Abstract
Heartwater, a tick-borne disease of domestic and wild ruminants, is caused by the intracellular rickettsia Ehrlichia ruminantium (previously known as Cowdria ruminantium). It is a major constraint to livestock production throughout subSaharan Africa, and it threatens to invade the Americas, yet there is no immediate prospect of an effective vaccine. A shotgun genome sequencing project was undertaken in the expectation that access to the complete protein coding repertoire of the organism will facilitate the search for vaccine candidate genes. We report here the complete 1,516,355-bp sequence of the type strain, the stock derived from the South African Welgevonden isolate. Only 62% of the genome is predicted to be coding sequence, encoding 888 proteins and 41 stable RNA species. The most striking feature is the large number of tandemly repeated and duplicated sequences, some of continuously variable copy number, which contributes to the low proportion of coding sequence. These repeats have mediated numerous translocation and inversion events that have resulted in the duplication and truncation of some genes and have also given rise to new genes. There are 32 predicted pseudogenes, most of which are truncated fragments of genes associated with repeats. Rather then being the result of the reductive evolution seen in other intracellular bacteria, these pseudogenes appear to be the product of ongoing sequence duplication events.
Keywords: gene duplication, bacterial genome, molecular sequence data, intracellular adaptation
Ehrlichia ruminantium (previously known as Cowdria ruminantium) is an obligate intracellular bacterium in the order Rickettsiales. Species in this order cause serious diseases in man and domestic animals throughout the world. E. ruminantium is transmitted by ticks of the genus Amblyomma and causes heartwater, a fatal and economically important disease of wild and domestic ruminants. The disease occurs throughout subSaharan Africa and on several Caribbean islands, from which it threatens to invade the Americas (1), but the existing immunization procedures are rudimentary and relatively ineffective (2). E. ruminantium is a fragile bacterium with exacting culture requirements in eukaryotic cell lines; genetic manipulation has not been attempted, and little is known about its mechanisms of virulence or pathogenesis. Heartwater affects all domestic ruminants, and 80-95% of naïve animals die within 3 weeks, but those that recover have a T cell-mediated immunity to subsequent homologous challenge (3). In the absence of any directed strategy to identify T cell-stimulatory proteins we sequenced the E. ruminantium genome in the expectation that access to the complete protein-coding repertoire of the organism would facilitate the search for vaccine candidate genes.
Materials and Methods
Genome Sequencing and Assembly. The genome sequence was obtained by whole-genome shotgun sequencing of clones from two small insert E. ruminantium (Welgevonden-type strain) genomic libraries. An existing library constructed in Lambda ZAP II (4) was used for initial sequencing. A second small insert library was constructed in pMOSBlue (Amersham Pharmacia) by using E. ruminantium genomic DNA prepared from the culture stock derived from the South African Welgevonden isolate (5) grown in a bovine aorta endothelial cell line (6). Genomic DNA was nebulized for 2 min at 100 kPa in a Medel (San Polo di Torrile, Italy) jet nebulizer reservoir, and fragments in the 600- to 1,500-bp range were cloned into the plasmid. All sequencing was done by using BigDye chemistry (Applied Biosystems) on ABI377 and ABI3100 sequencers. Sequences were assembled by using phrap (www.phrap.org) and gap4 (7) was used for manual checking and editing. Initial contig ordering was performed by comparing the positions of mapped genes and restriction sites to the physical map (8) and by exploiting synteny with the preliminary genomic sequence of Ehrlichia chaffeensis, the closest relative of E. ruminantium for which genomic data are available. The preliminary sequence was made available by The Institute for Genomic Research (www.tigr.org). Contig joining was performed by sequencing genomic PCR products spanning the gaps, and repeat regions and areas represented by single reads or clones were verified by the same means. The final assembly contains 25,648 reads with an average length of 569 bp, giving 9.6-fold coverage of the genome. The assembly was finally confirmed by comparison with the physical map (8).
Annotation and Analysis. Three gene modeling programs, genemarks (9), orpheus (10), and glimmer (11), were used to predict potential coding sequences (CDSs). The results were combined and checked manually and rbsfinder (www.tigr.org/software) was used to assist with the location of start codons. Proteins predicted from the revised gene model were searched against nonredundant protein databases by using fasta (12) and blastp (13). Domain analysis of predicted proteins was performed by searching pfam (14) and prosite (15). Transmembrane helices were identified by using tmhmm2.0 (16), and signalp (17) was used to detect putative signal peptides. Transfer RNAs were identified by trnascan-se (18). mreps (19) and tandem repeats finder (20) were used to detect tandem repeats. Genes that may have been acquired by horizontal gene transfer were identified by using sigi (21). The results of all searches were assembled, and predicted proteins were manually annotated in the artemis sequence viewer (22). Putative metabolic pathways were analyzed by using priam (23) and the online pathway tools on the Kyoto Encyclopedia of Genes and Genomes web site (24). A pathway/genome database was created, and a metabolic reconstruction was performed by using the pathologic component of the pathway tools software (25).
