Abstract
A fatal case associated with enterovirus D68 (EV-D68) infection affecting a 10-year-old boy was reported in Hong Kong in 2014. To examine if a new strain has emerged in Hong Kong, we sequenced the partial genome of the EV-D68 strain identified from the fatal case and the complete VP1, and partial 5′UTR and 2C sequences of nine additional EV-D68 strains isolated from patients in Hong Kong. Sequence analysis indicated that a cluster of strains including the previously recognized A2 strains should belong to a separate clade, clade D, which is further divided into subclades D1 and D2. Among the 10 EV-D68 strains, 7 (including the fatal case) belonged to the previously described, newly emerged subclade B3, 2 belonged to subclade B1, and 1 belonged to subclade D1. Three EV-D68 strains, each from subclades B1, B3, and D1, were selected for complete genome sequencing and recombination analysis. While no evidence of recombination was noted among local strains, interclade recombination was identified in subclade D2 strains detected in mainland China in 2008 with VP2 acquired from clade A. This study supports the reclassification of subclade A2 into clade D1, and demonstrates interclade recombination between clades A and D2 in EV-D68 strains from China.
Keywords: enterovirus D68, clade, recombination
1. Introduction
Enterovirus D68 (EV-D68) belongs to enterovirus species D (EV-D) in the genus Enterovirus of the family Picornaviridae. EV-D68 was first isolated from hospitalized children with lower respiratory illnesses in California in 1962 [1], and human rhinovirus 87 was later confirmed as an isolate of EV-D68 based on antigenic and genetic characterization and acid lability testing [2]. Sporadic cases of EV-D68 infection have been reported since 1970 [3], but there has been a significant increase in the number of reports of EV-D68 circulating globally in recent times [4,5,6,7,8,9,10,11]. In 2014, a nationwide outbreak of EV-D68 associated with severe respiratory disease in children and acute flaccid myelitis occurred in the United States (US), affecting over 1100 people in 49 states, 14 of whom died [12]. Most of the EV-D68 strains detected in the US outbreak belonged to clade B1, especially those sampled from acute flaccid myelitis (AFM) cases [13]. In our previous study, we examined the clinical and molecular epidemiology of EV-D68 among hospitalized patients with respiratory tract infections in Hong Kong [14]. Phylogenetic analysis of partial VP1, 2C, and 3D regions of EV-D68 strains in Hong Kong revealed four distinct lineages, clade A1, A2, B1, and the new clade, B3, with the elderly having severe lower respiratory illness exclusively caused by clade A2 [14]. Moreover, strains closely related to the US 2014 outbreak B1 strains were detected as early as 2011 in Hong Kong. These findings suggested that EV-D68 has been evolving into new lineages that might be emerging.
Mutations and recombinations are well-known phenomena in driving enterovirus evolution [15,16,17,18]. However, to date, only one study has demonstrated an intraclade recombination event within an EV-D68 strain US/KY/14-18951, which has arisen from recombination between subclades B1 and B2 [19]. In 2014, there was a fatal case associated with EV-D68 infection in a 10-year old boy in Hong Kong. To determine if a new strain has emerged in Hong Kong and if recombination events have played a role in the evolution of the emergence of new EV-D68 strains in Hong Kong, we sequenced the partial genome of the EV-D68 strain detected from the fatal case. We also randomly selected nine additional EV-D68 strains isolated from patients in Hong Kong during 2012 to 2014 and sequenced their complete VP1, partial 5′ untranslated region (5′UTR), and 2C region. We also determined the complete genome sequences of three EV-D68 strains belonging to three different clades, B1, B3, and D1.
2. Results
2.1. Clinical Characteristics of Patients with Enterovirus D68 (EV-D68) Infections
The fatal case occurred in a 10-year-old boy (case 1) with an α-thalassemia trait who was residing in mainland China and presented with acute encephalitis, neurogenic pulmonary edema with respiratory failure, autonomic dysfunction, and cardiac arrest requiring intensive care in August 2014 (Table 1). Due to the critical condition and coagulopathy state, lumbar puncture could not be performed. Therefore, he was started on broad-spectrum antimicrobial coverage for possible meningoencephalitis while awaiting investigation results. His condition was complicated by severe hypoxic ischemic encephalopathy and he died nine days after admission despite use of broad-spectrum antimicrobials including cefotaxime, tazobactam-piperacillin, vancomycin, levofloxacin, and acyclovir. Mycoplasma pneumoniae and EV-D68 nucleic acids were detected by PCR in his nasopharyngeal aspirate (NPA). The M. pneumoniae showed an A2063G mutation indicating macrolide resistance [20]. His peri-mortem cerebrospinal fluid (CSF) obtained after developing confusion and respiratory deterioration was negative for EV-D68 RNA.
Table 1.
Clinical characteristics of the 10 cases of enterovirus D68 (EV-D68) infections.
| Case | Strain | Specimen Type | Collection Date | Gender | Age | Underlying Diseases | Diagnosis | Clade |
|---|---|---|---|---|---|---|---|---|
| 1 | V14-8157864 | NPA | 16 August 2014 | M | 10 | α-thalassemia trait | Encephalitis, Mycoplasma pneumoniae co-infection | B3 # |
| 2 | V12-2240471 | NPS | 7 May 2012 | M | 5 | Allergic rhinitis | URTI, asthma | B1 |
| 3 | V12-2268728 * | NPA | 20 July 2012 | M | 2 | None | URTI, asthma | B1 |
| 4 | V13-2245157 * | NPS | 21 May 2013 | F | 94 | HT, CVA, gout, dementia | Pneumonia | D1 |
| 5 | V14-8133616 | NPA | 18 May 2014 | M | 4 | Allergic rhinitis, asthma | URTI, asthma | B3 |
| 6 | V14-8135150 | NPA | 23 May 2014 | F | 4 | MRSA skin infection | URTI, RSV co-infection | B3 |
| 7 | V14-8143594 | NPS | 21 June 2014 | F | 4 | Eczema | Wheezy bronchitis | B3 |
| 8 | V14-8143833 | NPS | 24 June 2014 | F | 4 | None | URTI, asthma | B3 |
| 9 | V14-8151518 | NPS | 23 July 2014 | M | 5 | α-thalassemia trait | URTI, Henoch Schonlein purpura | B3 |
| 10 | V14-8151546 * | NPS | 23 July 2014 | M | 5 | None | Febrile wheeze | B3 |
* EV-D68 strains selected for complete genome sequencing. # VP1 sequence not available; partial 5′UTR and 2C were sequenced (100% identical to those of EV-D68 strain V14-8151546; potential subclade B3). Abbreviations: CVA, cerebrovascular accident; HT, hypertension; MRSA, methicillin-resistant Staphylococcus aureus; NPA, nasopharyngeal aspirate; NPS, nasopharyngeal swab; RSV, respiratory syncytial virus; URTI, upper respiratory tract infection; M: male; F: female.
Nine other EV-D68 strains were isolated from the nasopharyngeal swabs (NPSs) or NPAs, obtained as part of the diagnostic workup of hospitalized patients from seven regional hospitals in Hong Kong from 2012 to 2014. Including the fatal case, most patients with EV-D68 infections were young children, while one was an elderly female patient (median age 4.5 years, range 2–94 years). Six were male and four were female. The clinical characteristics of the 10 patients with EV-D68 infections are summarized in Table 1. The 94-year-old woman with underlying medical illnesses presented with pneumonia. Six of the young children presented with upper respiratory tract infections (URTI), among which four were complicated by asthmatic exacerbation and one was complicated by Henoch Schonlein purpura. Two other children presented with wheezy bronchitis and febrile wheeze, respectively. Except for one fatal case, the other nine patients survived.
