Abstract
Purpose
Melanoma-associated retinopathy (MAR) is a paraneoplastic syndrome associated with malignant melanoma and the presence of anti-retinal autoantibodies, including autoantibodies against transient receptor potential melanopsin 1 (TRPM1), a cation channel expressed by both melanocytes and retinal bipolar cells. The goal of this study was to further map the antigenic epitope.
Methods
Patient sera were tested by immunofluorescence and Western blotting on HEK293 cells transfected with enhanced green fluorescent protein (EGFP)-TRPM1 fusion constructs and mouse retina sections.
Results
The epitope recognized by MAR patient sera was mapped to a region encoded by exons 9 and 10 of the human TRPM1 gene. This region of TRPM1 is highly conserved with TRPM3, and indeed MAR sera were found to cross-react with TRPM3, a closely related channel expressed in the retinal pigment epithelium (RPE).
Conclusions
These results indicate that TRPM1 autoantibodies in MAR patient sera recognize a short, intracellular segment of TRPM1. Cross-reactivity with TRPM3 in the RPE may account for other visual symptoms that are experienced by some MAR patients such as retinal and RPE detachments. We propose that TRPM1 autoantibodies are generated in response to abnormal TRPM1 polypeptides encoded by an alternate mRNA splice variant expressed by malignant melanocytes.
Keywords: TRPM1, MAR, TRPM3
Melanoma-associated retinopathy (MAR) is a paraneoplastic syndrome associated with cutaneous malignant melanoma. Visual deficits include flickering photopsias, sudden night blindness, and a generalized constriction of visual fields.1–3 MAR is believed to be caused by an autoimmune response to antigens expressed by the tumor that are also present in the retina. Electroretinogram (ERG) recordings from MAR patients point to a defect in ON-bipolar cell signaling.4 Furthermore, sera from MAR patients may contain autoantibodies that label retinal bipolar cells.1,2 These autoantibodies are not present in healthy individuals but may be present in patients with melanoma without reported visual problems.5 In at least some cases, the MAR antigen has been shown to be the TRPM1 cation channel, which is expressed under normal conditions by both bipolar cells and melanocytes.6–8
In the retina, transient receptor potential melastatin 1 (TRPM1) is required for the depolarizing light response of retinal ON-bipolar cells.9–12 TRPM1 is negatively coupled to the metabotropic glutamate receptor, mGluR6, via the heterotrimeric G-protein, Go.13,14 The light-induced decrease in glutamate release from photoreceptors results in the inactivation of mGluR6 and Go, relieving an inhibitory constraint on TRPM1, thus allowing the channel to open and depolarize the cell. This depolarization of the ON-bipolar cells gives rise to the b-wave in the ERG. TRPM1 is a major locus of mutations causing autosomal recessive type 1 congenital stationary night blindness (CSNB1) in humans.15–18 The ERG changes observed in CSNB1, absent b-wave with normal a-wave, are similar to those in MAR.19
The role of TRPM1 in melanocytes is less well understood, but it has been linked to melanin content. Oancea et al.20 described a nonselective cationic current in human melanocytes that they attributed to TRPM1 because it could be reduced by RNA interference-mediated knockdown of TRPM1 expression. They found that TRPM1 knockdown also correlated with reduced melanin content. Electrophysiological findings by Devi et al.21 confirm the presence of TRPM1 currents in melanocytes, and show further that, similar to retinal ON-bipolar cells, the TRPM1 current is controlled by endogenously expressed mGluR6. In melanocytes, however, mGluR6 activation increases the TRPM1 current (as opposed to reducing it in ON-bipolar cells), presumably due to mGluR6 coupling to different G-proteins in melanocytes and ON-bipolar cells. Devi et al.21 also showed that chronic stimulation of melanocytes with an mGluR6 agonist promoted melanin production and altered the cell morphology.
The occurrence of MAR is correlated with advanced-stage melanoma,5,22 and production of autoantibodies to TRPM1 is not known to occur in healthy individuals. Why is TRPM1 targeted by the immune system in melanoma patients? One possibility is that aberrant splicing of the TRPM1 mRNA, as has been shown to occur in malignant melanocytes,23,24 leads to the production of abnormal TRPM1 polypeptides that are seen as foreign by the immune system. Mapping the epitope or epitopes targeted by TRPM1 autoantibodies may yield insight into the mechanism by which TRPM1 becomes autoimmunogenic in melanoma, and may also provide insight into the array of visual symptoms associated with MAR. Previously, we have shown that TRPM1 autoantibodies in MAR patient sera bind to the intracellular, amino terminal domain of the channel. Here we show that MAR sera react not only with TRPM1, but also with the closely related family member TRPM3, which is expressed by the retinal pigment epithelium (RPE). Furthermore, we have narrowed the MAR epitope to amino acids encoded by exons 9 and 10 of human TRPM1, a region that is 82% identical in TRPM3.
