Abstract
Vaccine research in malaria has a high priority. However, identification of specific antigens as candidates for vaccines against asexual blood stages of malaria parasites has been based on largely circumstantial evidence. We describe here how genes encoding target antigens of strain-specific immunity in malaria can be directly located in the parasite's genome without prior information concerning their identity, by the method we call linkage group selection. Two genetically distinct clones of the rodent malaria parasite Plasmodium chabaudi chabaudi, each of which induces an immunity in laboratory mice that is more protective against challenge with itself than with the heterologous strain, were genetically crossed, and the uncloned cross progeny selected in mice that had been made partially immune by infection and drug cure with one or the other parental strain. Proportions of parental alleles in the selected and unselected cross progeny were compared by using quantitative genome-wide molecular markers. A small number, including groups of linked markers forming so-called selection valleys, were markedly reduced under strain-specific immune pressure. A very prominent selection valley was found to contain the gene for merozoite surface protein-1, a major candidate antigen for malaria vaccine development, at the locus at which the strongest reduction under strain-specific immune selection was detected. Closely linked to the merozoite surface protein-1 gene was a gene containing the signature motif of the ring-infected erythrocyte surface antigen family. Another affected locus, unlinked to this selection valley, contained a member of the serine repeat antigen gene family.
Keywords: amplified fragment length polymorphism, merozoite surface protein
Development of new vaccines to protect people from the most deleterious effects of malaria has been one of the main objectives of malaria research. As yet, no protective vaccine against the pathogenic blood stages of malaria exists, although trials in humans with candidate antigens encourage the view that such a vaccine is achievable (1). Most recently, a malaria vaccine based on a sporozoite and preerythrocytic stage antigen, the circumsporozoite protein, has shown convincing efficacy in a field trial (2). Among the antigens of the blood-stage malaria parasites that are considered to be candidates for a malaria vaccine are proteins of the merozoite surface protein (MSP) family such as MSP-1 (3, 4), MSP-2 (5), and MSP-3 (6); the apical membrane antigen 1 (AMA-1) (7, 8); the ring-infected erythrocyte surface antigen (RESA) (9, 10); and serine repeat antigen (11, 12).
The evidence upon which blood stage antigens of malaria parasites have been selected as vaccine candidates has previously been circumstantial and, therefore, usually uncertain in its predictive power. Such evidence could include the recognition of an antigen by antibodies in immune sera from human or animal hosts or its cellular location, such as on the surface of a merozoite or an infected erythrocyte (13). High levels of polymorphism in a protein of the blood-stage parasites, such as MSP-1 and AMA-1, is also considered to be a criterion for its candidacy as a target of protective immunity, as is evidence of preferential immunity against parasites carrying specific alleles of such antigens in malaria endemic human populations (4, 14-16). All such approaches require lengthy and complex experimentation to investigate the candidacy of an antigen as a target of effective protective immunity in malaria. With the completion of the Plasmodium falciparum Genome Project, bioinformatic approaches are now being used (17). Large numbers of sequences predicted as exons from Plasmodium genomic databases have been screened as DNA vaccine constructs (18).
We have developed a genetic method for identifying genes underlying any selectable phenotype in malaria parasites, including strain-specific protective immunity (SSPI). We call the method linkage group selection (LGS) (19) and apply this strategy to the present study. It allows, without prior information of any kind, the identification of loci containing genes that encode target antigens of protective immunity against the blood stages of a malaria parasite. Because our method is genetic, it depends on genetic differences between two parental malaria parasites. In the context of immunity, this procedure involves SSPI between the parasites. There is a small but persuasive literature that demonstrates the contribution that strain-specific immunity makes to protective immunity in human and primate malarias (20-22). SSPI has also been demonstrated in a rodent malaria parasite Plasmodium chabaudi chabaudi in laboratory mice (23) that is the system used in the present study.
Materials and Methods
The Principles of LGS. A more detailed description of the principles and validation of LGS is published elsewhere (19). In the context of malaria parasites, LGS is the application of a specific selection pressure against the recombinant progeny of a cross between two genetically different malaria parasite clones, one of which is susceptible to the selection pressure, whereas the other is resistant to it. In an organism such as Plasmodium, which is haploid for most of its life cycle, those recombinant haploid parasite progeny which carry alleles of genes conferring resistance to the specific selection pressure are most likely to survive, whereas those carrying susceptible alleles are most likely to be removed. Complications, such as dominance, do not arise in a haploid organism so that the relative probabilities of survival or elimination of parasites carrying alleles affected by a specific selection pressure will depend simply, and directly, on the strength of the selection pressure applied. A very strong selection pressure will lead to the total removal of parasites carrying a sensitive allele, as we have previously shown in a test of the principle of LGS by using a drug resistance/sensitivity phenotype as the target of selection (19). Genetic markers from the sensitive parent that are linked to the locus conferring sensitivity will also be reduced in intensity in relation to their distance from the target locus under selection. We define this relationship as a selection valley. Because these markers belong to a genetic linkage group containing the target locus, they serve to identify the genomic region within which the target locus lies, hence the name LGS.
