Abstract
Polyglutamine (polyQ) disorders, including Huntington's disease (HD), are caused by expansion of polyQ-encoding repeats within otherwise unrelated gene products. In polyQ diseases, the pathology and death of affected neurons are associated with the accumulation of mutant proteins in insoluble aggregates. Several studies implicate polyQ-dependent aggregation as a cause of neurodegeneration in HD, suggesting that inhibition of neuronal polyQ aggregation may be therapeutic in HD patients. We have used a yeast-based high-throughput screening assay to identify small-molecule inhibitors of polyQ aggregation. We validated the effects of four hit compounds in mammalian cell-based models of HD, optimized compound structures for potency, and then tested them in vitro in cultured brain slices from HD transgenic mice. These efforts identified a potent compound (IC50 = 10 nM) with long-term inhibitory effects on polyQ aggregation in HD neurons. Testing of this compound in a Drosophila HD model showed that it suppresses neurodegeneration in vivo, strongly suggesting an essential role for polyQ aggregation in HD pathology. The aggregation inhibitors identified in this screen represent four primary chemical scaffolds and are strong lead compounds for the development of therapeutics for human polyQ diseases.
Keywords: high-throughput screen, small-molecule therapeutics, Drosophila, R6/2 brain slices, genetic disease
At least nine inherited neurodegenerative diseases, including Huntington's disease (HD), are caused by expansion of polyglutamine (polyQ)-encoding repeats within otherwise unrelated proteins (1, 2). In HD, expansion of polyQ repeats within the huntingtin (Htt) protein causes an adult-onset neurodegenerative disease characterized by movement disorder, psychiatric symptoms, and cognitive dysfunction (3-5). As in several major neurological disorders, including Alzheimer's and Parkinson's diseases, the pathology and death of affected neurons in polyQ diseases are associated with accumulation of mutant polypeptides in insoluble aggregates (6-9). These polyQ-containing aggregates, or inclusions, have been found in the nuclei of affected neurons in postmortem patient tissues and brains from HD transgenic mice (10-12) and have emerged as a hallmark of HD pathology.
Mutant polypeptides with extended polyQ tracts aggregate in vitro and in vivo in a polyQ length-dependent manner, which closely correlates with the age of onset in HD and other polyQ-expansion diseases (2, 13-15). Although the precise role of neuronal aggregates in disease pathogenesis is not clear, therapeutic strategies aimed at inhibiting polyQ aggregation have shown some efficacy in vivo in both Drosophila and mouse models of HD (16, 17). These and other studies (18, 19) implicate polyQ-dependent aggregation as a cause of neurodegeneration in HD and suggest that inhibition of neuronal polyQ aggregation may be therapeutic in HD patients (8).
Chemical compounds that directly target polyQ aggregation have been identified in high-throughput screens using cell-free assays (20). As an alternative approach, we sought to identify small molecules that suppress polyQ aggregation by targeting cellular pathways. We described (21) a yeast model of polyQ aggregation and its associated toxicity; in this model, an N-terminal fragment of mutant Htt containing an extended polyQ tract (103Q) aggregates efficiently and is strongly cytotoxic, resulting in reduced yeast growth and reduced Htt-103Q expression levels. In contrast, a similar polypeptide with polyQ of normal length (25Q) does not aggregate and is not toxic in cells. We used this model to design a cell-based high-throughput screen for chemical compounds that inhibit polyQ aggregation, based on monitoring reversal of growth inhibition and increased overall Htt-103Q expression levels.
The yeast-based screening assay identified small molecules that potently inhibit polyQ aggregation in intact cells. Hits obtained from the primary yeast screen were tested subsequently in multiple secondary aggregation assays, including mammalian cell-based and in vitro assays (20, 22). Hit compounds were optimized for potency and then tested for activity in vitro in brain slices derived from HD transgenic mice (23, 24). These efforts identified a potent compound (IC50 = 10 nM) with long-term inhibitory effects on polyQ aggregation in neurons. Testing of this compound in a Drosophila HD model (22, 25) showed that it could suppress neurodegeneration in vivo, strongly suggesting an essential role for polyQ aggregation in HD pathology. The aggregation inhibitors identified in this screen represent four previously uncharacterized chemical scaffolds and are strong lead compounds for the development of therapeutics for HD.
Materials and Methods
Source of Compounds. The 16,000-compound collection (Diverse Set) for the primary screen in yeast and structural analogs of identified inhibitors were synthesized at and obtained from Chembridge (San Diego; hit2lead). Compounds C2-85B6 and C2-8C5 were synthesized at and obtained from TimTec (Newark, DE). For secondary assays, compounds were reobtained from vendors at a purity of >95%.