Methods
Expression Vectors
A series of deletion constructs were generated from the full-length mouse TRPM1 (GenBank NM_001039104) and inserted between the KpnI and SmaI sites of pEGFP-C3 (Clontech, Mountain View, CA, USA). Restriction enzymes were used to remove N-terminal– and C-terminal–encoding segments as follows: The pEGFP-C3-TRPM1 plasmid was digested with BamHI and recircularized to generate the M1-G799 (BamHI) segment. Similarly, digestion with AccI and AflII, followed by treatment with Klenow, and recircularization yielded the L1159 (AflII)-C1622 segment. The M1-G550 (ApaI) segment was generated by digesting the M1-G799 (BamHI) construct with ApaI and BamHI, T4 DNA polymerase treatment, and recircularization. The M1-I147 (EcoRV) segment was generated by digesting the pEGFP-C3-TRPM1 plasmid with EcoRV and recircularization. To generate the V149 (ApaLI)-V430 (AccI) construct, the 843-bp ApaLI-AccI fragment from M1-G550 (ApaI) was purified, treated with T4 DNA polymerase, and inserted into phosphatase-treated, SmaI-digested pEGFP-C3. Similarly the V149 (ApaLI)-Q283 (PstI) construct was generated by digesting the M1-G550 (ApaI) plasmid with ApaLI and PstI, T4 DNA polymerase treatment, and ligating the purified 399-bp fragment into SmaI-digested pEGFP-C3. The L282 (PstI)-G550 (ApaI) fragment was constructed from the M1-G550 (ApaI) plasmid, digested with PstI, and recircularized. Finally, digestion of the V149 (ApaLI)-V430 (AccI) construct with PstI followed by recircularization generated the L282 (PstI)-V430 (AccI) segment.
A human h109+TRPM1 cDNA plasmid (GenBank NM_001252020)20 was used as template to amplify small TRPM1-encoding fragments using the following PCR primers (restriction site linkers in lowercase):
5′-ATGCCTTGAAAGACCACTCCTC
5′-CCCTGCAGAAGATCAACACAAG
5′-gcgaattCTCGTGGGTCTCGTGGTG
5′-TTCTTCACAGTACTTGTGCGCAAAGG
5′-gcgaattcAGGCACAATCACATCAGC
5′-gcggatccGAGTTCTTTCTTCTTCATGC
5′-ccggatccGTTTGTTCCTTTCAGCAG
5′-ggtaccggtGGCCCAAAGACAAAGATCTG
5′-gtaccggtAGGAGCTTCACAAAGTCG
All PCR products were subcloned into pJET1.2 (Fermentas, Glen Burnie, MD, USA) and their nucleotide sequences were verified. The D189-E337 construct was amplified using primers 1 and 4. The resulting pJET1.2 plasmid was digested with XhoI and BglII, and the 465-bp fragment was subcloned into pEGFP-C3, digested with SalI and BamHI. The L275-L380 and L275-N407 constructs were prepared using primers 2 and 6, and 2 and 7, respectively. The PstI-BamHI fragments were subcloned into PstI-BamHI–digested pEGFP-C3. Similarly, the L288-L380 and L288-N407 constructs were prepared using primers 3 and 6, and 3 and 7, and the EcoRI-BamHI fragments were subcloned into EcoRI-BamHI–digested pEGFP-C3. The M372-P435 and M372-L519 constructs were amplified using primers 5 and 8, and 5 and 9, subcloned into pJET1.2 and the EcoRI-AgeI fragments inserted into EcoRI-AgeI–digested pEGFP-N1 (Clontech). For the K361-P435 plasmids, the same EcoRI-AgeI fragments were inserted into pEGFP-C3 digested with EcoRI and XmaI.
Animals
Adult mice of both sexes were used in this study. All mice were maintained on a 12-hour light/dark cycle and provided food and water ad libitum. Retina sections from C57BL6 mice were used for initial screening of MAR serum immunoreactivity. For identifying the target of labeling in RPE, targeted TRPM3 knockout mice (TRPM3tm1Lex; Texas Institute of Genomic Medicine, College Station, TX, USA) were used, and tissue from wild-type and knockout littermates was compared. Mice were maintained and used for experiments in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All animal procedures were approved by the Oregon Health & Science University (OHSU) Institutional Animal Care and Use Committee.
Patient Sera
This study was approved by the OHSU Institutional Review Board and adhered to the tenets of the Declaration of Helsinki. One MAR serum sample (no. 1) was from a patient diagnosed at the National Eye Institute25; the other serum sample (no. 2) was obtained from the Ocular Immunology Laboratory, OHSU.6 Thirty-eight control sera from normal healthy subjects were obtained from the Oregon Clinical and Translational Research Institute.