Results and Discussion
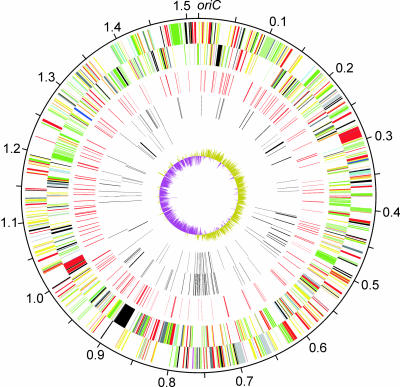
General Features of the Genome. The circular genome of the Welgevonden strain of E. ruminantium is 1,516,355 bp in length. As in the genomes of many other endosymbionts and intracellular pathogens, E. ruminantium has a low G + C content (27.5%). We identified 920 coding sequences with an average length of 1,032 bp, of which 32 (3.5%) probably represent pseudogenes (Fig. 1 and Table 1). The protein-coding capacity of the genome (62.0%) is even lower than that of Rickettsia prowazekii, which is 76% coding (26). We could assign functional information (including similarity to genes of known function, matches to pfam or prosite entries, or informative hydrophobicity plots) to 758 CDSs (82.8%). Of the CDSs, 50 (5.5%) were similar to conserved hypothetical genes of unknown function and 80 (8.7%) did not show any sequence similarity to known genes in other organisms, nor was any other functional information identified. The E. ruminantium genome is remarkable in possessing large numbers of repetitive sequences, which constitute 8.5% of the chromosome. The complete annotated sequence data have been submitted to European Molecular Biology Laboratory Bank under accession no. CR767821 and a full list of genes is given in Table 2, which is published as supporting information on the PNAS web site.
Fig. 1.
Circular representation of the genome of E. ruminantium (Welgevonden type strain). The outermost circle indicates the scale in megabases. The remaining concentric circles are described from outside to inside. The first and second circles represent the predicted coding sequences on the plus and minus strands, respectively, and are color-coded by function: dark blue, stable RNAs; black, chaperones and transporters; dark gray, energy metabolism; red, information transfer; yellow, central or intermediary metabolism; dark green, membrane proteins; cyan, degradation of large molecules; purple, degradation of small molecules; pale blue, regulators; orange, conserved hypothetical proteins; pink, phage and insertion sequence elements; brown, pseudogenes; pale green, unknown; light gray, miscellaneous. The third circle represents tandem repeats (shown in red). The fourth and fifth circles represent dispersed repeats (direct and inverted repeats, respectively), which are colored in black. The sixth circle represents G+C skew, with values greater than zero in olive and less than zero in magenta. A full-sized version of this figure is available as Fig. 4, which is published as supporting information on the PNAS web site.
Table 1. General features of the genome of the Welgevonden strain of E. ruminantium.
| Size, bp | 1,516,355 |
| G + C content, % | 27.5 |
| Protein coding regions,* % | 62.0 |
| CDSs total, n | 920 |
| Average length, bp | 1,032 |
| Probable pseudogenes, n (%) | 32 (3.5) |
| Average length, bp | 276 |
| Predicted protein CDSs, n (%) | 888 (96.5) |
| Average length, bp | 1,059 |
| CDSs with functional information,†n (%) | 758 (82.8) |
| Conserved hypothetical genes, n (%) | 50 (5.5) |
| Genes with no functional information, n (%) | 80 (8.7) |
| Stable RNAs | |
| rRNAs, n | 3 |
| tRNAs, n | 36 |
| Other RNAs (tmRNA/rnpB), n | 2 |
| Tandem repeats, bp (%) | 82,172 (5.4) |
| Dispersed repeats (direct and inverted), bp (%) | 43,976 (2.9) |
Not including pseudogenes.
Includes CDSs with database matches to genes of known function, matches to pfam or prosite entries, or informative hydrophobicity plots.
The real origin of replication (oriC) of the E. ruminantium genome was not experimentally determined, but we found a clear shift in GC-skew values, which frequently occurs close to the origin and termination of replication (27). There are many duplications and translocations in the area around the shift in GC-skew value between 740 and 770 kb (Fig. 1), suggesting that this region has a high rate of DNA reorganization. Comparisons of closely related bacteria have revealed a high frequency of recombination in the terminus region, which may be related to the mechanism of chromosome separation after replication (28). Therefore, an arbitrary position near the opposite transition in GC-skew values was chosen as base pair 1 of the genome (Fig. 1). In other bacteria, a conserved arrangement of 12 genes has been observed around oriC (29), which is often located close to the dnaA gene, but, in E. ruminantium, all 12 of these genes were scattered throughout the genome. The dnaA gene itself, which codes for chromosomal replication initiator protein, is located >200 kb away from the nearest transition in GC-skew values.
E. ruminantium has only a single rRNA operon, and the 16S gene is separated by 550 kb from the 23S-5S gene pair. Bacteria frequently have multiple rRNA operons (30), with the genes normally being arranged in the order 16S-23S-5S (31), but other Rickettsiales also have single rRNA operons in which the 16S gene is separated from the 23S-5S gene pair (32, 33). This atypical arrangement apparently arose in a common ancestor of the Rickettsiales after their divergence from the mitochondrial lineage (34), and it is a typical example of the scrambled gene organization that is a characteristic feature of intracellular bacteria (35). In E. ruminantium, in addition to the dispersion of the genes around oriC and the splitting of the rRNA operon, we found other genes that occur in operons in free-living bacteria to be dispersed throughout the genome. Examples include the ATP-synthase complex, the electron transfer complex, the NADH dehydrogenase complex, and the superribosomal protein gene operon (35). In the last mentioned case, the disrupted gene arrangement has been described for R. prowazekii, where it is thought to have arisen as the result of an intrachromosomal recombination event between two ancestral tuf genes, followed by the deletion of one copy of tuf (35). The disrupted arrangement of this operon is similar to that in R. prowazekii, but E. ruminantium has not lost the second copy of tuf.