Compound Screen in Yeast. For the primary screen, we engineered an erg6 yeast strain, which expresses Htt-103Q tagged with EGFP (Clontech) under the control of the GAL1 promoter. The erg6 mutation inhibits ergosterol biosynthesis, which enhances membrane fluidity, resulting in increased membrane permeability to various chemical compounds (26). The yeast culture was grown to midlogarithmic phase, shifted to galactose medium to induce Htt-103Q-EGFP expression, plated in 96-well plates, and supplemented with 10 μM compounds. We measured the OD600 to monitor yeast growth and EGFP fluorescence (excitation/emission, 485/520 nm) to monitor expression levels of 103Q-EGFP fusion proteins. EGFP fluorescence was assessed in samples before galactose induction (to select out autofluorescent compounds) and again after 20 h of treatment. We selected chemical compounds that caused at least a 25% increase in OD600 and/or EGFP fluorescence. The ability of each hit compound to suppress Htt-103Q-EGFP aggregation was then assessed microscopically.
Compound Tests in Mammalian Cell-Based Model of polyQ Aggregation. Quantification of polyQ aggregates in rat pheochromocytoma (PC12) cell lines that express the Htt-103Q-EGFP fusion protein in inducible fashion has been described (22). Previously, we isolated two subclones that produce visible aggregates in ≈50-80% of cells after 48 h of 5 μM muristerone A induction. PC12 cell lines were maintained with continued selection in DMEM (5% glucose) containing 10% horse serum, 5% FBS, 1% penicillin/streptomycin, 100 μg/ml G418, and 200 μg/ml zeocin.
Cells were plated on UV-treated coverslips, and Htt-103Q-EGFP expression was induced the next day by treatment with 1.25-5.0 μM muristerone A. Induced cells were exposed immediately to compounds dissolved in DMSO. Compounds were tested at concentration ranges of 1.0-10.0 μM in two cell lines. Subsequently, the compounds were retested at concentration ranges of 0.1-1.0 μM. The effects of compounds on polyQ aggregation were compared with control cells treated with the compound solvent (DMSO). After treatment, cells were fixed in 4% formaldehyde for 30 min. Visual counts of aggregates and EGFP-positive cells were performed by using fluorescence microscopy on an Axioplan II or Axiovert 25 (Zeiss). At least 300 cells were counted from five or six fields in two independent experiments for each data point (×10 magnification). Aggregation is expressed as the percentage of cells with aggregates versus the total number GFP-positive cells. The IC50 value for each compound is defined as the concentration of compound that was sufficient to suppress aggregation by 50% and was determined in duplicate in at least three independent experiments.
Filter-Trap Aggregation Assays in Vitro and in Cultured Cells. Compound tests using the modified filter-trap aggregation assays have been described (20, 27). For in vitro assays, 20 μl of compounds, which were diluted in buffer containing 150 mM NaCl, 20 mM Tris·HCl (pH 8.0), and 2 mM CaCl2, were mixed with an equal volume of recombinant HD Q51 fusion protein (25 ng/μl final, corresponding to 0.625 μM). Samples were incubated for 16 h at 37°C to allow aggregate formation. Reactions were stopped by addition of 40 μl of 4% SDS/100 mM DTT and boiling for 10 min. Aliquots corresponding to 125 ng of HDQ51 protein were filtered through a cellulose acetate membrane (0.2 μm, Schleicher & Schuell) by using a 96-well vacuum dot-blot apparatus, followed by two washing steps with 0.1% SDS.
To assess the effects of compounds on Htt aggregation in cultured cells, Cos1 cells were transiently transfected with the HD Q51 DNA construct. After incubation for 48 h, Cos1 cells were heat-denatured in 2% SDS/100 mM DDT and filtered through cellulose acetate membranes as described above. Membranes were blocked with 3% nonfat milk, and captured aggregates were detected by immunoblotting with a polyQ-specific antibody (CAG53b, 1:10,000 dilution; ref. 28), followed by incubation with alkaline phosphatase-conjugated anti-rabbit secondary antibody (1:10,000 dilution; Promega) and the fluorescent substrate AttoPhos (Promega). Signals corresponding to SDS-insoluble aggregates were quantified by using the aida 2.0 image analysis software (Raytest, Straubenhardt, Germany).
Compound Cytotoxicity and Compound-Mediated Cell Responses. Compounds were assessed for cytotoxic and other effects in PC12 cells by evaluating cell morphology, cell viability (Live/Dead kit, Molecular Probes), Htt-103Q-EGFP expression levels, and expression of several endogenous proteins including Hsp70 and p53 by Western blot analysis (data not shown).
The effects of compounds on the overall levels of Htt-103Q-EGFP fusion proteins were assessed in PC12 cells by measurement of total EGFP fluorescence. PC12 cells, in 96-well plates, were treated to induce Htt-103Q-GFP expression, exposed to compounds at a concentration range of 1.0-10.0 μM, and then incubated for 48 h and lysed in RIPA buffer (50 mM Tris, pH 8/150 mM NaCl/1% Triton X-100/10 mg/ml Na-deoxycholate/1 mg/ml SDS) with 5 mM EDTA for 30 min. EGFP fluorescence was measured by using a Victor2 V multilabel plate reader (PerkinElmer). The effect of each compound concentration on Htt-103Q-EGFP levels was determined in six independent experiments and the results were averaged.
