Abstract
Forkhead transcription factors FOXO1 (FKHR), FOXO3a (FKHRL1), and FOXO4 (AFX) play a pivotal role in tumor suppression by inducing growth arrest and apoptosis. Loss of function of these factors due to phosphorylation and proteasomal degradation has been implicated in cell transformation and malignancy. However, the ubiquitin ligase necessary for the ubiquitination of the FOXO factors and the relevance of this regulation to tumorigenesis have not been characterized. Here we demonstrate that Skp2, an oncogenic subunit of the Skp1/Cul1/F-box protein ubiquitin complex, interacts with, ubiquitinates, and promotes the degradation of FOXO1. This effect of Skp2 requires Akt-specific phosphorylation of FOXO1 at Ser-256. Moreover, expression of Skp2 inhibits transactivation of FOXO1 and abolishes the inhibitory effect of FOXO1 on cell proliferation and survival. Furthermore, expression of the FOXO1 protein is lost in a mouse lymphoma model, where Skp2 is overexpressed. These data suggest that the Skp2-promoted proteolysis of FOXO1 plays a key role in tumorigenesis.
Keywords: ubiquitin ligase, proteasomal degradation, cancer
Forkhead family members FOXO1, FOXO3a, and FOXO4 are multifunctional transcription factors, which regulate transcription of a number of genes that play critical roles in inducing either cell cycle arrest or apoptosis. Activation of each member of this family in transformed and nontransformed cells results in up-regulation of the cyclin-dependent kinase inhibitor p27KIP1 and/or down-regulation of d-type cyclins, thereby arresting cells at G1 (1, 2). Activated FOXO proteins also trigger apoptosis in many cancer cell lines through regulation of a number of proapoptotic proteins, including Fas ligand, TRAIL, and Bim (3–5). Knocking down the FOXO3a protein in human breast cancer cells or inhibition of the transcriptional activity of FOXO1 in chicken embryo fibroblasts promotes cell transformation and tumor progression (6, 7). Thus, it has been postulated that FOXO factors play a pivotal role in the inhibition of cell transformation and tumorigenesis.
The inhibitory function of FOXO proteins in cell proliferation and survival is often disrupted due to the overactivated phosphatidylinositol 3-kinase (PI3K)/Akt pathway in cancer cells. Activated Akt phosphorylates a wide range of downstream proapoptotic proteins, among which are the forkhead factors FOXO1, FOXO3a, and FOXO4. Phosphorylated forkhead proteins translocate from the nucleus to the cytoplasm where they are inactive (3, 8–10). Recently, another kinase, IκB kinase β, has been shown to phosphorylate and inactivate FOXO3a in breast cancer cells (6). The tumor suppressor gene PTEN encodes a lipid phosphatase that specifically dephosphorylates the D3 position of phosphatidylinositol 3,4,5-trisphosphate (11) and in so doing functionally antagonizes the PI3K pathway. Because of frequent deletions and mutations in the PTEN gene in human cancers, it is believed that protein phosphorylation is a key mechanism that inactivates the FOXO factors.
It has been demonstrated previously that a number of tumor suppressors (e.g., p53, RB, and p27KIP1) can be degraded by the ubiquitin pathway in human cancer (12). Indeed, several tumor suppressor proteins, including p27KIP1, p130, and p57KIP2, have been shown to be targeted by the F-box motif in Skp2 for degradation (13–16). Recently, ubiquitination and proteasome degradation of FOXO1 and FOXO3a have been reported (6, 7, 17, 18). However, the ubiquitin ligase necessary for this proteolysis has not been identified. In this study, we provide evidence that Skp2 interacts with and promotes the ubiquitin-dependent degradation of FOXO1, thereby inhibiting the tumor suppressor function of FOXO1.
Materials and Methods
Plasmids and Small Interfering RNAs (siRNAs). Plasmids for FLAG-tagged FOXO1 (FOXO1-WT and AAA, originally named FKHR-WT and FKHR-AAA), Skp2, and hemagglutinin (HA)-tagged ubiquitin were kindly provided by K. L. Guan (University of Michigan, Ann Arbor; ref. 8), H. Zhang (Yale University School of Medicine, New Haven, CT; ref. 19), and D. Bohmann (University of Rochester Medical Center, Rochester, NY; ref. 20), respectively. The luciferase reporter construct 3xIRS-Luc, which contains three copies of the FOXO responsive element from the IGFBP1 promoter, was a gift from K. L. Guan (8). The HA tag was integrated into pcDNA3.1 by PCR to make a HA-Skp2 expression vector. The expression vector for GST-Skp2 was constructed by cloning the full-length Skp2 into the pGEX-4T-1 plasmid (Amersham Pharmacia Biosciences). Plasmids for the C-terminal truncated and point-mutated FOXO1 proteins were generated by PCR-based mutagenesis (Stratagene). The myristoylated Akt expression vector has been described (21). The pEGFP vector was purchased from BD Biosciences (Clontech). A pool of siRNAs for the human Skp2 gene and nonspecific siRNAs was purchased from Dharmacon (Lafayette, CO).
Antibodies and Chemicals. The following antibodies were used: anti-FOXO1, anti-phospho-FOXO1 at Ser-256 (FOXO1-p), antiphospho-Akt at Ser-473 (Akt-p), anti-Akt (Cell Signaling Technology, Beverly, MA), anti-Skp2 monoclonal (Zymed), anti-Skp2 polyclonal (H-435, Santa Cruz Biotechnology), anti-FLAG (M2, Sigma), anti-HA (12CA5, Roche), anti-Erk2 (Santa Cruz Biotechnology), and anti-GST (Amersham Pharmacia Biosciences). Cycloheximide was purchased from Sigma, and MG132 and lactacystin were obtained from Calbiochem.
Tumor Samples and Cell Lines. Lymphomas in CBP–/– and CBP–/–p27KIP1+/– mice and thymocytes in control mice were collected, and protein samples were prepared for immunoblotting as described (22). LNCaP, 786-O, HepG2, Jurkat, NIH 3T3, and COS7 cells lines were purchased from the American Type Culture Collection.
