Abstract
Sterol regulatory element-binding proteins (SREBPs) are transcription factors central to the regulation of lipid metabolism. The SREBPs are synthesized as precursor proteins that require proteolytic processing to become transcriptionally active. Whereas the regulation of SREBP-1a and -2 cleavage by cellular sterol content is well defined, much less is known about the regulation of SREBP-1c, the predominant SREBP isoform in the liver. Both insulin and liver X receptor α (LXRα) induce SREBP-1c transcription; however, the respective roles of these factors and the mechanism responsible for proteolytic cleavage of this SREBP isoform are not known. In this study, we compare the effects of insulin and LXR agonist TO-901317 on SREBP-1c expression and transcriptional activity in isolated rat hepatocytes. We report that full induction of the mature and transcriptionally active form of SREBP-1c protein requires insulin. Although activation of LXR leads to the induction of SREBP-1c gene expression and precursor protein, it has a very poor effect in inducing the mature nuclear form of the transcription factor. This may be due to the induction of insulin-induced gene-2a mRNA and protein by LXR activation. The LXR-induced SREBP-1c precursor, however, is rapidly cleaved on acute exposure to insulin via a phosphatidylinositol 3-kinase-dependent mechanism. Finally, we show through experiments in suckling mice that this acute action of insulin to stimulate the proteolytic processing of SREBP-1c is functional in vivo.
Keywords: glucose homeostasis, cholesterol, lipogenesis, hepatocytes
The sterol regulatory element-binding proteins (SREBPs) are transcription factors integral to the maintenance of lipid homeostasis. The three SREBP isoforms (SREBP-1a, -1c, and -2) have overlapping target genes and show differential expression across tissues (1). SREBP-1c is the major isoform expressed in the liver and tissues involved in energy homeostasis (2). It regulates fatty acid synthesis through selective induction of hepatic glucokinase (GK) and an array of lipogenic genes (3-6). SREBP-2 is widely expressed and primarily regulates genes involved in cholesterol biosynthesis (7). The SREBP-1a isoform, which can transactivate both lipogenic and cholesterogenic genes, is highly expressed in cell lines but has very low expression in most organs in vivo (2).
The SREBPs are synthesized in the endoplasmic reticulum (ER) in the form of a precursor protein. To become transcriptionally active, the SREBP precursor must undergo proteolytic cleavage in the Golgi apparatus to liberate its N-terminal domain, which constitutes the mature transcription factor (1). Two proteins are essential to this cleavage process: SREBP cleavage-activating protein (SCAP) and insulin-induced gene (Insig). SCAP is a large integral membrane protein of the ER that interacts with newly synthesized SREBP precursor and escorts it to the Golgi apparatus (8, 9). However, SCAP can also interact with Insig, another ER protein that is deeply embedded in the membranes. Insig functions to retain the SCAP-SREBP complex within the ER (10-13).
The group of Brown and Goldstein has demonstrated that cellular sterol content can regulate SREBP processing (7). Sterols enhance the interaction between SCAP and Insig and thereby prevent the translocation of SCAP-SREBP to the Golgi compartment. In contrast, under the conditions of sterol depletion, SCAP undergoes a conformational change that prevents its association with Insig (10, 11, 14, 15). The SCAP-SREBP complex is thereby released from the ER, and the cleavage process is activated.
Importantly, these studies were performed in cell lines that predominantly express the SREBP-1a and -2 isoforms (2). Studies in vivo, however, indicate that sterol depletion does not regulate the proteolytic processing of SREBP-1c. Indeed, studies in hamsters showed that, in direct contrast to SREBP-2, hepatic expression of mature SREBP-1c was in fact decreased by sterol depletion (16). Furthermore, fasting/refeeding regimes in rodents showed that SREBP-1c is primarily regulated by changes in nutritional status that have little effect on SREBP-2 expression (17).
To date, the main regulation demonstrated for SREBP-1c is at the transcriptional level. Insulin induces the transcription of the SREBP-1c gene, and this leads to a parallel increase in both the membrane-bound precursor and the mature nuclear form (4, 5, 18). The transcription of SREBP-1c can also be induced by the activation of liver X receptor (LXR)α (19, 20). LXRα is a nuclear hormone receptor with high hepatic expression that is activated by oxysterols (intermediates of cholesterol metabolism) and induces the transcription of a range of genes involved in cholesterol efflux and clearance (21). Knockout studies and experiments in which animals have been fed LXR agonists identified a role for LXRα to induce lipogenic genes (19, 20, 22), mediated by both a direct action on the promoter of some of these genes including fatty acid synthase and acetyl-CoA carboxylase, as well as an indirect effect via the induction of SREBP-1c (21).
It is interesting that SREBP-1c is induced by two quite disparate stimuli: insulin, a hormone released in response to carbohydrate intake, and LXRα, a transcription factor that acts as a cholesterol sensor. The significance and respective roles of these two factors in the regulation of SREBP-1c, however, are largely unknown. Another unanswered question regarding the regulation of this SREBP isoform is how the SREBP-1c precursor is cleaved in the absence of a reduction in cellular sterol content. Is SREBP-1c cleavage simply a constitutive process that reflects the level of precursor protein, or does it require a specific stimulus equivalent to sterol depletion for the other SREBP isoforms? In this study, we compare the roles of insulin and LXRα activation in the control of SREBP-1c transcription and transcriptional activity.