Assessment of polyQ Aggregation in Organotypic Hippocampal Slice Cultures. Hippocampal slice cultures were prepared from P7-9 R6/2 neonate mouse pups and maintained in culture as described (23, 24). Compounds were dissolved in DMSO and 10-fold serial dilutions prepared to provide stocks spanning a four to five logconcentration range. The four hit compounds C1-C4 were tested at 0.1, 1, 10, and 100 μM, and their structural analogs C2-8, C3-6 and C4-7 were tested at concentrations of 0.001, 0.01, 0.1, 1, and 10 μM. In each case, the effect of the drug was compared with vehicle and no-drug controls. Compound dilutions were aliquoted and stored at -80°C, and then added as needed to fresh growth medium, which was prepared weekly. We prepared 60 slices (300 μm) from at least four independent mouse brains for each treatment group and incubated them with drug from the time at which they were first established. The medium was changed twice weekly, and 20 slices per treatment group were harvested and fixed after 2, 3, and 4 weeks in culture. Cell viability in control- and compound-treated slice cultures was assessed by propidium iodide uptake, as described (23). The slices were cut into 20-μm sections and immunoprobed by using the anti-Htt antibody S830 (29) and Alexa 488-conjugated anti-sheep secondary antibody (Molecular Probes) as described (23, 24). For any given compound, treatment groups harvested at all time points were immunoprobed in parallel to minimize staining variation.
Fluorescent images of aggregates in the CA1 region were captured at 2-μm intervals throughout the entire depth of the section by using a LSM150 confocal microscope (Zeiss) and compiled as a z stack. The z stack was collapsed and the aggregate load within the field was calculated by using a customized version of the ks-300 image-analysis program (Image Associates, Bicester, UK). The readout was generated in the form of the following three parameters: aggregate count (unit = aggregate number), total fluorescence intensity (unit = sum density), and aggregate area (unit = percentage of aggregate area of screen). Routinely, 20 sections were quantified for each treatment group, and the entire analysis for each compound was performed blindly. Data were analyzed by one-way ANOVA and two-way sample t tests and corrected for falsediscovery rate (30).
Testing Compound Efficacy in Drosophila. The compound test in Drosophila was performed as described (22, 31). Briefly, flies of the genotype p(elav-GAL4)/w; +/+; p(UAS-Htt93QP)/+ (females), which express the N-terminal fragment of the mutant Htt protein in all cells of the nervous system (25), were collected within 8 h of eclosion and placed in vials with food containing the indicated concentration of C2-8 (from a stock dissolved in DMSO), or containing solvent only. Flies from the same cross with the genotype w/Y; +/+; p(UAS-Htt93QP)/+, which do not express the Htt protein fragment, were also placed on food with and without drug and served as controls for general toxicity and other experimental conditions. Flies were placed on fresh food each day. The degree of neurodegeneration was assessed on day 7 in flies from each of the four vials by counting the number of photoreceptor cells present in each ommatidium.
Results
As described (21), aggregation of Htt-103Q-EGFP in yeast is cytotoxic; as a result, cells expressing this protein grow poorly and maintain the fusion protein at low levels. Genetic suppression of polyQ aggregation in this strain (e.g., by chaperone mutations, such as hsp104) leads to complete suppression of toxicity, restoration of normal growth, and elevation of 103Q expression levels (21). We used this assay to screen a library of 16,000 small chemical compounds (see Materials and Methods), and we identified nine compounds that result in increased yeast growth (at least a 25% increase in OD600) and/or increased Htt-103Q-EGFP expression levels (at least a 25% increase in EGFP fluorescence; Table 1 and Fig. 6, which is published as supporting information on the PNAS web site). The effects of compounds that met these criteria were then assessed microscopically (Fig. 1). Visual assessment confirmed that all of the nine compounds identified in the primary screening assay inhibit aggregation of Htt-103Q-EGFP in yeast.
Table 1. Identification of compounds causing increased growth (OD600) and fluorescence in the primary yeast screen.
| Hits | OD600 (fold change) | Fluorescence (fold change) |
|---|---|---|
| 1(C3) | 1.4 | 1.5 |
| 2(C1) | 1.4 | 1.0 |
| 3(C4) | 1.0 | 9.6 |
| 4(C2) | 1.0 | 1.3 |
| 5 | 1.3 | 1.4 |
| 6 | 5.6 | 7.0 |
| 7 | 1.0 | 4.0 |
| 8 | 1.0 | 4.0 |
| 9 | 1.0 | 3.0 |
The change in OD600 and EGFP fluorescence is expressed as fold change from control.
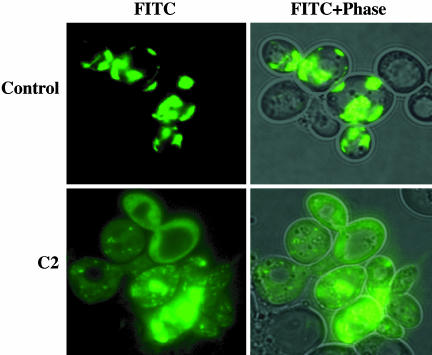
Fig. 1.