2.2. Complete VP1 Gene Sequence Analysis
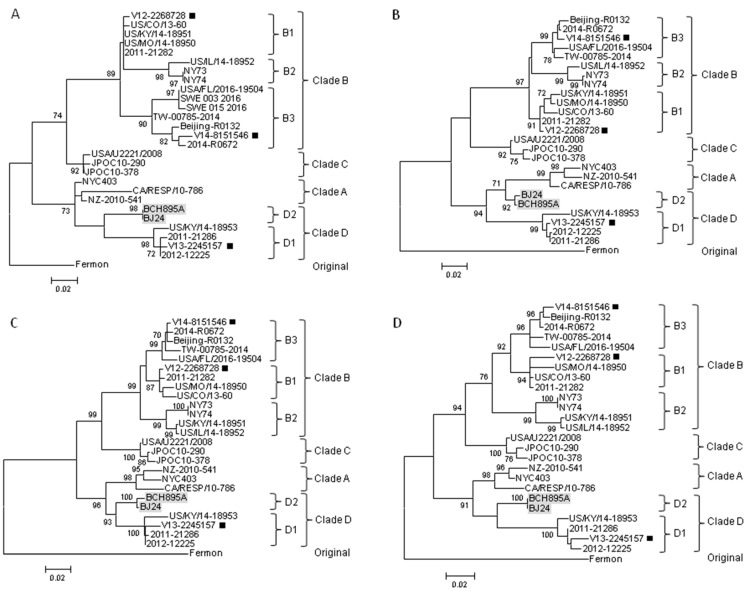
Since the strain V14-8157864 from the fatal case could not be grown in cell culture, only partial genomic sequences could be obtained and the complete VP1 sequence was not available. On the other hand, the complete VP1 sequences of the other nine EV-D68 virus isolates were successfully obtained. Phylogenetic analysis showed that they fell into three different subclades of EV-D68. Eight EV-D68 strains were clustered with other EV-D68 strains of clade B (Figure 1A). Among these eight strains, two strains from the year 2012 belonged to subclade B1 (nucleotide divergence of 0.2–3.5% for strains within B1), while six strains from the year 2014 belonged to subclade B3 (nucleotide divergence of 0.1–2.7% for strains within B3). The nucleotide divergence of 2.6–5.3% was observed between subclade B1 and subclade B3 strains (Table 2).
Figure 1.
Phylogenetic analysis using VP1, partial 5′UTR and 2C gene sequences of enterovirus D68 (EV-D68) strains.The trees were inferred from (A) VP1, (B) partial 5′UTR and (C) partial 2C sequence data by the maximum likelihood (ML) method, with bootstrap values calculated from 1000 trees. Sequences for 918 nucleotide positions in each VP1 region, 356 nucleotide positions in each 5′UTR region, and 531 nucleotide positions in each 2C region were included in the analysis. Bootstrap values expressed as a percentage are shown at the nodes and the scale reflects the number of nucleotide substitutions per site along the branches. Only bootstrap values >70% are shown. Black circles indicate EV-D68 strains collected for the present study.
Table 2.
Nucleotide sequence identities of VP1 genes between clades and subclades of EV-D68 strains.
| Strains * | Pairwise Identity (%) | Average Nucleotide Divergence (%) [Range] |
|---|---|---|
| Interclade | 10.8 [6.5–14.8] | |
| Prototype vs. A | 87–87.9 | |
| Prototype vs. B | 85.5–86.6 | |
| Prototype vs. C | 87.8–88 | |
| Prototype vs. D | 85.2–86.7 | |
| A vs. B | 86.5–90.4 | |
| A vs. C | 88.8–91.4 | |
| A vs. D | 90.2–93.5 | |
| B vs. C | 89.8–93.1 | |
| B vs. D | 86.4–89.7 | |
| C vs. D | 87.9–90.5 | |
| A vs. D1 | 90.2–91.7 | |
| A vs. D2 | 91.8–93.5 | |
| Intersubclade | 4.7 [2.6–7.1] | |
| B1 vs. B2 | 94–95.2 | |
| B1 vs. B3 | 94.7–97.4 | |
| B2 vs. B3 | 92.9–94.2 | |
| D1 vs. D2 | 94.1–95.6 | |
| Intrasubclade | 1.4 [0–3.5] | |
| B1 | 96.5–99.8 | |
| B2 | 98.9–100 | |
| B3 | 97.3–99.9 | |
| D1 | 97.5–99.7 | |
| D2 | 98.1–99.6 |
* VP1 sequences of the EV-D68 strains used for the analysis are shown in Table A1 in Appendix A.
One EV-D68 strain from the only elderly case (case 4) in this study was most closely related to subclade A2. However, recent studies demonstrated that the previously recognized A2 strains should be classified as clade D [21,22]. This was further supported by our in-silico analysis, in which (1) the nucleotide sequence divergence of 6.5–9.8% was observed between clade A and clade D strains (similar to that between other clades) (Table 2); (2) the strains in clade D formed a distinct cluster from clade A strains supported by a high bootstrap value of 97% (Figure 1A); and (3) amino acid residues in several positions of the VP1 of clade D strains were different from those of clade A strains (Table 3). Intriguingly, two distinct clusters were noted in clade D (Figure 1A), with nucleotide divergence of 4.4–5.9% between strains of the two clusters (Table 2). Moreover, amino acid residues in positions 5, 6, 103, and 279 of the VP1 were different between strains of these two clusters (Table 3). These findings suggested that these two clusters should be classified as subclade D1 and D2 (Figure 1A). Thus, the “A2” strain (case 4) in this study should also belong to subclade D1 (Figure 1A). This and other subclade D1 strains had a high degree of VP1 nucleotide sequence identity of 97.5–99.7% (Table 2).
Table 3.
Amino acid residues of VP1 between EV-D68 strains for clade A and D differentiation.
| Strains | Positions | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 5 | 6 | 92 | 103 | 131 | 140 | 142 | 147 | 177 | 186 | 279 | C-terminus | |
| Clade A | |||||||||||||
| CA/RESP/10-786 | D | D | A | A | F | V | S | S | T | A | I | R | VTT |
| NZ-2010-541 | D | D | A | A | F | V | S | S | T | A | I | R | VTT |
| NYC403 | D | D | A | A | F | V | S | S | T | A | I | R | VTT |
| TW-02378_2010 | D | D | A | A | F | V | S | S | T | A | I | R | |
| ITA/19391/12 | D | H | A | A | F | V | S | S | T | A | I | R | |
| HK2/2010 | D | D | A | A | F | V | S | S | T | A | I | R | VTT |
| Clade D | |||||||||||||
| Subclade D1: | |||||||||||||
| TW-02722_2013 | E | T | I | G | N | M | G | V | RLVNT | ||||
| US/KY/14-18953 | E | T | I | G | N | M | G | V | RLVNT | ||||
| 2011-21286 | E | T | I | G | N | M | G | V | RLVNT | ||||
| 2012-12225 | E | T | I | G | N | M | G | V | RLVNT | ||||
| 2013-1017-26 | E | T | I | G | N | M | G | V | RLVNT | ||||
| HK5/2011 | E | T | I | G | N | M | G | V | RLVNT | ||||
| HK13/2012 | E | T | I | G | N | M | G | V | RLVNT | ||||
| HK21/2013 | E | T | I | G | N | M | G | V | RLVNT | ||||
| V13-2245157 | E | T | I | G | N | M | G | V | RLVNT | ||||
| Subclade D2: | |||||||||||||
| BCH895A | E | D | A | T | F | I | G | N | M | G | V | R | RLVNT |
| BJ24 | E | D | A | T | F | I | G | N | M | G | V | R | RLVNT |
| TW-01166_2007 | E | D | A | T | F | I | G | N | M | G | V | R | |
| TW-11134_2009 | E | D | A | T | F | I | G | N | M | G | V | R | |
| 1737-Yamagata-2008 | E | D | A | T | F | I | G | N | M | G | V | R | RLVNT |
The VP1 amino acid residues of EV-D68 subclade D1 different from those of subclade D2 are highlighted in grey. Amino acid code: D, aspartic acid; A, alanine; F, phenylalanine; V, valine; S, serine; T, threonine; I, isoleucine; R, arginine; E, glutamic acid; H, histidine; Y, tyrosine; G, glycine; N, asparagine; M, methionine; K, lysine; L, leucine.