Central Metabolic Pathways. Reconstruction of the central metabolic pathways of E. ruminantium depicts an aerobic organism that probably does not ferment carbohydrates, such as glucose. Only a summary of the main points can be given here because of space constraints, a schematic overview is shown in Fig. 3, which is published as supporting information on the PNAS web site.
Many of the essential genes for the glycolytic pathway are absent; a glucose transport system was not found, nor were any enzymes for the Entner-Douderoff pathway. The primary carbon sources are likely to be proline and glutamate, and transporters for both were identified, together with enzymes for the conversion of proline to glutamate. This prediction is supported by the observation that the proline consumption of E. ruminantium-infected mammalian cells is increased in comparison with uninfected cells (36). All enzymes of the tricarboxylic acid pathway were found, and a putative glutamate dehydrogenase, which would enable transfer of glutamate into the tricarboxylic acid cycle. There is a partial gluconeogenesis pathway, terminating at fructose-6-phosphate, as well as a complete nonoxidative pentose-phosphate pathway.
Complete pathways for synthesis of purine and pyrimidine nucleotides were identified. These pathways also occur in other members of the family Anaplasmataceae, Wolbachia pipientis (37), and Anaplasma marginale (38). In contrast, R. prowazekii (26) and Rickettsia conorii (39) lack the ability to synthesize nucleotides. Rickettsia spp. grow freely in the cytoplasm of their host cells, where host nucleotides would be expected to be easily available, which may explain why these organisms have dispensed with the energy-intensive de novo synthesis of nucleotides. In contrast, organisms in the family Anaplasmataceae (Anaplasma, Ehrlichia, and Wolbachia) replicate in an intracellular vacuole, and they appear to have been obliged to retain their ability to synthesis nucleotides.
E. ruminantium has genes for the biosynthesis of five amino acids (arginine, lysine, proline, glutamate, and glutamine); hence, other amino acids are presumably obtained from the host cell, although specific amino acid transporters could not be identified. The situation is similar in A. marginale (38), which can apparently synthesize eight amino acids and yet has no specific transporters for the others. E. ruminantium has no gene for thymidylate synthase (thyA); instead, a gene for f lavin-dependent thymidylate synthase (thy1) was found, on which the organism must rely for thymidylate synthesis (40).
E. ruminantium has only two possible genes for enzymes involved in the metabolism of amino sugars (Erum5510 and Erum6670) and no genes for the biosynthesis of the cell wall components lipid A and murein sacculus. The inability to synthesize cell wall components is a common feature among the Anaplasmataceae, and the organism probably uses cholesterol from the host cell to compensate, as has been shown to occur in E. chaffeensis and Anaplasma phagocytophilum (41). A. marginale also has incomplete pathways for the synthesis of lipopolysaccharides, peptidoglycans, and murein sacculus, although it has more enzymes than E. ruminantium for the metabolism of amino sugars and peptidoglycans (38).
Energy Metabolism. E. ruminantium has genes for enzyme complexes typical of aerobic respiration, including the ATP synthase complex, a possible NADH dehydrogenase complex, the cytochrome bc1 reductase complex, the cytochrome oxidase complex, and a complete succinate dehydrogenase complex that allows linkage between the tricarboxylic acid cycle and the aerobic electron transport chain. No ATP/ADP translocases could be identified, indicating that E. ruminantium does not make use of ATP from the host cell, unlike many species of Rickettsia (42). The energy metabolism genes are listed in Table 2.
Information Transfer. E. ruminantium possesses genes for DNA replication and repair, although some genes that are present in free-living organisms are missing, as is the case with R. prowazekii (26). There are sets of genes for RNA synthesis, modification, and degradation, for tRNA synthesis and ribosomal protein synthesis (except for L30), for transcription and translation, and for homologous recombination. We also found a gene for tmRNA, which is responsible for tagging incomplete proteins on stalled ribosomes for proteolysis. The information transfer genes are listed in Table 2.
Transporters. The E. ruminantium genome sequence contains numerous orthologs involved in eubacterial membrane transport systems, several of which are ATP-binding cassette transporters and others that are involved in the import and efflux of cations. There are also two transporters putatively involved in multidrug efflux. E. ruminantium has the same basic secretion mechanisms that are found in free-living proteobacteria, including common chaperones, such as dnaK, dnaJ, hslU, hslV, groEL, groES, and htpG; genes of the secA-dependent secretion system; and the sec-independent secretion system tat. The transporter genes are listed in Table 2.
