) and hemodynamics (eg, vascular architectural imaging) (63). For these advanced methods, the DRO provides a tool with which to systematically investigate the sensitivity of DSC-MRI to such features and identify optimal acquisition protocols. Furthermore, the DRO can also be used to assess the accuracy of kinetic parameter estimates derived from newly developed pulse sequences, such as the recently proposed multiecho spin and gradient echo (SAGE) approach (64–69).
Although we trained the DRO with and validated it against in vivo data, any simulation approach that models complex biophysical phenomena has limitations. As described previously (22), the current computational approach does not consider the effects of arbitrary or heterogeneous CA distribution within a given tissue compartment such as the EES. The DRO could also be expanded to include the effects of transvascular water exchange rate, intravascular flow dynamics, atypical cellular geometries, and more heterogeneous vascular tree models. However, increasing the biological complexity of the input tissue structures also increases the number of unknown parameters that would need to be characterized.
The proposed DSC-MRI DRO provides a tool that can be leveraged by groups aiming to optimize and standardize acquisition and analysis methods for prospective clinical studies. It also enables the evaluation of bias and variance introduced by multisite data analysis. Such efforts are critical for establishing comparability of DSC-MRI data and interpreting multisite clinical trial data. To facilitate this effort, a range of DSC-MRI DROs is available for download from The Cancer Imaging Archive (www.cancerimagingarchive.net) under the collection name Barrow-DRO. The provided files contain multiple versions of the DRO, computed across a wide range of pulse sequence parameters and preload dosing schemes, all saved in Digital Imaging and Communications in Medicine (DICOM) and Matlab formats. Table 2 summarizes the range of pulse sequence parameters and CA dosing schemes that, when combined, yield 360 different acquisition methods. Each DRO file is a DSC-MRI time series data set similar to what would be acquired clinically and includes predefined regions of interest for the AIF, normal tissue and tumor voxels. Accordingly, these data may be processed using commercial or customized DSC-MRI analysis packages. The data set summary page details the organization of the files, the regions of interest, and the instructions for use.
Table 2.
Summary of Pulse Sequence Parameter Values and CA Dosing Schemes
| TR (ms) | FA (°) | TE (ms) | B0 (T) | Preload + Bolus |
|---|---|---|---|---|
| (1000, 1500, 2000) | (30, 60, 90) | (20, 30, 40, 50) | (1.5, 3) | (0 + 1, 1/4 + 3/4, 1/2 + 1/2, 1/2 + 1, 1 + 1) |
All possible combinations yielded 360 different acquisition methods. Dosing schemes are presented as a fraction of a standard 0.1 mmol/kg dose.
Acknowledgments
This work was performed at the Barrow Neurological Institute, with support from R01 CA158079. We thank Dr. Kathleen Schmainda (Medical College of Wisconsin) for access to dual-echo data used for validation.
Disclosures: No disclosures to report.
Conflict of Interest: None reported.
Footnotes
- DSC
- Dynamic susceptibility contrast
- MRI
- magnetic resonance imaging
- DRO
- digital reference object
- CBF
- cerebral blood flow
- CBV
- cerebral blood volume
- CA
- contrast agent
- TR
- repetition time
- TE
- echo time
- TCIA
- The Cancer Imaging Archive
- FA
- flip angle
- 3D
- 3-dimensional
- FPFDM
- finite perturber finite difference method
- EES
- extravascular extracellular space
- Gd-DTPA
- gadopentetate dimeglumine
- AIF
- arterial input function
- PSR
- percent signal recovery
References
- 1. Aronen HJ, Gazit IE, Louis DN, Buchbinder BR, Pardo FS, Weisskoff RM, Harsh GR, Cosgrove GR, Halpern EF, Hochberg FH. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology. 1994;191(1):41–51. [DOI] [PubMed] [Google Scholar]
- 2. Cha S, Knopp EA, Johnson G, Litt A, Glass J, Gruber ML, Lu S, Zagzag D. Dynamic contrast-enhanced T2-weighted MR imaging of recurrent malignant gliomas treated with thalidomide and carboplatin. AJNR Am J Neuroradiol. 2000;21(5):881–890. [PMC free article] [PubMed] [Google Scholar]