2.3. Partial 5′UTR and 2C Sequence Analysis
Although the VP1 of the strain V14-8157864 from the fatal case (case 1) could not be amplified, the partial 5′UTR and 2C gene region of this strain and the other nine EV-D68 strains from Hong Kong were all successfully amplified and sequenced. Phylogenetic analysis using the partial 5′UTR and 2C sequences (Figure 1B,C) showed similar topologies to those using VP1 sequences. The strain V14-8157864 from the fatal case was clustered with B3 strains.
2.4. Complete Genome Analysis of Local and Regional EV-D68 Strains
To look for potential mutation and recombination events among EV-D68 strains in Hong Kong, one strain belonging to each subclade (B1, B3, and D1) were selected for complete genome sequencing (Table 1). The three genomes are around 7.3 kbp in length, after excluding the polyadenylated tract, and the G + C content is 42%. Two deletions (CCTCAAAACCTCCAGTACATAAC and AAACTTATTTAT corresponding to positions 682–704 and 718–729 of the prototype strain Fermon, respectively) in the spacer region of 5′UTR between the end of the internal ribosome entry site and the start codon of the open reading frame were noted in strains belonging to clades B and C when compared to the prototype strain Fermon [23], while only the first deletion (CCTCAAAACCTCCAGTACATAAC) was observed in strains belonging to clades A and D (Figure 2).
Figure 2.
Deletion blocks in 5′UTR of EV-D68 strains. Nucleotides 672–742 (based on the prototype strain with GenBank accession no. AY426531) of the 5′UTR are shown. EV-D68 representative strains of clade A, B, C, D, and the prototype strain Fermon are included.
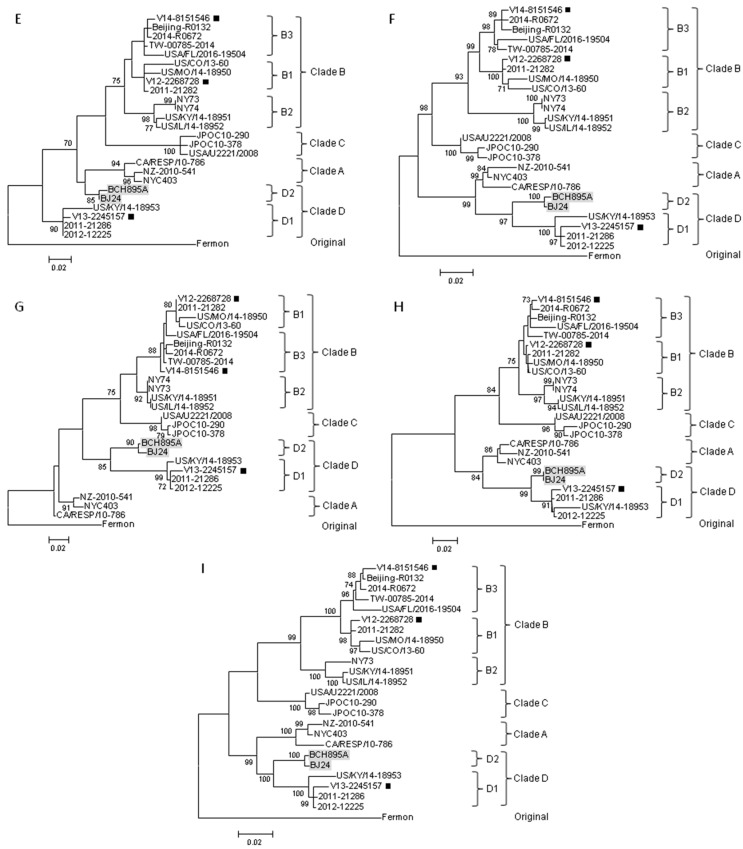
To examine for incongruent tree topologies which may suggest potential recombination events, phylogenetic trees using nucleotide sequences of VP4 to VP1, 2A to 2C, and 3A to 3D regions of three EV-D68 strains from Hong Kong and other EV-D68 strains with complete genome sequences available in the GenBank were constructed (Figure 1A and Figure 3). The results did not reveal potential recombination in the three Hong Kong strains, and confirmed that V13-2245157 belonged to clade D1, while V12-2268728 and V14-8151546 belonged to clades B1 and B3, respectively (Figure 1A and Figure 3). A study showed that the majority of the outbreak strains belonging to subclade B1 in the US in 2014 differed from others, with signature mutations M291T, V341A, T860N, D927N, S1108G, and R2005K [13,23,24], which were also observed in the isolate V12-2268728 in Hong Kong. The genome sequence of this isolate shared 98% nucleotide identity to those of outbreak strains in the U.S. (subclade B1), supporting the idea that the EV-D68 epidemic in the US may have originated from strains circulating in Asia a few years ago [14].
Figure 3.
Phylogenetic analysis using various gene sequences of EV-D68 strains. The trees were constructed using the ML method, with bootstrap values calculated from 1000 trees. Sequences for 207 nucleotide positions in each VP4 region (A), 744 nucleotide positions in each VP2 region (B), 705 nucleotide positions in each VP3 region (C), 441 nucleotide positions in each 2A region (D), 297 nucleotide positions in each 2B region (E), 990 nucleotide positions in each 2C region (F), 333 nucleotide positions in each 3A–3B region (G), 549 nucleotide positions in each 3C region (H) and 1371 nucleotide positions in each 3D region (I) were included in the analysis. Bootstrap values expressed as percentages are shown at the nodes and the scale reflects the number of nucleotide substitutions per site along the branches. Only bootstrap values >70% are shown. Black squares indicate the three EV-D68 strains subjected to complete genome sequencing in the present study. The interclade recombinants from mainland China are highlighted in grey.