Hit compounds inhibit Htt-103Q-EGFP aggregation in Saccharomyces cerevisiae. The effect of each hit compound on aggregation was assessed microscopically. A representative compound, C2, inhibits polyQ aggregation in yeast.
Next, we investigated the effects of these nine inhibitors on polyQ aggregation in mammalian cells by using a PC12 cell-based model (22) (see Materials and Methods). Four compounds (C1, C2, C3, and C4; Fig. 2A) inhibited Htt-103Q-EGFP aggregation in two independent PC12 clones (Fig. 2B Middle and data not shown). The IC50 values for compounds in this assay were 10 (C1), 5 (C2 and C3), and 2.5 μM (C4). Except for C2, compounds were nontoxic up to 10 μM; C2 was toxic above 7.5 μM. The efficiencies with which these compounds suppressed polyQ aggregation in PC12 cells were compared with that of Congo red (Fig. 2B Bottom), known to be a direct blocker of polyQ aggregation (17, 32). The IC50 of Congo red in this assay was 10 μM (Fig. 2B Bottom, and data not shown).
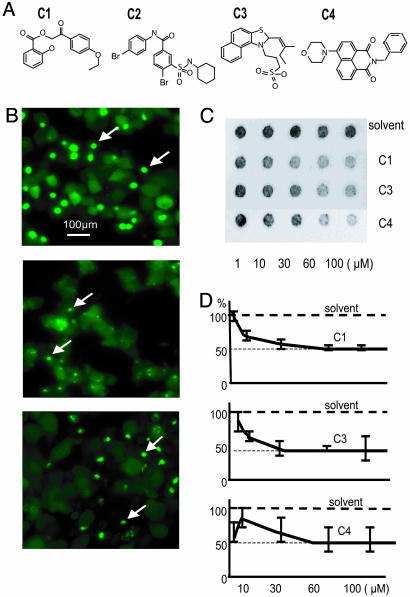
Fig. 2.
Hit compounds from the yeast screen inhibit polyQ aggregation in mammalian cells. (A) Structures of four hits from the yeast primary screen (C1-C4) that inhibit polyQ aggregation in PC12 cells. (B) Fluorescence microscopic assessment of Htt-103Q-EGFP aggregation in control- or compound-treated PC12 cells. Cells were treated with DMSO (control, Top), 2.5 μM C4 (Middle), or 10 μM Congo red (a known aggregation inhibitor, Bottom). Aggregates are indicated by arrows. (C and D) Inhibition of HD 51Q aggregation in Cos1 cells by compounds C1, C3, and C4, assessed using the filter-trap assay. (C) Immunodetection of insoluble (aggregated) HD 51Q trapped on the filter. (D) Quantification of results shown in C. Signal intensity from the sample incubated in the presence of the solvent alone was used as reference and set at 100%.
The inhibitory effects of the four compounds on polyQ-mediated aggregation were confirmed and quantified in African green monkey kidney cells (Cos1) by using a modified filter-trap method to detect polyQ aggregates. In this assay, soluble proteins pass through a cellulose acetate membrane, whereas insoluble (aggregated) proteins are retained; aggregated proteins can then be quantified by immunoblotting of the membrane (27). Cos1 cells were transiently transfected with a construct encoding an N-terminal fragment of mutant Htt (HD Q51, see Materials and Methods) to ensure high-level expression of the polyQ protein and, subsequently, a high rate of polyQ aggregation. Aggregates formed by HD Q51 in Cos1 cells are not disrupted when cell lysates are denatured in SDS, and thus, they are retained on cellulose acetate membranes (Fig. 2C). However, denatured lysates from transfected Cos1 cells treated with C1, C3, or C4 contained lower levels of insoluble HD Q51 (Fig. 2C). The IC50 values (Fig. 2D) for compounds in this assay were 60 μM for C1, 30 μM for C3, and 50 μM for C4. Because compound C2 was toxic for Cos1 cells, its IC50 value was not determined. The higher IC50 values obtained for compounds in the Cos1 cell assay, compared with the PC12 cell assay, probably reflect the significantly higher expression of polyQ-containing proteins in these cells relative to the PC12 system.
To obtain aggregation inhibitors with greater potency, we assembled four compound libraries consisting of chemical entities with >70% structural similarity to C1-C4. A total of 24, 28, 24, and 53 structural analogs of C1, C2, C3, and C4, respectively, were tested for their effects on polyQ aggregation in PC12 cells at concentrations of 0.025-5.0 μM. An example of these structural analog studies is shown for the C2 series in Fig. 7, which is published as supporting information on the PNAS web site. Three compounds from these analog libraries potently inhibited polyQ aggregation in the PC12 cell system, with IC50 values of 0.05-0.1 μM (Fig. 3 A and B). These compounds demonstrated no effects on overall expression levels of Htt-103Q-EGFP (data not shown) and no toxicity at the tested concentrations. No high-potency inhibitors of polyQ aggregation were identified among the C1 derivatives.
