Abstract
Five years after the completion of the sequence of the Drosophila melanogaster genome, the number of protein-coding genes it contains remains a matter of debate; the number of computational gene predictions greatly exceeds the number of validated gene annotations. We have assembled a collection of >10,000 gene predictions that do not overlap existing gene annotations and have developed a process for their validation that allows us to efficiently prioritize and experimentally validate predictions from various sources by sequencing RT-PCR products to confirm gene structures. Our data provide experimental evidence for 122 protein-coding genes. Our analyses suggest that the entire collection of predictions contains only ≈700 additional protein-coding genes. Although we cannot rule out the discovery of genes with unusual features that make them refractory to existing methods, our results suggest that the D. melanogaster genome contains ≈14,000 protein-coding genes.
Keywords: gene number, validation, genome annotation
The total number of protein-coding genes in the Drosophila melanogaster genome remains a subject of debate. Whereas those who curated the D. melanogaster genome concluded that the annotated 13,659 genes in the 3.1 release likely constitute 95% of all protein-coding genes (1), others researchers have concluded that many, possibly thousands, of protein-coding genes remain unannotated (2). Two issues have fueled the debate surrounding gene number in D. melanogaster: the large numbers of computational gene predictions located within intergenic regions and varying standards of experimental evidence for concluding that a gene prediction corresponds to a real gene.
As of release 3.1, ≈50% of the D. melanogaster genome is intergenic. Running the gene prediction program genscan (3) on every intergenic region in the D. melanogaster genome results in 10,644 gene predictions spread amongst 62 megabases (Mb) of annotation-free sequence. Surely some of these predictions are real, but how many? The best way to answer this question is to subject a representative sample of the gene predictions to some validation procedure.
The design and interpretation of experiments intended to assay expression of genes that have been predicted computationally have become controversial. One approach is to rely on hybridization to microarrays or RT-PCR assays for transcript expression (2), with the detection of a product by agarose gel electrophoresis taken as confirmation of the corresponding gene prediction. However, as our results show, unless the diagnostic PCR product includes a splice junction, amplification of residual genomic DNA and detection of unprocessed transcripts may lead to false verifications of gene predictions. It has also been critical to determine the sequence, and not just the size, of the PCR products (4).
One way to obtain spliced cDNAs for sequencing is to perform RT-PCR with a 3′-oligo(dT) primer and an upstream PCR primer located in the prediction's 5′-most exon. The advantages of this approach are that it requires only a single, prediction-specific oligonucleotide (oligo) and that the sequenced PCR product provides much useful information about transcript structure. A disadvantage is that the limited processivity of reverse transcriptase can make it difficult to obtain products from long transcripts. Another approach (5–7), the one that we have pursued, involves designing two PCR primers to a pair of flanking exons. This approach circumvents the problems associated with obtaining PCR products from long transcripts, but it provides less information about transcript structure.
One drawback to all of these approaches is that they are labor intensive. Microarray-based validation assays offer a possible alternative in this regard. Hild et al. (2) recently reported the identification and validation of 2,636 previously unrecognized D. melanogaster genes, based on a microarray-based approach that involved hybridizing randomly primed cDNA against probes corresponding to a large set of fgenesh predictions. Microarray-based approaches scale well when large numbers of gene predictions need to be verified. As our results show, however, one drawback of this approach is that it is unable to distinguish “background” transcription from the generation of properly processed, discrete transcripts.
We have constructed a pipeline for validation of gene predictions, no matter what their source (computational gene-finder, human annotation, etc.) that allows us to subject every potential gene to the same procedures of oligo design and validation. This approach produces consistent and standardized results and makes possible more accurate estimates of how many genes in D. melanogaster remain unannotated.
Because testing every prediction in our collection would be very labor intensive, we also sought to develop the means to identify and prioritize for validation those predictions most likely to test true. Toward this end, we explored both homology and gene structure as means to prioritize predictions for validation. Below, we describe our results to date on a collection of genscan and fgenesh predictions, existing genome annotations, and gene predictions recently “confirmed” by a microarray-based validation approach (2). We conclude that our collection of gene predictions contains only ≈700 additional protein-coding genes.