- 3. Knopp EA, Cha S, Johnson G, Mazumdar A, Golfinos JG, Zagzag D, Miller DC, Kelly PJ, Kricheff II. Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology. 1999;211(3):791–798. [DOI] [PubMed] [Google Scholar]
- 4. Law M, Yang S, Babb JS, Knopp EA, Golfinos JG, Zagzag D, Johnson G. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol. 2004;25(5):746–755. [PMC free article] [PubMed] [Google Scholar]
- 5. Law M, Yang S, Wang H, Babb JS, Johnson G, Cha S, Knopp EA, Zagzag D. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol. 2003;24(10):1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 6. Lev MH, Ozsunar Y, Henson JW, Rasheed AA, Barest GD, Harsh GR 4th, Fitzek MM, Chiocca EA, Rabinov JD, Csavoy AN, Rosen BR, Hochberg FH, Schaefer PW, Gonzalez RG. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected]. AJNR Am J Neuroradiol. 2004;25(2):214–221. [PMC free article] [PubMed] [Google Scholar]
- 7. Lev MH, Rosen BR. Clinical applications of intracranial perfusion MR imaging. Neuroimaging Clin N Am. 1999;9(2):309–331. [PubMed] [Google Scholar]
- 8. Rosen BR, Belliveau JW, Vevea JM, Brady TJ. Perfusion imaging with NMR contrast agents. Magn Reson Med. 1990;14(2):249–265. [DOI] [PubMed] [Google Scholar]
- 9. Sugahara T, Korogi Y, Kochi M, Ikushima I, Hirai T, Okuda T, Shigematsu Y, Liang L, Ge Y, Ushio Y, Takahashi M. Correlation of MR imaging-determined cerebral blood volume maps with histologic and angiographic determination of vascularity of gliomas. AJR Am J Roentgenol. 1998;171(6):1479–1486. [DOI] [PubMed] [Google Scholar]
- 10. Sugahara T, Korogi Y, Shigematsu Y, Hirai T, Ikushima I, Liang L, Ushio Y, Takahashi M. Perfusion-sensitive MRI of cerebral lymphomas: a preliminary report. J Comput Assist Tomogr. 1999;23(2):232–237. [DOI] [PubMed] [Google Scholar]
- 11. Sugahara T, Korogi Y, Shigematsu Y, Liang L, Yoshizumi K, Kitajima M, Takahashi M. Value of dynamic susceptibility contrast magnetic resonance imaging in the evaluation of intracranial tumors. Top Magn Reson Imaging. 1999;10(2):114–124. [DOI] [PubMed] [Google Scholar]
- 12. Villringer A, Rosen BR, Belliveau JW, Ackerman JL, Lauffer RB, Buxton RB, Chao YS, Wedeen VJ, Brady TJ. Dynamic imaging with lanthanide chelates in normal brain: contrast due to magnetic susceptibility effects. Magn Reson Med. 1988;6(2):164–174. [DOI] [PubMed] [Google Scholar]
- 13. Boxerman JL, Ellingson BM. Response assessment and magnetic resonance imaging issues for clinical trials involving high-grade gliomas. Top Magn Reson Imaging. 2015;24(3):127–136. [DOI] [PubMed] [Google Scholar]
- 14. Boxerman JL, Ellingson BM, Jeyapalan S, Elinzano H, Harris RJ, Rogg JM, Pope WB, Safran H. Longitudinal DSC-MRI for distinguishing tumor recurrence from pseudoprogression in patients with a high-grade glioma. Am J Clin Oncol, 2014. [Epub ahead of print]. doi: 10.1097/COC.0000000000000156 [DOI] [PubMed] [Google Scholar]
- 15. Boxerman JL, Zhang Z, Safriel Y, Larvie M, Snyder BS, Jain R, Chi TL, Sorensen AG, Gilbert MR, Barboriak DP. Early post-bevacizumab progression on contrast-enhanced MRI as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 Central Reader Study. Neuro Oncol. 2013;15(7):945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raghavan D, Boxerman J, Rogg J, Cosgrove R. Glioblastoma multiforme: utilization of advanced MRI techniques for preoperative planning. Med Health R I. 2012;95(2):42–43. [PubMed] [Google Scholar]
- 17. Schmainda KM, Zhang Z, Prah M, Snyder BS, Gilbert MR, Sorensen AG, Barboriak DP, Boxerman JL. Dynamic susceptibility contrast MRI measures of relative cerebral blood volume as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 multicenter trial. Neuro Oncol. 2015;17(8):1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shiroishi MS, Boxerman JL, Pope WB. Physiologic MRI for assessment of response to therapy and prognosis in glioblastoma. Neuro Oncol. 