Immunization of Mice and Challenge Infections with P. c. chabaudi. Inbred female CBA/CA mice and genetically distinct cloned lines of P. c. chabaudi, AS-PYR1 and CB, were used in these experiments. These clones were derived from two different lines, or strains, of P. c. chabaudi (AS and CB) isolated from thicket rats, Thamnomys rutilans, in the Central African Republic (24). The clones were derived by limiting dilution of blood-stage parasites. AS was subsequently subjected to selection for pyrimethamine resistance to produce clone AS-PYR1 (25).
To induce SSPI, 6- to 8-week-old female CBA/CA mice were infected by i.p. inoculation with 1 × 106 parasitized erythrocytes of AS-PYR1 or CB. Infections were terminated by mefloquine treatment before the peak of parasitemia at 5 days after inoculation. Mice were, thereafter, regularly screened for parasites by microscopic analysis of Giemsa-stained blood smears. Three weeks after the initial mefloquine treatment, mice were reinfected with the same clone (AS-PYR1 or CB), the infections were grown for 5 days, and were then treated with mefloquine as before, to produce mice that were AS-PYR1- or CB-immune, respectively. Six weeks after the second mefloquine treatment, the same mice were challenged with either a mixture of clones AS-PYR1 and CB, to verify the existence of SSPI between these two parasite strains, or with the uncloned progeny of a genetic cross between AS-PYR1 and CB in the subsequent LGS analysis (see below). All of the challenge inocula contained 5 × 106 parasitized erythrocytes.
Genetic Crosses Between Clones of P. c. chabaudi. To conduct a genetic cross between the two clones AS-PYR1 and CB of P. c. chabaudi, 2 × 105 blood-stage parasites of CB and 2 × 106 blood-stage parasites of AS-PYR1 (the differences in the numbers of each clone being to allow for their difference in growth rate, with CB being the faster growing of the two) were inoculated into two, 6- to 8-week-old female CBA mice. Six days later, when the parasitemias in the mice were 16% and 26%, and gametocytes of both sexes were present (approximately four female gametocytes to one male gametocyte) at gametocytemias of 0.02% and 0.04% respectively, the mice were used to infect Anopheles stephensi female mosquitoes from a laboratory colony. A cage of ≈200 female mosquitoes that had emerged from pupae 6 days previously were allowed to feed on each mouse. The mice were anesthetized and placed on top of the mosquito cages, and the mosquitoes were allowed to feed for 30 min. The mice were humanely killed thereafter, while still under anesthesia. To verify the presence of oocysts, six mosquitoes were dissected from each cage, 8 days after infection; the mean number of oocysts per mosquito for the two cages was 15. Sixteen days after infection with the mixed clone infections, sporozoites were dissected from the salivary glands of a total of 278 of these mosquitoes and injected i.p. into three nonimmune CBA mice in 0.1-ml volumes of a 50:50 FCS:Ringer's solution mixture to grow up the cross progeny for the LGS experiments described in the next section.
Selection of Cross Progeny in Immune Mice for LGS Analysis. Five days after inoculation of the sporozoites from the genetic cross of AS-PYR1 × CB, two each of AS-PYR1- or CB-immunized CBA female mice (whose course of strain-specific immunizations, by infection and drug cure had been completed 6 weeks previously, see above) were inoculated i.p. with 5 × 106 blood-stage parasites from these sporozoite-induced infections to subject the cross progeny to strain-specific immune selection for the LGS analysis. A total of 2 × 106 blood-stage parasites from the sporozoite-induced cross progeny infections were inoculated into nonimmunized CBA female mice, for the LGS control. The cross progeny were grown for 6 days in these mice (both the immune and nonimmune control mice) to allow strain-specific immune selection to take place in the AS-PYR1- or CB-immunized mice. To grow parasites in sufficient quantities to prepare DNA for subsequent analyses, blood was thereupon collected, and 1 × 106 blood-stage parasites from each mouse (immune or nonimmune) were used to initiate infections in four nonimmune mice, and the parasites were grown in these mice for 5 days before harvesting for DNA extraction as described (26).
The Use of Amplified Fragment Length Polymorphism (AFLP) to Identify Selection Valleys. DNA was extracted from the uncloned progeny of the genetic cross, with and without immune selection as described above, and also from equivalent preparations of blood-stage parasites of the parental clones (AS-PYR1 or CB) and screened for genetic markers between AS-PYR1 and CB by using the AFLP method as described (26). AFLP reactions were analyzed on polyacrylamide gels, and AS-PYR1 or CB-specific marker band intensities were measured with both PhosphorImager (Molecular Dynamics) and imagequant software (Molecular Dynamics), and the values were converted to relative intensity indices (RIIs).