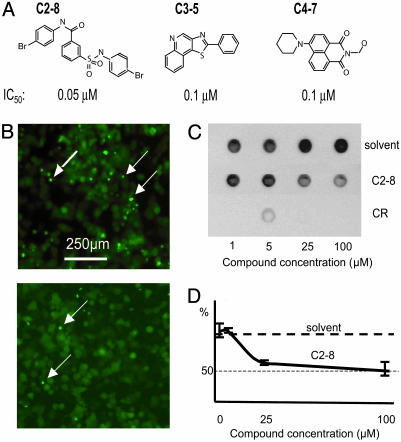
Fig. 3.
Identification of a potent polyQ-aggregation inhibitor, C2-8. (A) Structures of three potent analogs (C2-8, C3-5, and C4-7) derived from focused libraries of compounds with structural similarity to primary hits. The IC50 value of each compound in the PC12 cell assay (B) is noted. No cytotoxicity was observed using compound concentrations up to 10 μM. (B) C2-8, at 100 nM, (Lower) inhibits polyQ aggregation in 103Q-EGFP PC12 cells, as shown by fluorescence microscopy. Upper shows control (DMSO-treated) cells. (C) Immunodetection of insoluble recombinant HD 51Q trapped on the filter. Purified HF 51Q was incubated with solvent, C2-8, or Congo red (CR). (D) Quantification of results shown in C. Signal intensity from the sample incubated in the presence of the solvent alone was used as reference and set at 100%.
To test whether the selected compounds can interfere directly with polyQ aggregation, we used the filter-trap assay described above to assess aggregation in a cell-free system (28). Surprisingly, all of the compounds identified in the initial screen (C1-C4) failed to block aggregation of polyQ in vitro even at 100 μM concentrations (data not shown), suggesting that these compounds do not target soluble or aggregated polyQ directly. However, the analog C2-8 inhibited aggregation both in PC12 cells (Fig. 3B) and in vitro (Fig. 3 C and D). Although C2-8 strongly inhibits aggregation in PC12 cells (Fig. 3B), it does not affect the overall level of Htt103Q-EGFP protein in these experiments (data not shown). C2-8 efficiently inhibits aggregation in PC12 cells (IC50 = 50 nM, Fig. 3 A and B), but is effective in vitro only at very high concentrations (IC50 = 25 μM, Fig. 3 C and D). In contrast, Congo red, which is known to directly block aggregate formation (32), was a more efficient inhibitor in the cell-free system than in live cells (data not shown).
Because the ultimate goal of this study was to identify small molecules that inhibit polyQ-dependent aggregation in neurons, the effects of these high-potency analogs and of the original four compounds were assessed in brain-slice cultures from the R6/2 transgenic mouse model of HD. R6/2 mice ubiquitously express human Htt exon 1 containing >150 glutamines (HD Q150), and they develop a neurological phenotype with many similarities to the human disease (33-35). PolyQ aggregates are detectable in the CA1 hippocampal neurons of 3-week-old mice, before the appearance of behavioral changes (23, 36). Organotypic hippocampal slices can be established at P7-P9 and maintained in culture for up to 3 months (23). Aggregates form within the hippocampal neurons at the same rate and in the same sequence as in the R6/2 brain in vivo and can be detected in CA1 neurons after 2 weeks in culture. Thus, the potency of aggregation inhibitors can be assessed in cultured brain slices, which allow access to neurons in a largely normal physiological context without requiring transport of drugs across the blood-brain barrier (23, 24).
The effects of compounds on aggregation were assessed in the slice cultures over a 4-week period (see Materials and Methods). Among the four primary hit compounds (C1-C4), C1 and C2 were toxic at 100 μM after 2 weeks in culture and C4 was toxic at 10 and 100 μM (Fig. 8, which is published as supporting information on the PNAS web site, and data not shown). Compounds C1 and C2 showed evidence of aggregate inhibition at 2 weeks, but this effect had been lost by 4 weeks (shown for C2 in Fig. 8). At 4 weeks, there was no significant difference in the aggregate load between slices treated with the various drug concentrations and those receiving vehicle, as determined by any of the measurement parameters used (Fig. 8 and data not shown).
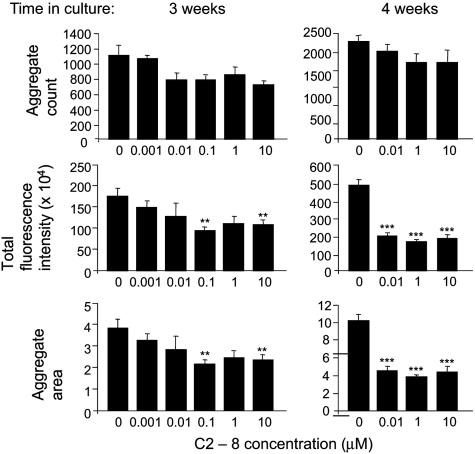
The analogs C2-8, C3-6, and C4-7 were tested at concentrations ranging from 0.001-10 μM. Compounds C3-6 and C4-7 showed no dose-responsive inhibitory effects on aggregation and no consistent inhibitory effects over the course of the experiment (data not shown). However, compound C2-8 had a pronounced inhibitory effect on polyQ aggregation in CA1 neurons in the hippocampal slices (Fig. 4). After 3 weeks, aggregate load as determined by total fluorescence intensity and the total aggregate area was reduced significantly in slices exposed to 0.1 and 10 μM C2-8. After 4 weeks, a highly significant reduction in total fluorescence intensity and the total aggregate-area inhibition (one-way ANOVA with falsediscovery-rate correction; P < 10-4) was found at concentrations ranging from 0.01-10 μM. In contrast, there was no significant reduction in the number of aggregates that had formed in the CA1 region in all C2-8 treatment groups and at all time points. These results were replicated in a second independent trial. C2-8 had no detrimental effects on slice culture viability over the course of the experiment (see Materials and Methods).