2016;18(4):467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ellingson BM, Bendszus M, Boxerman J, Barboriak D, Erickson BJ, Smits M, Nelson SJ, Gerstner E, Alexander B, Goldmacher G, Wick W, Vogelbaum M, Weller M, Galanis E, Kalpathy-Cramer J, Shankar L, Jacobs P, Pope WB, Yang D, Chung C, Knopp MV, Cha S, van den Bent MJ, Chang S, Yung WK, Cloughesy TF, Wen PY, Gilbert MR; Jumpstarting Brain Tumor Drug Development Coalition Imaging Standardization Steering Committee. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol. 2015;17(9):1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldmacher GV, Ellingson BM, Boxerman J, Barboriak D, Pope WB, Gilbert M. Standardized brain tumor imaging protocol for clinical trials. AJNR Am J Neuroradiol. 2015;36(10):E65–E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bosca RJ. Jackson EF. Creating an anthropomorphic digital MR phantom–an extensible tool for comparing and evaluating quantitative imaging algorithms. Phys Med Biol. 2016;61(2):974–982. [DOI] [PubMed] [Google Scholar]
- 22. Semmineh NB, Semmineh NB, Xu J, Boxerman JL, Delaney GW, Cleary PW, Gore JC, Quarles CC. An efficient computational approach to characterize DSC-MRI signals arising from three-dimensional heterogeneous tissue structures. PLoS One. 2014;9(1):e84764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Semmineh NB, Xu J, Skinner JT, Xie J, Li H, Ayers G, Quarles CC. Assessing tumor cytoarchitecture using multiecho DSC-MRI derived measures of the transverse relaxivity at tracer equilibrium (TRATE). Magn Reson Med. 2015; 74(3):772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brix G, Bahner ML, Hoffmann U, Horvath A, Schreiber W. Regional blood flow, capillary permeability, and compartmental volumes: measurement with dynamic CT–initial experience. Radiology. 1999;210(1):269–276. [DOI] [PubMed] [Google Scholar]
- 25. Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 26. Liu HL, Pu Y, Liu Y, Nickerson L, Andrews T, Fox PT, Gao JH. Cerebral blood flow measurement by dynamic contrast MRI using singular value decomposition with an adaptive threshold. Magn Reson Med. 1999;42(1):167–172. [DOI] [PubMed] [Google Scholar]
- 27. Carroll TJ, Rowley HA, Haughton VM. Automatic calculation of the arterial input function for cerebral perfusion imaging with MR imaging. Radiology. 2003;227(2):593–600. [DOI] [PubMed] [Google Scholar]
- 28. Wu O, Østergaard L, Weisskoff RM, Benner T, Rosen BR, Sorensen AG. Tracer arrival timing-insensitive technique for estimating flow in MR perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med. 2003;50(1):164–174. [DOI] [PubMed] [Google Scholar]
- 29. Newton AT, Pruthi S, Stokes AM, Skinner JT, Quarles CC. Improving perfusion measurement in DSC-MR imaging with multiecho information for arterial input function determination. AJNR Am J Neuroradiol. 2016;37(7):1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paulson ES, Prah DE, Schmainda KM. Spiral Perfusion Imaging with Consecutive Echoes (SPICE™) for the simultaneous mapping of DSC- and DCE-MRI parameters in brain tumor patients: theory and initial feasibility. Tomography, 2016;2(4):295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schramm P, Xyda A, Klotz E, Tronnier V, Knauth M, Hartmann M. Dynamic CT perfusion imaging of intra-axial brain tumours: differentiation of high-grade gliomas from primary CNS lymphomas. Eur Radiol. 2010;20(10):2482–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Donahue KM, Weisskoff RM, Burstein D. Water diffusion and exchange as they influence contrast enhancement. J Magn Reson Imaging. 1997;7(1):102–110. [DOI] [PubMed] [Google Scholar]
- 33. Landis CS, Li X, Telang FW, Coderre JA, Micca PL, Rooney WD, Latour LL, Vétek G, Pályka I, Springer CS Jr. Determination of the MRI contrast agent concentration time course in vivo following bolus injection: effect of equilibrium transcytolemmal water exchange. Magn Reson Med. 2000;44(4):563–574. [DOI] [PubMed] [Google Scholar]
- 34. Quarles CC, Gochberg DF, Gore JC, Yankeelov TE. A theoretical framework to model DSC-MRI data acquired in the presence of contrast agent extravasation. Phys Med Biol. 2009;54(19):5749–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delaney GW, Cleary PW. The packing properties of superellipsoids. Europhys Lett 2010;89(3):34002. [Google Scholar]