Materials and Methods
Priority Scores and Primer Design. Our validation strategy required us to identify the best pair of exons within which to locate primers. We developed a fuzzy logic (8) algorithm to accomplish this task. Fuzzy-logic algorithms have been used with success in a variety of problems, from antiskid braking systems for Boeing aircraft to genomic analysis (9). The approach is especially well suited to situations wherein little training data exists but considerable expert knowledge is available (see the supporting information, which is published on the PNAS web site). We used primer3 (10) to pick primers that were within 50 bp of the predicted exon splice junction.
Partitioning Subsets of Predictions for Verification. For purposes of our analyses, we defined intergenic regions as those regions between two genes on either strand. Introns, even if they contained a gene, were classified as genic regions. genscan (3) was run with the Arabidopsis parameter file. The genscan and fgenesh predictions were loaded into a release 3.1 D. melanogaster annotation gadfly database (11) as putative annotations. The Heidelberg predictions were obtained as gene finding format (GFF) files (M. Hild, personal communication). Using a bioperl script based on BioModel::IntersectionGraph (12), we identified genscan and fgenesh predictions that did not overlap any release 3.1 genes or repeatmasker results (www.repeatmasker.org), then identified Heidelberg predictions that did not overlap any of the genscan or fgenesh predictions that were being tested or release 3.1 genes or transposons. Predictions with conserved splice junctions were identified by using cgl, a software library designed to facilitate such comparisons (unpublished work).
RNA Isolation. RNA was isolated with RNAwiz reagent (Ambion, Austin, TX) from the following developmental stages from an isogenic y; cn bw sp strain: 0- to 24-h embryo (derived from 4- to 6-h subcollections), late third-instar larva (L3), mixed-stage pupa, and mixed-age adult. Poly(A)+ RNA was selected by using Poly(A)+ Purist kits (Ambion).
See the supporting information for RT-PCR and DNA sequencing conditions.
Alignment of Oligos and Sequenced RT-PCR Products to the Genome. RT-PCR product sequence reads were quality-trimmed as described in ref. 14. Short (<160 bp) and poorer-quality sequence traces were read manually. For other low-quality reads, we used phred to call bases and score quality. RT-PCR and oligo sequences were aligned to the genomic sequence by using sim4wrap (11). Matches were filtered by using the berkeley output parser (11). The control, genscan, fgenesh, and Heidelberg (2) predictions and associated oligos were loaded into a modified release 3.1 gadfly database, and each prediction was visualized with aligned RT-PCR products and oligos by using the apollo genome annotation browser and editor (15). In some cases, poor RT-PCR product sequence quality required manual National Center for Biotechnology Information blastn (16) comparison against the release 3 genomic sequence. When the sequence corresponded to a spliced mRNA transcript, the prediction was scored as “sequence validated” and checked against release 3.2 of the annotated genome in the FlyBase database (17). For those validated genes not annotated in release 3.2, a gene model was curated and communicated to FlyBase (17). See the supporting information for details of replication of RT-PCRs described by Hild et al. (2) and analysis of RT-PCR products of unexpected size.
Results and Discussion
A Collection of >10,000 Nonoverlapping Gene Models Representing Potentially Unannotated Genes. Because our pipeline for gene prediction validation is independent of any particular source of predictions, we assembled a large collection of potentially unannotated genes derived from multiple sources. Our primary source consisted of 10,644 genscan predictions lying within intergenic regions in the D. melanogaster 3.1 release. These predictions were obtained by running genscan over the complete set of intergenic regions whose repeats had been masked by using repeatmasker (www.repeatmasker.org). Excluding all single exon predictions (because these are unsuitable substrates for our validation procedure) resulted in a set of 9,811 multiexon genscan predictions. We then sought additional predictions that did not overlap any of our genscan predictions from a set of 1,167 fgenesh predictions produced by using a modified version of the program especially trained for use on D. melanogaster (18), which gave an additional 325 predictions, for a total of 10,136. To these we added 1,266 multiexon gene models located in intergenic regions reported by Hild et al. (2) to consist of previously unrecognized protein-coding genes that had been validated by the microarray-based approach; however, all but 37 of these overlapped one of the 10,136 predictions generated by genscan or fgenesh.