Immunochemistry
HEK293 cells were seeded onto polylysine-coated coverslips in 12- or 24-well tissue culture plates, transfected with pEGFP-C3 and -N1 expression plasmids encoding fragments of TRPM1 fused at the C- or N-terminus of enhanced green fluorescent protein (EGFP), using Effectene (Qiagen, Valencia, CA, USA), LipoJet (SignaGen, Rockville, MD, USA), or calcium phosphate coprecipitation. CHO-K1 cells were transfected with mouse TRPM3 (GenBank AEE80504.1) in pcDNA3, using TransIT-CHO Transfection Kit (Mirus, Madison, WI, USA). Twenty-four to 36 hours after transfection, coverslips were removed from the wells, cells were fixed for 10 minutes by immersion in cold 4% paraformaldehyde, and then processed for immunofluorescence as follows.
Freshly dissected mouse eyes were hemisected and the front of the eye and lens discarded. The remaining eyecup containing the retina was fixed by immersion in ice-cold 4% paraformaldehyde for 20 minutes, washed in ice-cold PBS, then cryoprotected by consecutive incubations in ice-cold 10%, 20%, and 30% sucrose. Vertical sections, 16 μm, were cut on a cryostat, air dried, and then stored at −80°C until use.
Transfected cells on coverslips and thawed retinal sections were processed for immunofluorescence confocal microscopy as described previously,10 with dilutions (1:100–1:2000) of the MAR serum instead of primary antibodies and anti-human IgG conjugated to Alexa Fluor 594 (1:1000; Invitrogen, Carlsbad, CA, USA) as the secondary antibody. Fluorescence images were acquired with an Olympus FluoView FV1000 confocal microscope (Olympus, Waltham, MA, USA) using a ×60/1.42 oil immersion objective. Image brightness and contrast were enhanced using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA, USA).
Western Blot Analysis
TRPM1-transfected HEK cells were collected in radio-immunoprecipitation assay (RIPA) buffer with protease inhibitor cocktail (Cell Signaling Technology, Danvers, MA, USA) for Western blot analysis. Lysates were electrophoresed on precast 4% to 12% polyacrylamide gradient gels (Novex; Invitrogen). The separated proteins were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes, which were then probed with human sera (1:1000), goat anti-GFP (1:1000), or sheep antibodies to mouse TRPM1 (amino acids 1423–1622). Anti-human IRDye 680CW, anti-sheep IRDye 680W, and anti-goat IRDye 800CW secondary antibodies (Li-Cor, Lincoln, NE, USA) were used at a dilution of 1:10,000 and visualized with an Odyssey infrared imaging system (Li-Cor).
Results
The Epitope for TRPM1 Autoantibodies Is Located Near the Middle of the Cytoplasmic N-Terminal Domain
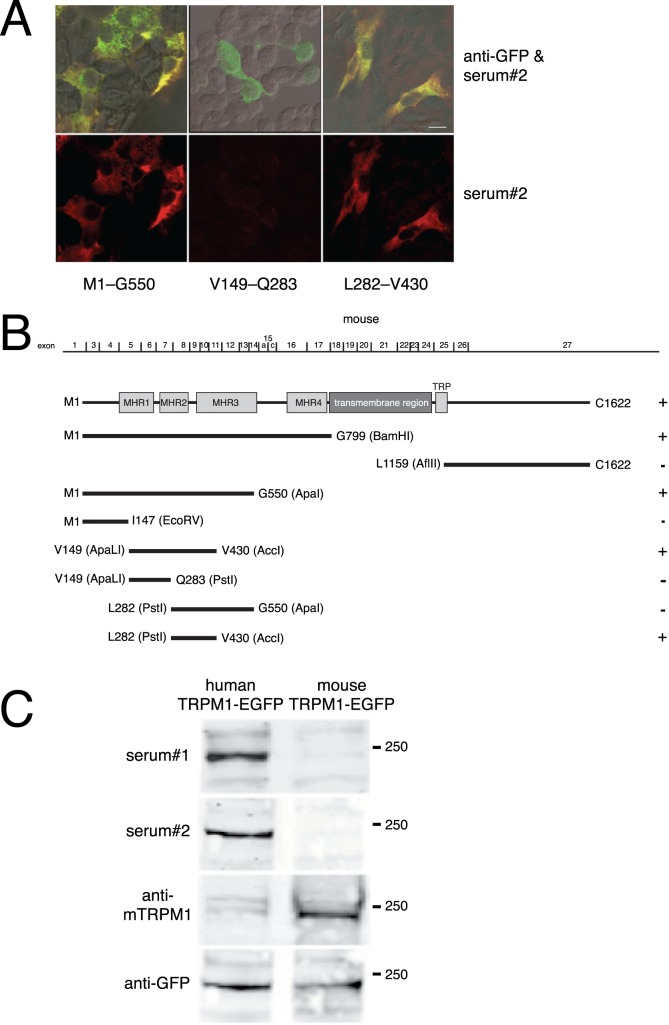
We previously found that MAR autoantibodies bind to a site within the predicted intracellular N-terminal domain of TRPM1.6 To localize the MAR epitope further, we subcloned successively smaller mouse TRPM1 cDNA restriction fragments and expressed them as EGFP fusion proteins in transfected HEK293 cells (Fig. 1). Transfected cells were then fixed and immunostained with two MAR antisera we previously described.6,25 Then, a fluorescent anti-human IgG was used to detect human autoantibodies. Following the transfection procedure, approximately 20% to 50% of the cells expressed the TRPM1–EGFP fusion proteins, as determined by the detection of EGFP (green) fluorescence. The untransfected cells (no EGFP fluorescence) served as a control for nonspecific immunofluorescence with the human sera (Fig. 1A). The two MAR sera showed similar patterns of immunoreactivity toward the TRPM1 fragments (not shown). The smallest immunoreactive fragment spans amino acids L282 to V430 (Fig. 1B). This region comprises the C-terminal 