Cell Culture, Transfection, and Luciferase Assays. NIH 3T3, HepG2, and COS7 cells were grown in DMEM supplemented with 10% FBS. LNCaP and 786-O cells were grown in RPMI medium 1640. Cycloheximide, MG132, or lactacystin was added at a final concentration of 30 μg/ml, 10 μM, or 10 μM, respectively. siRNAs were used at a final concentration of 100 nM. Transfections were performed with lipofectamine 2000 (Invitrogen) for COS7 cells and by electroporation for LNCaP, 786-O, NIH 3T3, and Jurkat cells, as described (21). Transfection efficiencies of ≈60–90% in LNCaP, 786-O, NIH 3T3, and COS7 cells were achieved. The luciferase reporter construct 3xIRS-Luc was cotransfected with FOXO1 expression vectors into NIH 3T3 cells or with Skp2 siRNAs into Jurkat cells, and luciferase activity was measured as described (23).
Cell Sorting and Cell Cycle Analysis. 786-O Cells were cotransfected with pEGFP, Skp2, and FOXO1. For cell cycle analysis, cells were fixed briefly with 1% formaldehyde for 10 min at room temperature. For Western blot analysis, GFP-positive cells were sorted with a FACSVantage SE (Becton Dickinson). Cell cycle analysis was performed as described (21). Cell cycle distributions were determined by a FACSCalibur flow cytometer (Becton Dickinson) and analyzed with modfit II software (Verity Software House, Topsham, ME).
Immunoprecipitation, Immunoblotting Analysis, GST Fusion Protein Purification, and GST Pull-Down Assays. Immunoprecipitations were performed by using an immunoprecipitation kit (Roche Applied Science). Immunoblotting was performed as described (23). GST and GST-Skp2 fusion proteins were purified from the BL21 Star (DE3) Escherichia coli strain (Invitrogen). GST pull-down assays were performed as described (24). Briefly, NIH 3T3 cells were transfected with a FLAG-FOXO1 expression plasmid for 36 h, and cells were lysed for 30 min in lysis buffer containing 50 mM Tris, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 10 mM NaF, and protease inhibitors. Cell debris was removed by centrifugation. Lysates were precleared with GST beads for 1 h and incubated with GST or GST-Skp2 fusion proteins overnight at 4°C. Protein-bound GST beads were washed four times with lysis buffer and eluted in SDS/PAGE sample buffer. Eluted proteins were analyzed by immunoblotting.
In Vitro Protein-Binding and in Vitro Ubiquitination Assays. Bacterially expressed GST or GST-Skp2 proteins were incubated with immobilized FOXO1 peptides in binding buffer (50 mM Tris, pH 7.4/150 mM NaCl/5 mM MgCl2/1 mM EDTA/0.15% N-P40/5 mM DTT/10 mM NaF/protease inhibitors) for 5 h at 4°C. Bound proteins were washed three times with binding buffer and analyzed by SDS/PAGE. In vitro ubiquitination assays using rabbit reticulocyte lysate were performed as described (13, 25).
Further Details. For further details, please see Supporting Text, which is published as supporting information on the PNAS web site.
Results
Skp2 Promotes Ubiquitination and Degradation of the FOXO1 Protein. It has been shown recently that the FOXO1 protein is targeted for proteasomal degradation in FL5.12 murine lymphocytes and HepG2 cells (17, 18). Because the expression pattern of FOXO1 protein is inversely correlated with the Skp1/Cul1/F-box protein subunit Skp2 during the cell cycle of NIH 3T3 cells (data not shown), we sought to determine whether Skp2 is involved in the degradation of FOXO1. Three sublines of LNCaP cells were generated that stably express Skp2. Elevated levels of Skp2 were found to correlate with decreased expression of endogenous FOXO1 protein in all three clones, although to varying degrees (Fig. 1A). Moreover, transient expression of Skp2 resulted in a reduction of FLAG-FOXO1 protein in LNCaP cells (Fig. 1B), as well as in NIH 3T3 and COS7 cells (Fig. 4A and Fig. 7A, which is published as supporting information on the PNAS web site). In contrast, knockdown of endogenous Skp2 by siRNA resulted in an increase in the level of endogenous FOXO1 protein (Fig. 1C), whereas nonspecific siRNAs had no effect. Moreover, expression of the adenoviral protein E1A resulted in an increase in the level of Skp2 in androgen-treated LNCaP cells but caused a decrease in the FOXO1 protein (Fig. 8A, which is published as supporting information on the PNAS web site). Similar to the findings reported in Rat-6 and IMR-90 fibroblasts (26), expression of Skp2 is temporally up-regulated in NIH 3T3 cells in an adhesion-dependent manner (Fig. 8B). In contrast, the level of FOXO1 is decreased in adherent cells but not in those cultured in suspension. To elucidate the underlying mechanism, we examined whether Skp2 regulates the expression of FOXO1 at the messenger level by using RT-PCR. No effect of Skp2 was observed on the expression of FOXO1 mRNA in LNCaP cells that were transfected with Skp2 either transiently or stably (Fig. 9A, which is published as supporting information on the PNAS web site). Taken together, these findings suggest that Skp2 negatively regulates the expression of FOXO1 at the protein level.
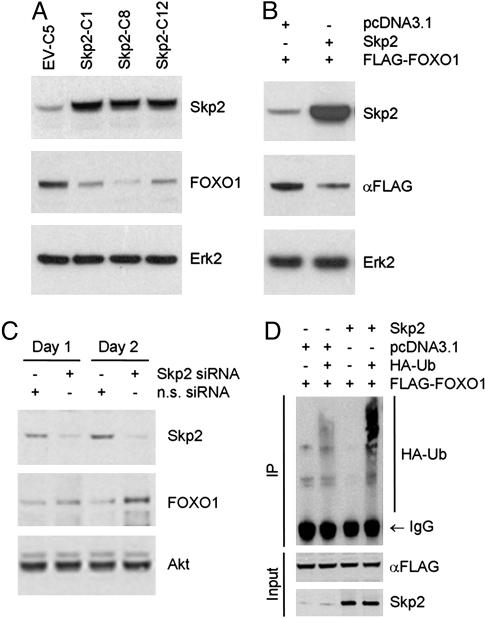
Fig. 1.