Experimental Strategy. Our experimental approach required that we design two exon-specific primers for each gene model, but to which pair of exons? We reasoned that regardless of the particulars of the gene-finder that produced the prediction, the longer the open-reading frame, the less likely it is to occur by chance, and, therefore, the more likely it is to actually encode a protein. The fact that experimentally confirmed exons tend to be longer than genscan-predicted exons in gene-containing regions and much longer than the genscan-predicted exons in intergenic regions (data not shown) supports this hypothesis. If the gene model is the product of human annotation, its longer exons are still the better choice, because they afford more leeway for optimal primer placement with regard to sequence complexity and melting temperatures. We also sought whenever possible to design primers to exon pairs that flanked introns whose length was as close as possible to the modal intron length in D. melanogaster, because these exon pairs, we reasoned, comprise the portion of the prediction most likely to be correct. Finally, given a prediction with two otherwise identical sets of exon pairs flanking introns of the same length, we selected the 3′-most exon pair, which would be relevant if using oligo(dT) to prime reverse transcription reactions. However, because we used gene-specific primers for both the RT and PCR steps, this criterion was not pertinent to the experiments described here. Given these three criteria (exon lengths, intron lengths, and distance of the exon pair from the prediction's 3′-end) we were then faced with the issue of how to weigh each criterion when choosing which exons to use for primer design. To address this problem, we developed a fuzzy-logic (8) algorithm that considers ideal two exons, both ≈300 nt or longer, that flank an intron of >70 but <200 nt in length. The algorithm considers each exon pair and scores it relative to this ideal. We then used primer3 (10) to design a matched pair of primers to the high-scoring pair of exons. One consequence of this approach is that each gene model in our collection now had a score associated with its potential PCR product (see the supporting information) independent of the original gene prediction program. If our reasoning as to what constituted a likely pair of real exons was correct, then, on average, those gene models whose PCR products were assigned a high score would be more likely to validate than would those gene models whose PCR products had received a low score. Thus, we were able to investigate whether these scores would prove useful not only in estimating the number of true positives in a collection of gene models, but also for prioritizing predictions for validation; hence, we dubbed them “priority scores.”
Our validation procedure entailed an RT-PCR assay with these gene-specific primers to amplify a portion of a predicted transcript. Both poly(A)+ and total RNA isolated from whole animals representing a broad range of Drosophila stages served as templates for the RT-PCRs. Any RT-PCR product whose size corresponded to the respective prediction was gel-purified where necessary and then sequenced and aligned to the genome to verify its identity and exon structure. In addition, all unique RT-PCR products, regardless of relation to predicted size, were analyzed at this level. In a pilot experiment, 105 of the RT-PCRs that produced no band were sequenced; of these, none verified the tested prediction, so we decided not to sequence reactions that did not produce a band on an agarose gel. Only PCR products reflecting transcript splicing were considered to verify gene predictions. If a reaction did not yield specific products, that negative result was only included in our analysis in cases where the corresponding primers were shown to produce the expected amplicon from genomic DNA in a control PCR (see the supporting information). We are aware that this rigorous requirement will result in our discarding a significant number of true negatives and thus result in an overestimate of the percentage of gene models that validate, because many of the primer pairs will fail to amplify genomic DNA simply because the lengths of intervening introns make the amplicons prohibitively large. In fact, the median expected size for genomic products for those reactions that failed this test (2,824 bp) was >5 times that of those that passed (510 bp).
The Collection Was Partitioned into Four Subsets for Validation Purposes. Because we originally had no data as to how indicative our priority scores might be of a gene prediction's likelihood to be experimentally validated, we subdivided our working prediction collection on the basis of sequence homology rather than priority score, hypothesizing that frequency of experimental validation would also correlate with extent of interspecies sequence conservation (Table 1). To identify genes with sequence conservation, we first searched each of the 9,811 genscan and 375 fgenesh predictions in our collection against the Drosophila pseudoobscura genome by using tblastn in an effort to identify those predictions having at least two conserved exons. We identified 559 predictions satisfying this criterion.