- 36. Ogawa S, Lee TM. Magnetic resonance imaging of blood vessels at high fields: in vivo and in vitro measurements and image simulation. Magn Reson Med. 1990;16(1):9–18. [DOI] [PubMed] [Google Scholar]
- 37. Fisel CR, Ackerman JL, Buxton RB, Garrido L, Belliveau JW, Rosen BR, Brady TJ. MR contrast due to microscopically heterogeneous magnetic susceptibility: numerical simulations and applications to cerebral physiology. Magn Reson Med. 1991;17(2):336–347. [DOI] [PubMed] [Google Scholar]
- 38. Kennan RP, Zhong J, Gore JC. Intravascular susceptibility contrast mechanisms in tissues. Magn Reson Med. 1994;31(1):9–21. [DOI] [PubMed] [Google Scholar]
- 39. Weisskoff RM, Zuo CS, Boxerman JL, Rosen BR. Microscopic susceptibility variation and transverse relaxation: theory and experiment. Magn Reson Med. 1994;31(6):601–610. [DOI] [PubMed] [Google Scholar]
- 40. Yablonskiy DA, Haacke EM. Theory of NMR signal behavior in magnetically inhomogeneous tissues: the static dephasing regime. Magn Reson Med. 1994;32(6):749–763. [DOI] [PubMed] [Google Scholar]
- 41. Boxerman JL, Bandettini PA, Kwong KK, Baker JR, Davis TL, Rosen BR, Weisskoff RM. The intravascular contribution to fMRI signal change: Monte Carlo modeling and diffusion-weighted studies in vivo. Magn Reson Med. 1995;34(1):4–10. [DOI] [PubMed] [Google Scholar]
- 42. Boxerman JL, Hamberg LM, Rosen BR, Weisskoff RM. MR contrast due to intravascular magnetic susceptibility perturbations. Magn Reson Med. 1995b;34(4):555–566. [DOI] [PubMed] [Google Scholar]
- 43. Stables LA, Kennan RP, Gore JC. Asymmetric spin-echo imaging of magnetically inhomogeneous systems: theory, experiment, and numerical studies. Magn Reson Med. 1998;40(3):432–442. [DOI] [PubMed] [Google Scholar]
- 44. Christen T, Pannetier NA, Ni WW, Qiu D, Moseley ME, Schuff N, Zaharchuk G. MR vascular fingerprinting: A new approach to compute cerebral blood volume, mean vessel radius, and oxygenation maps in the human brain. Neuroimage. 2014;89:262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tropres I, Pannetier N, Grand S, Lemasson B, Moisan A, Péoc'h M, Rémy C, Barbier EL. Imaging the microvessel caliber and density: principles and applications of microvascular MRI. Magn Reson Med. 2015;73(1):325–341. [DOI] [PubMed] [Google Scholar]
- 46. Hosokawa M, Kenmotsu H, Koh Y, Yoshino T, Yoshikawa T, Naito T, Takahashi T, Murakami H, Nakamura Y, Tsuya A, Shukuya T, Ono A, Akamatsu H, Watanabe R, Ono S, Mori K, Kanbara H, Yamaguchi K, Tanaka T, Matsunaga T, Yamamoto N. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One. 2013;8(6):e67466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jain RK. Transport of molecules in the tumor interstitium: a review. Cancer Res. 1987;47(12):3039–3051. [PubMed] [Google Scholar]
- 48. Weisskoff RM, Kiihne S. MRI susceptometry: image-based measurement of absolute susceptibility of MR contrast agents and human blood. Magn Reson Med. 1992;24(2):375–383. [DOI] [PubMed] [Google Scholar]
- 49. Le Bihan D. [Diffusion, perfusion and functional magnetic resonance imaging]. J Mal Vasc. 1995;20(3):203–214. [Article in English, French]. [PubMed] [Google Scholar]
- 50. Paulson ES, Schmainda KM. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology. 2008;249(2):601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bjornerud A, Sorensen AG, Mouridsen K, Emblem KE. T1- and T2*-dominant extravasation correction in DSC-MRI: part I–theoretical considerations and implications for assessment of tumor hemodynamic properties. J Cereb Blood Flow Metab. 2011;31(10):2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heiland S, Benner T, Debus J, Rempp K, Reith W, Sartor K. Simultaneous assessment of cerebral hemodynamics and contrast agent uptake in lesions with disrupted blood-brain-barrier. Magn Reson Imaging. 1999;17(1):21–27. [DOI] [PubMed] [Google Scholar]
- 53. Johnson G, Wetzel SG, Cha S, Babb J, Tofts PS. Measuring blood volume and vascular transfer constant from dynamic, T(2)*-weighted contrast-enhanced MRI. Magn Reson Med. 2004;51(5):961–968. [DOI] [PubMed] [Google Scholar]