58 amino acids from the melastatin homology region (MHR)-2,26 66 amino acids from the N-terminal part of MHR3, and a less conserved segment linking these two regions. This region of TRPM1 is 91% identical between the mouse and human sequences, and we have previously demonstrated that human MAR sera react with mouse TRPM1 by immunofluorescent labeling of mouse retina sections and CHO cells transfected with mouse TRPM1.6,25 Though both MAR sera react well with mouse TRPM1 by immunofluorescence, both sera reacted more strongly with human TRPM1 than mouse TRPM1 by Western blotting (Fig. 1C). This suggests that the sequence differences between human and mouse TRPM1 are not significant for binding of the autoantibodies to TRPM1 in its native conformation (i.e., immunofluorescent labeling of fixed cells or tissue), but do affect the binding affinity of the autoantibodies when TRPM1 is in a denatured state (i.e., Western blotting).
Figure 1.
The MAR epitope is encoded by mouse TRPM1 exons 8 through 11. To map the MAR epitope, HEK cells were transfected with a series of EGFP–mouse TRPM1 deletion constructs and tested for immunofluorescence with MAR serum. (A) Top row: superimposition of GFP (green) and MAR serum 2 (red) immunofluorescence, with colocalization appearing yellow. Bottom row: MAR serum immunofluorescence alone. Scale bar: 10 μm. (B) Diagram of the mouse TRPM1 cDNA deletion constructs. Exon 2, which is alternatively spliced and encodes an alternative N-terminus, is not present in the plasmid constructs used. The first and last amino acids encoded by each construct are indicated. Positive immunofluorescence with MAR serum was graded as positive (+), or negative (−). MHR: TRPM homology regions.26 (C) HEK293 cells were transfected with plasmids encoding either mouse TRPM1-EGFP or human TRPM1-EGFP and then Western blotted with either MAR serum 1, MAR serum 2, an antibody to mouse TRPM1, or an antibody to GFP.
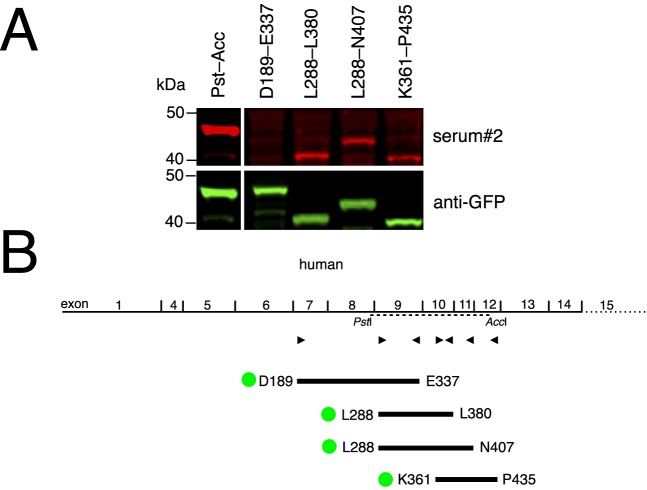
To further narrow the region of TRPM1 recognized by MAR autoantibodies, we used a human TRPM1 cDNA20 as template for PCR to generate a series of overlapping TRPM1–EGFP fusion constructs, spanning amino acids D189 to L519. This region is encoded by human exons 7 through 14 and encompasses the region corresponding to amino acids L282 to V430 of the mouse fragment identified in Figure 1. Protein extracts from HEK293 cells transfected with these plasmids were used for Western blotting with the two MAR sera. Using the Li-Cor Odyssey imaging system, it is possible to double-label the blot with an antibody against EGFP and the MAR serum. Results using MAR serum 2 are shown in Figure 2, but no specific labeling with MAR serum 1 was observed (not shown). Constructs containing amino acids L282 to L380 (encoded by exons 9–10) and overlapping amino acids K361 to P435 (encoded by exons 10–12) are the smallest segments reacting with MAR serum 2.
Figure 2.
Western blotting identifies human TRPM1 exons 9 and 10 as encoding the MAR epitope. To further define the MAR epitope, HEK cells were transfected with a series of EGFP–human–TRPM1 constructs and protein extracts were used in Western blotting. (A) Western blotting of TRPM1–EGFP fusion proteins using MAR serum. (B) Diagram of human TRPM1 constructs. Exons 2 and 3 are alternatively spliced and not present in the template cDNA used. Triangles indicate the position of the PCR primers used to generate the TRPM1 fragments. A dotted line shows the region corresponding to the mouse TRPM1 PstI-AccI construct (Fig. 1). For each fragment, the first and last encoded amino acids are indicated and the position of the EGFP fusion is indicated by a green disk.