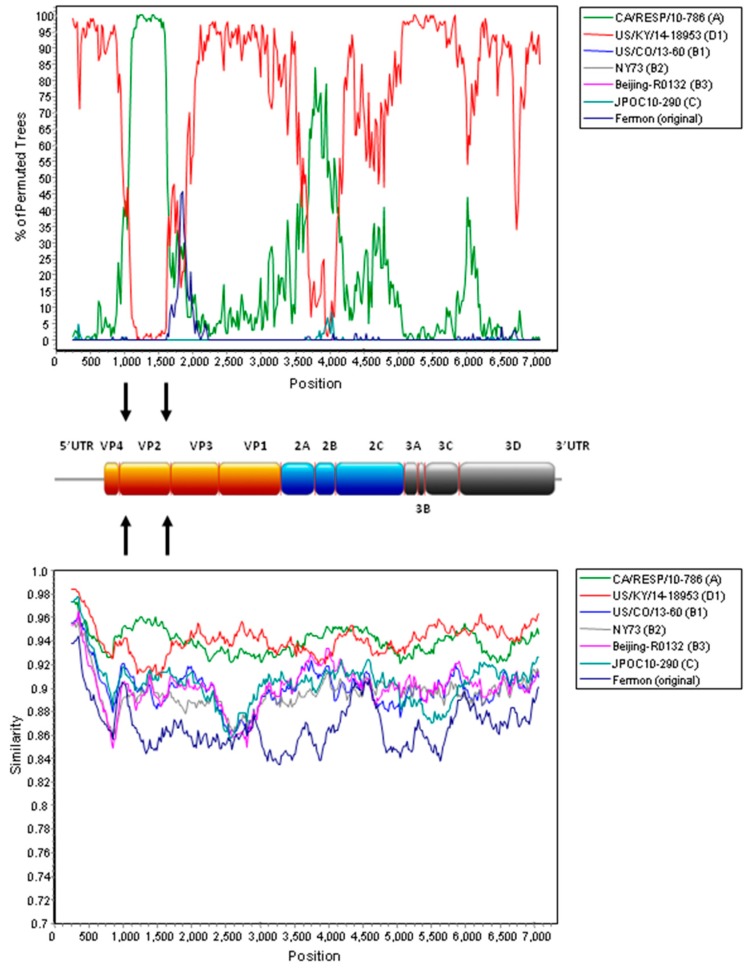
On the other hand, potential recombination events were identified in two EV-D68 subclade D2 strains from mainland China, as retrieved from the GenBank search, during our analysis. Examination of the tree topologies of the other EV-D68 strains revealed that the EV-D68 subclade D2 strains BCH895A and BJ24 from mainland China were most closely related to the strains of clade A for VP2 and 2B, while they were most closely related to subclade D1 strains for other gene regions (Figure 1A and Figure 3), indicating that recombination events might have occurred in the two subclade D2 strains. Similarity plot and bootscan analyses, using the BCH895A strain as a query sequence, were performed to identify potential recombination sites (Figure 4). The analyses demonstrated that four recombination sites were probably located at VP4–VP2 (between nucleotide positions 900 and 1300), VP2–VP3 (between nucleotide positions 1500 and 1800), 2A–2B (between nucleotide positions 3500 and 3800), and 2B–2C (between positions 4000 and 4300). The region between the VP4–VP2 and VP2–VP3 junctions and the region between the 2A–2B and 2B–2C junctions of the D2 strain were highly similar to the clade A strain, while other regions were most closely related to the D1 strain. Potential recombination within the genome sequences was also examined by the Recombination Detection Program (RDP4). Recombination events were detected in the genome sequence of the strains BCH895A and BJ24, supported by p values of less than 0.05 in the GENECONV, Bootscan, and SiScan methods. Two significant recombination breakpoints were identified in the VP2 gene at nucleotide positions 1297 and 1506. While the similarity plot and bootscan analysis supported the recombination event in the 2B region, this lacked statistical support by RDP4. The above findings suggested that the two EV-D68 D2 strains BCH895A and BJ24 identified in mainland China in 2008 represent interclade recombinants with the VP2 acquired from clade A.
Figure 4.
Recombination analysis of the complete genome of EV-D68. Bootscan analysis (upper panel) and similarity plot (lower panel) were conducted with SimPlot version 3.5.1 (Kimura distance model; window size, 500 bp; step, 20 bp) on a gapless nucleotide alignment, generated with Clustal X version 2.0 (University College Dublin, Dublin, Ireland), with the genome sequence of the EV-D68 strain BCH895A as a query sequence. The red line denotes EV-D68 subclade D1 strain US/KY/14-18953 and the green line denotes EV-D68 clade A strain CA/RESP/10-786. Arrows indicate the gene regions in which potential recombination breakpoints are located.
3. Discussion
This study reported the first fatal case associated with EV-D68 infection in Hong Kong. The 10-year-old boy (case 1) presented with acute encephalitis and deteriorated rapidly after admission. Although co-infection by M. pneumoniae might have contributed to the encephalitis, the role of EV-D68 in his illness or exacerbating the mycoplasma infection cannot be excluded. Evidence of neurological complications associated with EV-D68 infection is increasing in recent epidemiological investigations [25,26,27], and the absence of EV-D68 in CSF in the present case may be due to delayed specimen collection and was consistent with previous reports [26,27]. The failure to detect EV-D68 in CSF is not surprising as other known neurotropic enteroviruses, like EV-A71 and poliovirus, were not always detected from CSF specimens of patients with neurological complications, while other specimen types such as throat swabs, rectal swabs, or stool specimens gave higher diagnostic yields [13,28,29,30,31]. Another possible reason is that the neurological symptoms are caused by an aberrant immune response to EV-D68 infection [13]. On the other hand, the detection of EV-D68 in CSF from patients with neurological complications has been reported in two cases [32,33], suggesting the possibility of direct viral neuroinvasion. The latest study demonstrated that EV-D68 strains from the US 2014 outbreak could cause motor neuron death and produce paralytic disease in mice resembling human acute flaccid myelitis (AFM), supporting the causative role of EV-D68 in neurological diseases [34]. Further investigation is therefore needed to delineate whether an indirect immune effect and/or direct damage to the neuron is the key mechanism of causing neurological symptoms by EV-D68 infection. Nevertheless, the two pathogenic mechanisms may not contradict each other, as both processes may co-exist to cause pathologies, as in many other pathogens. Recent studies have shown that neurological symptoms were associated with EV-D68 subclade B3 strains. An outbreak of EV-D68 subclade B3 mainly involving children was reported in Sweden, in which some patients had severe respiratory or neurological symptoms and one died [35]. In another study describing an upsurge of EV-D68 in the Netherlands, some infected children required intensive care unit admission due to respiratory insufficiency and one had AFM that were associated with EV-D68 subclade B3 strains [36]. In Italy, an AFM case that was probably caused by the EV-D68 B3 strain was also reported recently [37]. Unfortunately, the VP1 gene sequence of the EV-D68 strain (V14-8157864) from the present fatal case could not be amplified probably due to low viral load. We have attempted to propagate this strain in cell culture, but were unsuccessful. Yet, the partial 5′UTR and 2C sequences of this strain were 100% identical to those of the subclade B3 strain V14-8151546 (case 10). Interestingly, these two strains, together with the strains identified in the other five young children (cases 5–9), were detected during the summer of 2014, suggesting that similar B3 strains may have been predominating in our population. This is in line with our previous study showing that EV-D68 subclade B3 was the predominant lineage circulating in Hong Kong since 2014 [14]. In fact, subclade B3 strains were also detected in Taiwan in 2014 [22]. It is likely that the recent outbreaks of EV-D68 due to subclade B3 in Europe may have originated from strains that emerged in Asia in 2014. Clinicians should be aware of the possible neurological manifestations of EV-D68 infection, which may help better understand the pathogenicity of the emerging subclade B3 strains. The six mutations (M291T, V341A, T860N, D927N, S1108G, and R2005K) associated with neurovirulence in the US outbreak strains (subclade B1 strains in 2014) [13,23,24] were noted in the B1 strain, but not in the B3 strain in the present study. Further studies are required to examine the role of these mutations in the pathogenicity caused by EV-D68.