Skp2 induces ubiquitination and degradation of FOXO1. (A) Stable clones of LNCaP cells expressing Skp2 (Skp2-C1, Skp2-C8, and Skp2-C12) or empty vector (EV-C5) were established, and cell extracts were prepared for Western blot analysis. (B) A FLAG-tagged FOXO1 expression vector was cotransfected transiently with Skp2 or an empty vector (pcDNA3.1) into LNCaP cells, and whole cell lysates were prepared for Western bot analysis. (C) HepG2 cells were transfected with a pool of nonspecific (n.s.) siRNAs or siRNAs for Skp2, and cell extracts were prepared at the indicated times for Western blot analysis. (D) LNCaP cells were transfected with the indicated plasmids including HA-tagged ubiquitin, and cell extracts were either immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-HA antibody or immunoblotted directly with antibodies for the FLAG tag or Skp2. Samples were analyzed by Western blots.
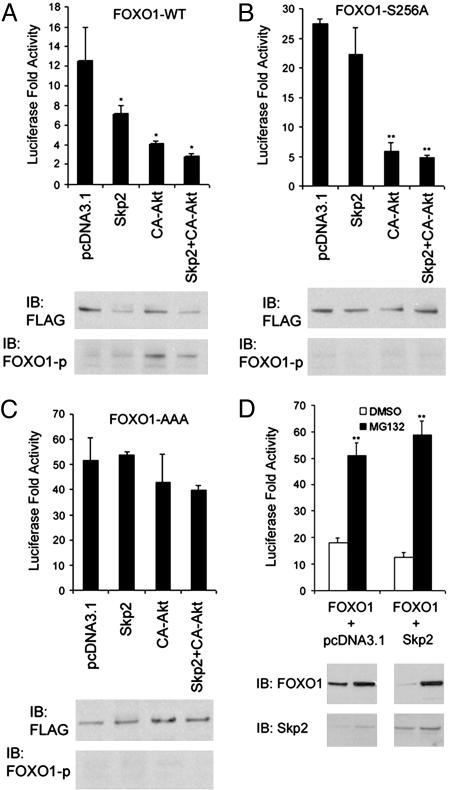
Fig. 4.
Effect of Skp2 on transactivation of FOXO1. (A) NIH 3T3 cells were transfected with luciferase reporter 3xIRS-Luc and the wild-type FOXO1. At 36 h after transfection, luciferase activity, levels of FOXO1 and Skp2 proteins as well as the status of phosphorylated FOXO1 were analyzed. Activity of luciferase was determined by normalizing the measured light units of firefly luciferase with the measured Renilla luciferase activity. *, P < 0.05 comparing the effect of Skp2 and Akt with that of the empty vector pcDNA3.1. (B) NIH 3T3 cells were transfected with luciferase reporter 3xIRS-Luc and FOXO1-S256A. Luciferase analysis and immunoblotting were performed as in A. **, P < 0.01 comparing the effect of Akt and Akt plus Skp2 with that of the empty vector pcDNA3.1. (C) NIH 3T3 cells were transfected with luciferase reporter 3xIRS-Luc and FOXO1-AAA. Luciferase analysis and immunoblotting were performed as in A.(D) NIH 3T3 cells were transfected with luciferase reporter 3xIRS-Luc and FOXO1. At 24 h after transfection, cells were treated with 15μM of MG132 or DMSO for 12 h. Luciferase analysis and immunoblotting were performed as in A. **, P < 0.01 comparing the effect of MG132 with the vehicle.
To determine whether the effect of Skp2 on the FOXO1 protein is mediated through protein degradation, we compared the half-life of FOXO1 protein in cells transfected with Skp2 or empty vector. Ectopic expression of Skp2 resulted in a rapid decrease in the FOXO1 protein (Fig. 10A, which is published as supporting information on the PNAS web site). Importantly, this effect was abolished completely by the proteasome inhibitor lactacystin (Fig. 10A). In contrast, FOXO1 protein was degraded much more slowly in mock-transfected than in Skp2-transfected cells, and lactacystin reduced the rate of degradation even further (Fig. 10B). A similar result was obtained in a Skp2-stable clone, Skp2-C8, vs. the emptyvector control, EV-C5 (data not shown). These findings indicate that Skp2 targets FOXO1 protein for proteasome degradation. Because FOXO1 has been identified as a ubiquitination target in HepG2 cells (18), we examined whether expression of Skp2 affects ubiquitination of FOXO1 in LNCaP cells. Indeed, ubiquitination of FOXO1 was confirmed in this prostate cancer cell line (Fig. 1D), and importantly, the ubiquitination of FOXO1 was enhanced markedly by the transfection of Skp2 (Fig. 1D). Moreover, this effect was enhanced further by MG132 (Fig. 10C). These findings demonstrate that Skp2 promotes the ubiquitination and proteasome degradation of FOXO1.