The RII of an AFLP marker in a genetically mixed population of parasites is defined as the intensity of the AFLP marker band in the mixture divided by the intensity of a designated nonpolymorphic band in the same mixed parasite sample on the same AFLP gel (its intensity index), divided by the equivalent ratio (intensity index) of the same two bands (AFLP and nonpolymorphic band) when measured in a sample of the pure parental strain (27). For the purpose of determining an intensity index, we always chose nonpolymorphic reference bands that were close to the polymorphic AFLP bands to minimize error due to any variation in gel or image quality, as previously discussed (26). Comparative Intensity (CI) is defined as the RII of an AFLP marker in the cross progeny selected in the immunized mice (RIIi), divided by the RII of the marker of the cross progeny grown in parallel in nonimmunized mice (RIIni), and expressed as a percentage, i.e., CI = (RIIi/RIIni) × 100.
Assignment of AFLP Markers to Locations in Plasmodium Genomes. AFLP markers that showed a significant reduction in CI in immune mice (relative to nonimmune mice) were located on a genetic linkage map of P. c. chabaudi (28) obtained from previous genetic crosses between AS-derived clones and AJ [another strain of P. c. chabaudi from the same African location as AS and CB (24)]. Some of these AFLP markers were also physically mapped to locations in the P. falciparum (sequence data for P. falciparum were obtained from the Sanger Centre web site, which can be accessed at www.sanger.ac.uk/Projects/P_falciparum) and/or Plasmodium yoelii yoelii (sequence data for P. y. yoelii were obtained from The Institute for Genomic Research web site, which can be accessed at www.tigr.org/tdb/e2k1/pya1) genomes. The markers were eluted from the gel, amplified, sequenced, and their homologues in the P. falciparum and/or P. y. yoelii genomes identified by blast searches (29).
Real-Time Quantitative PCR (RTQ-PCR) Measurement of P. c. chabaudi msp1. RTQ-PCRs were performed on a LightCycler instrument (Roche Diagnostics) as described (30), with calibrated mixtures of parasite DNA by using an assay that quantitatively discriminates between clones carrying the AS-PYR1 or CB allele of msp1. The msp1 allele-specific primers used in this assay were: AS-PYR1 forward primer 5′-CCACAACACCTGAAACCAC-3′ with AS-PYR1 reverse 5′-CTGGTGCTGCTGGGACA-3′; and the CB forward primer 5′-CTGTTACAACCCAAACC-3′ with CB reverse primer 5′-AGTTGTTCCTGTGGCAG-3′. The actual DNA concentrations measured were converted to show the relative proportions of msp1 alleles of AS-PYR1 and CB present in each sample analyzed.
The RTQ-PCR assay has been calibrated for alleles of msp1 by using rigorously prepared and quantified mixtures of blood-stage parasites of AS-PYR1 and CB and shown to measure the proportions of parasites in a mixture to an accuracy of <1% (30).
Results
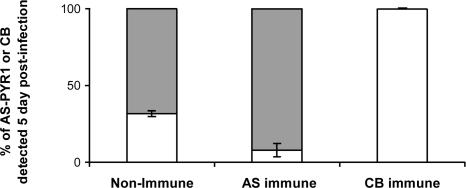
Demonstration of SSPI Between Cloned Strains AS-PYR1 and CB of P. c. chabaudi. In a preliminary experiment, we verified the existence of SSPI generated by, and between, P. c. chabaudi-cloned strains AS-PYR1 and CB. This procedure was performed by inoculating mixtures of equal proportions of blood-stage parasites of AS-PYR1 and CB into mice made immune to one or the other strain (see Materials and Methods). The proportions of each strain in the subsequent infections were followed by RTQ-PCR of the AS-PYR1 and CB alleles of msp1 (see Materials and Methods). The results showed that protective immunity generated by each of these cloned strains was highly strain-specific and resulted in the almost complete removal (AS-PYR1 clone), or the complete removal (CB clone), of the immunizing strain (Fig. 1). This level of SSPI between AS-PYR1 and CB, and especially that induced by CB, was considered to be sufficiently strong to be used in LGS experiments to identify loci containing genes for the target antigens of SSPI in P. c. chabaudi.
Fig. 1.
The proportion of parasites carrying the AS-PYR1 or CB allele of P. c. chabaudi msp1 measured by RTQ-PCR analysis of DNA samples obtained from mixed-strain infections in two nonimmune mice, two AS-PYR1-immune mice, and two CB-immune mice at 5 days after inoculation. The DNA concentrations of each msp1 allele (AS-PYR1 or CB) as measured by RTQ-PCR, are given as a ratio to show their relative proportions in each sample analyzed. The bars represent the SD of AS-PYR1 and CB measurements between mice. Gray bars represent the proportions of CB parasites in a mixture, and white bars represent the proportions of AS-PYR1 parasites in a mixture.