The previously recognized subclade A2 strains should be re-designated as subclade D1 within clade D. Current taxonomy and classification of picornaviruses, including enteroviruses, are based on the capsid region, particular VP1 [38,39,40]. In the present study, we have determined the pairwise nucleotide identities using VP1 gene sequences of EV-D68 strains, with the results showing that the average nucleotide sequence divergences between clades and between subclades were 10.8% (6.5–14.8%) and 4.7% (2.6–7.1%), respectively (Table 2). As the average nucleotide divergence between clade A and “A2” strains was 8.4%, together with the evidence from the VP1 sequence and phylogenetic analyses (Figure 1A and Figure 3), all these findings suggested that the “A2” strains should belong to clade D, which has been proposed by other authors [21,22]. Among the clade D strains, two distinct clusters were shown in the phylogenetic tree of VP1 (Figure 1A) and they had an average nucleotide divergence of 5.1% (Table 2). Furthermore, amino acid differences were noted in the VP1 of the strains between these two clusters (Table 3). Therefore, the strains of clade D should be further divided into subclades D1 and D2. In this study, one subclade D1 strain was detected in the elderly (case 4) with pneumonia, which is in line with our previous study showing that adults/the elderly in Hong Kong were exclusively infected by subclade D1 (“A2”) [14]. Although subclade D1 strains have been circulating globally [22,36,41], the clinical characteristics of patients infected by these strains were not reported in other studies. Further epidemiological investigations on subclade D1 strains are needed to determine if this lineage also has a predilection for infecting adults/the elderly in other countries or continents.
We described the first evidence for interclade recombination in EV-D68. Recombination is a well-known phenomenon in enterovirus evolution and recombination breakpoints have been frequently detected in non-structural regions of enterovirus genomes [18,42,43,44]. This is possibly because the non-structural genes are highly conserved at certain regions among the same enterovirus species when compared to structural (capsid) genes. Nevertheless, natural recombination in the capsid region of polioviruses has also been reported [45,46]. To date, only one study has demonstrated recombination between subclades B1 and B2 in an EV-D68 strain from the US during the 2014 outbreak, with a breakpoint in VP2 [19]. In the present study, recombination in capsid region was identified in two EV-D68 strains (BCH895A and BJ24) belonging to subclade D2 from mainland China in 2008. Both phylogenetic and recombination analyses showed that the nucleotide sequences of the two subclade D2 strains from mainland China were most closely related to those of clade A strains in VP2, but to those of clade D strains in other genomic regions, suggesting that interclade recombination had occurred between clade A and D strains of EV-D68. Such recombination is not unexpected since the capsid regions are sufficiently conserved among different clades of a single enterovirus genotype. Though the association between natural enterovirus recombination and pathogenicity remains unclear, an in vitro study showed that replacing a structural region of a slow-growth EV-A71 strain with that of a fast-growth EV-A71 strain could generate a recombinant virus with growth improvement and larger plaque phenotypes [47]. Further studies are warranted to investigate the role of recombination in driving the evolution of EV-D68.
In conclusion, we described the molecular epidemiology of EV-D68 in Hong Kong and presented further evidence for taxonomic reclassification of subclade A2 into a separate clade D1. We also showed for the first time that recent subclade D2 strains from mainland China probably originated from an interclade recombination event between clade A and clade D strains. Further evolutionary analysis of currently circulating EV-D68 strains in East Asia is warranted. Continuous epidemiological and laboratory surveillance is required to detect novel variants with epidemic potential.
4. Materials and Methods
4.1. Clinical Specimens
NPAs or NPSs were collected from 10 hospitalized patients from seven regional hospitals in Hong Kong during the years from 2012–2014 (Table 1). EV-D68 detection and isolation from NPAs or NPSs was performed at the Public Health Laboratory Centre, Centre for Health Protection, Department of Health, Hong Kong. Detection of EV-D68 RNA from clinical specimens was performed using real-time RT-PCR targeted to the 5′UTR region and sequencing using a previously described protocol [48]. Isolation of EV-D68 was performed using the human embryonal rhabdomyosarcoma (RD) cell line. The clinical features, laboratory results, and outcome of illness of these patients were analyzed. The ethical approval was obtained from the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 16-365; approval date: 20 July 2016) for this study.
4.2. RNA Extraction
The clinical specimens (200 µL each) were subjected to total nucleic acid extraction by the EZ1 Virus Mini Kit v2.0 (QIAGEN, Hilden, Germany), with the elution volume of 60 µL. The eluate was used as a template for reverse-transcription polymerase chain reaction (RT-PCR).
4.3. Reverse-Transcription Polymerase Chain Reaction (RT-PCR) and Sequencing of VP1 of EV-D68 for Clade Determination
RT was performed using random hexamers and a SuperScript III Kit (Invitrogen, Carlsbad, CA, USA) as described previously [14,18]. The VP1 of EV-D68 strains detected from the clinical specimens were amplified and sequenced using the primers and the strategy described in our previous publication [14]. Both strands of PCR products were sequenced twice with an ABI 3130xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using the PCR primers. A phylogenetic tree of the VP1 was constructed using maximum likelihood (ML) method in MEGA6 with the model T92 + I [49], with bootstrap analysis of 1000 replicates. The percentage of VP1 nucleotide sequence identity was determined by MatGAT2.01 software (Montclair State University, Montclair, NJ, USA) [50].
4.4. RT-PCR and Sequencing of Partial 5′UTR and 2C of EV-D68 and Phylogenetic Analysis
Due to the unsuccessful amplification of the VP1 of the EV-D68 strain V14-8157864 from the fatal case, we attempted to amplify and sequence the partial 5′UTR and 2C of this strain, together with the EV-D68 strains detected from the other 9 patients using the primers and the strategy described in our previous publication [14]. Phylogenetic trees of each region were constructed using ML method in MEGA6 with the model K2 + G for partial 5′UTR and T92 + I for partial 2C [49], with bootstrap analysis of 1000 replicates.
4.5. Complete Genome Sequencing of EV-D68
The complete genomes of three EV-D68 strains (V12-2268728, V13-2245157, and V14-8151546) were amplified and sequenced using the strategy described in our previous publications [18,51]. The cDNA was synthesized using the viral RNA by a combined random-priming and oligo(dT) priming strategy. PCR amplification was achieved by degenerate primers designed by multiple alignments of the EV-D68 genomes and other primers designed based on the results of the first and subsequent rounds of sequencing. The primer sequences are available upon request. The 5′ ends of the EV-D68 genomes were confirmed by rapid amplification of cDNA ends (RACE) using Terminal Deoxynucleotidyl Transferase, recombinant (Invitrogen, Carlsbad, CA, USA). Sequences were assembled and manually edited to produce final sequences of the viral genomes by BioEdit version 7.2.5 (NC State University, Raleigh, NC, USA).
4.6. Genome Analysis
The nucleotide sequences of various genomic regions of the EV-D68 strains detected in Hong Kong were compared to those of EV-D68 strains with available genome sequences in GenBank (Table A1). Phylogenetic tree construction was performed using the ML method in MEGA6 with the models T92 + G for VP4, VP2, 2B, 3A-3B, and 3C, and T92 + I for VP3, VP1, 2A, 2C, and 3D (Figure 1A and Figure 3), with bootstrap values calculated from 1000 trees [49]. Recombination analysis was conducted using a nucleotide alignment of the genome sequences of the EV-D68 strains in this study, EV-D68 prototype strain Fermon (original), strain CA/RESP/10-786 (clade A), strain US/KY/14-18953 (clade D1), strain US/CO/13-60 (clade B1), strain NY73 (clade B2), strain Beijing-R0132 (clade B3), and strain JPOC10-290 (clade C) was generated by ClustalX version 2.0 [52], and edited manually. Once aligned, similarity plot and bootscan analyses were performed using SimPlot version 3.5.1 (National Institute of Virology, Pune, India) (window size, 500 bp; step, 20 bp) [53]. Potential recombination within the genome sequences of EV-D68 was also examined by the Recombination Detection Program version 4.46 (RDP4) (University of Cape Town, Cape Town, South Africa) [54]. Phylogenetic incongruence between different regions and with p values of less than 0.05 in the methods further supported the presence of recombination events.