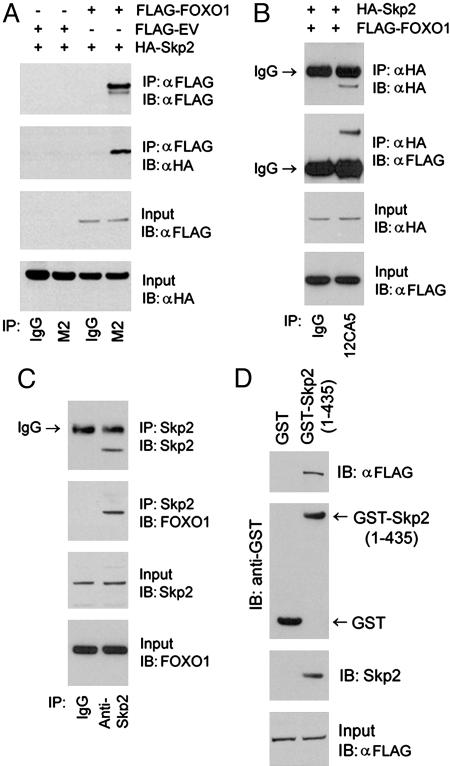
Skp2 Interacts with FOXO1 in Vivo and in Vitro. Next we assessed whether Skp2 interacts with FOXO1 by coimmunoprecipitation assays (Fig. 2 A–C). FLAG-tagged FOXO1 and HA-tagged Skp2 expression vectors were transfected either alone or together into LNCaP cells. As demonstrated in Fig. 2A, HA-Skp2 was detected in the FLAG-FOXO1 immune complex, whereas no HA-Skp2 was found in the immune complex precipitated with a nonspecific mouse IgG (Fig. 2A). Also, no HA-Skp2 was immunoprecipitated in cells transfected with empty vector (Fig. 2A). Conversely, FLAG-FOXO1 was found in the HA-Skp2 immune complex (Fig. 2B). Next we sought to determine whether endogenous FOXO1 interacts with Skp2 in LNCaP and NIH 3T3 cells. The FOXO1 protein was detected in the immune complex precipitated with anti-Skp2 antibody in both cell lines (Fig. 2C and Fig. 11, which is published as supporting information on the PNAS web site). Also, FLAG-FOXO1 in NIH 3T3 cells was pulled down by GST-Skp2 fusion protein purified from bacteria but not GST alone (Fig. 2D). Further immunoprecipitation analyses demonstrated that no interaction was detected between FOXO1 and the other F-box protein hCDC4/Fbw7 in LNCaP cells. Neither was there evidence of interaction between Skp2 and other FOXO proteins, including FOXO3a and FOXO4 in NIH 3T3 cells (Fig. 12, which is published as supporting information on the PNAS web site). Thus, Skp2 forms a complex specifically with FOXO1 in vivo and in vitro.
Fig. 2.
Skp2 interacts with FOXO1 in vivo and in vitro. LNCaP cells were transfected with the indicated plasmids, and protein lysates were immunoprecipitated (IP) with anti-FLAG antibody (M2) (A) or an anti-HA antibody (12CA5) (B). A mouse IgG was used as a negative control. Immunoprecipitates were analyzed by immunoblotted (IB) with anti-FLAG and anti-HA antibodies. (C) Protein lysates from NIH 3T3 cells were immunoprecipitated with an anti-Skp2 antibody and immunoblotted with antibodies for FOXO1 and Skp2. (D) Lysates of NIH 3T3 cells transfected with FLAG-FOXO1 were subjected to GST pull-down by GST-Skp2 purified from bacteria.
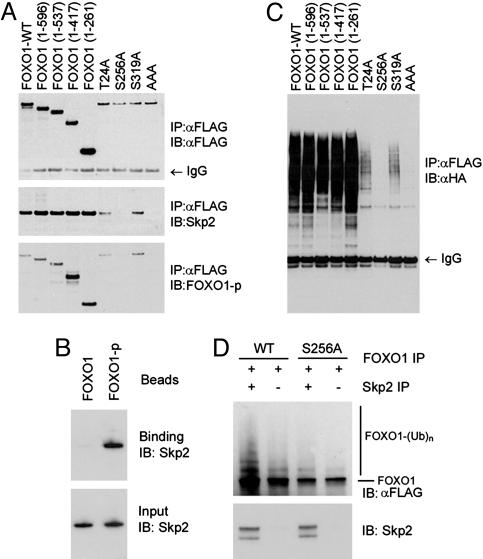
Interaction and Degradation of FOXO1 by Skp2 Require Phosphorylation of FOXO1 at Ser-256. To identify the domain of the FOXO1 protein that mediates the interaction with Skp2, we constructed a series of C-terminal truncation mutants of FOXO1. These mutant constructs were transfected into LNCaP cells, and cell lysates were precipitated with an anti-FLAG antibody. Each of these mutants formed a complex with the Skp2 protein (Fig. 3A), suggesting that the N-terminal 261 amino acids of FOXO1 contain the Skp2-interaction domain. Because Akt-dependent phosphorylation plays a key role in the proteasomal degradation of FOXO1 (7, 17, 18), and the F-box proteins in Skp1/Cul1/F-box protein ligases preferentially recognize phosphorylated substrates (27), we assessed whether mutation of the Akt-phosphorylation sites Thr-24, Ser-256, and/or Ser-319 to alanine affects the interaction of FOXO1 with Skp2. As demonstrated in Fig. 3A, all of the mutant forms of FOXO1 were expressed at comparable levels in LNCaP cells. Both the T24A and S319A mutants were able to form a complex with Skp2 (Fig. 3A). However, the interaction between FOXO1 and Skp2 was abolished by a point mutation at Ser-256 or triple mutations at Thr-24, Ser-256, and Ser-319 (Fig. 3A). As expected, no phosphorylation was detected at Ser-256 in these two mutants (Fig. 3A). These findings prompted us to determine whether a FOXO1 phosphopeptide, with a phosphoserine at position 256, would bind to Skp2 in a phosphorylation-dependent manner. The phospho- and nonphosphopeptides of FOXO1 were immobilized on agarose beads and incubated with bacterially produced GST-Skp2. The recombinant Skp2 bound to the phosphopeptide of FOXO1 significantly better than the nonphosphorylated peptide (Fig. 3B). GST alone showed no binding to these peptides (data not shown). Thus, these findings suggest that phosphorylation at Ser-256 in FOXO1 is required for its interaction with Skp2.
Fig. 3.