Fig. 4.
Analysis of C2-8 effects on polyQ aggregation in the hippocampal slice culture assay. The effect of treating slice cultures with 0.001-10 μM compound C2-8 after 3 and 4 weeks in culture is shown. Slices treated with 0.001 μM and 0.1 μM C2-8 are missing from the 4-week analysis because they had been lost to contamination. Aggregate formation in the slices treated with vehicle was comparable for the assessment of C2-8 and three additional compounds that were tested in parallel. Error bars represent standard errors of the mean. **, P < 0.01; ***, P < 0.0001.
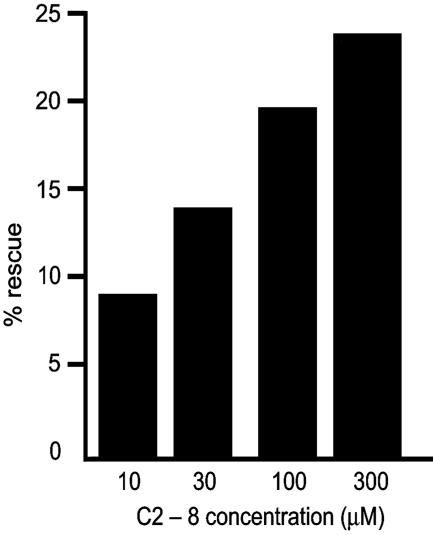
To assess the efficacy of C2-8 in relieving the progressive neurodegeneration typical of polyQ diseases, we tested whether C2-8 could reduce this degeneration in vivo in a Drosophila model of HD. In this model, Drosophila express the human pathogenic peptide Httex1 93QP, which is similar to that produced in the R6/2 transgenic mice (25). Drosophila expressing Httex1 93QP emerge (eclose) as adults with ongoing neurodegeneration seen most readily in the photoreceptor cells of the compound eye (22, 25, 31). When fed on food containing C2-8 from the first 8 h after eclosion onwards, the progressive degeneration seen in controls is significantly relieved in a dose-responsive manner (Fig. 5). The percentage of rescue is underestimated by ≈40% because, by the time emerging adults are first exposed to compound in the food, a certain amount of photoreceptor degeneration has already taken place (25). These data show that C2-8, a potent small-molecule inhibitor of aggregation in vitro, also rescues polyQ-dependent neurodegeneration in a dose-dependent manner in vivo.
Fig. 5.
C2-8 relieves neurodegeneration in a dose-dependent manner in Drosophila. The average number of rhabdomeres/ommatidium for flies fed the drug continuously from eclosion (emergence as adults) was compared with the average for flies fed control food. The percentage of rescue expressed as the increase in the average number of surviving rhabdomeres when drug is present compared with the maximum increase possible (i.e., to seven rhabdomeres) is shown. Percentage of rescue is calculated by the (number of photoreceptors with drug minus the number of photoreceptors without drug) divided by (seven minus the number of photoreceptors without drug).
Discussion
In this study, we describe a cell-based high-throughput screening approach that permits rapid and straightforward identification of pharmacological polyQ-aggregation inhibitors. A key feature of this process is that hits obtained from the primary screen were subsequently validated in multiple in vitro, cell-based, and in vivo secondary assays. By using this approach, we have identified four classes of chemical compounds that inhibit aggregation in intact cells, without significant cytotoxicity. These efforts have yielded a highly potent analog, C2-8, that inhibits polyQ aggregation in cultured cells and intact neurons and can rescue polyQ-mediated neurodegeneration in vivo.
The contribution of polyQ-containing aggregates to disease pathogenesis remains controversial. Although some studies have suggested a causal association between intranuclear inclusions and toxicity (15, 18), others imply that aggregate formation could be unrelated to disease pathogenesis or could represent a cellular neuroprotective mechanism (37, 38). Nonetheless, several reports have demonstrated that aggregation inhibitors can exert beneficial effects in vivo, in Drosophila and in mouse models of HD (16, 17, 19). These studies, together with the results presented here, suggest a key role for polyQ aggregation in neurodegeneration in HD and validate inhibition of aggregation as a useful pharmacological target.