Pathogenicity-Associated Genes. A type IV secretion system was identified that contains homologues of the virB gene operon, a system that has been well characterized in Agrobacterium tumefaciens (43, 44). E. ruminantium possesses two clusters of virB genes (virD4/virB8/virB9/virB10/virB11 and virB3/virB4/virB6) and three additional large genes (Erum5210, Erum5220, and Erum5230), which probably encode type IV secretion proteins. We did not find the virB1, virB2, virB5, and virB7 genes, genes for the effector proteins VirD2, VirE2 and VirF, or genes for the regulatory proteins VirA and VirG. A putative trbG gene was located that is involved in conjugal transfer of T-DNA in A. tumefaciens. The arrangement of virB genes in E. ruminantium is similar to that in A. phagocytophilum and E. chaffeensis (45), and, in view of the dispersed nature of many Ehrlichia and Anaplasma genes, this conservation of operon structure may have considerable significance. The system is known to be essential for the survival and/or virulence of several intracellular bacterial pathogens of plants and animals (46-48) and could well be involved in pathogenesis in E. ruminantium.
Membrane Protein Genes. There are many possible membrane proteins in the E. ruminantium genome: 28% (247) of all CDSs, other than pseudogenes, are predicted to contain at least one transmembrane helix, 197 of which begin within the first 10 aa of the protein. Signal peptides were predicted for 66 CDS, of which 53 contained no predicted transmembrane helix. It is difficult to differentiate between N-terminal signal peptides and N-terminal transmembrane helices (49), so many of the 197 predicted N-terminal transmembrane domains could, in fact, be signal sequences. This situation allows for up to 263 potential signal sequences and, hence, the possibility that 29.6% of all CDSs code for membrane proteins. There are 37 CDSs containing 1-20 transmembrane helices, but no likely signal peptide, of which 3 are pseudogenes and 24 are unknown, whereas 10 have homologues in other bacteria.
Several families of hypothetical membrane protein genes were identified. We assigned genes to a family if they were predicted to code for proteins of similar lengths, had similar features, and had a mean of all pairwise identities that did not fall below the 15-25% “twilight zone,” below which a common origin is unlikely (50). One of these, the E. ruminantium map1 multigene family of outer-membrane proteins, consists of 16 paralogs (51), having identities ranging from 13.3% to 66.5%, with a mean of 35.1%. Homologues of the map1 family occur in Ehrlichia canis (52), E. chaffeensis (53), A. phagocytophilum (54), and A. marginale (55). These immunodominant proteins are some of the first proteins to have been identified for each of the organisms mentioned, but they have not been found to be good vaccine candidates. It has been suggested that, in the case of E. ruminantium, the function of the most immunodominant of these outer-membrane proteins, MAP1, is to block an immune response to paralogs that are important to the survival of the organism (51).
Two other potentially interesting membrane protein families were identified of 14 and 10 members neither of which has homologues in other sequenced genomes. The 14-member family (Erum2400-Erum2240) contains paralogs having lengths of 914-1126 bp and identities ranging from 10.8% to 46.6%, with a mean of 18.3%. Seven adjacent members of this family were predicted by sigi to be of alien origin based on codon usage analysis. The 10-member family is split between two loci (Erum2750-Erum2800 and Erum3600-Erum3630). The paralogs have lengths of 1,514-1,962 bp and identities ranging from 18.3% to 36.9%, with a mean of 26.3%.
Fourteen of the 16 proteins of the MAP1 family are predicted to have signal peptides, and all members of the 10- and 14-member membrane protein families are predicted to have either signal peptides or N-terminal transmembrane domains that could be signal peptides. All of these terminal peptides are predicted to be noncleavable (56), which would suggest that the proteins are located on the bacterial inner membrane. It has been experimentally demonstrated, however, that MAP1 is expressed on the parasite surface (57); hence, any or all of the members of these membrane protein families could be located on the parasite surface, although no firm conclusions can be drawn without experimental evidence.
Repeats. A striking feature of the E. ruminantium genome is the large number of repetitive sequences, constituting 8.3% of the chromosome, basically of three sorts: simple sequence repeats (SSRs), which include homopolymeric tracts and short repeats of 2-5 bp; large tandem repeats (LTRs) of 6 bp and up to several hundred bp; and dispersed repeats, which include direct and inverted repeats. The repeats constitute 8.5% of the chromosome and contribute to the high proportion of noncoding sequence, which results in a 20% larger size for the E. ruminantium genome than for R. conorii (1.27 megabases), the next largest member of the Rickettsiales that has been completely sequenced. The smaller W. pipientis genome (1.27 megabases) also contains very high levels of repetitive DNA and mobile elements (37), and it has an irregular GC-skew pattern that has been attributed to intragenomic rearrangements associated with the repeat elements. In the repeat-rich E. ruminantium genome, however, the GC-skew transitions at the origin and termination of replication are maintained.
The E. ruminantium genome contains 126 SSRs, including four homopolymeric tracts of G/C nucleotides and many homopolymeric tracts of A/T nucleotides. In other bacteria, SSRs, including hypervariable homopolymeric tracts (usually of G/C bases), have been implicated in phase variation of surface-associated proteins (58). In the E. ruminantium genome, only one of the homopolymeric G/C tracts is variable, being either 11 or 12 bp long. It is located in a noncoding region and so is unlikely to have any effect on surface-associated protein phase variation. Thirteen SSRs are located within promoter regions upstream of the predicted start codons of genes, and three are located within ORFs close to the start codon. It is possible that these SSRs play a role in promoter regulation or phase variation in the E. ruminantium genome.