Table 1. Sequence validation rates of predictions.
| Gene prediction set | Total no. of predictions* | No. of predictions tested | No. sequence validated (%)† |
|---|---|---|---|
| sjc set | 197 | 171 | 64 (37.4) |
| Heidelberg set‡ | 1,266 | 160 | 18 (11.3) |
| Homol-2 set | 362 | 209 | 28 (13.4) |
| Homol-0 set§ | 9,577 | 204 | 12 (5.9) |
| Total predictions | 11,402 | 744 | 122 (16.4) |
| Controls | 159 | 159 | 154 (96.9) |
See Table 2 for specific numbers from genscan vs. fgenesh predictions
Gene predictions were considered validated if the aligned sequence of the PCR product was consistent with a spliced gene model in the region of the prediction
Only 1,266 multiexon predictions from the 2,636 predictions described by Hild et al. (2) were considered for analysis, and, of these, we tested only the 160 with the highest priority scores that did not overlap any genscan or fgenesh predictions tested in the other sets
The homol-0 set was selected to be representative of the full range of priority scores
Next we subjected each of these predictions to a more stringent analysis: testing for evidence of conserved splice junctions at the same position in the D. pseudoobscura genome. We termed those that met this criterion the “sjc set” (“splice-junction conserved”) because each has at least two adjacent exons, both with homology to D. pseudoobscura, flanking conserved donor and acceptor sites. We tested 171 (Table 1). Those predictions with two conserved exons but without evidence for a conserved splice junction were called the “homol-2 set,” and we tested 209. For both the sjc set and homol-2 set, we forced our exon selector algorithm to design oligos to the best-conserved pair of exons and kept track of their priority scores.
The remaining 9,577 genscan and fgenesh predictions with at most a single exon with D. pseudoobscura conservation constitute the “homol-0 set.” We selected predictions such that one-quarter had priority scores >75, one-quarter had scores between 75 and 50, one-quarter had scores between 50 and 25, and one-quarter had scores <25. We tested a total of 204 predictions.
We then tested 160 genes from the 1,266 previously unrecognized multiexon genes reported by Hild et al. (2) to have been verified based on expression detected by microarrays. We chose those with the highest priority scores that did not overlap any of the gene predictions for testing from the sjc, homol-0, or homol-2 datasets; we termed this the “Heidelberg set.”
Finally, we collected a set of 159 existing genome annotations for which we were particularly certain of the gene structures. All have at least one published cDNA sequence confirming their annotated transcript's intron–exon structure and no conflicting EST or cDNA sequence evidence. These genes constituted our control set.
Validation Results. The results in Table 1 show that whereas gene models with homology in D. pseudoobscura are more likely to test true, simple sequence conservation (across the evolutionary distance separating D. melanogaster from D. pseudoobscura) is not as useful in determining that a gene prediction corresponds to a real gene as exon–intron structure conservation. Gene predictions having at least one conserved splice junction in D. pseudoobscura (sjc set) were >2.5 times as likely to validate as those lacking a conserved splice junction but having two (partially) conserved exons (homol-2 set). In this regard, our results accord well with previous analyses of human and rodent predictions (4), which also found conservation of gene structure to be the most powerful in silico diagnostic of how likely a gene prediction was to reflect the structure of a real gene.
Only 11% of the Heidelberg set was validated by our criteria. We do not believe this low rate of validation reflects limitations of our experimental approach because we were able to confirm the existence of nearly all (154/159) of the genome annotations contained in our control set. Based on EST frequency data (35 of the 159 have no associated ESTs), we know that not all of the genes in our control set were highly expressed genes, yet they were still detected in our assay. We also observed a relatively high percentage of verified genes, 37.4%, in the sjc set. To verify the sensitivity of our RT-PCR conditions, we replicated the secondary RT-PCR validation protocol described by Hild et al. (2) for 22 gene models for which they reported positive RT-PCR results; we also were able to obtain positive results for 21 of 22 (95%) of these, confirming that our assay conditions are able to detect previously unrecognized Drosophila transcripts at a comparable rate, and none of these produced a band in control reactions lacking reverse transcriptase. Thus, although we were able to confirm that a high percentage of the Heidelberg gene models overlap transcribed regions in the genome, our results suggest that only a small fraction of these transcribed regions give rise to spliced mRNAs. Therefore, we do not dispute the data reported by Hild et al. (2), only their interpretation that a high percentage of these gene models represent previously unrecognized protein-coding genes.
The distributions of priority scores associated with the four different sets also suggest that the Heidelberg set contains few previously unannotated protein-coding genes. As can be seen in Fig. 1, the priority score distribution associated with our control set is very different from that of the 9,811 multiexon genscan predictions included in our collection. The mode for the priority score distribution for the control set was 80, twice that of the genscan predictions; likewise, validated predictions (sjc set, homol-2 set, and homol-0 set) also tend to have higher priority scores (Fig. 1B). On the other hand, the distribution of priority scores for the Heidelberg set (Fig. 1C) is nearly indistinguishable from that of the 9,811 genscan predictions. Thus, the data shown in Fig. 1 provide additional support for our conclusion that the Heidelberg set contains few genes that produce spliced mRNAs. Taken together, these facts suggest that the microarray-based validation procedure used by Hild et al. (2) has a high false positive rate and that priority scores and homology, especially conservation of a splice junction, are better indicators that a gene prediction produces a spliced mRNA.