Transfected HEK293 cells were also seeded onto polylysine-coated coverslips, which were then fixed and immunostained using MAR sera 1 and 2. For both sera, the smallest immunopositive segment spanned human TRPM1 amino acids L288 to L380 (Fig. 3). Interestingly, MAR serum 1 did not react with the EGFP fusion construct containing amino acids K361 to P435 (Fig. 3A), whereas MAR serum 2 was positive for this construct (Fig. 3B). As negative control, sera from 38 healthy subjects were not reactive in these assays (not shown).
Figure 3.
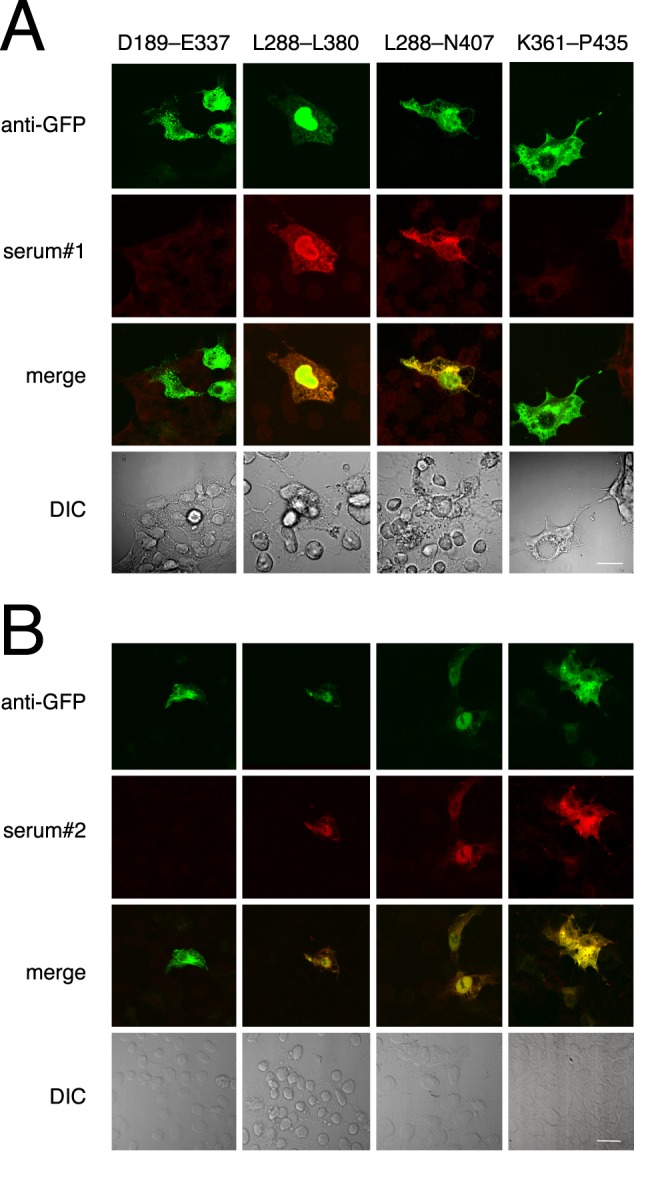
The MAR epitope is encoded by human TRPM1 exons 9 and 10. HEK cells, transfected with a series of EGFP–human TRPM1 constructs, were tested for immunofluorescence with both MAR sera. (A) Top row: EGFP immunofluorescence showing transfected cells. Second row: MAR serum 1 immunofluorescence. Third row: superimposition of EGFP (green) and MAR serum (red) immunofluorescence, with colocalization appearing yellow. Bottom row: differential interference contrast (DIC) image of the coverslip, showing TRPM1-transfected, and untransfected control cells. (B) Immunofluorescence analysis of HEK transfected cells using the same constructs as in (A), but with MAR serum 2. Serum 2 reacts with the K361-P435 fragment, whereas serum 1 does not. Scale bars: 10 μm.