We identified 158 LTRs. Fifty (31.6%) occur within a total of 31 genes, whereas 85 (53.8%) are located in noncoding regions. Twenty-three LTRs overlap genes: 3 at the 5′ end, 18 at the 3′ end, and 2 at the 3′ end of one gene and the 5′ end of the following gene. In four cases, LTRs overlap and triplicate the beginning or end of a gene, thereby producing eight pseudogenes, amounting to 25.0% of the pseudogenes identified. Of the 31 CDSs containing LTRs, 27 (87.1%) are genes whose products are predicted to be membrane-associated or are genes unique to E. ruminantium (Table 3, which is published as supporting information on the PNAS web site). Two of these genes (Erum3750 and Erum3980) contain ankyrin repeat domains, which are important in many different types of protein-protein interactions in eukaryotes. The few examples that are found in prokaryotes are thought to have been acquired by horizontal gene transfer from eukaryotic hosts (59). Four E. ruminantium CDSs contain ankyrin repeats, but none of them was predicted by sigi to be of alien origin.
At four sites, the numbers of repeats were found to be variable, and three of these are tandem repeats of different 7-bp motifs, all with highly variable (16.7-38.7, 4-80, 7-88) numbers of the repeated sequence motif. The fourth repeat is 122 bp and occurs with continuously variable frequency from 1.5-7.5 times. Amplification across these regions yielded amplicons of different lengths and, when several clones covering the four repeat regions were sequenced, different clones were found to contain different repeat frequencies. It is not possible to obtain enough E. ruminantium DNA from a single tissue culture flask to generate a genomic library; hence, the sequencing libraries contained DNA pooled from several culture passages, representing many generations of the organism. At four specific repetitive loci, therefore, the repeat frequencies vary dynamically between generations. No additional sequence variations, other than occasional single nucleotide polymorphisms, were found between different clones of any other repetitive or nonrepetitive region; hence, the process that generates the variability is specific to just four repeat regions. Possible mechanisms for the generation of repeats are slipped-strand mispairing (60, 61) and secondary structure formation (62). All three 7-bp tandem repeat regions have a higher G+C content than the rest of the genome and exhibit strand asymmetry (one strand contains predominantly either G or C). High G+C sequences tend to form secondary structures, which can cause the DNA polymerase to pause, resulting in the rapid generation of tandem repeats (62).
There are 75 dispersed repeats (duplicated sequences) in the genome, including 52 direct repeats and 40 inverted repeats, the majority of which occur twice (Table 4, which is published as supporting information on the PNAS web site). Of the 32 putative pseudogenes in the genome, 21 (65.6%) appear to have arisen as a result of translocation and inversion events. Of five large (>1 kb) duplications identified, four are associated with genes and only one is located in an intergenic region.
Duplications Appear to Generate New Genes. Of the 32 predicted pseudogenes in the genome, 29 (90.6%) are associated with repeats and 25 (78.1%) appear to be truncated fragments of other genes. Rather then being the result of the reductive evolution seen in other intracellular bacteria, most of the pseudogenes in E. ruminantium appear to be the product of sequence duplication events.
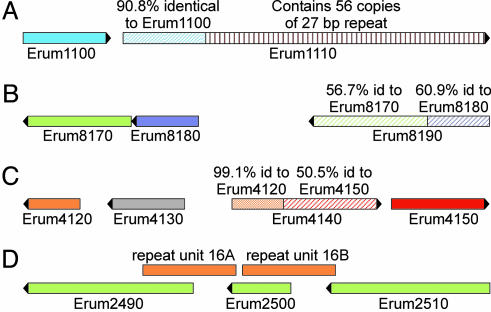
In addition to the generation of pseudogenes, duplications also appear to have resulted in the formation of new genes, and four examples are shown in Fig. 2. The first example (Fig. 2 A) concerns gene Erum1110, which contains an unremarkable 421-bp 5′ region, followed by a 1,511-bp region that consists of a 27-bp motif repeated 56 times. The 5′ region of Erum1110, plus an additional 65 bp upstream of the start codon, has a duplicate with >90% identity, which forms the adjacent gene, Erum1100. Erum1100 terminates where the 27-bp tandem repeat starts in Erum1110. An ortholog of Erum1110 containing 21.7 copies of the 27-bp motif has been identified in E. ruminantium (Highway) (63). It is not possible to distinguish between two possibilities that could have given rise to the current situation: that Erum1100 was duplicated and subsequently became fused with the tandem repeat or that the nonrepetitive part of gene Erum1110 was duplicated and became an independent gene.
Fig. 2.
Schematic representation of E. ruminantium genes that appear to have arisen through duplication events. id, identical.
The second example (Fig. 2B) shows two sections of gene Erum8190, each of which has approximately the same level of identity to one of the two adjacent genes, Erum8170 and Erum8180. These two latter genes would form a single ORF if it were not for the existence of the single stop codon that separates them. It appears likely that Erum8190 was duplicated and that the paralog acquired mutations that eventually led to the appearance of a stop codon and the creation of two new genes. However, without performing transcription and translation studies, it is not possible to be certain that these two genes are expressed.