Methods
Animals. Procedures were carried out according to French guidelines (Institut Nationale de la Santé et de la Recherche Médicale office of animal experimentation, Paris). Female Wistar rats (≈250 g) and C3H mice (Charles River Breeding Laboratories) were housed in a controlled environment (12-h light/12-h dark cycle) and fed ad lib with a laboratory-chow diet and free access to water. Suckling mice (13-d-old) were administered insulin i.p. (0.4 units in 100 μl of saline; Novo-Nordisk, Copenhagen) or vehicle. After 20 min, mice were killed by cervical dislocation and livers harvested. For LXR agonist gavage experiments, 10 week-old fed mice were force-fed the synthetic LXR agonist T0-901317 (Sigma; 50 mg/kg in 1% carboxymethylcellulose) or vehicle and the liver collected 12 h later.
Primary Hepatocyte Isolation and Culture. Hepatocytes were isolated and cultured as described (6). After 16 h in basal medium (M199, GIBCO/BRL, plus 100 nM Dexamethasone, Sigma), cells were treated with fresh basal medium supplemented with 100 nM insulin/10 μM TO-901317 (20)/50 μg/ml ALLN (N-acetyl-Leu-Leu-norleu-al) calpain-inhibitor 1 (Sigma) (23) and/or 100 nM wortmannin (Sigma), as indicated.
Isolation of Total RNA and Real-Time RT-PCR. Total RNA was isolated according to ref. 24. Real-time quantitative RT-PCR analyses were performed with 50 ng of cDNA/3 mM MgCl2/250 nM sense and antisense primers (Proligo, Boulder, CO) in a final reaction volume of 25 μl by using the qPCR TM Core Kit (Eurogentec, Brussels) and the MyiQ real-time PCR detection system (Bio-Rad). Specific primers are provided in Table 1, which is published as supporting information on the PNAS web site. Relative quantitation of each gene was calculated after normalization to 18S ribosomal RNA by using the comparative CT method.
Isolation of Microsomal Membranes and Nuclear Extracts. Microsomal membranes were isolated according to ref. 25. Nuclear extracts from primary hepatocytes were prepared according to refs. 26 and 27. Extracts from mouse liver were prepared as described (28).
Western Blotting and EMSA. For detection of SREBP-1, 50-70 μg of proteins from microsomal membranes or 20-40 μg of protein from nuclear extracts was used. For detection of Insig-2, 40 μg of proteins from microsomal membranes was used and denatured as described (12). SREBP-1c was detected by using a mouse monoclonal antibody (IgG 2A4; NeoMarkers, Fremont, CA). Insig-2 was detected by using the antibody described below. Calnexin antibody (BD Biosciences) was used as a loading control for microsomal membrane preparations from mouse liver. EMSA was performed using 4 μg of nuclear protein, as described (28).
Development of an Antibody Against Insig-2a. The antibody was developed by Eurogentec (Brussels) by injecting rabbits with two peptides specific for the rat/mouse Insig-2 isoform (N-AEGETESPRPKKRGPC; N-CKVIAEKSHQE) and the serum collected 2 months later. Antibody specificity was checked by Western blotting by using microsomal membranes isolated from hepatocytes infected with an adenovirus expressing rat Insig-2 (data not shown). Furthermore, Insig-2 was up-regulated in the liver of fasted rats, as described (12).
Statistical Analyses. Statistical analysis was performed by using a commercial software package (prism, GraphPad, San Diego). Comparisons were performed by one-way ANOVA incorporating a Newman-Keuls multiple comparison posthoc test. Results are presented as means ± SEM.
Results
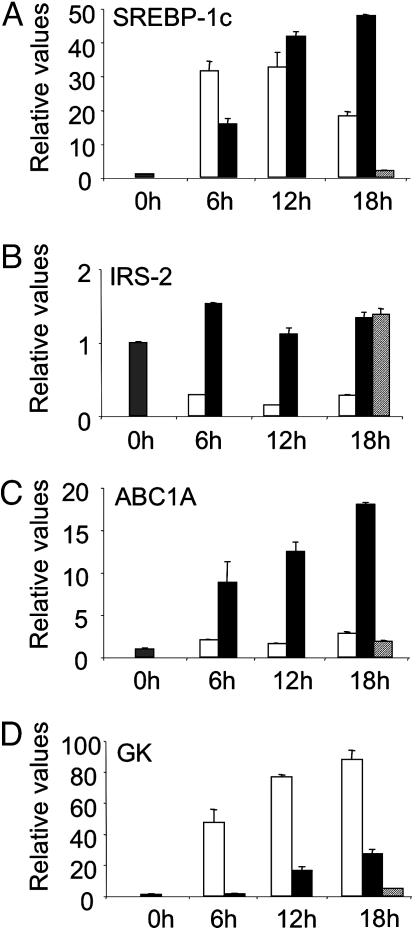
Differential Induction of SREBP-1c and GK Expression by Insulin and LXR Activation. We first performed studies in isolated rat hepatocytes to compare the ability of insulin and LXR activation to induce SREBP-1c gene expression and the expression of a target gene of SREBP-1c, GK. Incubation of hepatocytes with insulin induces the expression of SREBP-1c mRNA (Fig. 1A). This induction tapers off after 12 h, for which we have no clear explanation. It does not appear to be due to a decrease in insulin efficiency, because IRS-2 expression remained inhibited throughout the treatment period (Fig. 1B). Activation of LXR by using the specific agonist TO-901317 induces the expression of SREBP-1c, as well as another LXR target gene, ATP-binding cassette A1 (ABCA1), in a time-dependent fashion (Fig. 1 A and C).