The Application of LGS to the Location of Genes for the Target Antigens of SSPI in P. c. chabaudi. The uncloned progeny of a genetic cross between AS-PYR1 and CB were subjected to strain-specific immune selection by growth in mice immunized with one or other of the parental strains (see Materials and Methods). The cross progeny were also grown in nonimmune mice. DNA prepared from parasites derived from these infections was analyzed by AFLP. A total of 481 AFLP markers that distinguished AS-PYR1 and CB were identified; 206 of these markers were specific to AS-PYR1, and 275 markers were specific to CB. To evaluate whether each marker is linked to a locus under selection, the RIIs for each AFLP marker from immune and nonimmune mice were compared, and a CI was obtained for each marker (see Materials and Methods for definitions of RII and CI).
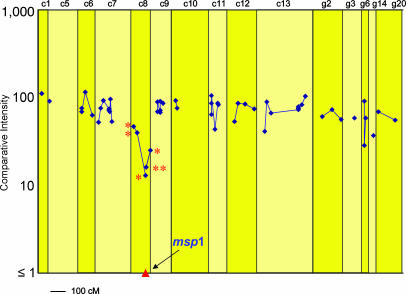
Of the 275 CB-specific markers, 44 showed CIs of <50% under CB-specific immune selection, suggesting that they may be linked to loci affected by CB-specific protective immunity. We wished to locate these markers on a P. c. chabaudi genetic linkage map that had previously been made from the progeny of a genetic cross between strains AJ and AS (28). The effects of CB-specific immune selection across the P. c. chabaudi genome is illustrated in Fig. 2 in relation to a total of 56 CB AFLP markers that we were able to locate on the P. c. chabaudi genetic linkage map. Of the CB AFLP markers whose CIs were considered to be significantly reduced after selection, 10 appeared to be identical to markers of AJ (based on their size and on the selective bases used for their amplification), and could, therefore, be located on the AJ × AS genetic linkage map (28) (Fig. 2). Six of these markers mapped to P. c. chabaudi chromosome 8, which contains the msp1 gene in this species, and one each to chromosomes 11 and 13, and to the unassigned linkage groups g6 and g14 (Table 1).
Fig. 2.
The CIs after selection in CB-immunized mice, of those CB-specific AFLP markers that could be located on a P. c. chabaudi genetic linkage map (see text). Because the genetic linkage map was generated by using strains AS and AJ, only 56 of the CB markers of the total 275 CB markers could be assigned (see text). Genetic distances are represented in centimorgans. The proportion of the progeny carrying the CB allele at the msp1 locus was measured by RTQ-PCR (see also Table 1 and Figs. 3 and 4). A selection valley is clearly evident on chromosome 8 with the msp1 locus at its lowest point. The AFLP markers represented here on P. c. chabaudi chromosome 8 are also represented in Table 1 and Fig. 3. These markers are indicated by an asterisk.
Table 1. The genetic and genomic locations of markers of P. c. chabaudi strain CB affected by CB-specific immune selection in mice.
| Chromosomal mapping
|
||||
|---|---|---|---|---|
|
P. chabaudi*
|
P. falciparum
|
|||
| Name of marker | Cl in CB-immune mice, % | Chromosome no. | Chromosome no. | Approximate position, kb |
| CBCA01CT | 10 | ND | 1 | pf1-0288 |
| CBGA01AG | 32 | ND | 2 | pf2-0310 |
| CBCA07AT | 18 | ND | 5 | pf5-0285 |
| CBGT02AC | 36 | ND | 5 | pf5-1013 |
| CBAC03AT | 46 | 8 | ND | ND |
| CBAC01AG | 40 | 8 | 9 | pf9-0742 |
| CBAT02TA | 39 | ND | 9 | pf9-0910 |
| CBGT01GT | 28 | ND | 9 | pf9-1007 |
| CBAT01AG | 30 | ND | 9 | pf9-1029 |
| CBTT04AT | 13 | 8 | 9 | pf9-1040 |
| CBTC02TT | 18 | ND | 9 | pf9-1164 |
| CBGT06AT | 8 | ND | 9 | pf9-1168 |
| CB msp1† | 1 | 8 | 9 | pf9-1203 |
| CBAG05AT | 12 | 8 | 9 | pf9-1263 |
| CBAT01TA | 16 | 8 | ND | ND |
| CBAG02AG | 26 | 8 | 9 | pf9-1372 |
| CBCT01TC | 44 | 11 | ND | ND |
| CBAC01GA | 34 | ND | 12 | pf12-1490 |
| CBAC04AT | 41 | 13 | ND | ND |
| CBTA01GA | 45 | ND | 14 | pf14-1621 |
| CBTT01CA | 37 | ND | 14 | pf14-1892 |
| CBTC05AG | 45 | ND | 14 | pf14-2037 |
| CBTT01TG | 28 | g6 | ND | ND |
| CBTT06TT | 37 | g14 | ND | ND |
CB markers that were strongly reduced (i.e., with a Cl of <50%) in the uncloned progeny of the genetic cross between P. c. chabaudi AS-PYR1 and CB after selection in CB immunized mice are shown. Physical distances in kilobase pairs for P. falciparum are from the P. falciparum genome database. Genetic distances for P. c. chabaudi markers are shown in Figs. 2 and 3 in centimorgans. The largest group of linked markers under selection (in bold) form the selection valley around the msp1 locus (underlined because it is not an AFLP marker) on chromosome 8 of P. c. chabaudi and chromosome 9 of P. falciparum (see also Fig. 3). ND, not determined.