The third example (Fig. 2C) shows two sections of gene Erum4140, each of which has a very different level of identity to one of the two adjacent genes, Erum4120 and Erum4150. These two latter genes each have homologues in other bacteria, and for this reason and because of their very different levels of identity with Erum4140, they are unlikely to be the products of the duplication of that gene. It appears that, in this case, Erum4150 was duplicated and that the paralog acquired mutations to attain its present level of identity at 50.5%. Much later, Erum4120 was duplicated, and the new paralog fused with the older one to generate the new gene, Erum4140. Erum4120 is believed to code for an iron-sulfur cofactor synthesis protein, and Erum4150 is believed to code for a cysteine desulfurase. If these functions are preserved in the new composite gene, then the product will be a bifunctional “Rosetta Stone” protein (64). A similar bifunctional protein occurs in a Parachlamydia sp. (TrEMBL accession no. Q6MEW3).
The fourth example (Fig. 2D) shows a direct repeat (repeat units 16A and 16B) that appears to have resulted in the duplication of the 3′ end of Erum2490 to create the new gene, Erum2500. We searched for this new gene in 11 other stocks of E. ruminantium by performing PCR amplifications of genomic DNA by using primers at the 5′ and 3′ ends of the gene (data not shown). The gene was found in six stocks that fall phylogenetically into the “southern” African clade of E. ruminantium stocks (Blaauwkrans, Ball 3, Kwanyanga, Gardel, Mara 87/7, and Vosloo) but not in five stocks in the “western” African clade (Pokoase, Sankat, Senegal, Mali, and Kümm 1) (65). These findings suggest that the duplication event that gave rise to Erum2500 occurred in southern Africa some time after E. ruminantium had spread out from southern Africa, where the species is believed to have originated (65).
Adaptation in the Intracellular Environment. It is well established that free-living bacteria readily acquire alien genes by horizontal gene transfer and that this is a major mechanism in the evolution of bacterial pathogenesis and symbiosis (66). Many pathogenic bacteria appear to have acquired 10-15% of their genes by this means (21). The figure predicted by sigi is much lower in the case of E. ruminantium, where 3% of the genes are predicted to be of alien origin. The corresponding figure for R. prowazekii is 0.9%. In general, only one species of intracellular parasite inhabits a host cell, which restricts the parasite's access to new genes (35), so the small number of genes acquired by horizontal gene transfer is to be expected.
Genome reduction in bacteria appears to occur by way of intrachromosomal recombination events at repeated sequences that lead to deletions (61). Intracellular bacteria being unable to regain the lost sequences from other bacterial species through horizontal transfer thus suffer a loss of genes whose products must then be obtained from the host. This process leads to the reductive evolution frequently seen for these organisms (35). The active duplication of tandemly repeated sequences in E. ruminantium, however, appears to be operating to counter reductive evolution and to create new genes, and this may be an alternative mechanism for adaptation to the host by increasing antigenic diversity. Support for this idea lies in the observation that orthologs of the duplicated genes represented in Fig. 2 have been identified in E. ruminantium (Highway) by screening of an expression library with immune serum (63). This indicates that the products of these genes are readily visible to the immune system of the host and that they therefore play a role in immune recognition. Recombinant proteins from these genes, however, did not stimulate lymphoproliferation of mononuclear cells from E. ruminantium-immune animals (63), suggesting that the genes are not likely to be good vaccine candidates.
There is other evidence that, although immune recognition proteins may play an essential survival role, their genes may not be useful as vaccine candidates. In Mycoplasma hyorhinis, for example, there appears to be positive selection for surface lipoproteins containing large numbers of tandem repeats, and it has been suggested that these act as a protective shield for other surface proteins that are less free to change (67). A similar suggestion has been made in respect of the map1 gene of E. ruminantium (51).
This genome sequencing project was undertaken to facilitate the search for vaccine candidate genes. The criteria for identifying such genes from their sequences are not well formulated, and although one may speculate that secreted proteins or outer-membrane proteins might be good candidates, there is little experimental evidence on which to base such a conclusion. It may be possible to identify peptide motifs that are likely to be recognized by T cells from immunized ruminants (68), but this approach currently suffers from a lack of extensive ruminant MHC data. Access to the complete coding repertoire of the organism will, however, enable the design of experiments to screen genes for possible use in a vaccine formulation, either as the genes themselves (in a DNA vaccine vector) or as recombinant proteins.
Supplementary Material
Acknowledgments
We thank Sharen Bowman, Julian Parkhill, Kim Rutherford, Lee Murphy, Rob Davies, Matthew Berriman, and the staff of the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute for assistance with sequencing and data processing; Rainer Merkl (Institut für Mikrobiologie und Genetic, Göttingen, Germany) for performing the sigi analysis; and Ian Paulsen (Institute for Genomic Research, Rock-ville, MD) for performing a transporter prediction analysis. This research was sponsored by the Agricultural Research Council of South Africa. The majority of the financial support came from the LEAD Programmes Fund of the Department of Science and Technology of South Africa. We also received partial support through a grant from the Netherlands Organization for Scientific Research/Netherlands Foundation for the Advancement of Tropical Research to the University of Utrecht. The preliminary sequence of E. chaffeensis was supported by National Institutes of Health Grant R01 AI47885 (to Y. Rikihisa, Ohio State University, Columbus).