Fig. 1.
Relative expression of SREBP-1c, IRS-2, ABCA1, and GK mRNA in primary hepatocytes treated with insulin or the LXR agonist TO-901317. Hepatocytes were maintained overnight in basal medium before treatment with insulin (100 nM; open bars), TO-901317 (10 μM; black bars), or DMSO (vehicle control; hatched bars) for the time period indicated. Total RNA from triplicate plates of hepatocytes was extracted and analyzed for SREBP-1c (A), IRS-2 (B), ABCA1 (C), and GK (D). Data are means ± SEM. Representative of three independent experiments.
Insulin treatment strongly induces the expression of GK (Fig. 1D). SREBP-1c induces GK gene transcription through binding to SREs in the GK promoter (6, 29). That GK mRNA continues to increase at 18 h despite a decline in SREBP-1c mRNA expression may indicate supplementary posttranslational effects of insulin to regulate the mature SREBP-1c protein. In contrast to insulin, and despite a clear induction of SREBP-1c mRNA, TO-901317 treatment had no effect on GK expression at 6 h and thereafter induced GK in a comparatively weak manner (Fig. 1D).
To understand this discrepancy between SREBP-1c gene induction and GK expression, we measured SREBP-1c protein levels. Although the antibody used to recognize SREBP-1 recognizes both SREBP-1a and -1c isoforms, SREBP-1c is the predominant isoform expressed in liver (2), and only the SREBP-1c gene is induced by insulin (4) and LXR activation (19).
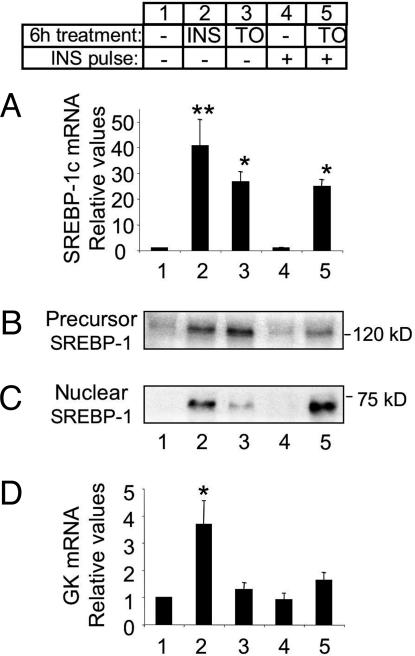
The induction of SREBP-1c mRNA by a 6-h incubation with either insulin or TO-901317 leads to an increase in SREBP-1c precursor protein in the microsomal membranes (Fig. 2 A and B, lanes 1-3). Measurement of SREBP-1c expression in the nucleus shows that insulin also causes a strong induction of mature SREBP-1c (Fig. 2C, lanes 1-2). In contrast, cells treated with TO-901317 show only very weak expression of the nuclear SREBP-1c protein (Fig. 2C, lane 3). This poor expression of the mature transcription factor in cells treated with TO-901317 would explain the lack of GK induction by the LXR agonist (Fig. 2D), despite strong SREBP-1c mRNA and precursor expression. Note that fold induction of GK expression shown here is substantially lower than in Fig. 1. This is due to variations in the basal expression of GK, which is extremely low and sometimes at the limit of detection.
Fig. 2.
Differential induction of nuclear SREBP-1c by insulin and TO-901317. After 16 h in basal medium, hepatocytes were incubated for 6 h in either fresh basal medium or basal medium supplemented with insulin (100 nM; INS) or TO-901317 (10 μM; TO). Subsets of hepatocytes cultured in basal medium or with TO-901317 were treated with insulin (100 nM) for the final 30 min of this 6-h treatment period (INS pulse; lanes 4 and 5). (A) SREBP-1c mRNA expression. (B) Immunoblot of SREBP-1c precursor expressed in the microsomal fraction of hepatocytes. (C) Immunoblot of mature SREBP-1c in the nuclear extracts of hepatocytes. (D) GK mRNA expression. mRNA values represent the mean ± SEM of three independent experiments performed in triplicate. Immunoblots are representative of three independent experiments. *, P < 0.01; **, P < 0.001 compared with untreated control (lane 1).
This result also indicates that, in addition to simply increasing SREBP-1c gene expression, insulin may have a complementary action on SREBP-1c processing to increase the amount of the mature protein in the nucleus. Alternatively, this result may indicate an effect of LXR activation to induce a factor that inhibits the cleavage of SREBP-1c precursor.