Interaction and ubiquitination of FOXO1 by Skp2 depend on phosphorylation of FOXO1 at Ser-256. (A) FOXO1 plasmids were cotransfected with Skp2 into LNCaP cells, and cell extracts were prepared for immunoprecipitation with an anti-FLAG antibody and immunoblotted with antibodies for FLAG, Skp2, or phospho-Ser-256 FOXO1 (FOXO1-p). (B) LNCaP cells were transfected with the indicated FOXO1, Skp2, and HA-tagged ubiquitin plasmids, and cell extracts were immunoprecipitated with the anti-FLAG antibody and immunoblotted with the anti-HA antibody. (C) Bacterially expressed GST-Skp2 was incubated in vitro with beads coupled to the FOXO1 phosphopeptide GKSPRRRAApSMDNNSKFAKS (FOXO1-p), which contains a phosphoserine at position 256. A corresponding nonphosphopeptide (FOXO1) was included as a control. (D) In vitro ubiquitination of FOXO1. Anti-FLAG immunoprecipitates immobilized on beads from LNCaP cells transfected with either wild-type (WT) or mutant (S256A) FLAG-FOXO1 were incubated with Skp2 proteins immunoprecipitated from Skp2-stable LNCaP cell lines (see Fig. 1) in a reaction system that contained 20 μM MG132 and rabbit reticulocyte lysate, which was precleared with anti-Skp2 antibody. Reactions were terminated by washing the pellets three times in RIPA buffer, and samples were analyzed by SDS/PAGE and immunoblotting.
Next we sought to determine whether phosphorylation of FOXO1 at Ser-256 is required for the Skp2-mediated ubiquitination of FOXO1 in LNCaP cells. Each of the C-terminal-truncated FOXO1 proteins was found to be ubiquitinated similarly to the wild-type FOXO1 (Fig. 3C). Moreover, ubiquitination was observed on the FOXO1 protein mutated at either Thr-24 or Ser-319, although to a lesser extent than that of wild-type FOXO1 (Fig. 3B). However, ubiquitination of FOXO1 was abolished by the single mutation at Ser-256 or the triple mutations (Fig. 3B). Furthermore, in vitro ubiquitination assays demonstrated that the phosphorylation of FOXO1 at Ser-256 is required for the Skp2-mediated ubiquitination of FOXO1 (Fig. 3D). These findings demonstrate that phosphorylation of FOXO1 at Ser-256 is required for its interaction with Skp2 and subsequent ubiquitination. The reduced ubiquitination of the T24A and S319A mutants could be due to several factors. One possibility is their nuclear localization (9), because ubiquitination of FOXO1 has been shown to be affected by its cellular localization (18). Another possibility is that the mutated proteins are less well tolerated than the wild-type in LNCaP cells (Fig. 3A) (23, 28).
To further examine whether phosphorylation of FOXO1 at Ser-256 plays a critical role in Skp2-mediated degradation of FOXO1, Skp2 was cotransfected with a constitutively active form of Akt (CA-Akt) and wild-type or mutated FOXO1 (S256A) into COS7 cells. Ectopic expression of Akt resulted in a decrease in levels of FOXO1 (Fig. 7A). However, this effect was abolished by the mutation at Ser-256 (Fig. 7B). Moreover, pulse–chase analyses were performed to determine the stability of the wild-type and S256A mutated FOXO1. The results demonstrated that the half-life of the S256A mutant was much longer than that of the wild-type protein (Fig. 7C), thus confirming that phosphorylation of FOXO1 on Ser-256 is required for Skp2-mediated degradation.
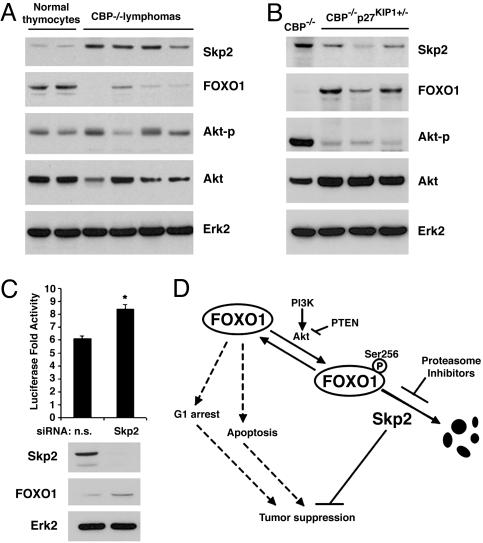
Skp2 Inhibits Transactivation of FOXO1. Next we sought to determine whether the Skp2- and Akt-promoted degradation of FOXO1 affects its transactivational activity. Expression vectors for wild-type and degradation-resistant mutants of FOXO1 were transfected into NIH 3T3 cells together with the luciferase reporter 3xIRS-luc and vectors for Skp2, CA-Akt, or both. Ectopic expression of Skp2 induced an ≈50% decrease in its transactivation activity (Fig. 4A) and a marked decrease in the level of wild-type FLAG-FOXO1 protein (Fig. 4A). Expression of CA-Akt resulted in a more robust decrease in the transactivation activity of FOXO1 (Fig. 4A). This effect of Akt is very likely mediated by the phosphorylation and/or degradation of FOXO1 (Fig. 4A). The most significant inhibition of the transactivation of FLAG-FOXO1 was obtained in cells transfected with both Skp2 and CA-Akt. This correlated with a decreased expression and an increased phosphorylation of FOXO1 protein (Fig. 4A). The single mutation of FOXO1 at Ser-256 resulted in a higher transactivation activity of FOXO1 (Fig. 4B), similar to previous findings in 293T cells (8). In contrast to the wild-type FOXO1, there was no effect of Skp2 expression on either protein levels or transcriptional activity of FOXO1 (Fig. 4B). As expected, expression of CA-Akt resulted in a marked decrease in the activity of FOXO1-S256A, but no effect on protein levels (Fig. 4B). This is presumably due to Akt-mediated phosphorylation of FOXO1 at Thr-24 and Ser-319 (9). Expression of the triple mutant FOXO1-AAA resulted in an ≈6-fold increase in its transactivation when compared with the wild-type protein (Fig. 4C). Like the S256A mutant, the triple mutant was resistant to Skp2-induced degradation and decreased transactivation (Fig. 4). No inhibitory effect of Akt was obtained on the transactivation of the triple mutant control (Fig. 4C). Thus, Skp2 inhibits transcriptional activity of FOXO1 via protein degradation. Next, we sought to determine the effect of the proteasome inhibitor MG132 on the regulation of FOXO1 transactivation by Skp2. Treatment of NIH 3T3 cells with MG132 resulted in a marked increase in the transactivation of FOXO1, with a concomitant increase in FOXO1 protein (Fig. 4D). Although expression of Skp2 inhibited the transcriptional activity of FOXO1, this effect was abolished by MG132 treatment (Fig. 4D), suggesting that the inhibitory effect of Skp2 on transactivation of FOXO1 can be completely reversed by inhibition of proteasome activity. Moreover, knockdown of endogenous Skp2 with siRNA resulted in an increase in the transcriptional activity of FOXO1 in Jurkat cells (Fig. 6C). Taken together, these data suggest that Skp2 negatively regulates the transactivation activity of FOXO1 by inducing its degradation.