4.7. Nucleotide Sequence Accession Numbers
The complete genome sequences of the three EV-D68 strains and the VP1, partial 5′UTR, and 2C sequences of the EV-D68 strains (Table 1) have been deposited in the GenBank database under accession numbers KY767820 to KY767842.
Acknowledgments
We are grateful to the generous support of Carol Yu, Richard Yu, Hui Hoy, and Hui Ming in the genomic sequencing platform. This work is partly supported by the Health and Medical Research Fund (HMRF), Food and Health Bureau, Hong Kong SAR Government (Ref. No. HKM-15-M02 and HKM-15-M04); National Science and Technology Major Project of China (grant number 2012ZX10004213); University Research Committee, the University of Hong Kong and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health. Views expressed in this paper are those of the authors only, and may not represent the opinion of the Government of the HKSAR.
Abbreviations
| AFM | Acute flaccid myelitis |
| CVA | Cerebrovascular accident |
| CSF | Cerebrospinal fluid |
| EV-D | Enterovirus species D |
| HT | Hypertension |
| kbp | Kilobase pairs |
| ML | Maximum likelihood |
| MRMP | Macrolide-resistant Mycoplasma pneumoniae |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| NPA | Nasopharyngeal aspirate |
| NPS | Nasopharyngeal swab |
| RACE | Rapid amplification of cDNA ends |
| RD | Rhabdomyosarcoma |
| RDP | Recombination Detection Program |
| RSV | Respiratory syncytial virus |
| RT-PCR | Reverse transcription polymerase chain reaction |
| URTI | Upper respiratory tract infection |
| US | United States |
| UTR | Untranslated region |
Appendix A
Table A1.
List of EV-D68 strains used for analysis in the present study.
| Strain | Year of Isolation | Place of Isolation | Clade * | Source | GenBank Accession No. |
|---|---|---|---|---|---|
| Fermon (prototype) | 1962 | USA | Original | GenBank | AY426531 |
| CA/RESP/10-786 | 2013 | USA | A | GenBank | KM892500 |
| NYC403 | 2009 | USA | A | GenBank | JX101846 |
| NZ-2010-541 | 2010 | NZ | A | GenBank | JX070222 |
| HK2/2010 | 2010 | HK | A | GenBank | KT959174 (VP1), |
| KT959114 (2C) | |||||
| TW-02378_2010 | 2010 | TW | A | GenBank | KP657713 (VP1) |
| ITA/19391/12 | 2012 | Italy | A | GenBank | KC763159 (VP1) |
| 2011-21282 | 2011 | China | B1 | GenBank | KT285320 |
| HK8/2011 | 2011 | HK | B1 | GenBank | KT959180 (VP1), |
| KT959120 (2C) | |||||
| HK14/2012 | 2012 | HK | B1 | GenBank | KT959186 (VP1) |
| KT959126 (2C) | |||||
| HK26/2014 | 2014 | HK | B1 | GenBank | KT959198 (VP1), |
| KT959138 (2C) | |||||
| NY316 | 2014 | USA | B1 | GenBank | KP745764 |
| STL_2014_12 | 2014 | USA | B1 | GenBank | KM881710 |
| US/CO/13-60 | 2013 | USA | B1 | GenBank | KP100794 |
| US/CO/14-93 | 2014 | USA | B1 | GenBank | KP126911 |
| US/MO/14-18950 | 2014 | USA | B1 | GenBank | KM851228 |
| NY73 | 2014 | USA | B2 | GenBank | KP745768 |
| NY74 | 2014 | USA | B2 | GenBank | KP745769 |
| US/KY/14-18951 | 2014 | USA | B2 | GenBank | KM851229 |
| US/IL/14-18952 | 2014 | USA | B2 | GenBank | KM851230 |
| 2014-R0672 | 2014 | China | B3 | GenBank | KT280500 |
| Beijing-R0132 | 2014 | China | B3 | GenBank | KP240936 |
| HK25/2014 | 2014 | HK | B3 | GenBank | KT959197 (VP1), |
| KT959137 (2C) | |||||
| HK28/2014 | 2014 | HK | B3 | GenBank | KT959200 (VP1), |
| KT959140 (2C) | |||||
| TW-00932-2014 | 2014 | TW | B3 | GenBank | KT711081 |
| TW-00785-2014 | 2014 | TW | B3 | GenBank | KT711083 |
| NY10_16 | 2016 | USA | B3 | GenBank | KX957754 |
| USA/FL/2016-19504 | 2016 | USA | B3 | GenBank | KX675261 |
| SWE_003_2016 | 2016 | SWE | B3 | GenBank | KY215829 (VP4) |
| SWE_015_2016 | 2016 | SWE | B3 | GenBank | KY215841 (VP4) |
| USA/U2221/2008 | 2008 | USA | C | GenBank | KX255371 |
| USA/MO93/2009 | 2009 | USA | C | GenBank | KX261814 |
| JPOC10-290 | 2010 | Japan | C | GenBank | AB601882 |
| JPOC10-378 | 2010 | Japan | C | GenBank | AB601883 |
| 2011-21286 | 2011 | China | D1 | GenBank | KT306743 |
| 2012-12225 | 2012 | China | D1 | GenBank | KT285319 |
| 2013-1017-26 | 2013 | China | D1 | GenBank | KT280501 |
| HK5/2011 | 2011 | HK | D1 | GenBank | KT959177 (VP1), |
| KT959117 (2C) | |||||
| HK13/2012 | 2012 | HK | D1 | GenBank | KT959185 (VP1), |
| KT959125 (2C) | |||||
| HK21/2013 | 2013 | HK | D1 | GenBank | KT959193 (VP1), |
| KT959133 (2C) | |||||
| US/KY/14-18953 | 2014 | USA | D1 | GenBank | KM851231 |
| TW-02722_2013 | 2013 | TW | D1 | GenBank | KP657720 (VP1) |
| BCH895A | 2008 | China | D2 | GenBank | KF726085 |
| BJ24 | 2008 | China | D2 | GenBank | KU242683 |
| 1737-Yamagata-2008 | 2008 | Japan | D2 | GenBank | AB667899 (VP1) |
| TW-01166_2007 | 2007 | TW | D2 | GenBank | KP657701 (VP1) |
| TW-11134_2009 | 2009 | TW | D2 | GenBank | KP657708 (VP1) |
| V12-2240471 | 2012 | HK | B1 | This study | KY767823 (VP1), |
| KY767829 (5′UTR), | |||||
| KY767836 (2C) | |||||
| V12-2268728 | 2012 | HK | B1 | This study | KY767820 |
| V13-2245157 | 2013 | HK | D1 | This study | KY767821 |
| V14-8133616 | 2014 | HK | B3 | This study | KY767824 (VP1), |
| KY767830 (5′UTR), | |||||
| KY767837 (2C) | |||||
| V14-8135150 | 2014 | HK | B3 | This study | KY767825 (VP1), |
| KY767831 (5′UTR), | |||||
| KY767838 (2C) | |||||
| V14-8143594 | 2014 | HK | B3 | This study | KY767826 (VP1), |
| KY767832 (5′UTR), | |||||
| KY767839 (2C) | |||||
| V14-8143833 | 2014 | HK | B3 | This study | KY767827 (VP1), |
| KY767833 (5′UTR), | |||||
| KY767840 (2C) | |||||
| V14-8151518 | 2014 | HK | B3 | This study | KY767828 (VP1), |
| KY767834 (5′UTR), | |||||
| KY767841 (2C) | |||||
| V14-8151546 | 2014 | HK | B3 | This study | KY767822 |
| V14-8157864 | 2014 | HK | B3 # | This study | KY767835 (5′UTR), |
| KY767842 (2C) |
# VP1 sequence not available; partial 5’UTR and 2C were sequenced (100% identical to those of EV-D68 strain V14-8151546; potential subclade B3). Abbreviations: HK, Hong Kong; NZ, New Zealand; SWE, Sweden; TW, Taiwan; USA, the United States of America.