Fig. 1.
Priority scores have predictive value. (A) The priority score distribution for all genscan predictions in our collection (gray bars) vs. the priority score distribution of the control set (black bars). (B) All genscan predictions (gray bars) vs. all validated genscan predictions (black bars). (C) All genscan predictions (gray bars) vs. the Heidelberg set (black bars).
Overall, homol-0 set gene models had the lowest validation rate (5.9%). Recall that for testing, we randomly selected predictions such that each quartile had scores of 0–25, 25–50, 50–75, or 75–100, and we tested a total of 204 predictions. None of the predictions having priority scores <25 validated; 3.3% of the predictions with priority scores between 25 and 50 validated; 10% of the predictions with priority scores >50 but <75 validated; and 10.2% of those with scores >75 validated. Overall, the validation rate of the homol-0 set was half that of the Heidelberg set. We chose for inclusion in the Heidelberg set gene models from Hild et al. (2) having high priority scores; thus, the higher percentage of verified genes in the Heidelberg set reflects this selection bias, because it is similar to the validation rate of the highest scoring homol-0 predictions.
Estimating the Number of Protein-Coding Genes That Remain Unannotated. We have experimentally tested 744 gene models: ≈7% of our total collection. We can estimate the total number of protein-coding genes contained in our collection by using these results (Table 1) to estimate what percentage of the 9,373 homol-0 set, 153 homol-2 set, and 26 sjc set predictions, which were not directly tested, correspond to real genes. These extrapolations suggest that 553 homol-0, 21 homol-2, and 8 sjc gene predictions (a total of 582) would validate by our approach. Including the 122 experimentally validated gene models gives an estimate that 705 protein-coding genes remained unannotated at the time of the 3.1 release. (Ninety-seven percent of the Heidelberg predictions overlap one of our gene prediction sets and are thus included in the above estimate.)
As Fig. 1 demonstrates, priority scores do indeed indicate how likely a gene model is to be validated, regardless of the means used to produce it. To obtain a second estimate of the remaining number of unannotated genes, we investigated the use of priority scores to estimate the number of real genes contained in a collection of predictions. To do so, we first partitioned the remaining untested 9,552 genscan and fgenesh gene predictions and the remaining untested 1,106 gene models from Hild et al. (2) into four bins based on their priority scores: <25, 25–50, 50–75, and 75–100. We then multiplied the number of genes in each bin by the observed frequency of validation for our 204 tested homol-0 genes: 0 for the first quartile, 0.033 for the second, 0.1 for the third, and 0.102 for the fourth. This approach gave an estimate of 404 additional real genes among the untested genscan and fgenesh predictions; 20 additional genes are predicted from the Heidelberg set, but, as mentioned above, these are likely to be redundant of those in the genscan and fgenesh predictions. Adding the 122 experimentally validated gene models places the total number of unannotated genes at ≈572, a 20% decrease compared with the number of 705 derived by extrapolation of the data in Table 1 as described above. Nevertheless, because we wished to obtain a reasonable upper bound for the number of genes our collection might contain, we choose 705 as a working estimate for the additional considerations presented below.
A factor not addressed by any of our analyses is how many single-exon genes remain unannotated. Because these are not substrates of our validation procedure, none were tested. Running genscan over all 62 Mb of intergenic sequence produced 833 single exon genes. By definition, these single exon genes fall into the homol-0 class. Because there is no evidence that single exon gene predictions are more accurate than multiexon predictions, the validation rate (5.9%) of the homol-0 genes would give 49 additional genes, to bring the estimated number of unannotated protein-coding genes in the 3.1 release to ≈750.