Fig. 6.
The FOXO1 proteins are lost in lymphomas in CBP knockout mice. Representative immunoblot analyses of Skp2, FOXO1, phosphorylated Akt at Ser-473 (Akt-p), and Akt in normal mouse thymocytes (n = 4), CBP–/– lymphomas (n = 8) (A), or CBP–/–p27KIP1+/– lymphomas (n = 3) (B). Erk2 was used as a loading control. (C) Jurkat cells were transfected with luciferase reporter 3xIRS-Luc and Skp2 siRNAs. At 36 h after transfection, luciferase activity, levels of FOXO1 and Skp2 proteins were analyzed. *, P < 0.05. (D) A diagram depicts the role of Skp2 in tumorigenesis via promoting degradation of FOXO1. Dashed lines indicate the tumor suppression functions of FOXO1 in the absence of interference of Skp2.
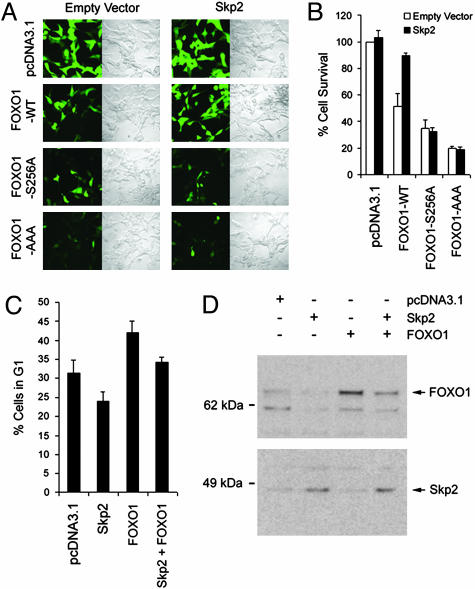
Skp2 Inhibits the Tumor Suppression Function of FOXO1. It has been shown that FOXO1 possesses a tumor suppressor function by inhibiting cell proliferation and survival (2, 8, 23, 28). Similar to previous findings (23, 28), expression of wild-type and constitutively active forms of FOXO1 resulted in a significant loss of viability in LNCaP cells (Fig. 5 A and B). Moreover, the inhibitory effect of the wild-type FOXO1 was almost completely abolished by cotransfection of Skp2 into LNCaP cells. In control cells, no effect on viability was observed when cells were cotransfected with Skp2 and mutants of FOXO1 (Fig. 5 A and B). A similar result was obtained for the single mutant S256A (Fig. 5 A and B). Similar to a previous report (28), expression of FOXO1 in 786-O cells resulted in G1 arrest (Fig. 5C). This effect was inhibited by Skp2 expression. Consistent with this result, transfection of Skp2 into 786-O cells resulted in a decrease in the level of total FOXO1 proteins (Fig. 5D). Thus, our findings demonstrate that Skp2 inhibits the tumor suppressor activity of FOXO1.
Fig. 5.
Skp2 antagonizes the tumor suppression function of FOXO1. (A and B) LNCaP cells were cotransfected with pEGFP vector along with control vector pcDNA3.1, wild-type FOXO1, or two mutants FOXO1-S256A and FOXO1-AAA in the presence or absence of Skp2. At 48 h after transfection, transfected viable cells were photographed under both UV and transmitted light (A) and quantified by using trypan blue (B). (C and D) 786-O cells were cotransfected with pEGFP vector along with control vector pcDNA3.1, wild-type FOXO1, or Skp2. At 48 h after transfection, cells were fixed briefly with formaldehyde and subjected to two-color FACS analysis for cell cycle distribution (C) and lysed for immunoblotting with FOXO1 and Skp2 antibodies (D).
Loss of the FOXO1 Protein Is Inversely Correlated with Skp2 Overexpression in Mouse Lymphomas. The Skp2 protein is overexpressed in high-grade lymphomas in the human (29). Skp2 is consistently up-regulated in T-cell lymphomas that develop in CBP knockout mice. However, disruption of one allele of p27KIP1 in this lymphoma model alleviates the need for Skp2 up-regulation (22). Consistent with the idea that Skp2 up-regulation causes down-regulation of FOXO1, we found that CBP null lymphomas with higher Skp2 have lower FOXO1 (Fig. 6A), whereas lymphomas with lower Skp2 had higher FOXO1 (Fig. 6B). No consistent increase in phosphorylation of Akt was detected in CBP null lymphomas (Fig. 6A). To further examine the causal effect of Skp2 on the low level of FOXO1 protein in T cells, a pool of Skp2 siRNAs was transfected into Jurkat malignant lymphoblasts. Knockdown of Skp2 by siRNAs resulted in an increase of the endogenous FOXO1 protein (Fig. 6C). These findings suggest that elevated Skp2 is a major factor responsible for the degradation of FOXO1 in lymphomagenesis.