These markers were genetically mapped onto their chromosomes.
Measured by RTQ-PCR assay.
We also wished to map markers with reduced intensity onto the P. falciparum genome by blast searching. Of the 44 CB markers that were reduced under selection, 17 were sequenced, and their homologues were successfully located in P. falciparum. Nine of these markers mapped to P. falciparum chromosome 9, on which msp1 is located in this species (Table 1). The other eight markers mapped to positions on P. falciparum chromosomes 1, 2, and 12 (one marker each), 5 (two markers each) and 14 (three markers each) (Table 1).
Note that a total of 11 CB markers that were reduced under CB-immune selection were mapped to positions linked to msp1, either genetically in P. c. chabaudi, or physically in P. falciparum, or, in the case of four of these markers (CBAC01AG, CBTT04AT, CBAG05AT, and CBAG02AG), in both (in bold in Table 1).
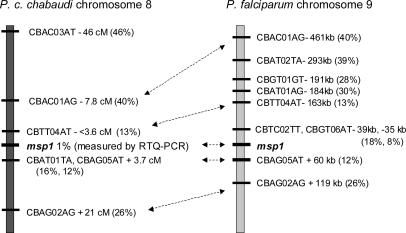
CB markers generally decreased in CI with decreasing physical (P. falciparum) or genetic (P. c. chabaudi) distance from the msp1 locus (Table 1 and Figs. 2 and 3), thereby defining a selection valley pointing toward the msp1 locus. The four AFLP markers that showed the strongest intensity reduction under CB-immune selection, and which could also be assigned physical locations in the P. falciparum genome, having CIs of 8%, 12%, 13%, and 18%, respectively, lay 35, 60, 163, and 39 kb, respectively, from the P. falciparum msp1 locus (Fig. 3). Three CB markers whose CIs were reduced to <20% under the selection, and which could be located on the P. c. chabaudi genetic linkage map, all lay within 4 cM, predicted to be equivalent to ≈80 kb in P. c. chabaudi (31), from the msp1 locus (Fig. 3).
Fig. 3.
CB AFLP markers linked to msp1 in P. c. chabaudi and P. falciparum. The distances of CB markers from the msp1 locus on P. c. chabaudi chromosome 8 are shown in centimorgans. Distances from the msp1 locus of the homologues of the affected CB AFLP markers along P. falciparum chromosome 9 are shown in base pairs. Those AFLP markers that could be located in both species are indicated by double-headed arrows. The CIs of the CB AFLP markers, and the proportion of parasites carrying the CB allele at the msp1 locus, are indicated by percentages.
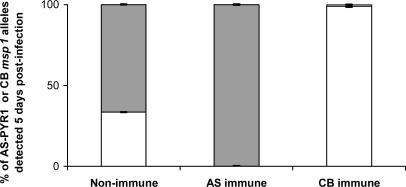
To quantify, as accurately as possible, the proportions of parasites carrying the AS-PYR1 and CB alleles of msp1, we used RTQ-PCR (30) based on the extensive polymorphism between the AS-PYR1 and CB alleles at the msp1 locus. This procedure showed that the CB msp1 allele was reduced to ≈1% in the CB-immune-selected population (Fig. 4), below the reduction in CI of the two AFLP CB markers closest to msp1 (Figs. 2 and 3). The locus containing msp1 lies, therefore, at the lowest point that we have yet detected in the selection valley. Marker CBAG05AT (Table 1 and Fig. 3), which is among those most closely linked to the msp1 gene in both P. chabaudi and P. falciparum, and is, likewise, one of the markers under the strongest detected reduction under CB-immune selection, showed homology to a gene in P. y. yoelii with the motif for RESA.
Fig. 4.
The relative proportions of AS-PYR1 and CB alleles of msp1 measured by RTQ-PCR in the progeny of the genetic cross between AS-PYR1- and CB-cloned strains of P. c. chabaudi. Data represent the mean values and SE obtained from five independent measurements of AS-PYR1 or CB allelic variants of msp1. Gray bars represent the proportions of parasites carrying the CB allele of msp1, and white bars represent the proportions of parasites carrying the AS-PYR1 allele of msp1.
As shown in Table 1, 12 other markers that showed a CI of <50% in the CB-immune mice relative to nonimmune mice, were not associated with the msp1 selection valley and were distributed elsewhere in the Plasmodium genome. We have yet to confirm or characterize selection valleys associated with any of these other markers under apparent CB-immune selection. One of the markers, CBGA01AG, showed homology to a member of the serine repeat antigen gene family on chromosome 2 of P. falciparum. Of the other affected markers outside the msp1 selection valley, one had a homologue on P. falciparum chromosome 1, two on chromosome 5, one on chromosome 12, and three on chromosome 14 (Table 1). Four other markers were located on P. c. chabaudi chromosomes 11 and 13, and on unassigned linkage groups g6 and g14 (Table 1 and Fig. 2).