We also considered the possibility that some additional genes might reside among the unvalidated sjc predictions, but we do not believe this to be true. To address this possibility, however, we searched every sjc prediction's predicted protein against the GenBank nonredundant protein database. Ninety percent of the validated sjc predictions have a significant hit (expect <1e-4; default wu-blastp parameters) to a non-Drosophila protein, whereas only 21% of the unverified sjc predictions do. Thus, the validated sjc predictions are highly enriched for protein homology relative to the unvalidated remainder; this finding suggests that much of the homology associated with the unvalidated sjc predictions is noncoding. Moreover, given the extensive genomic sequence conservation between the two Drosophila species and the large numbers of false positives in our collection of gene models, we find it unsurprising that 1% (133/10,136) of the genscan and fgenesh predictions would have significant, noncoding homology to D. pseudoobscura, the details of which were consistent with conserved GT and AG dinucleotides at predicted exon borders. Thus, we believe that most of the unvalidated sjc predictions are indeed true negatives, although we cannot rule out the possibility that some fraction of them may correspond to real genes whose expression is spatially or temporally very limited.
A final issue to consider is how many of these 750 or so genes represent previously unrecognized genes as opposed to merely unannotated 5′ and 3′ exons of existing annotations. It seems reasonable to assume that some of our validated predictions are, in fact, unannotated portions of known genes. Assuming that 10% are missed exons rather than genes, a conservative estimate, considering that Stolc et al. (13) have concluded that the number may be as high as 32% (because 369 of 1,155 expressed genscan exons they tested belonged to existing annotations), would bring the number of genes in our extrapolation to 675.
Priority Scores Provide a Means to Manage a Validation Pipeline in a Cost-Effective Manner. The large numbers of predictions in our collection relative to the small number of real genes contained within it means that identifying which of the remaining 10,658 untested predictions is real is potentially an inefficient and labor intensive task. In principle, the scores provided by each gene-finder might be used to prioritize predictions for validation, but if the models are derived from several gene-finders, relating individual gene-finder scores to one another becomes a complex problem. We therefore examined the utility of the priority scores assigned by our primer-design algorithm as a general method to score gene models independent of their original source. As Fig. 2 shows, 50% of all true positives could be obtained from our collection of 10,136 genscan and fgenesh predictions in the first 1,000 PCRs by using priority scores; choosing models randomly would require 5,068 PCRs to obtain the same number of true positives. Thus, our approach can be used to prioritize predictions for validation in a cost-effective manner such that a greater number of previously unrecognized genes can be identified with fewer PCRs.
Fig. 2.
Priority scores can be used to prioritize gene predictions for validation in a cost-effective manner. Fraction of all validated gene predictions obtained (y axis) vs. the number of PCRs performed (x axis) if choosing randomly (black line) or choosing on the basis of priority score (black diamonds) from the collection of 10,136 genscan and fgenesh predictions.
Conclusions
Our goal was to obtain an accurate estimate of the number of predicted protein-coding genes left unannotated in the D. melanogaster genome as of release 3.1. Toward this end, we assembled a large collection of putative unannotated, multiexon protein-coding genes located within intergenic regions: 9,811 genscan predictions, 375 fgenesh predictions, and 1,266 purportedly previously unrecognized protein-coding genes that had been validated by a microarray-based approach (2). To consider a prediction experimentally verified, we required that the sequence of a corresponding RT-PCR product align specifically to the prediction and reveal evidence of transcript splicing.
One drawback to our validation approach is that it is labor intensive. Our collection of gene predictions was large and, we suspected, contained few true positives. These issues made highly desirable any in silico method capable of identifying gene predictions likely to be real, because it would allow us to test these predictions first. Accordingly, we explored several such measures: homology, conservation of gene structure, and priority scores. Overall, those predictions having at least one conserved splice junction when compared with D. pseudoobscura validated at the highest rate, 37.4% (Table 1). We found simple sequence conservation to be a much less reliable in silico indicator of gene predictions' validation rates than conservation of gene structure. Only 13.4% of gene models with two partially conserved exons but lacking evidence of a conserved splice site in D. pseudoobscura verified. Expression of spliced transcripts was detected for only 11.3% of the predictions represented by the Heidelberg set. It appears that our requirement that diagnostic RT-PCR products represent spliced transcripts accounts for the difference in our validation rate from that of the Heidelberg group. Although those predictions may overlap transcribed regions of the genome, our evidence suggests that only ≈11% of those predictions actually correspond to mRNAs. The generally low priority scores assigned to the Heidelberg set by our methods are further consistent with our verification rate.