Discussion
The molecular mechanisms by which FOXO1 and its related proteins FOXO3a and FOXO4 participate in tumorigenesis are not fully elucidated. Chromosomal translocation of FOXO1 (FKHR) with PAX3 has been demonstrated in human rhabdomyosarcoma (30). Moreover, activation of the protein kinases Akt and IκB kinase β in cancer cells promotes phosphorylation and nuclear exclusion of the FOXO proteins, thereby inhibiting their tumor suppressor function (3, 6). In the present study, we provide evidence that the cellular level of the FOXO1 protein is tightly controlled by the Skp1/Cul1/F-box protein component Skp2 and that the tumor suppressor activity of FOXO1 is abolished by the increased expression of Skp2 in cancer cells. We further demonstrate that the FOXO1 proteins are lost in mouse primary lymphomas and inversely correlated with Skp2 overexpression. Thus, Skp2-mediated inhibition of FOXO1 represents a previously uncharacterized mechanism of inactivation of this tumor suppressor protein during tumorigenesis. Support of this concept can also be found in the literature, where rhabdomyosarcoma cell lines that contain the PAX3-FKHR translocation express no FKHR protein (30). Moreover, cell transformation by PAX3-FKHR results in an increase in the cellular level of Skp2 (31). Recent transgenic studies suggest that loss of one allele of the FOXO1 gene is indeed a pivotal event in the genesis of this tumor (32). Therefore, it is conceivable that PAX3-FKHR translocation may trigger the complete loss of the FOXO1 protein by means of the Skp2-mediated degradation.
Overexpression or amplification of Skp2 has been documented in a large number of human cancers, including prostate cancer, breast cancer, lymphoma, small cell lung cancer, oral squamous cell carcinoma, and colorectal carcinoma. Transgenic expression of Skp2 in mice leads to tumor formation in a variety of tissues, further suggesting that Skp2 is oncogenic. However, the signaling pathways that mediate this oncogenic effect are not fully understood. p27KIP1 is a putative tumor suppressor gene and is targeted by Skp2 for degradation. The p27KIP1 protein has been shown to be consistently lost in the spontaneous lymphomas of CBP knockout mice (22). Indeed, loss of p27KIP1 is a key player in this model, because deletion of p27KIP1 accelerates the rate of tumor formation in CBP–/– mice. The mechanism of p27KIP1 reduction in these tumors involves decreased gene transcription and increased protein degradation (22). Increased degradation of p27KIP1 can be attributed to the elevated levels of Skp2 in tumors (22). Because p27KIP1 is a direct transactivation target of FOXO1 (28), it is conceivable that decreased transcription of p27KIP1 results from a loss of FOXO1 protein. Transgenic expression of Skp2 induces carcinoma in the mouse prostate (33). However, mice deficient in p27KIP1 do not develop prostate cancer (34). These findings suggest a p27KIP1-independent mechanism acting downstream of Skp2 in the prostate. We demonstrate that overexpression of Skp2 results in a decrease in the level of FOXO1 protein in the prostate cancer cell line LNCaP. Importantly, the FOXO1-induced loss of viability of LNCaP cells was abolished by Skp2 expression. Together with the observation that Skp2 proteins are elevated in human primary prostate tumors (26, 35), our findings suggest that Skp2 may play a key role in human prostate cancer by promoting the degradation of the FOXO1 protein.
Another important component of our findings is that Skp2-mediated degradation of FOXO1 requires its phosphorylation at Ser-256 by Akt. We provide additional evidence that FOXO1 degradation is under the dual control of Akt and Skp2. Thus, it is not surprising that increased levels of Skp2, activated Akt, or both can accelerate the degradation of FOXO1. Indeed, it has been reported that activation of Akt promotes the degradation of FOXO1 in HepG2 cells, FL5.12 murine pro-B lymphocytic cells, and chicken embryo fibroblasts, although the E3 ligase responsible the Akt-promoted degradation of FOXO1 was unknown (7, 17, 18). Importantly, we demonstrate that increased expression of Skp2 alone can result in a marked decrease in the levels of FOXO1 protein, thus abrogating a requirement for the gain of function of Akt. A similar scenario may exist in lymphomas triggered by CBP knockout in mice, because no increase in Akt phosphorylation was observed in those tumors compared with normal tissues. Thus, we envisage a model whereby Skp2 promotes tumorigenesis via the degradation of FOXO1 (Fig. 6D). In normal nonmalignant cells, Akt activity is finely controlled by the two antagonizing signals phosphatidylinositol 3-kinase and PTEN, where a balance between phosphorylated and unphosphorylated forms of FOXO1 is maintained. During tumor progression with the elevated cellular level of Skp2, the phosphorylated form of FOXO1 starts to be degraded. This triggers new rounds of phosphorylation and degradation of FOXO1 and eventually leads to a crash in the protein, thereby eliminating the tumor suppressor function of FOXO1 and promoting tumor formation. Thus, our findings suggest that Skp2 promotes the degradation of FOXO1 independently of the gain of function of Akt. This represents a pathway by which FOXO1 may be inactivated during tumorigenesis.
Conclusion
Our studies demonstrate that Skp2 interacts with and promotes degradation of FOXO1. Through this mechanism, the tumor suppressor function of FOXO1, including induction of G1 arrest and triggering of cell death, is abolished by Skp2 expression. We also demonstrate that loss of FOXO1 protein is inversely correlated with gain of Skp2 protein in a mouse lymphoma model. The finding that Skp2-mediated degradation and loss of function of FOXO1 can be reversed by proteasome inhibitors, even in the presence of overexpressed Skp2, suggests that this signaling network is a viable therapeutic target in human cancers, especially those with high levels of Skp2.
Supplementary Material
Acknowledgments
We thank K. L. Guan, H. Zhang, and D. Bohmann for plasmids. This work was supported by National Institutes of Health Grants DK65236, DK60920, and CA91956 and the T. J. Martell Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: siRNA, small interfering RNA; HA, hemagglutinin.