Insulin Acts Acutely to Induce the Cleavage of SREBP-1c Precursor in Isolated Hepatocytes. To test the hypothesis that insulin can induce SREBP-1c cleavage, we examined the effect of a short “pulse” of insulin on the precursor and nuclear forms of SREBP-1c in isolated hepatocytes. A 30-min incubation with insulin alone has no effect in increasing SREBP-1c mRNA or protein expression in cells cultured in basal medium (Fig. 2 A-C, lane 1 vs. 4). However, in cells in which the SREBP-1c precursor is already induced by pretreatment with TO-901317, a 30-min insulin pulse greatly augments the presence of mature SREBP-1c in the nucleus (Fig. 2C, lane 3 vs. 5) and decreases the abundance of the SREBP-1c precursor (Fig. 2B, lane 3 vs. 5). The rapid induction of mature SREBP-1c has no effect on GK expression, probably due to the short period (30 min) involved (Fig. 2D, lane 3 vs. 5).
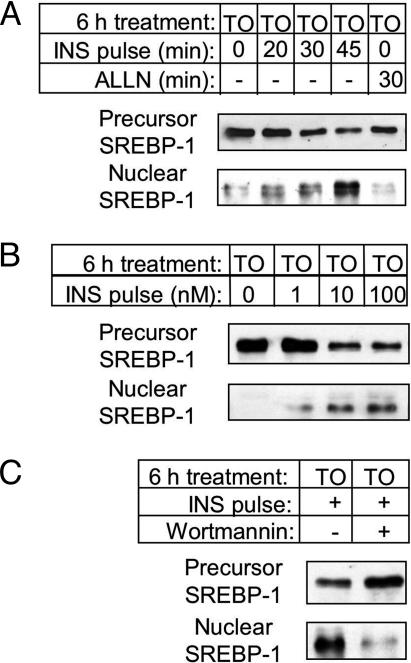
This effect of insulin in inducing nuclear SREBP-1c is both time- and dose-dependent. In cells pretreated with LXR agonist, an increase in nuclear SREBP-1c is evident after only 20-min incubation with insulin and continues to accumulate with increasing exposure to the hormone (Fig. 3A). Furthermore, insulin is effective at a dose of as low as 1 nM (Fig. 3B). Accompanying this accumulation of nuclear SREBP-1c, we see a corresponding decrease in membrane-bound SREBP-1c precursor (Figs. 2 and 3). Over seven separate experiments, the 30-min insulin pulse caused a 34 ± 5% (P < 0.001) decrease in SREBP-1c precursor, measured by densitometry. These data indicate that insulin has an acute action to stimulate the cleavage of SREBP-1c precursor protein.
Fig. 3.
Characterization of the acute accumulation of nuclear SREBP-1c with insulin. After 16 h in basal medium, hepatocytes were incubated for 6 h in basal medium supplemented with TO-901317 (10 μM) with additional treatments as detailed. (A) Time-course and effect of ALLN: cells were treated with insulin (100 nM; INS pulse) for the indicated time, or the calpain-1 inhibitor, ALLN (50 μg/ml; 30 min). (B) Dose-response to insulin: cells were treated with 0, 1, 10, or 100 nM insulin, as indicated (30 min; INS pulse). (C) Inhibition by wortmannin: wortmannin (100 nM) or vehicle was added to plates 15 min before the insulin pulse (100 nM, 30 min; INS pulse). At the end of the 6-h treatment period, cells were collected for preparation of microsomal membranes and nuclear extracts. Each blot is representative of two independent experiments.
An effect of insulin to increase the stability of mature SREBP-1c could also contribute to its accumulation in the nucleus. It has been shown that nuclear SREBP-1 is degraded by calpain-1 type enzymes, and inhibition of this proteolytic action by treatment with ALLN causes the transcription factor to accumulate over 4-5 h (23, 30). If stabilization of mature SREBP-1c does indeed contribute to its accumulation on acute exposure to insulin, we may expect that inhibition of SREBP-1c degradation by ALLN should similarly cause the protein to accumulate in the nucleus over the same time period. In hepatocytes in which the SREBP-1c precursor was induced by pretreatment with the LXR agonist, however, a 30-min insulin pulse induces nuclear SREBP-1c, whereas 30-min exposure to ALLN has no such effect (Fig. 3A). This suggests that stabilization of mature SREBP-1c may not be a major contributor to the rapid accumulation of nuclear SREBP-1c induced by acute exposure to insulin.
It has been shown (18, 31) that insulin regulation of SREBP-1c gene expression involves a phosphatidylinositol 3-kinase-dependent (PI3-kinase) pathway (18, 31). We therefore tested whether this signaling pathway is also implicated in the acute action of insulin on SREBP-1c cleavage. Indeed, in cells in which the SREBP-1c precursor is induced by TO-901317, the presence of wortmannin (an inhibitor of PI3-kinase) greatly diminishes the effect of an insulin pulse to induce mature SREBP-1c (Fig. 3C). Thus insulin has an acute effect in stimulating the cleavage of the SREBP-1c precursor via a PI3-kinase-dependent mechanism.