We also analyzed the intensities of AS-PYR1 AFLP markers in the reciprocal experiment where mice were immunized with the AS-PYR1 strain and then challenged with the AS-PYR1 × CB cross progeny. In general, the results were less clear than those given by CB-immune selection on the CB-specific AFLP markers. The main reason for this conclusion was that in the unselected (control) progeny of the AS-PYR1 × CB cross, the intensities of many of the AS-PYR1-specific AFLP markers appeared very faint, especially when compared with those of the CB markers. This result was probably due to the overrepresentation of parental CB (because of its faster growth rate) in the cross progeny and it would also account for the fact that the CIs of CB markers in the CB-immune-selected progeny are generally less than one (see Fig. 2). This finding is because the more parental CB is present in the unselected progeny the greater must be the relative reduction of all CB markers under any CB-specific selection. Nevertheless, despite this overrepresentation of parental CB, we were able, by using the RTQ-PCR method, to determine that the proportion of the AS-PYR1 allele of msp1 was reduced to <1% in the AS-PYR1-immune-selected population (Fig. 4). This result is well below the general intensity of most other AS-PYR1 markers in the AS-PYR1-immune-selected progeny. The result is consistent with the AS-PYR1 allele of msp1, or of a gene closely linked to it, coding for a target of SSPI in AS-PYR1.
Discussion
Our results show how LGS can be used to locate regions in a Plasmodium genome that contains genes encoding target antigens of strain-specific immunity in malaria. Thus, the progeny of a genetic cross between two strains of P. c. chabaudi, AS-PYR1, and CB, were subjected to AS-PYR1 or CB strain-specific immune selection and analyzed in a genome-wide scan by AFLP. Several AFLP markers that were strongly affected by CB-specific immune selection pressure were located on P. c. chabaudi chromosome 8 forming a so-called selection valley. After genome database mining, homologues of CB markers in this selection valley were located to a region on P. falciparum chromosome 9, close to the gene encoding the MSP-1 antigen, a major candidate for a target of SSPI in malaria. In the P. c. chabaudi AS-PYR1 × CB selection valley under CB-induced immune pressure, the CB allele of msp1 marked the locus under the strongest detected reduction, as measured by RTQ-PCR, as did the AS-PYR1 allele of msp1 under AS-PYR1-induced immune pressure. Unfortunately, poor general representation of the AS-PYR1 AFLP markers in this cross progeny prevented the identification of other AS-PYR1 markers associated with selection around this locus.
This demonstration that alleles at the msp1 locus in P. c. chabaudi are strongly affected by strain-specific immune selection in two reciprocal LGS experiments (CB- or AS-PYR1-immune selection), is very strong evidence that msp1, or a gene very closely linked to it, encodes a major target antigen of SSPI in this malaria parasite. Although the msp1 gene is itself the most likely candidate in the region under selection, our analysis has revealed that the affected linkage group also contains a gene encoding a protein with a RESA signature motif in P. y. yoelii, a rodent malaria parasite that is phylogenetically closely related to P. chabaudi. RESA is a protein expressed in the membrane of erythrocytes infected with ring stage malaria parasites and is a candidate antigen as a target of protective immunity in malaria (9, 10). A RESA-related protein could likewise be a potential target of protective immunity. In addition, there are several predicted proteins of unknown function located within 60 kb of the msp1 gene in the P. falciparum genome (the region corresponding to the most intense selection in P. c. chabaudi) whose candidacy will need to be tested.
Evaluating the candidacy of each of the expressed proteins within this otherwise tightly defined region in the parasite genome could involve the sequencing of all such genes in P. c. chabaudi AS-PYR1 and CB. This process would reveal whether, and to what extent, the genes for the individual proteins are polymorphic and, therefore, whether they could, in principle, elicit, and be targeted by, SSPI responses. Target antigens of strain-specific immunity are expected to have a large number of polymorphic sites. Sites with at least three consecutive SNPs would be suitable for generating primers that could be used for RTQ-PCR. Such very accurately quantifiable markers would lead to a more accurate characterization of the slopes and dimensions of the selection valley around the msp1 locus and would allow us to determine whether the msp1 locus, or another locus, lay at the point of strongest selection within the valley. Should the msp1 locus be the only significantly polymorphic locus in the region of P. c. chabaudi chromosome 8 under strong strain-specific immune selection, this would identify msp1 itself as the only likely candidate antigen for strain-specific immunity in the region. If necessary, backcrossing can be used to increase the resolution of an LGS analysis. A single back-cross to the sensitive parasite reduces the width of a selection valley and increases the resolution of any markers within it. Bioinformatic analysis, such as prediction of membrane location and stage-specific expression profile, may also be used to evaluate the likely involvement of a candidate locus in a protective immune response. Together, these approaches should allow us to identify the gene, or genes, if more than one, that are the targets of SSPI in the selection valley associated with msp1.