Author Contributions
Cyril C. Y. Yip, Patrick C. Y. Woo, Kwok-Yung Yuen, and Susanna K. P. Lau designed the study. Janice Y. C. Lo, Kwok-Hung Chan, Jasper F. W. Chan, Vincent C. C. Cheng, and Kwok-Yung Yuen provided virus isolates. Janice Y. C. Lo, Siddharth Sridhar, David C. Lung, Shik Luk, and Susanna K. P. Lau provided clinical data. Cyril C. Y. Yip conducted experiments. Cyril C. Y. Yip and Susanna K. P. Lau performed data analysis. Cyril C. Y. Yip and Susanna K. P. Lau wrote the manuscript. Janice Y. C. Lo, Kwok-Hung Chan, Patrick C. Y. Woo, and Kwok-Yung Yuen corrected the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Schieble J.H., Fox V.L., Lennette E.H. A probable new human picornavirus associated with respiratory diseases. Am. J. Epidemiol. 1967;85:297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- 2.Blomqvist S., Savolainen C., Raman L., Roivainen M., Hovi T. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 2002;40:4218–4223. doi: 10.1128/JCM.40.11.4218-4223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khetsuriani N., Lamonte-Fowlkes A., Oberst S., Pallansch M.A. Centers for Disease Control and Prevention. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill. Summ. 2006;55:1–20. [PubMed] [Google Scholar]
- 4.Ikeda T., Mizuta K., Abiko C., Aoki Y., Itagaki T., Katsushima F., Katsushima Y., Matsuzaki Y., Fuji N., Imamura T., et al. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol. Immunol. 2012;56:139–143. doi: 10.1111/j.1348-0421.2012.00411.x. [DOI] [PubMed] [Google Scholar]
- 5.Imamura T., Fuji N., Suzuki A., Tamaki R., Saito M., Aniceto R., Galang H., Sombrero L., Lupisan S., Oshitani H. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg. Infect. Dis. 2011;17:1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linsuwanon P., Puenpa J., Suwannakarn K., Auksornkitti V., Vichiwattana P., Korkong S., Theamboonlers A., Poovorawan Y. Molecular epidemiology and evolution of human enterovirus serotype 68 in Thailand, 2006–2011. PLoS ONE. 2012;7:e35190. doi: 10.1371/journal.pone.0035190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Q.B., Wo Y., Wang H.Y., Wei M.T., Zhang L., Yang H., Liu E.M., Li T.Y., Zhao Z.T., Liu W., et al. Detection of enterovirus 68 as one of the commonest types of enterovirus found in patients with acute respiratory tract infection in China. J. Med. Microbiol. 2014;63:408–414. doi: 10.1099/jmm.0.068247-0. [DOI] [PubMed] [Google Scholar]
- 8.Meijer A., van der Sanden S., Snijders B.E., Jaramillo-Gutierrez G., Bont L., van der Ent C.K., Overduin P., Jenny S.L., Jusic E., van der Avoort H.G., et al. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology. 2012;423:49–57. doi: 10.1016/j.virol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 9.Opanda S.M., Wamunyokoli F., Khamadi S., Coldren R., Bulimo W.D. Genetic diversity of human enterovirus 68 strains isolated in Kenya using the hypervariable 3′ end of VP1 gene. PLoS ONE. 2014;9:e102866. doi: 10.1371/journal.pone.0102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piralla A., Girello A., Grignani M., Gozalo-Marguello M., Marchi A., Marseglia G., Baldanti F. Phylogenetic characterization of enterovirus 68 strains in patients with respiratory syndromes in Italy. J. Med. Virol. 2014;86:1590–1593. doi: 10.1002/jmv.23821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokarz R., Firth C., Madhi S.A., Howie S.R., Wu W., Sall A.A., Haq S., Briese T., Lipkin W.I. Worldwide emergence of multiple clades of enterovirus 68. J. Gen. Virol. 2012;93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enterovirus D68. [(accessed on 13 March 2017)]; Available online: https://www.cdc.gov/non-polio-enterovirus/about/ev-d68.html.
- 13.Greninger A.L., Naccache S.N., Messacar K., Clayton A., Yu G., Somasekar S., Federman S., Stryke D., Anderson C., Yagi S., et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012–14): A retrospective cohort study. Lancet Infect. Dis. 2015;15:671–682. doi: 10.1016/S1473-3099(15)70093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau S.K., Yip C.C., Zhao P.S., Chow W.N., To K.K., Wu A.K., Yuen K.Y., Woo P.C. Enterovirus D68 infections associated with severe respiratory illness in elderly patients and emergence of a novel clade in Hong Kong. Sci. Rep. 2016;6:25147. doi: 10.1038/srep25147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis T.C., Kirkegaard K. Poliovirus RNA recombination: Mechanistic studies in the absence of selection. EMBO J. 1992;11:3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashev A.N., Lashkevich V.A., Ivanova O.E., Koroleva G.A., Hinkkanen A.E., Ilonen J. Recombination in circulating enteroviruses. J. Virol. 2003;77:10423–10431. doi: 10.1128/JVI.77.19.10423-10431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yip C.C., Lau S.K., Lo J.Y., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of EV71 isolates from 2004 to 2010 reveals predominance and persistent circulation of the newly proposed genotype D and recent emergence of a distinct lineage of subgenotype C2 in Hong Kong. Virol. J. 2013;10:222. doi: 10.1186/1743-422X-10-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Y., Hassan F., Schuster J.E., Simenauer A., Selvarangan R., Halpin R.A., Lin X., Fedorova N., Stockwell T.B., Lam T.T., et al. Molecular Evolution and Intraclade Recombination of Enterovirus D68 during the 2014 Outbreak in the United States. J. Virol. 2015;90:1997–2007. doi: 10.1128/JVI.02418-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Ye X., Zhang H., Xu X., Li W., Zhu D., Wang M. Antimicrobial susceptibility of Mycoplasma pneumoniae isolates and molecular analysis of macrolide-resistant strains from Shanghai, China. Antimicrob. Agents Chemother. 2009;53:2160–2162. doi: 10.1128/AAC.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du J., Zheng B., Zheng W., Li P., Kang J., Hou J., Markham R., Zhao K., Yu X.F. Analysis of Enterovirus 68 strains from the 2014 north american outbreak reveals a new clade, indicating viral evolution. PLoS ONE. 2015;10:e0144208. doi: 10.1371/journal.pone.0144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Y.N., Yang S.L., Shih S.R., Huang Y.C., Chang P.Y., Huang C.G., Kao K.C., Hu H.C., Liu Y.C., Tsao K.C. Molecular evolution and the global reemergence of enterovirus D68 by genome-wide analysis. Medicine (Baltimore) 2016;95:e4416. doi: 10.1097/MD.0000000000004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pabbaraju K., Wong S., Drews S.J., Tipples G., Tellier R. Full genome analysis of enterovirus D-68 strains circulating in Alberta, Canada. J. Med. Virol. 2016;88:1194–1203. doi: 10.1002/jmv.24444. [DOI] [PubMed] [Google Scholar]