Insulin Rapidly Induces the Cleavage of SREBP-1c Precursor in Vivo. The above findings in isolated hepatocytes are reminiscent of results recently reported in studies of suckling rodents. The suckling period is characterized by low levels of plasma insulin, which is a consequence of the low carbohydrate content of the milk diet (32). Despite the extremely low insulinemia, SREBP-1c mRNA and precursor protein are highly expressed in suckling rats (25) and mice (28). This induction of the SREBP-1c gene may be mediated by LXRα, which is induced and activated during the suckling period (28), possibly due to the abundance of oxysterols in milk. Despite the expression of SREBP-1c mRNA and precursor protein, however, the mature form of SREBP-1c is absent from the liver of suckling rats (25) and mice (28), and SREBP-1c target genes are not expressed (25, 28, 33). This in vivo situation is comparable to our results shown above in hepatocytes cultured with LXR agonist in the absence of insulin.
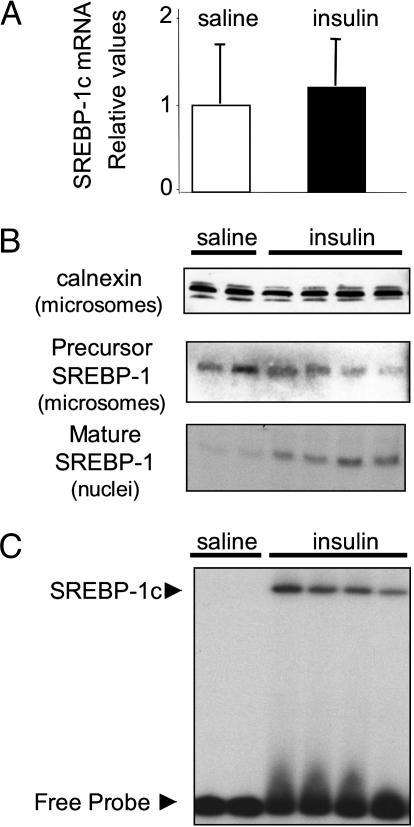
As for our experiments in hepatocytes, we wanted to investigate the ability of insulin to acutely induce the cleavage of the SREBP-1c precursor in this in vivo model. We therefore injected 13-day-old suckling mice with a bolus of insulin and collected the liver 20 min later. This short exposure to insulin has no effect on SREBP-1c mRNA expression (Fig. 4A). As seen in Fig. 4B, suckling mice injected with saline express SREBP-1c precursor in the microsomal membranes, but the nuclear form of the transcription factor is absent. Acute insulin exposure, however, rapidly induces the appearance of mature SREBP-1c in the liver of suckling mice (Fig. 4B). Thus the acute action of insulin to stimulate the proteolytic processing of SREBP-1c is also functional in vivo. Moreover, in an EMSA, we show that mature SREBP-1c induced by the insulin pulse is able to bind to the fatty acid synthase SRE sequence (Fig. 4C), indicating transcriptional potential. The decrease in SREBP-1c precursor that accompanies the appearance of nuclear SREBP-1c in the experiments in hepatocytes was not apparent in these in vivo experiments, possibly being masked by variability in protein expression among individual mice.
Fig. 4.
Acute induction of SREBP-1c cleavage by insulin in liver of suckling mice. Suckling mice were administered 0.4 units of insulin or saline by i.p. injection. After 20 min, mice were killed and livers collected. (A) SREBP-1c mRNA expression. (B) Immunoblot of calnexin (90 kD) is provided as a loading control for microsomal membrane proteins. Immunoblot of SREBP-1c in hepatic microsomal membranes (SREBP-1c precursor) and nuclear extracts (mature SREBP-1). (C) EMSA performed with hepatic nuclear extracts from suckling mice. The position of the SREBP-1c specific complex is indicated. Each lane represents an individual animal.
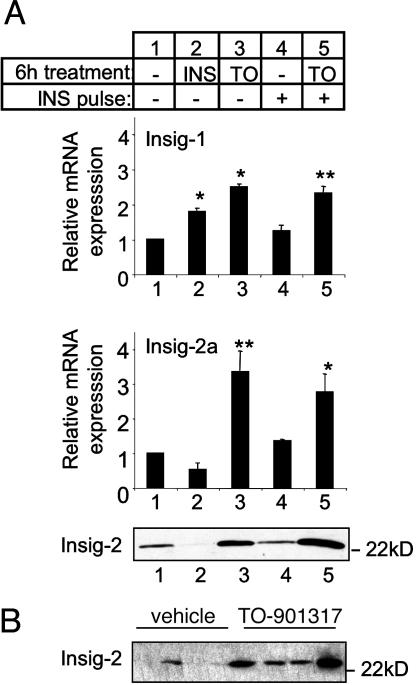
Differential Effects of Insulin and LXR Activation on Insig Gene Expression. To test the hypothesis that LXR activation may induce a factor that inhibits the proteolytic processing of SREBP-1c, we measured the expression of genes that code for the Insig proteins. There are two isoforms of Insig protein (Insig-1 and -2) encoded by separate genes (10, 11). Through the use of different promoters, the Insig-2 gene produces two transcripts, designated Insig-2a and -2b, which are independently regulated (34). The Insig-2a transcript is of particular interest in regard to the regulation of SREBP-1c, because it shows liver-specific expression and is selectively down-regulated by insulin. By reducing the expression of the Insig-2 protein, it has been proposed that insulin, in the long term, could promote SREBP-1c cleavage (34).