References
- 1.Medema, R. H., Kops, G. J., Bos, J. L. & Burgering, B. M. (2000) Nature 404, 782–787. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy, S., Nakamura, N., Sansal, I., Bergeron, L. & Sellers, W. R. (2002) Cancer Cell 2, 81–91. [DOI] [PubMed] [Google Scholar]
- 3.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 4.Modur, V., Nagarajan, R., Evers, B. M. & Milbrandt, J. (2002) J. Biol. Chem. 277, 47928–47937. [DOI] [PubMed] [Google Scholar]
- 5.Gilley, J., Coffer, P. J. & Ham, J. (2003) J. Cell Biol. 162, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu, M. C., Lee, D. F., Xia, W., Golfman, L. S., Ou-Yang, F., Yang, J. Y., Zou, Y., Bao, S., Hanada, N., Saso, H., Kobayashi, R. & Hung, M. C. (2004) Cell 117, 225–237. [DOI] [PubMed] [Google Scholar]
- 7.Aoki, M., Jiang, H. & Vogt, P. K. (2004) Proc. Natl. Acad. Sci. USA 101, 13613–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang, E. D., Nunez, G., Barr, F. G. & Guan, K. L. (1999) J. Biol. Chem. 274, 16741–16746. [DOI] [PubMed] [Google Scholar]
- 9.Biggs, W. H., III, Meisenhelder, J., Hunter, T., Cavenee, W. K. & Arden, K. C. (1999) Proc. Natl. Acad. Sci. USA 96, 7421–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kops, G. J., de Ruiter, N. D., De Vries-Smits, A. M., Powell, D. R., Bos, J. L. & Burgering, B. M. (1999) Nature 398, 630–634. [DOI] [PubMed] [Google Scholar]
- 11.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 12.Scheffner, M., Werness, B. A., Huibregtse, J. M., Levine, A. J. & Howley, P. M. (1990) Cell 63, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 13.Carrano, A. C., Eytan, E., Hershko, A. & Pagano, M. (1999) Nat. Cell Biol. 1, 193–199. [DOI] [PubMed] [Google Scholar]
- 14.Tedesco, D., Lukas, J. & Reed, S. I. (2002) Genes Dev. 16, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamura, T., Hara, T., Kotoshiba, S., Yada, M., Ishida, N., Imaki, H., Hatakeyama, S., Nakayama, K. & Nakayama, K. I. (2003) Proc. Natl. Acad. Sci. USA 100, 10231–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- 17.Plas, D. R. & Thompson, C. B. (2003) J. Biol. Chem. 278, 12361–12366. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzaki, H., Daitoku, H., Hatta, M., Tanaka, K. & Fukamizu, A. (2003) Proc. Natl. Acad. Sci. USA 100, 11285–11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mamillapalli, R., Gavrilova, N., Mihaylova, V. T., Tsvetkov, L. M., Wu, H., Zhang, H. & Sun, H. (2001) Curr. Biol. 11, 263–267. [DOI] [PubMed] [Google Scholar]
- 20.Musti, A. M., Treier, M. & Bohmann, D. (1997) Science 275, 400–402. [DOI] [PubMed] [Google Scholar]
- 21.Huang, H., Cheville, J. C., Pan, Y., Roche, P. C., Schmidt, L. J. & Tindall, D. J. (2001) J. Biol. Chem. 276, 38830–38836. [DOI] [PubMed] [Google Scholar]
- 22.Kang-Decker, N., Tong, C., Boussouar, F., Baker, D. J., Xu, W., Leontovich, A. A., Taylor, W. R., Brindle, P. K. & van Deursen, J. M. (2004) Cancer Cell 5, 177–189. [DOI] [PubMed] [Google Scholar]
- 23.Huang, H., Muddiman, D. C. & Tindall, D. J. (2004) J. Biol. Chem. 279, 13866–13877. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. R., Ramos, S. M., Ko, A., Masiello, D., Swanson, K. D., Lu, M. L. & Balk, S. P. (2002) Mol. Endocrinol. 16, 85–99. [DOI] [PubMed] [Google Scholar]
- 25.Siegel, D., Anwar, A., Winski, S. L., Kepa, J. K., Zolman, K. L. & Ross, D. (2001) Mol. Pharmacol. 59, 263–268. [DOI] [PubMed] [Google Scholar]
- 26.Carrano, A. C. & Pagano, M. (2001) J. Cell Biol. 153, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura, N., Ramaswamy, S., Vazquez, F., Signoretti, S., Loda, M. & Sellers, W. R. (2000) Mol. Cell. Biol. 20, 8969–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latres, E., Chiarle, R., Schulman, B. A., Pavletich, N. P., Pellicer, A., Inghirami, G. & Pagano, M. (2001) Proc. Natl. Acad. Sci. USA 98, 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galili, N., Davis, R. J., Fredericks, W. J., Mukhopadhyay, S., Rauscher, F. J., III, Emanuel, B. S., Rovera, G. & Barr, F. G. (1993) Nat. Genet. 5, 230–235. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L. & Wang, C. (2003) J. Biol. Chem. 278, 27–36. [DOI] [PubMed] [Google Scholar]
- 32.Accili, D. & Arden, K. C. (2004) Cell 117, 421–426. [DOI] [PubMed] [Google Scholar]
- 33.Shim, E. H., Johnson, L., Noh, H. L., Kim, Y. J., Sun, H., Zeiss, C. & Zhang, H. (2003) Cancer Res. 63, 1583–1588. [PubMed] [Google Scholar]
- 34.Fero, M. L., Rivkin, M., Tasch, M., Porter, P., Carow, C. E., Firpo, E., Polyak, K., Tsai, L. H., Broudy, V., Perlmutter, R. M., et al. (1996) Cell 85, 733–744. [DOI] [PubMed] [Google Scholar]
- 35.Yang, G., Ayala, G., De Marzo, A., Tian, W., Frolov, A., Wheeler, T. M., Thompson, T. C. & Harper, J. W. (2002) Clin. Cancer Res. 8, 3419–3426. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.