Overall, 16.4% of the gene predictions tested proved valid. We believe that these 122 previously unrecognized transcripts comprise ≈18% of all real protein-coding genes within our collection of gene models. Our reasons for believing so are twofold. First, the high rates of validation associated with the sjc and homol-2 sets suggest that homology, especially conservation of gene structure, is a good indicator that a gene prediction actually reflects a real gene. We tested the majority of such predictions; therefore, the 9,552 predictions that remain untested are probably greatly depleted for true positives. Second, the low rate of validation (5.9%) associated with the homol-0 set independently suggests the same conclusion. Extrapolating from the validation rate of the homol-0 set or using the more sophisticated method that makes use of priority scores gives similar results; both suggest that our collection of gene predictions contains ≈700 protein-coding genes. Analysis of our findings compared with the more recent FlyBase annotation release 3.2 (17) suggested that 34% of our 122 previously unrecognized genes were included in this release. Thus, we conclude that our collection of gene predictions provides no evidence to support the notion that D. melanogaster has more than ≈14,000 protein-coding genes.
We discovered that sequencing was crucial to our verification procedure. For a small fraction of our predictions that gave an expected RT-PCR product size on an agarose gel, the sequenced products aligned not to the predictions but to other regions of the genome, usually highly expressed, previously annotated genes, suggesting that these products were due to mispriming on abundantly expressed genes. Another 153 RT-PCRs yielded unique products of unexpected size (see the supporting information). Sequencing indicated that >40% of these products aligned to a different part of the genome, and another 12% verified a spliced transcript, but the details of exon–intron structure were incorrect in the original prediction. Thus, one must use caution when interpreting the results of studies examining expressed regions of the genome by hybridization to microarrays or sizing RT-PCR products. Without a sequenced product or sufficient microarray hybridization specificity, one might interpret a positive signal as a previously unrecognized gene, when it is actually cross-hybridization to, or mispriming from, an already annotated gene.
How justified are we in concluding that 95% of all D. melanogaster protein-coding genes have been identified and at least provisionally annotated? Certainly, there is no shortage of untested predictions. However, if our estimates are correct, experimentally testing all 9,552 remaining gene predictions would identify only about another 500 genes. Moreover, implicit in our approach is the assumption that demonstrating the existence of a spliced transcript that has the capacity to encode a polypeptide constitutes proof of the existence of a gene, which may not always be the case. We believe that our results allow us to conclude that only a few hundred D. melanogaster protein-coding genes that are identifiable by using homology or commonly used gene-finders remain unannotated. Thus, we believe that the results presented here put to rest arguments that would claim that some massive, previously unrecognized subset of genscan or fgenesh gene predictions is real. No doubt there are still more D. melanogaster genes to be discovered, but these genes probably don't look like the traditional protein-coding genes found by genscan and fgenesh. Some, for example, may be nested among the introns of existing annotations; our approach would have missed these from the outset. Finding the remaining protein-coding genes will require new strategies, and it seems likely that these new strategies will entail searching very large collections of gene models for a small number of additional genes. Thus, the approach to large-scale prediction-validation that we have presented here should provide an effective infrastructure for the next round of gene identification, as well. In the quest to identify previously unrecognized genes, however, it should not be forgotten that the details of most genome annotations remain provisional, awaiting verification by cDNA sequencing. Clearly, another way forward is to employ a pipeline such as we describe for the directed, high-throughput confirmation of the details of a genome's annotations.
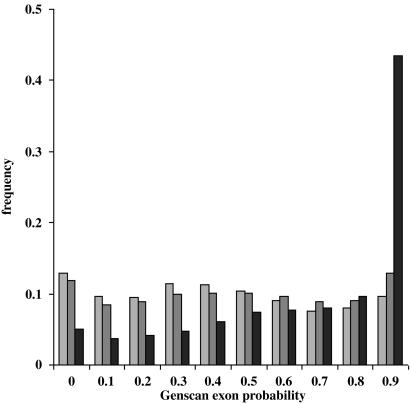
We believe our results also speak to the ongoing debate about estimating gene numbers in other metazoan organisms. The distribution of genscan exon probabilities within intergenic regions of the D. melanogaster genome closely approximates those that we obtained by performing the same analysis on random sequence (Fig. 3); the short exons and lax requirements for splicing, characteristic of most large eukaryotic genomes, mean that gene-like structures will occur in any sufficiently large intergenic region simply by random chance. Thus, the existence of large numbers of computational gene predictions does not make a compelling case for the presence of additional genes. Moreover, in humans (19) and Drosophila (13), a much larger proportion of the genome is transcribed than previously appreciated, which means that spurious gene predictions will often fall within transcribed regions simply by chance; thus, showing that a gene prediction is transcribed is not sufficient to validate a gene model. Gene predictions must be subjected to rigorous validation before being promoted to the status of annotations. Finally, our observations may also suggest a mechanism for the evolution of new genes. Gene-like structures occur by chance in DNA, and transcription is promiscuous, raising the possibility that, on an evolutionary time scale, new protein-coding genes may be produced “spontaneously” within genomes.