Fig. 5A shows the expression of Insig-1 and -2a mRNAs corresponding to the isolated hepatocyte experiments described in Fig. 2. Insig-1 and -2a were the predominant mRNA detected in isolated hepatocytes. Insig-2b was poorly expressed in the liver and showed only a small induction with 6 h of insulin and no change with TO-901317 treatment (data not shown). Confirming previous studies (11, 34), 6-h insulin treatment induces the expression of Insig-1 and tends to down-regulate the Insig-2a transcript (P < 0.06; Fig. 5A, lane 1 vs. 2). Similar to the effect of insulin, a 6-h treatment with TO-901317 induces Insig-1 mRNA expression in hepatocytes (Fig. 5A lane 1 vs. 3).
Fig. 5.
Differential regulation of Insig expression by insulin and TO-901317. (A) Hepatocytes were treated as described in Fig. 2. Insig-1 and -2a mRNA were measured by RT-PCR; each value represents the mean ± SEM of three independent experiments performed in triplicate. *, P < 0.05; **, P < 0.01 compared with untreated control (lane 1). Protein expression of Insig-2 in hepatocyte microsomal membranes was measured by Western blot, as described in Methods. (B) Insig-2 protein in liver of adult mice force-fed with TO-901317 or vehicle, detected by Western blot. Each lane represents one individual animal.
Strikingly, however, LXR activation also produces a marked increase in Insig-2a mRNA. This is in direct contrast to the effect of insulin on this transcript (Fig. 5A, lanes 1-3). This opposing regulation of Insig-2a mRNA expression by insulin and LXR is reflected at the protein level (Fig. 5A, lanes 1-3). The induction of Insig-2 protein by LXR activation shown here in hepatocytes confirms suspicions raised in our recent in vivo study in mice during development (28). In that study, we showed that, compared with other periods of development, suckling mice express high levels of Insig-2a mRNA, which we proposed could be mediated by LXR (28). The ability of LXR activation to induce Insig-2 in vivo is shown here by the induction of Insig-2 protein in adult mice when force-fed the LXR agonist (Fig. 5B). This induction of Insig-2 would be expected to retain SREBP-1c within the ER (10, 13) and could contribute to the relative lack of nuclear SREBP-1c evident with LXR activation, despite strong induction of both SREBP-1c mRNA and precursor protein (Fig. 2).
Importantly, although long-term (6-h) insulin treatment causes a decrease in Insig-2 mRNA and protein (Fig. 5A, lane 1 vs. 2), the 30-min insulin pulse, which induces the cleavage of the SREBP-1c precursor, has no effect in decreasing either Insig-2a mRNA transcript or Insig-2 protein expression (Fig. 5A, lane 3 vs. 5). Thus the cleavage induced by a 30-min insulin pulse occurs despite high levels of Insig expression. This means that the short-term (≈30-min) effects of insulin to stimulate SREBP-1c cleavage must therefore be mediated by an acute mechanism that is distinct from its long-term effect to decrease the expression of the Insig-2 retention factor.
Discussion
In these studies, we describe distinct roles of insulin and LXRα in the regulation of SREBP-1c. Activation of LXR by TO-901317 strongly induces SREBP-1c mRNA and precursor protein but is not sufficient for full induction of the mature form of the transcription factor. The SREBP-1c precursor induced by LXR activation, however, is rapidly cleaved with acute exposure to insulin, revealing a previously undescribed role of this hormone to stimulate the proteolytic processing of SREBP-1c. After demonstrating this effect of insulin in isolated hepatocytes, we show that it is also functional in vivo by injecting the hormone into suckling mice, which exhibit activated LXR, extremely low endogenous insulin levels, and high expression of SREBP-1c precursor but negligible levels of nuclear SREBP-1c.
It has been known for some time that insulin can induce SREBP-1c gene expression, and that this leads to a parallel increase in the mature transcription factor (4, 5, 18). However, until now, it was not known that insulin also had an active role in stimulating the proteolytic processing of SREBP-1c. The current study shows that insulin induces a marked accumulation of nuclear SREBP-1 in <30 min. This is associated with a decrease in the SREBP-1c precursor, indicating that the accumulation of the mature transcription factor is due to an increase in SREBP-1c precursor cleavage.
It has been proposed (34) that insulin may promote the cleavage of SREBP-1c through the selective down-regulation of the Insig-2 protein. Importantly, in the current study, we show that the acute (<30-min) action of insulin to induce SREBP-1c cleavage occurs despite the strong expression of Insig-2, which is induced by LXR activation (Fig. 5). We propose that in the short term, insulin acts via an acute mechanism analogous to that of sterol depletion for the cleavage of SREBP-1a and -2. This may involve disruption of the interaction between SCAP and Insig-2 through a conformational change or rapid degradation of a protein implicated in the SREBP-processing pathway. This acute action of insulin may be reinforced in the longer term by the decrease in Insig-2a expression that occurs with more prolonged exposure to insulin (ref. 34 and Fig. 5). Hepatic overexpression of Insig-1 (12, 13) or Insig-2 (13) in rodents diminishes nuclear SREBP-1c expression and the lipogenic response to insulin. This suggests that either or both the acute and long-term mechanisms of insulin to induce nuclear SREBP-1c can be blocked when Insig proteins are expressed at supraphysiological levels.