Author contributions: N.E.C., E.P.d.V., K.A.B., and B.A.A. designed research; N.E.C., J.L., E.P.d.V., K.A.B., E.L., A.P., F.E.F., H.v.H., A.J., H.C.S., F.v.S., E.Z., J.C.M., D.B., M.T.A., and B.A.A. performed research; N.E.C., J.L., E.P.d.V., E.L., A.L.P., M.v.K., M.B., F. Joubert, C.H.C., N.R.T., M.T.A., and B.A.A. analyzed data; and N.E.C., J.L., K.A.B., M.T.A., and B.A.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CDS, coding sequence; SSR, simple sequence repeat; LTR, large tandem repeat.
Data deposition: The sequence data have been deposited in the European Molecular Biology Laboratory database (EMBL accession no. CR767821).
References
- 1.Provost, A. & Bezuidenhout, J. D. (1987) Onderstepoort J. Vet. Res. 54, 165-169. [PubMed] [Google Scholar]
- 2.Oberem, P. T. & Bezuidenhout, J. D. (1987) Onderstepoort J. Vet. Res. 54, 485-488. [PubMed] [Google Scholar]
- 3.Totte, P., McKeever, D., Martinez, D. & Bensaid, A. (1997) Infect. Immun. 65, 236-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brayton, K. A., Fehrsen, J., de Villiers, E. P., van Kleef, M. & Allsopp, B. A. (1997) Vet. Parasitol. 72, 185-199. [DOI] [PubMed] [Google Scholar]
- 5.Du Plessis, J. L. (1985) Onderstepoort J. Vet. Res. 52, 55-61. [PubMed] [Google Scholar]
- 6.Zweygarth, E., Josemans, A. I. & Horn, E. (1998) Ann. N. Y. Acad. Sci. 849, 307-312. [DOI] [PubMed] [Google Scholar]
- 7.Bonfield, J. K., Smith, K. & Staden, R. (1995) Nucleic Acids Res. 23, 4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Villiers, E. P., Brayton, K. A., Zweygarth, E. & Allsopp, B. A. (2000) Microbiology 146, 2627-2634. [DOI] [PubMed] [Google Scholar]
- 9.Besemer, J., Lomsadze, A. & Borodovsky, M. (2001) Nucleic Acids Res. 29, 2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frishman, D., Mironov, A., Mewes, H. W. & Gelfand, M. (1998) Nucleic Acids Res. 26, 2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson, W. R. (2000) Methods Mol. Biol. 132, 185-219. [DOI] [PubMed] [Google Scholar]
- 13.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E. L., et al. (2004) Nucleic Acids Res. 32, D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigrist, C. J., Cerutti, L., Hulo, N., Gattiker, A., Falquet, L., Pagni, M., Bairoch, A. & Bucher, P. (2002) Brief Bioinform. 3, 265-274. [DOI] [PubMed] [Google Scholar]
- 16.Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001) J. Mol. Biol. 305, 567-580. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Int. J. Neural Syst. 8, 581-599. [DOI] [PubMed] [Google Scholar]
- 18.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolpakov, R., Bana, G. & Kucherov, G. (2003) Nucleic Acids Res. 31, 3672-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson, G. (1999) Nucleic Acids Res. 27, 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkl, R. (2004) BMC Bioinformatics 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944-945. [DOI] [PubMed] [Google Scholar]
- 23.Claudel-Renard, C., Chevalet, C., Faraut, T. & Kahn, D. (2003) Nucleic Acids Res. 31, 6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H. & Kanehisa, M. (1999) Nucleic Acids Res. 27, 29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karp, P. D., Paley, S. & Romero, P. (2002) Bioinformatics 18, Suppl. 1, 225-232. [DOI] [PubMed] [Google Scholar]
- 26.Andersson, S. G., Zomorodipour, A., Andersson, J. O., Sicheritz-Ponten, T., Alsmark, U. C., Podowski, R. M., Naslund, A. K., Eriksson, A. S., Winkler, H. H. & Kurland, C. G. (1998) Nature 396, 133-140. [DOI] [PubMed] [Google Scholar]
- 27.Lobry, J. R. (1996) Mol. Biol. Evol. 13, 660-665. [DOI] [PubMed] [Google Scholar]
- 28.Hughes, D. (2000) Genome Biol. 1, REVIEWS0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogasawara, N. & Yoshikawa, H. (1992) Mol. Microbiol. 6, 629-634. [DOI] [PubMed] [Google Scholar]
- 30.Acinas, S. G., Marcelino, L. A., Klepac-Ceraj, V. & Polz, M. F. (2004) J. Bacteriol. 186, 2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krawiec, S. & Riley, M. (1990) Microbiol. Rev. 54, 502-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson, S. G., Stothard, D. R., Fuerst, P. & Kurland, C. G. (1999) Mol. Biol. Evol. 16, 987-995. [DOI] [PubMed] [Google Scholar]
- 33.Rurangirwa, F. R., Brayton, K. A., McGuire, T. C., Knowles, D. P. & Palmer, G. H. (2002) Int. J. Syst. Evol. Microbiol. 52, 1405-1409. [DOI] [PubMed] [Google Scholar]
- 34.Lang, B. F., Burger, G., O'Kelly, C. J., Cedergren, R., Golding, G. B., Lemieux, C., Sankoff, D., Turmel, M. & Gray, M. W. (1997) Nature 387, 493-497. [DOI] [PubMed] [Google Scholar]