TRPM1-Positive MAR Autoantibodies Cross-React With TRPM3
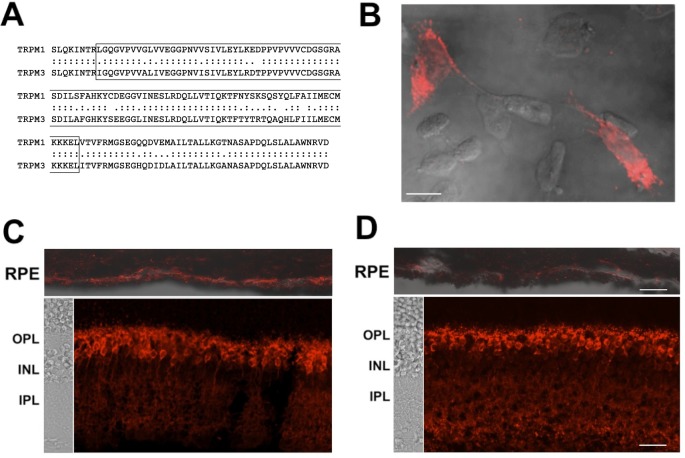
The antigenic region of TRPM1 (human amino acids L288–L380) is 82% identical between TRPM1 and TRPM3 (Fig. 4A), suggesting that the MAR sera may react with both TRP channels. Labeling TRPM3-transfected cells with both MAR sera revealed that they indeed react with TRPM3 (Fig. 4B). TRPM3 mRNA has been shown to be abundantly expressed by the RPE.27 We detected MAR serum immunofluorescence in the RPE of wild-type mice that was markedly reduced in TRPM3 knockout mice, whereas immunofluorescence in the ON-bipolar cells, which express TRPM1, was unaffected in the TRPM3 knockout retina (Figs. 4C, 4D).
Figure 4.
MAR IgG cross-react with TRPM3. (A) Sequence alignment of the TRPM1 epitope and the corresponding region of TRPM3. Identical amino acids are indicated by colons, similar amino acids by periods. The minimal MAR epitope is boxed. (B) Immunofluorescent labeling of TRPM3-transfected CHO cells with MAR serum 2. (C, D) MAR serum 2 gave rise to strong immunofluorescence over the RPE in sections of TRPM3+/+ mouse eyecups ([C] upper image), but markedly reduced fluorescence over the RPE in TRPM3−/− eyecups ([D] upper image), whereas bipolar cell staining was strong in both TRPM3+/+ and TRPM3−/− retinas ([C, D] lower images). RPE immunostaining is overlaid onto DIC images ([C, D] upper images). DIC images of the retina layers are shown to the left of the retina immunostaining ([C, D] lower images). The scale bars represent 10 μm in (B) and 20 μm in (D) (applies to both [C] and [D]). OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer.
Discussion
TRPM1 is expressed by both melanocytes and retinal bipolar cells,12,28 and it is downregulated in metastatic melanoma.29,30 Sera from melanoma patients diagnosed with MAR have been shown to react with TRPM1.6–8 The visual symptoms associated with MAR suggest that the TRPM1 autoantibodies inhibit channel function upon binding. Here, we mapped the immunogenic region of TRPM1 to a segment encoded by exons 9 and 10 of the human gene (corresponding to exons 7 and 8 in mouse), which is located in the cytoplasmic N-terminal domain of TRPM1. This region is 82% identical in TRPM3, a closely related channel that is expressed at high levels by the RPE in the eye,31 and is also expressed in pancreatic β-cells,32 dorsal root ganglia,33 and vascular smooth muscle cells.34 Indeed, we found that the MAR autoantibodies labeled TRPM3-expressing CHO cells, as well as the RPE in wild-type but not TRPM3 knockout mice. While inactivation of TRPM1 channels in the ON-bipolar cells by MAR autoantibodies is likely to account for the suppression of the ERG b-wave, cross-reactivity with TRPM3 may explain additional eye-related deficits seen in some cases of MAR, such as vitelliform lesions,35–37 characterized by multiple sites of retinal detachment from the RPE. In these cases, it is possible that TRPM3 may be the primary autoantigen rather than TRPM1, particularly in patients with paraneoplastic vitelliform lesions but no classical MAR symptoms.37
It is noteworthy that both MAR sera reacted with the same small region of TRPM1, raising the question of what makes this region autoimmunogenic. Interestingly, a tumor suppressor microRNA, miR-211, is encoded within intron 8 of the TRPM1 gene (i.e., the intron between exons 8 and 9 of the human sequence).38–41 This microRNA is generated during processing of the TRPM1 pre-mRNA42 and is downregulated in advanced melanoma by an unknown mechanism. It is intriguing that intron 8 is located close to the TRPM1 region recognized by the MAR autoantibodies. A possible mechanism by which downregulation might occur is by alternative splicing of the TRPM1 mRNA in a manner that prevents the generation of the microRNA. This aberrant mRNA splicing could also result in truncated TRPM1 polypeptides that may be recognized as non-self by the immune system and trigger an autoimmune response. Indeed, alternate TRPM1 transcripts encoding truncated N-terminal TRPM1 polypeptides are more abundant than the transcript encoding full-length TRPM1 in pigmented metastatic melanoma cells.23 This is consistent with an RNA seq analysis of TRPM1 transcripts in the melanoma cell line SK-Mel-30, which reveals much higher levels of transcripts encoding exons 2 to 11, than downstream exons.43 These shorter transcripts may include an alternative exon 9 donor site and encode a truncated protein. Examples of such cDNA clones and EST sequences are deposited in GenBank (e.g., BC033627). Indeed, the presence of truncated TRPM1 polypeptides has been reported in malignant melanocytes.24 We thus propose that such polypeptides are autoimmunogenic and cause MAR.