The majority of prior studies that have identified a role for LXRα in inducing hepatic SREBP-1c, and its target genes have been performed in vivo, under conditions where endogenous insulin could stimulate the cleavage of SREBP-1c precursor (19, 20). Similarly, the only study in which LXR agonists are shown to induce mature SREBP-1c in isolated hepatocytes was performed in the presence of insulin (35).
Whereas the ability of insulin to regulate SREBP-1c, and thereby lipogenesis, is central to energy homeostasis, the induction of SREBP-1c by LXRα may be more closely linked to cholesterol balance. It has been proposed that LXRα induces SREBP-1c to generate fatty acids needed for the formation of cholesterol esters, which buffer the free cholesterol concentration (36). The current studies show that the regulation of SREBP-1c by LXR differs from the regulation by insulin in two important aspects: not only does LXR lack the effect of insulin to stimulate the proteolytic processing of SREBP-1c (Figs. 2 and 3), but also, in direct contrast to insulin, LXR induces the expression of Insig-2 (Fig. 5). This effect of LXR to induce Insig-2 would be expected to inhibit the cleavage of the SREBP-1c precursor by retaining the SCAP-SREBP complex in the ER. This prompts the question of why LXR activation would stimulate the expression of the SREBP-1c precursor but, paradoxically, simultaneously induce a gene that functions to retain the SREBP precursor in the inactive form. One possible explanation could be that Insig-2 is induced as a safety mechanism. Stimulation of SREBP-1c-mediated lipogenesis by LXR during periods of low glucose availability would be detrimental to glucose homeostasis. The concurrent induction of Insig-2a and the requirement of insulin for cleavage of SREBP-1c would ensure that lipogenesis occurs only when glucose is abundant. The need for such a mechanism is well illustrated by the suckling period, in which active glucose production is necessary due to the high-fat low-carbohydrate diet. Unregulated utilization of this glucose for lipogenesis could be not only futile but also lifethreatening. LXR may also have a role in priming the SREBP-1c system. By inducing the precursor protein but inhibiting the cleavage process via induction of Insig-2, LXR activation may act to provide SREBP-1c precursor that is ready for immediate cleavage on the appearance of insulin. This would allow for a rapid transcriptional response in SREBP-1c-mediated insulin target genes, avoiding the delays inherent in gene transcription and translation. This could be relevant to the rapid adaptations required to respond to everyday feeding cycles.
Supplementary Material
Acknowledgments
We thank S. Lambot, F.Diot-Dupuy, M. Daval, F. Darakhshan, and D. Eberlé for experimental assistance and helpful discussions. B.D.H. is supported by a fellowship from the National Health and Medical Research Council of Australia (no. 188833). A.B. is the recipient of a scholarship from the Ministère de l'Enseignement Supérieur et de la Recherche, France.
Author contributions: B.D.H., A.B., P.F., P.B., and F.F. designed research; B.D.H., A.B., I.H., and P.B. performed research; B.D.H., A.B., P.F., P.B., and F.F. analyzed data; and B.D.H., P.F., and F.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; GK, glucokinase; LXR, liver X receptor; SREBP, sterol regulatory element-binding protein; SCAP, SREBP cleavage activating protein; Insig, insulin-induced gene.