- 35.Andersson, S. G. & Kurland, C. G. (1998) Trends Microbiol. 6, 263-268. [DOI] [PubMed] [Google Scholar]
- 36.Josemans, A. I. & Zweygarth, E. (2002) Ann. N.Y. Acad. Sci. 969, 141-146. [DOI] [PubMed] [Google Scholar]
- 37.Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., McGraw, E. A., Martin, W., Esser, C., Ahmadinejad, N., et al. (2004) PLoS Biol. 2, E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brayton, K. A., Kappmeyer, L. S., Herndon, D. R., Dark, M. J., Tibbals, D. L., Palmer, G. H., McGuire, T. C. & Knowles, D. P., Jr. (2005) Proc. Natl. Acad. Sci. USA 102, 844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogata, H., Audic, S., Renesto-Audiffren, P., Fournier, P. E., Barbe, V., Samson, D., Roux, V., Cossart, P., Weissenbach, J., Claverie, J. M., et al. (2001) Science 293, 2093-2098. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal, N., Lesley, S. A., Kuhn, P. & Kohen, A. (2004) Biochemistry 43, 10295-10301. [DOI] [PubMed] [Google Scholar]
- 41.Lin, M. & Rikihisa, Y. (2003) Infect. Immun. 71, 5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greub, G. & Raoult, D. (2003) Appl. Environ. Microbiol. 69, 5530-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, D. V., Melchers, L. S., Idler, K. B., Schilperoort, R. A. & Hooykaas, P. J. (1988) Nucleic Acids Res. 16, 4621-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das, A. & Pazour, G. J. (1989) Nucleic Acids Res. 17, 4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohashi, N., Zhi, N., Lin, Q. & Rikihisa, Y. (2002) Infect. Immun. 70, 2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmid, M. C., Schulein, R., Dehio, M., Denecker, G., Carena, I. & Dehio, C. (2004) Mol. Microbiol. 52, 81-92. [DOI] [PubMed] [Google Scholar]
- 47.Celli, J. & Gorvel, J. P. (2004) Curr. Opin. Microbiol. 7, 93-97. [DOI] [PubMed] [Google Scholar]
- 48.Christie, P. J. (2001) Mol. Microbiol. 40, 294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, Z., Davis, M. J., Zhang, F. & Teasdale, R. D. (2003) Biochem. Biophys. Res. Commun. 312, 1278-1283. [DOI] [PubMed] [Google Scholar]
- 50.Doolittle, R. F. (1987) Of URFs and ORFs (Univ. Sci. Books, Sausalito, CA).
- 51.Van Heerden, H., Collins, N. E., Brayton, K. A., Rademeyer, C. & Allsopp, B. A. (2004) Gene 330, 159-168. [DOI] [PubMed] [Google Scholar]
- 52.Ohashi, N., Unver, A., Zhi, N. & Rikihisa, Y. (1998) J. Clin. Microbiol. 36, 2671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohashi, N., Zhi, N., Zhang, Y. & Rikihisa, Y. (1998) Infect. Immun. 66, 132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhi, N., Ohashi, N. & Rikihisa, Y. (1999) J. Biol. Chem. 274, 17828-17836. [DOI] [PubMed] [Google Scholar]
- 55.Palmer, G. H., Eid, G., Barbet, A. F., McGuire, T. C. & McElwain, T. F. (1994) Infect. Immun. 62, 3808-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Heijne, G. (1986) Nucleic Acids Res. 14, 4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jongejan, F., Thielemans, M. J., De Groot, M., van Kooten, P. J. & van der Zeijst, B. A. (1991) Vet. Microbiol. 28, 199-211. [DOI] [PubMed] [Google Scholar]
- 58.Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T., Holroyd, S., et al. (2000) Nature 403, 665-668. [DOI] [PubMed] [Google Scholar]
- 59.Bork, P. (1993) Proteins 17, 363-374. [DOI] [PubMed] [Google Scholar]
- 60.Levinson, G. & Gutman, G. A. (1987) Mol. Biol. Evol. 4, 203-221. [DOI] [PubMed] [Google Scholar]
- 61.Rocha, E. P. (2003) Genome Res. 13, 1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weitzmann, M. N., Woodford, K. J. & Usdin, K. (1997) J. Biol. Chem. 272, 9517-9523. [DOI] [PubMed] [Google Scholar]
- 63.Barbet, A. F., Whitmire, W. M., Kamper, S. M., Simbi, B. H., Ganta, R. R., Moreland, A. L., Mwangi, D. M., McGuire, T. C. & Mahan, S. M. (2001) Gene 275, 287-298. [DOI] [PubMed] [Google Scholar]
- 64.Marcotte, C. J. & Marcotte, E. M. (2002) Appl. Bioinform. 1, 93-100. [PubMed] [Google Scholar]
- 65.Allsopp, M. T., Van Heerden, H., Steyn, H. C. & Allsopp, B. A. (2003) Ann. N. Y. Acad. Sci. 990, 685-691. [DOI] [PubMed] [Google Scholar]
- 66.Ochman, H. & Moran, N. A. (2001) Science 292, 1096-1099. [DOI] [PubMed] [Google Scholar]
- 67.Citti, C., Kim, M. F. & Wise, K. S. (1997) Infect. Immun. 65, 1773-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vordermeier, M., Whelan, A. O. & Hewinson, R. G. (2003) Infect. Immun. 71, 1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.