Acknowledgments
The authors thank Elena Oancea (Brown University, Providence, RI, USA) for supplying human TRPM1 cDNA, Kirill Martemayanov (Scripps Research Institute, Jupiter, FL, USA) for a gift of sheep TRPM1 antiserum, and Nida Sen (National Eye Institute, Bethesda, MD, USA) and Grazyna Adamus (Oregon Health & Science University, Portland, OR, USA) for patient antiserum.
Supported by National Institutes of Health (NIH) Grants EY022369 (CWM) and EY009534 and EY019907 (RMD), by the Oregon Clinical and Translational Research Institute (CWM and RMD), which is supported by NIH Grant UL1TR000128 and the Medical Research Foundation of Oregon (CWM).
Disclosure: R.M. Duvoisin, None; T.L. Haley, None; G. Ren, None; I. Strycharska-Orczyk, None; J.P. Bonaparte, None; C.W. Morgans, None
References
- 1. Milam AH,, Saari JC,, Jacobson SG,, Lubinski WP,, Feun LG,, Alexander KR. Autoantibodies against retinal bipolar cells in cutaneous melanoma-associated retinopathy. Invest Ophthalmol Vis Sci. 1993; 34: 91–100. [PubMed] [Google Scholar]
- 2. Keltner JL,, Thirkill CE,, Yip PT. Clinical and immunologic characteristics of melanoma-associated retinopathy syndrome: eleven new cases and a review of 51 previously published cases. J Neuroophthalmol. 2001; 21: 173–187. [DOI] [PubMed] [Google Scholar]
- 3. Lu Y,, Jia L,, He S,, et al. Melanoma-associated retinopathy: a paraneoplastic autoimmune complication. Arch Ophthalmol. 2009; 127: 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexander KR,, Fishman GA,, Peachey NS,, Marchese AL,, Tso MO. “On” response defect in paraneoplastic night blindness with cutaneous malignant melanoma. Invest Ophthalmol Vis Sci. 1992; 33: 477–483. [PubMed] [Google Scholar]
- 5. Ladewig G,, Reinhold U,, Thirkill CE,, Kerber A,, Tilgen W,, Pföhler C. Incidence of antiretinal antibodies in melanoma: screening of 77 serum samples from 51 patients with American Joint Committee on Cancer stage I-IV. Br J Dermatol. 2005; 152: 931–938. [DOI] [PubMed] [Google Scholar]
- 6. Xiong W-H,, Duvoisin RM,, Adamus G,, Jeffrey BG,, Gellman C,, Morgans CW. Serum TRPM1 autoantibodies from melanoma associated retinopathy patients enter retinal on-bipolar cells and attenuate the electroretinogram in mice. PLoS One. 2013; 8: e69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhingra A,, Fina ME,, Neinstein A,, et al. Autoantibodies in melanoma-associated retinopathy target TRPM1 cation channels of retinal ON bipolar cells. J Neurosci. 2011; 31: 3962–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kondo M,, Sanuki R,, Ueno S,, et al. Identification of autoantibodies against TRPM1 in patients with paraneoplastic retinopathy associated with ON bipolar cell dysfunction. PLoS One. 2011; 6: e19911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen Y,, Heimel JA,, Kamermans M,, Peachey NS,, Gregg RG,, Nawy S. A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J Neurosci. 2009; 29: 6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morgans CW,, Zhang J,, Jeffrey BG,, et al. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc Natl Acad Sci U S A. 2009; 106: 19174–19178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koike C,, Obara T,, Uriu Y,, et al. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc Natl Acad Sci U S A. 2010; 107: 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morgans CW,, Brown RL,, Duvoisin RM. TRPM1: the endpoint of the mGluR6 signal transduction cascade in retinal ON-bipolar cells. Bioessays. 2010; 32: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dhingra A,, Lyubarsky A,, Jiang M,, et al. The light response of ON bipolar neurons requires G[alpha]o. J Neurosci. 2000; 20: 9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duvoisin RM,, Morgans CW,, Taylor WR. The mGluR6 receptors in the retina: analysis of a unique G-protein signaling pathway. Cell Sci Rev. 2005; 2: 18. [Google Scholar]
- 15. Li Z,, Sergouniotis PI,, Michaelides M,, et al. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet. 2009; 85: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Audo I,, Kohl S,, Leroy BP,, et al. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2009; 85: 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Genderen MM,, Bijveld MMC,, Claassen YB,, et al. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet. 2009; 85: 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakamura M,, Sanuki R,, Yasuma TR,, et al. TRPM1 mutations are associated with the complete form of congenital stationary night blindness. Mol Vis. 2010; 16: 425–437. [PMC free article] [PubMed] [Google Scholar]
- 19. Koh AH,, Hogg CR,, Holder GE. The incidence of negative ERG in clinical practice. Doc Ophthalmol. 2001; 102: 19–30. [DOI] [PubMed] [Google Scholar]