References
- 1.Horton, J. D., Goldstein, J. L. & Brown, M. S. (2002) J. Clin. Invest. 109, 1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimomura, I., Shimano, H., Horton, J. D., Goldstein, J. L. & Brown, M. S. (1997) J. Clin. Invest. 99, 838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimano, H., Yahagi, N., Amemiya-Kudo, M., Hasty, A. H., Osuga, J., Tamura, Y., Shionoiri, F., Iizuka, Y., Ohashi, K., Harada, K., et al. (1999) J. Biol. Chem. 274, 35832-35839. [DOI] [PubMed] [Google Scholar]
- 4.Shimomura, I., Bashmakov, Y., Ikemoto, S., Horton, J. D., Brown, M. S. & Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 13656-13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foretz, M., Pacot, C., Dugail, I., Lemarchand, P., Guichard, C., Le Liepvre, X., Berthelier-Lubrano, C., Spiegelman, B., Kim, J. B., Ferre, P. & Foufelle, F. (1999) Mol. Cell. Biol. 19, 3760-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foretz, M., Guichard, C., Ferre, P. & Foufelle, F. (1999) Proc. Natl. Acad. Sci. USA 96, 12737-12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton, J. D., Shimomura, I., Brown, M. S., Hammer, R. E., Goldstein, J. L. & Shimano, H. (1998) J. Clin. Invest. 101, 2331-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakai, J., Nohturfft, A., Cheng, D., Ho, Y. K., Brown, M. S. & Goldstein, J. L. (1997) J. Biol. Chem. 272, 20213-20221. [DOI] [PubMed] [Google Scholar]
- 9.Nohturfft, A., DeBose-Boyd, R. A., Scheek, S., Goldstein, J. L. & Brown, M. S. (1999) Proc. Natl. Acad. Sci. USA 96, 11235-11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, T., Espenshade, P. J., Wright, M. E., Yabe, D., Gong, Y., Aebersold, R., Goldstein, J. L. & Brown, M. S. (2002) Cell 110, 489-500. [DOI] [PubMed] [Google Scholar]
- 11.Yabe, D., Brown, M. S. & Goldstein, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 12753-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelking, L. J., Kuriyama, H., Hammer, R. E., Horton, J. D., Brown, M. S., Goldstein, J. L. & Liang, G. (2004) J. Clin. Invest. 113, 1168-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaishi, K., Duplomb, L., Wang, M. Y., Li, J. & Unger, R. H. (2004) Proc. Natl. Acad. Sci. USA 101, 7106-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yabe, D., Xia, Z. P., Adams, C. M. & Rawson, R. B. (2002) Proc. Natl. Acad. Sci. USA 99, 16672-16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams, C. M., Goldstein, J. L. & Brown, M. S. (2003) Proc. Natl. Acad. Sci. USA 100, 10647-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng, Z., Otani, H., Brown, M. S. & Goldstein, J. L. (1995) Proc. Natl. Acad. Sci. USA 92, 935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton, J. D., Bashmakov, Y., Shimomura, I. & Shimano, H. (1998) Proc. Natl. Acad. Sci. USA 95, 5987-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzout-Marniche, D., Becard, D., Guichard, C., Foretz, M., Ferre, P. & Foufelle, F. (2000) Biochem. J. 350, 389-393. [PMC free article] [PubMed] [Google Scholar]
- 19.Repa, J. J., Liang, G., Ou, J., Bashmakov, Y., Lobaccaro, J. M., Shimomura, I., Shan, B., Brown, M. S., Goldstein, J. L. & Mangelsdorf, D. J. (2000) Genes Dev. 14, 2819-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz, J. R., Tu, H., Luk, A., Repa, J. J., Medina, J. C., Li, L., Schwendner, S., Wang, S., Thoolen, M., Mangelsdorf, D. J., Lustig, K. D. & Shan, B. (2000) Genes Dev. 14, 2831-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffensen, K. R. & Gustafsson, J. A. (2004) Diabetes 53, S36-S42. [DOI] [PubMed] [Google Scholar]
- 22.Peet, D. J., Turley, S. D., Ma, W., Janowski, B. A., Lobaccaro, J. M., Hammer, R. E. & Mangelsdorf, D. J. (1998) Cell 93, 693-704. [DOI] [PubMed] [Google Scholar]
- 23.Wang, X., Sato, R., Brown, M. S., Hua, X. & Goldstein, J. L. (1994) Cell 77, 53-62. [DOI] [PubMed] [Google Scholar]
- 24.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 25.Botolin, D. & Jump, D. B. (2003) J. Biol. Chem. 278, 6959-6962. [DOI] [PubMed] [Google Scholar]
- 26.Parker, C. S. & Topol, J. (1984) Cell 36, 357-369. [DOI] [PubMed] [Google Scholar]
- 27.Gorski, K., Carneiro, M. & Schibler, U. (1986) Cell 47, 767-776. [DOI] [PubMed] [Google Scholar]
- 28.Bobard, A., Hainault, I., Ferré, P., Foufelle, F. & Bossard, P. (October 27, 2004) J. Biol. Chem., 10.1074/jbc.M406522200.
- 29.Kim, S. Y., Kim, H. I., Kim, T. H., Im, S. S., Park, S. K., Lee, I. K., Kim, K. S. & Ahn, Y. H. (2004) J. Biol. Chem. 279, 30823-30829. [DOI] [PubMed] [Google Scholar]
- 30.Hirano, Y., Yoshida, M., Shimizu, M. & Sato, R. (2001) J. Biol. Chem. 276, 36431-36437. [DOI] [PubMed] [Google Scholar]
- 31.Fleischmann, M. & Iynedjian, P. B. (2000) Biochem. J. 349, 13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girard, J. R., Ferre, P., Pegorier, J. P., Turlan, P., El Manoubi, L. & Callikan, S. (1981) Biochem. Soc. Trans. 9, 369-370. [DOI] [PubMed] [Google Scholar]
- 33.Perdereau, D., Narkewicz, M., Coupe, C., Ferre, P. & Girard, J. (1990) Adv. Enzyme Regul. 30, 91-108. [DOI] [PubMed] [Google Scholar]
- 34.Yabe, D., Komuro, R., Liang, G., Goldstein, J. L. & Brown, M. S. (2003) Proc. Natl. Acad. Sci. USA 100, 3155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawar, A., Botolin, D., Mangelsdorf, D. J. & Jump, D. B. (2003) J. Biol. Chem. 278, 40736-40743. [DOI] [PubMed] [Google Scholar]
- 36.Tontonoz, P. & Mangelsdorf, D. J. (2003) Mol. Endocrinol. 17, 985-993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.