We noted several regions on other chromosomes that may be selected by strain-specific immunity (Table 1) and that could, therefore, indicate the presence of other selection valleys containing genes for targets of strain-specific immunity. However, unlike the region around msp1, these contain only one to three affected markers. This number of markers is too few with which to identify clear selection valleys at this stage. One such affected region, on P. falciparum chromosome 14, contains three markers spanning 416 kb (Table 1), or ≈20 cM of genetic distance in this species (32). As none of the markers was reduced <37%, this region could contain a gene that, compared with the gene within the selection valley associated with msp1, has a relatively small contribution to strain-specific immunity. Alternatively, these three markers could belong to one edge of a deeper selection valley. If this is the case, markers within the deeper region of the valley remain to be identified. Another affected marker, located on P. falciparum chromosome 2, represents a gene encoding a protein related to serine repeat antigen, a candidate for a target of protective immunity in malaria (11, 12).
It is noteworthy that the gene for the polymorphic malaria vaccine candidate antigen, AMA-1 (8), which is located on chromosome 9 of P. c. chabaudi, is not found at any of the genomic locations implicated in strain-specific immunity in this LGS study (Table 1). This outcome, however, was expected because, most unusually, there are no polymorphisms in ama-1 between P. c. chabaudi clones AS-PYR1 and CB in the cell surface, ectodomain-coding regions of the gene where they have otherwise been detected (S.C. and R. Carter, unpublished data). By contrast, comparison of the AS-PYR1 and CB alleles of msp1, which was strongly implicated as a target of strain-specific immunity in this study, reveals a large amount of sequence polymorphism between them, with sequence identity being only ≈85% (S.C. and R. Carter, unpublished data).
In conclusion, LGS has enabled us to locate regions in the genome of the malaria parasite P. c. chabaudi, which contains genes controlling strain-specific immunity against the disease-causing blood stages of the parasites. These regions include a well defined selection valley whose focus lies on, or very close to, the gene encoding the malaria vaccine candidate antigen, MSP-1. Together with our identification of the locus (containing dhfr) underlying pyrimethamine resistance in P. c. chabaudi (19), our present results demonstrate the power and versatility of LGS for the location of genes of malaria parasites that encode selectable phenotypes of these parasites. Because of the equivalence of the genetic features of their life cycles, LGS will be equally applicable to other Apicomplexan parasites, including such important pathogens as the Theilerias, Babesias, and Eimerian parasites of domestic animals.
Acknowledgments
We thank Les Steven for technical assistance, Jacobus de Roode (University of Edinburgh) for assistance in the preparation of material for RTQ-PCR, Deborah Charlesworth for critical reading the manuscript, and Danilo Martinelli for the financial support of A.M. This work was supported by the Wellcome Trust and the Medical Research Council of the United Kingdom.
Author contributions: R. Carter and M.M. designed research; A.M., S.C., R. Culleton, A.R., and R. Carter performed research; A.M., S.C., and P.H. analyzed data; and A.M., P.H., and R. Carter wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LGS, linkage group selection; SSPI, strain-specific protective immunity; MSP, merozoite surface protein; AMA-1, apical membrane antigen 1; AFLP, amplified fragment length polymorphism; RQT-PCR, real-time quantitative PCR; RESA, ring-infected erythrocyte surface antigen; RII, relative intensity index; CI, comparative intensity.
References
- 1.Carvalho, L. J., Daniel-Ribeiro, C. T. & Goto, H. (2002) Scand. J. Immunol. 56, 327-343. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, P. L., Sacarlal, J., Aponte, J. J., Leach, A., Macete, E., Milman, J., Mandomando, I., Spiessens, B., Guinovart, C., Espasa, M., et al. (2004) Lancet 364, 1411-1420. [DOI] [PubMed] [Google Scholar]