Although the precise mechanism by which C2-8 inhibits polyQ aggregation is not yet clear, our results provide some intriguing clues. The process of polyQ aggregation proceeds in two phases (14, 15, 22, 39). The slow rate-limiting nucleation step, or seeding, is followed by polymerization, or aggregate growth, which occurs rapidly in cell culture models. Notably, C2-8 did not significantly reduce aggregate number in brain slices, but it decreased aggregate size and density (Fig. 4). These data suggest that C2-8 does not affect the nucleation step of aggregation but instead blocks the polymerization step of the process. C2-8 cannot disrupt preformed aggregates in vitro (data not shown); however, we found that C2-8, unlike C2, inhibits polyQ aggregation in a cell-free system, although only at very high concentrations (IC50 = 25 μM, data not shown). Thus, it appears that at least some activity of C2-8 does not depend upon cellular factors or pathways, although cellular processes may increase its potency. Our data suggest either that C2-8 is metabolically converted by cells into a highly potent inhibitor or that it affects cellular factors involved in the regulation of protein aggregation.
The drug-discovery pipeline described here can be extended to screen large compound collections to yield additional drug candidates. This yeast-based screen has yielded four previously uncharacterized scaffolds of aggregation inhibitors, including sulfobenzoic acid, naphthalimide, annelated thiazole, and oxoethyl salicylate derivatives. Compound C2-8, N-(4-bromophenyl)-3-{[(4-bromophenyl)amino]sulfonyl}benzamide, is a sulfobenzoic acid derivative that potently inhibits polyQ aggregation in brain slice cultures and in cell-based models. These results, together with the in vivo suppression of pathogenesis by C2-8 in Drosophila, make it a strong lead compound for human drug development. The scaffolds identified in this study will require further medicinal chemistry optimization for preclinical testing in HD mouse models, with the ultimate goal of developing effective therapies for HD patients.
Supplementary Material
Acknowledgments
We thank Susan Lindquist, Sylvia Krobitsch, Valerie Berthelier, Ronald Wetzel, Pamela McLean, and Bradley Hyman for testing primary hits in their experimental HD models; Ben Woodman and Anj Mahal for the maintenance and genotyping of the R6/2 mouse colony; Emma Hockly for advice on statistics; Alexander McCampbell and Brigid Davis for critical review of the manuscript; and Robert Chrovat for providing medicinal chemistry expertise. This work was supported by Wellcome Trust Grants 051897, 060360, and 066270 (to G.P.B.), the Huntington's Disease Society of America Coalition for the Cure program (G.P.B., L.M.T., and E.E.W.), the Hereditary Disease Foundation Cure Huntington's Disease Initiative (G.P.B., L.M.T., M.Y.S., and A.B.Y.), Human Frontiers Science Program Grant RG0132 (to G.P.B., L.M.T., and E.E.W.), the Federal Ministry for Education, Science, Research, and Technology Grant 0311853 (to S.E. and E.E.W.), the Sierra Foundation (A.G.K. and D.E.H.), and National Institutes of Health Grants HD36081 and HD36049 (to J.L.M.).
Author contributions: J.L.M., L.M.T., E.E.W., A.B.Y., D.E.H., G.P.B., M.Y.S., and A.G.K. designed research;
Abbreviations: HD, Huntington's disease; Htt, huntingtin; polyQ, polyglutamine.
References
- 1.Gusella, J. F. & MacDonald, M. E. (2000) Nat. Rev. Neurosci. 1, 109-115. [DOI] [PubMed] [Google Scholar]
- 2.Zoghbi, H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23, 217-247. [DOI] [PubMed] [Google Scholar]
- 3.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 4.Myers, R. H., Marans, K. S. & MacDonald, M. E. (1998) in Genetic Instabilities and Hereditary Neurological Diseases, eds. Wells, R. D. & Warren, S. T. (Academic, San Diego), pp. 301-323.
- 5.Bates, G., Harper, P. S. & Jones, L. (2002) Huntington's Disease (Oxford Univ. Press, New York).