Fig. 3.
The distribution of genscan exon probabilities for predicted genes within D. melanogaster intergenic regions closely approximates that of random sequence. Distribution of exon probabilities assigned by genscan to predictions lying within 2 Mb of random sequence 0.25 G:A:T:C (light gray bars), 62 Mb of D. melanogaster release 3.1 intergenic regions (gray bars), and the first 2 Mb of the D. melanogaster chromosome arm 2L (black bars).
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Joe Carlson, Suzanna Lewis, Kenneth Wan, Chris Mungall, Chris Smith, and Gina Dailey. We also thank Marc Hild and Victor Solovyev for providing datasets. This work was funded in part by National Human Genome Research Institute Grant HG002673 (to S.E.C.), National Institutes of Health Grant HG00750 (to G.M.R.), and the Howard Hughes Medical Institute (M.Y. and G.M.R.).
Abbreviations: Mb, megabases; oligo, oligonucleotide; sjc, splice-junction conserved.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CX309415–CX309654).
References
- 1.Misra, S., Crosby, M. A., Mungall, C. J., Matthews, B. B., Campbell, K. S., Hradecky, P., Huang, Y., Kaminker, J. S., Millburn, G. H., Prochnik, S. E., et al. (2002) Genome Biol. 3, research0083.1-0083.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hild, M., Beckmann, B., Haas, S. A., Koch, B., Solovyev, V., Busold, C., Fellenberg, K., Boutros, M., Vingron, M., Sauer, F., et al. (2003) Genome Biol. 5, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 4.Wu, J. Q., Garcia, A. M., Hulyk, S., Sneed, A., Kowis, C., Yuan, Y., Steffen, D., McPherson, J. D., Gunaratne, P. H. & Gibbs, R. A. (2004) Biotechniques 36, 690-696, 698-700. [DOI] [PubMed] [Google Scholar]
- 5.Flicek, P., Keibler, E., Hu, P., Korf, I. & Brent, M. R. (2003) Genome Res. 13, 46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guigo, R., Dermitzakis, E. T., Agarwal, P., Ponting, C. P., Parra, G., Reymond, A., Abril, J. F., Keibler, E., Lyle, R., Ucla, C., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenney, A. E., Brown, R. H., Vaske, C., Lodge, J. K., Doering, T. L. & Brent, M. R. (2004) Genome Res. 14, 2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zadeh, L. A. (1965) Inf. Control 8, 338-353. [Google Scholar]
- 9.Sadegh-Zadeh, K. (2001) Medizintheorie 1, 41-82. [Google Scholar]
- 10.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- 11.Mungall, C. J., Misra, S., Berman, B. P., Carlson, J., Frise, E., Harris, N., Marshall, B., Shu, S., Kaminker, J. S., Prochnik, S. E., et al. (2002) Genome Biol. 3, research0081.1-0081.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stajich, J. E., Block, D., Boulez, K., Brenner, S. E., Chervitz, S. A., Dagdigian, C., Fuellen, G., Gilbert, J. G., Korf, I., Lapp, H., et al. (2002) Genome Res. 12, 1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M. F., Rifkin, S. A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P. E., et al. (2004) Science 306, 655-660. [DOI] [PubMed] [Google Scholar]
- 14.Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. (2002) Genome Res. 12, 1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis, S. E., Searle, S. M. J., Harris, N., Gibson, M., Iyer, V., Richter, J., Wiel, C., Bayraktaroglu, L., Birney, E., Crosby, M. et al. (2002) Genome Biol. 3, research0082.1-0082.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 17.FlyBase Consortium (2002) Nucleic Acids Res. 30, 106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salamov, A. A. & Solovyev, V. V. (2000) Genome Res. 10, 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916-919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.