- 24.Huang W., Wang G., Zhuge J., Nolan S.M., Dimitrova N., Fallon J.T. Whole-genome sequence analysis reveals the enterovirus D68 isolates during the United States 2014 outbreak mainly belong to a novel clade. Sci. Rep. 2015;5:15223. doi: 10.1038/srep15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aliabadi N., Messacar K., Pastula D.M., Robinson C.C., Leshem E., Sejvar J.J., Nix W.A., Oberste M.S., Feikin D.R., Dominguez S.R. Enterovirus D68 infection in children with acute flaccid myelitis, Colorado, USA, 2014. Emerg. Infect. Dis. 2016;22:1387–1394. doi: 10.3201/eid2208.151949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messacar K., Schreiner T.L., van Haren K., Yang M., Glaser C.A., Tyler K.L., Dominguez S.R. Acute flaccid myelitis: A clinical review of US cases 2012–2015. Ann. Neurol. 2016;80:326–338. doi: 10.1002/ana.24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sejvar J.J., Lopez A.S., Cortese M.M., Leshem E., Pastula D.M., Miller L., Glaser C., Kambhampati A., Shioda K., Aliabadi N., et al. Acute flaccid myelitis in the United States, August–December 2014: Results of nationwide surveillance. Clin. Infect. Dis. 2016;63:737–745. doi: 10.1093/cid/ciw372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grard G., Drexler J.F., Lekana-Douki S., Caron M., Lukashev A., Nkoghe D., Gonzalez J.P., Drosten C., Leroy E. Type 1 wild poliovirus and putative enterovirus 109 in an outbreak of acute flaccid paralysis in Congo, October–November 2010. Euro Surveill. 2010;15:pii:19723. doi: 10.2807/ese.15.47.19723-en. [DOI] [PubMed] [Google Scholar]
- 29.Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Velez C.M., Anderson M.S., Robinson C.C., McFarland E.J., Nix W.A., Pallansch M.A., Oberste M.S., Glodé M.P. Outbreak of neurologic enterovirus type 71 disease: A diagnostic challenge. Clin. Infect. Dis. 2007;45:950–957. doi: 10.1086/521895. [DOI] [PubMed] [Google Scholar]
- 31.Teoh H.L., Mohammad S.S., Britton P.N., Kandula T., Lorentzos M.S., Booy R., Jones C.A., Rawlinson W., Ramachandran V., Rodriguez M.L., et al. Clinical characteristics and functional motor outcomes of enterovirus 71 neurological disease in children. JAMA Neurol. 2016;73:300–307. doi: 10.1001/jamaneurol.2015.4388. [DOI] [PubMed] [Google Scholar]
- 32.Kreuter J.D., Barnes A., McCarthy J.E., Schwartzman J.D., Oberste M.S., Rhodes C.H., Modlin J.F., Wright P.F. A fatal central nervous system enterovirus 68 infection. Arch. Pathol. Lab. Med. 2011;135:793–796. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 33.Levy A., Roberts J., Lang J., Tempone S., Kesson A., Dofai A., Daley A.J., Thorley B., Speers D.J. Enterovirus D68 disease and molecular epidemiology in Australia. J. Clin. Virol. 2015;69:117–121. doi: 10.1016/j.jcv.2015.06.079. [DOI] [PubMed] [Google Scholar]
- 34.Hixon A.M., Yu G., Leser J.S., Yagi S., Clarke P., Chiu C.Y., Tyler K.L. A mouse model of paralytic myelitis caused by enterovirus D68. PLoS Pathog. 2017;13:e1006199. doi: 10.1371/journal.ppat.1006199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyrdak R., Grabbe M., Hammas B., Ekwall J., Hansson K.E., Luthander J., Naucler P., Reinius H., Rotzén-Östlund M., Albert J. Outbreak of enterovirus D68 of the new B3 lineage in Stockholm, Sweden, August to September 2016. Euro Surveill. 2016;21:pii:30403. doi: 10.2807/1560-7917.ES.2016.21.46.30403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoester M., Schölvinck E.H., Poelman R., Smit S., Vermont C.L., Niesters H.G., van Leer-Buter C.C. Upsurge of enterovirus D68, the Netherlands, 2016. Emerg. Infect. Dis. 2017;23:140–143. doi: 10.3201/eid2301.161313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esposito S., Chidini G., Cinnante C., Napolitano L., Giannini A., Terranova L., Niesters H., Principi N., Calderini E. Acute flaccid myelitis associated with enterovirus-D68 infection in an otherwise healthy child. Virol. J. 2017;14:4. doi: 10.1186/s12985-016-0678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan Y.F., Sam I.C., AbuBakar S. Phylogenetic designation of enterovirus 71 genotypes and subgenotypes using complete genome sequences. Infect. Genet. Evol. 2010;10:404–412. doi: 10.1016/j.meegid.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Oberste M.S., Maher K., Kilpatrick D.R., Flemister M.R., Brown B.A., Pallansch M.A. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 1999;37:1288–1293. doi: 10.1128/jcm.37.5.1288-1293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberste M.S., Maher K., Kilpatrick D.R., Pallansch M.A. Molecular evolution of the human enteroviruses: Correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown B.A., Nix W.A., Sheth M., Frace M., Oberste M.S. Seven strains of enterovirus D68 Detected in the United States during the 2014 severe respiratory disease outbreak. Genome Announc. 2014;2:pii:e01201-14. doi: 10.1128/genomeA.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson P., Edman K., Lindberg A.M. Molecular analysis of the echovirus 18 prototype: Evidence of interserotypic recombination with echovirus 9. Virus Res. 2002;85:71–83. doi: 10.1016/S0168-1702(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 43.Lindberg A.M., Andersson P., Savolainen C., Mulders M.N., Hovi T. Evolution of the genome of Human enterovirus B: Incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 2003;84:1223–1235. doi: 10.1099/vir.0.18971-0. [DOI] [PubMed] [Google Scholar]
- 44.Lukashev A.N., Lashkevich V.A., Koroleva G.A., Ilonen J., Hinkkanen A.E. Recombination in uveitis-causing enterovirus strains. J. Gen. Virol. 2004;85:463–470. doi: 10.1099/vir.0.19469-0. [DOI] [PubMed] [Google Scholar]
- 45.Blomqvist S., Savolainen-Kopra C., Paananen A., El Bassioni L., El Maamoon Nasr E.M., Firstova L., Zamiatina N., Kutateladze T., Roivainen M. Recurrent isolation of poliovirus 3 strains with chimeric capsid protein Vp1 suggests a recombination hot-spot site in Vp1. Virus Res. 2010;151:246–251. doi: 10.1016/j.virusres.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Martín J., Samoilovich E., Dunn G., Lackenby A., Feldman E., Heath A., Svirchevskaya E., Cooper G., Yermalovich M., Minor P.D. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J. Virol. 2002;76:10921–10928. doi: 10.1128/JVI.76.21.10921-10928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phuektes P., Chua B.H., Sanders S., Bek E.J., Kok C.C., McMinn P.C. Mapping genetic determinants of the cell-culture growth phenotype of enterovirus 71. J. Gen. Virol. 2011;92:1380–1390. doi: 10.1099/vir.0.029371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X., Holloway B., Dare R.K., Kuypers J., Yagi S., Williams J.V., Hall C.B., Erdman D.D. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campanella J.J., Bitincka L., Smalley J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003;4:29. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yip C.C., Lau S.K., Woo P.C., Wong S.S., Tsang T.H., Lo J.Y., Lam W.K., Tsang C.C., Chan K.H., Yuen K.Y. Recombinant coxsackievirus A2 and deaths of children, Hong Kong, 2012. Emerg. Infect. Dis. 2013;19:1285–1288. doi: 10.3201/eid1908.121498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]