- 54. Quarles CC, Ward BD, Schmainda KM. Improving the reliability of obtaining tumor hemodynamic parameters in the presence of contrast agent extravasation. Magn Reson Med. 2005;53(6):1307–1316. [DOI] [PubMed] [Google Scholar]
- 55. Uematsu H, Maeda M, Sadato N, Matsuda T, Ishimori Y, Koshimoto Y, Kimura H, Yamada H, Kawamura Y, Yonekura Y, Itoh H. Blood volume of gliomas determined by double-echo dynamic perfusion-weighted MR imaging: a preliminary study. AJNR Am J Neuroradiol. 2001;22(10):1915–1919. [PMC free article] [PubMed] [Google Scholar]
- 56. Uematsu H, Maeda M, Sadato N, Matsuda T, Ishimori Y, Koshimoto Y, Yamada H, Kimura H, Kawamura Y, Matsuda T, Hayashi N, Yonekura Y, Ishii Y. Vascular permeability: quantitative measurement with double-echo dynamic MR imaging–theory and clinical application. Radiology. 2000;214(3):912–917. [DOI] [PubMed] [Google Scholar]
- 57. Vonken EJ, van Osch MJ, Bakker CJ, Viergever MA. Measurement of cerebral perfusion with dual-echo multi-slice quantitative dynamic susceptibility contrast MRI. J Magn Reson Imaging. 1999;10(2):109–117. [DOI] [PubMed] [Google Scholar]
- 58. Leu K, Boxerman JL, Cloughesy TF, Lai A, Nghiemphu PL, Liau LM, Pope WB, Ellingson BM. Improved leakage correction for single-echo dynamic susceptibility contrast perfusion MRI estimates of relative cerebral blood volume in high-grade gliomas by accounting for bidirectional contrast agent exchange. AJNR Am J Neuroradiol. 2016;37(8):1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stokes AM, Semmineh N, Quarles CC. Validation of a T1 and T2* leakage correction method based on multiecho dynamic susceptibility contrast MRI using MION as a reference standard. Magn Reson Med. 2016;76(2):613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mangla R, Kolar B, Zhu T, Zhong J, Almast J, Ekholm S. Percentage signal recovery derived from MR dynamic susceptibility contrast imaging is useful to differentiate common enhancing malignant lesions of the brain. AJNR Am J Neuroradiol. 2011;32(6):1004–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sourbron S, Heilmann M, Walczak C, Vautier J, Schad LR, Volk A. T2*-relaxivity contrast imaging: first results. Magn Reson Med. 2013;69(5):1430–1437. [DOI] [PubMed] [Google Scholar]
- 62. Tropres I, Grimault S, Vaeth A, Grillon E, Julien C, Payen JF, Lamalle L, Décorps M. Vessel size imaging. Magn Reson Med. 2001;45(3):397–408. [DOI] [PubMed] [Google Scholar]
- 63. Emblem KE, Mouridsen K, Bjornerud A, Farrar CT, Jennings D, Borra RJ, Wen PY, Ivy P, Batchelor TT, Rosen BR, Jain RK, Sorensen AG. Vessel architectural imaging identifies cancer patient responders to anti-angiogenic therapy. Nat Med. 2013;19(9):1178–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schmiedeskamp H, Andre JB, Straka M, Christen T, Nagpal S, Recht L, Thomas RP, Zaharchuk G, Bammer R. Simultaneous perfusion and permeability measurements using combined spin- and gradient-echo MRI. J Cereb Blood Flow Metab. 2013;33(5):732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schmiedeskamp H, Straka M, Newbould RD, Zaharchuk G, Andre JB, Olivot JM, Moseley ME, Albers GW, Bammer R. Combined spin- and gradient-echo perfusion-weighted imaging. Magn Reson Med. 2012;68(1):30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Skinner JT, Robison RK, Elder CP, Newton AT, Damon BM, Quarles CC. Evaluation of a multiple spin- and gradient-echo (SAGE) EPI acquisition with SENSE acceleration: applications for perfusion imaging in and outside the brain. Magn Reson Imaging. 2014;32(10):1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stokes AM, Quarles CC. A simplified spin and gradient echo approach for brain tumor perfusion imaging. Magn Reson Med. 2016;75(1):356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stokes AM, Skinner JT, Quarles CC. Assessment of a combined spin- and gradient-echo (SAGE) DSC-MRI method for preclinical neuroimaging. Magn Reson Imaging. 2014;32(10):1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stokes AM, Skinner JT, Yankeelov T, Quarles CC. Assessment of a simplified spin and gradient echo (sSAGE) approach for human brain tumor perfusion imaging. Magn Reson Imaging. 2016;34(9):1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]