- 6.Paulson, H. L. (1999) Am. J. Hum. Genet. 64, 339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey, B. & Lansbury, P. T. (2003) Annu. Rev. Neurosci. 26, 267-298. [DOI] [PubMed] [Google Scholar]
- 8.Bates, G. (2003) Lancet 361, 1642-1644. [DOI] [PubMed] [Google Scholar]
- 9.Ross, C. A. & Poirier, M. A. (2004) Nat. Med. 10, Suppl., S10-S17. [DOI] [PubMed] [Google Scholar]
- 10.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537-548. [DOI] [PubMed] [Google Scholar]
- 11.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277, 1990-1993. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto, A., Lucas, J. J. & Hen, R. (2000) Cell 101, 57-66. [DOI] [PubMed] [Google Scholar]
- 13.Kazantsev, A., Preisinger, E., Dranovsky, A., Goldgaber, D. & Housman, D. (1999) Proc. Natl. Acad. Sci. USA 96, 11404-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherzinger, E., Sittler, A., Schweiger, K., Heiser, V., Lurz, R., Hasenbank, R., Bates, G. P., Lehrach, H. & Wanker, E. E. (1999) Proc. Natl. Acad. Sci. USA 96, 4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, S., Ferrone, F. A. & Wetzel, R. (2002) Proc. Natl. Acad. Sci. USA 99, 11884-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazantsev, A., Walker, H. A., Slepko, N., Bear, J. E., Preisinger, E., Steffan, J. S., Zhu, Y. Z., Gertler, F. B., Housman, D. E., Marsh, J. L. & Thompson, L. M. (2002) Nat. Genet. 30, 367-376. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez, I., Mahlke, C. & Yuan, J. (2003) Nature 421, 373-379. [DOI] [PubMed] [Google Scholar]
- 18.Yang, W., Dunlap, J. R., Andrews, R. B. & Wetzel, R. (2002) Hum. Mol. Genet. 11, 2905-2917. [DOI] [PubMed] [Google Scholar]
- 19.Pollitt, S. K., Pallos, J., Shao, J., Desai, U. A., Ma, A. A., Thompson, L. M., Marsh, J. L. & Diamond, M. I. (2003) Neuron 40, 685-694. [DOI] [PubMed] [Google Scholar]
- 20.Heiser, V., Engemann, S., Brocker, W., Dunkel, I., Boeddrich, A., Waelter, S., Nordhoff, E., Lurz, R., Schugardt, N., Rautenberg, S., et al. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16400-16406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meriin, A. B., Zhang, X., He, X., Newnam, G. P., Chernoff, Y. O. & Sherman, M. Y. (2002) J. Cell. Biol. 157, 997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apostol, B. L., Kazantsev, A., Raffioni, S., Illes, K., Pallos, J., Bodai, L., Slepko, N., Bear, J. E., Gertler, F. B., Hersch, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5950-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, D. L., Portier, R., Woodman, B., Hockly, E., Mahal, A., Klunk, W. E., Li, X. J., Wanker, E., Murray, K. D. & Bates, G. P. (2001) Neurobiol. Dis. 8, 1017-1026. [DOI] [PubMed] [Google Scholar]
- 24.Smith, D. L., Woodman, B., Mahal, A., Sathasivam, K., Ghazi-Noori, S., Lowden, P. A., Bates, G. P. & Hockly, E. (2003) Ann. Neurol. 54, 186-196. [DOI] [PubMed] [Google Scholar]
- 25.Marsh, J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J. & Thompson, L. M. (2000) Hum. Mol. Genet. 9, 13-25. [DOI] [PubMed] [Google Scholar]
- 26.Emter, R., Heese-Peck, A. & Kralli, A. (2002) FEBS Lett. 521, 57-61. [DOI] [PubMed] [Google Scholar]
- 27.Wanker, E. E., Scherzinger, E., Heiser, V., Sittler, A., Eickhoff, H. & Lehrach, H. (1999) Methods Enzymol. 309, 375-386. [DOI] [PubMed] [Google Scholar]
- 28.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. & Wanker, E. E. (1997) Cell 90, 549-558. [DOI] [PubMed] [Google Scholar]
- 29.Sathasivam, K., Woodman, B., Mahal, A., Bertaux, F., Wanker, E. E., Shima, D. T. & Bates, G. P. (2001) Hum. Mol. Genet. 10, 2425-2435. [DOI] [PubMed] [Google Scholar]
- 30.Hochberg, Y. & Benjamini, Y. (1990) Stat. Med. 9, 811-818. [DOI] [PubMed] [Google Scholar]
- 31.Steffan, J. S., Bodai, L., Pallos, J., Poelman, M., McCampbell, A., Apostol, B. L., Kazantsev, A., Schmidt, E., Zhu, Y. Z., Greenwald, M., et al. (2001) Nature 413, 739-743. [DOI] [PubMed] [Google Scholar]
- 32.Heiser, V., Scherzinger, E., Boeddrich, A., Nordhoff, E., Lurz, R., Schugardt, N., Lehrach, H. & Wanker, E. E. (2000) Proc. Natl. Acad. Sci. USA 97, 6739-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W. & Bates, G. P. (1996) Cell 87, 493-506. [DOI] [PubMed] [Google Scholar]
- 34.Carter, R. J., Lione, L. A., Humby, T., Mangiarini, L., Mahal, A., Bates, G. P., Dunnett, S. B. & Morton, A. J. (1999) J. Neurosci. 19, 3248-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lione, L. A., Carter, R. J., Hunt, M. J., Bates, G. P., Morton, A. J. & Dunnett, S. B. (1999) J. Neurosci. 19, 10428-10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morton, A. J., Lagan, M. A., Skepper, J. N. & Dunnett, S. B. (2000) J. Neurocytol. 29, 679-702. [DOI] [PubMed] [Google Scholar]
- 37.Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. (1998) Cell 95, 55-66. [DOI] [PubMed] [Google Scholar]
- 38.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805-810. [DOI] [PubMed] [Google Scholar]
- 39.Perutz, M. F., Pope, B. J., Owen, D., Wanker, E. E. & Scherzinger, E. (2002) Proc. Natl. Acad. Sci. USA 99, 5596-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.