- 20. Oancea E,, Vriens J,, Brauchi S,, Jun J,, Splawski I,, Clapham DE. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci Signal. 2009;2:ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Devi S,, Markandeya Y,, Maddodi N,, et al. Metabotropic glutamate receptor 6 signaling enhances TRPM1 calcium channel function and increases melanin content in human melanocytes. Pigment Cell Melanoma Res. 2013; 26: 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pföhler C,, Haus A,, Palmowski A,, et al. Melanoma-associated retinopathy: high frequency of subclinical findings in patients with melanoma. Br J Dermatol. 2003; 149: 74–78. [DOI] [PubMed] [Google Scholar]
- 23. Fang D,, Setaluri V. Expression and up-regulation of alternatively spliced transcripts of melastatin, a melanoma metastasis-related gene, in human melanoma cells. Biochem Biophys Res Commun. 2000; 279: 53–61. [DOI] [PubMed] [Google Scholar]
- 24. Zhiqi S,, Soltani MH,, Bhat KMR,, et al. Human melastatin 1 (TRPM1) is regulated by MITF and produces multiple polypeptide isoforms in melanocytes and melanoma. Melanoma Res. 2004; 14: 509–516. [DOI] [PubMed] [Google Scholar]
- 25. Dalal MD,, Morgans CW,, Duvoisin RM,, et al. Diagnosis of occult melanoma using transient receptor potential melastatin 1 (TRPM1) autoantibody testing: a novel approach. Ophthalmology. 2013; 120: 2560–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fleig A,, Penner R. The TRPM ion channel subfamily: molecular, biophysical and functional features. Trends Pharmacol Sci. 2004; 25: 633–639. [DOI] [PubMed] [Google Scholar]
- 27. Gilliam JC,, Wensel TG. TRP channel gene expression in the mouse retina. Vision Res. 2011; 51: 2440–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oancea E,, Wicks NL. TRPM1: new trends for an old TRP. Adv Exp Med Biol. 2011; 704: 135–145. [DOI] [PubMed] [Google Scholar]
- 29. Deeds J,, Cronin F,, Duncan LM. Patterns of melastatin mRNA expression in melanocytic tumors. Hum Pathol. 2000; 31: 1346–1356. [PubMed] [Google Scholar]
- 30. Erickson LA,, Letts GA,, Shah SM,, Shackelton JB,, Duncan LM. TRPM1 (Melastatin-1/MLSN1) mRNA expression in Spitz nevi and nodular melanomas. Mod Pathol. 2009; 22: 969–976. [DOI] [PubMed] [Google Scholar]
- 31. Brown RL,, Weei-Hong X,, Peters JH,, et al. TRPM3 expression in mouse retina. PLoS One. 2015; 10: e0117615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner TFJ,, Loch S,, Lambert S,, et al. Transient receptor potential M3 channels are ionotropic steroid receptors in pancreatic beta cells. Nat Cell Biol. 2008; 10: 1421–1430. [DOI] [PubMed] [Google Scholar]
- 33. Vriens J,, Owsianik G,, Hofmann T,, et al. TRPM3 is a nociceptor channel involved in the detection of noxious heat. Neuron. 2011; 70: 482–494. [DOI] [PubMed] [Google Scholar]
- 34. Naylor J,, Li J,, Milligan CJ,, et al. Pregnenolone sulphate- and cholesterol-regulated TRPM3 channels coupled to vascular smooth muscle secretion and contraction. Circul Res. 2010; 106: 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sotodeh M,, Paridaens D,, Keunen J,, Schooneveld MV,, Adamus G,, Baarsma S. Paraneoplastic vitelliform retinopathy associated with cutaneous or uveal melanoma and metastases. Klin Monatsbl Augenheilkd. 2005; 222: 910–914. [DOI] [PubMed] [Google Scholar]
- 36. Javaheri M,, Khurana RN,, Bhatti RA,, Lim JI. Optical coherence tomography findings in paraneoplastic pseudovitelliform lesions in melanoma-associated retinopathy. Clin Ophthalmol. 2008; 2: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lincoff N,, Nadeem M,, Younus Z,, Thirkill CE. Exudative polymorphous vitelliform retinopathy: importance of early recognition of the condition in patients with metastatic melanoma. Ophthalmol Ther. 2016; 5: 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levy C,, Khaled M,, Iliopoulos D,, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010; 40: 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mazar J,, DeYoung K,, Khaitan D,, et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One. 2010; 5: e13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boyle GM,, Woods SL,, Bonazzi VF,, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011; 24: 525–537. [DOI] [PubMed] [Google Scholar]
- 41. Xu Y,, Brenn T,, Brown ERS,, Doherty V,, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012; 106: 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janas MM,, Khaled M,, Schubert S,, et al. Feed-forward microprocessing and splicing activities at a microRNA–containing intron. PLoS Genet. 2011; 7: e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klijn C,, Durinck S,, Stawiski EW,, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2014; 33: 306–312. [DOI] [PubMed] [Google Scholar]