- 3.Siddiqui, W. A., Tam, L. Q., Kramer, K. J., Hui, G. S., Case, S. E., Yamaga, K.M., Chang, S. P., Chan, E. B. & Kan, S. C. (1987) Proc. Natl. Acad. Sci. USA 84, 3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conway, D. J., Cavanagh, D. R., Tanabe, K., Roper, C., Mikes, Z. S., Sakihama, N., Bojang, K. A., Oduola, A. M., Kremsner, P. G., Arnot, D. E., et al. (2000) Nat. Med. 6, 689-692. [DOI] [PubMed] [Google Scholar]
- 5.Saul, A., Lord, R., Jones, G. L. & Spencer, L. (1992) J. Immunol. 148, 208-211. [PubMed] [Google Scholar]
- 6.Carvalho, L. J., Oliveira, S. G., Theisen, M., Alves, F. A., Andrade, M. C., Zanini, G. M., Oeuvray, C., Povova, M. M., Muniz, J. A., Druilhe, P., et al. (2004) Scand. J. Immunol. 59, 363-372. [DOI] [PubMed] [Google Scholar]
- 7.Collins, W. E., Pye, D., Crewther, P. E., Vanderbergh, K .L., Galland, G. G., Sulzer, A. J., Kemp, D. J., Edwards, S. J., Coppel, R. L., Sullivan, J. S., et al. (1994) Am. J. Trop. Med. Hyg. 51, 711-719. [DOI] [PubMed] [Google Scholar]
- 8.Anders, R. F., Crewther, P. E., Edwards, S., Margetts, M., Matthew, M .L., Pollock, B. & Pye, D. (1998) Vaccine 16, 240-247. [DOI] [PubMed] [Google Scholar]
- 9.Collins, W. E., Anders, R. F., Ruebush, T. I., Kemp, D .J., Woodrow, G. C., Campbell, G. H., Brown, G. V., Irving, D. O., Gloss, N., Filipski, V. K., et al. (1991) Am. J. Trop. Med. Hyg. 44, 34-41. [DOI] [PubMed] [Google Scholar]
- 10.Berzins, K., Perlmann, H., Wahlin, B., Ekre, H. P., Hogh, B., Petersen, E., Wellde, B., Schoenbechler, M., William, J., Chulay, J., et al. (1991) Infect. Immun. 59, 1500-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin, L. H., Ramirez, E., Lambert, P. H. & Miescher, P. A. (1981) Nature 298, 301-303. [DOI] [PubMed] [Google Scholar]
- 12.Inselburg, J., Bathurst, I. C., Kansopon, J., Barr, P. F. & Rossan, R. N. (1993) Infect. Immun. 61, 2048-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good, M. F. (2001) Nat. Rev. Immunol. 1, 117-125. [DOI] [PubMed] [Google Scholar]
- 14.Conway, D. J. (1997) Parasitol. Today 13, 26-29. [DOI] [PubMed] [Google Scholar]
- 15.Conway, D. J. & Polley, S. D. (2002) Parasitology 125, S3-S16. [DOI] [PubMed] [Google Scholar]
- 16.Polley, S. D., Chokejindachai, W. & Conway, D. J. (2003) Genetics 165, 5555-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doolan, D. L., Aguiar, J. C., Weiss, W. R., Sette, A., Felger, P. L., Regis, D. P., Quinones-Casa, P., Yates, J. R. III, Blair, P. L., Richie, T .L., et al. (2003) J. Exp. Biol. 206, 3789-3802. [DOI] [PubMed] [Google Scholar]
- 18.Haddad, D., Bilcikova, E., Witney, A. A., Carlton, J. M., White, C. E., Blair, P. L., Chattopadhyay R., Russell, J., Abot, E., Charoenvit, Y., et al. (2004) Infect. Immun. 72, 1594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culleton, R., Martinelli, A., Hunt, P. & Carter, R. (2005) Genome Res. 15, 92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciuca, M., Balli, L. & Chelarescu, V. M. (1934) Trans. R. Soc. Trop. Med. Hyg. 27, 619-622. [Google Scholar]
- 21.Jeffrey, G. M. (1966) Bull. W. H. O. 35, 873-882. [PMC free article] [PubMed] [Google Scholar]
- 22.Powell, R. D., McNamara, J. V. & Rieckmann, K. H. (1972) Proc. Helminthol. Soc. Wash. 39, 51-66. [Google Scholar]
- 23.Jarra, W. & Brown, K. N. (1985) Parasite Immunol. (Oxf.) 7, 595-606. [DOI] [PubMed] [Google Scholar]
- 24.Carter, R. & Walliker, D. (1975) Ann. Trop. Med. Parasitol. 69, 187-196. [DOI] [PubMed] [Google Scholar]
- 25.Walliker, D., Carter R. & Sanderson, A. (1975) Parasitology 70, 19-24. [DOI] [PubMed] [Google Scholar]
- 26.Grech, K., Martinelli, A., Pathirana, S., Walliker, D., Hunt, P. & Carter, R. (2002) Mol. Biochem. Parasitol. 123, 95-104. [DOI] [PubMed] [Google Scholar]
- 27.Martinelli, A., Hunt, P., Cheesman, S J. & Carter, R. (2004) Mol. Biochem. Parasitol. 136, 117-122. [DOI] [PubMed] [Google Scholar]
- 28.Martinelli, A. (2003) Ph.D. thesis (Univ. of Edinburgh, Edinburgh, U.K.).
- 29.Hunt, P., Martinelli, A., Fawcett, R., Carlton, J., Carter, R. & Walliker, D. (2004) Mol. Biochem. Parasitol. 136, 157-164. [DOI] [PubMed] [Google Scholar]
- 30.Cheesman, S. J., de Roode, J. C., Read, A. F. & Carter, R. (2003) Mol. Biochem. Parasitol. 131, 83-91. [DOI] [PubMed] [Google Scholar]
- 31.Carlton, J. (1995) Ph.D. thesis (Univ. of Edinburgh, Edinburgh, U.K.).
- 32.Walker-Jonah, A., Dolan, S. A., Gwadz, R. W., Panton, L. J. & Wellems, T. E. (1992) Mol. Biochem. Parasitol. 51, 313-320. [DOI] [PubMed] [Google Scholar]