Abstract
Estrogen signaling occurs through at least two distinct molecular pathways: (i) direct binding of liganded estrogen receptors (ERs) to estrogen-responsive DNA elements (EREs) (the “ER/ERE pathway”) and (ii) indirect recruitment of liganded ERs to activating protein-1 (AP-1)-responsive DNA elements via heterodimers of Fos and Jun (the “ER/AP-1 pathway”). We have developed a biochemical assay for examining ligand-regulated transcription by ERs in the ER/AP-1 pathway. This assay recapitulates the altered (i.e., agonistic) pharmacology of selective estrogen receptor modulator drugs in this pathway reported previously by using various cell-based assays. We used our biochemical assay to examine the detailed mechanisms of ER/AP-1-dependent transcription. Our studies indicate that (i) ERα/AP-1 complexes play a critical role in promoting the formation of stable RNA polymerase II preinitiation complexes leading to transcription initiation, (ii) chromatin is a key determinant of estrogen and selective estrogen receptor modulator signaling in the ERα/AP-1 pathway, (iii) distinct domains of ERα are required for recruitment to DNA-bound Fos/Jun heterodimers and transcriptional activation at AP-1 sites, and (iv) different enhancer/activator combinations in the ERα and AP-1 pathways use coactivators in distinct ways. These studies have increased our understanding of the molecular mechanisms underlying ligand-dependent signaling in the ER/AP-1 pathway and demonstrate the usefulness of this biochemical approach.
Keywords: chromatin, enhancer, Fos/Jun heterodimers, histone acetyltransferase, selective estrogen receptor modulator
Estrogens play crucial roles in the normal physiology of a variety of tissues, including those of the mammary glands, reproductive tract, central nervous system, and skeleton (1–3). Selective estrogen receptor (ER) modulators (SERMs), pharmacologic agents that act as estrogen antagonists or, in some cases, agonists depending on the target tissue, are used in the treatment of estrogen-related diseases, such as mammary cancers (4, 5). The actions of estrogens and SERMs are mediated by two ER isoforms, ERα and ERβ, which function as ligand-regulated, DNA-binding transcriptional activators (1–3). The ERs contain conserved DNA- and ligand-binding domains (DBD and LBD, respectively), as well as two activation functions (AFs), AF-1 in the N-terminal A/B region and AF-2 in the LBD (2).
Cellular signaling by estrogens occurs through at least two distinct pathways: (i) direct binding of liganded ERs to estrogen response elements (EREs) (the “ER/ERE pathway”) and (ii) indirect recruitment of liganded ERs to AP-1-responsive elements via heterodimers of the b-zip transcription factors c-Fos and c-Jun that comprise AP-1 (the “ER/AP-1 pathway”) (6). An interesting aspect of the ER/AP-1 pathway is that, under certain cell type and promoter contexts, some SERMs that were originally defined as classical antagonists in the ER/ERE pathway can function as agonists (6–8). Although the molecular details of the ERE pathway are well characterized, our understanding of the ER/AP-1 pathway is limited, especially with regard to the mechanisms of altered pharmacology. Expression microarray studies have shown that many genes are regulated by ERs, but it is unclear what percentage of these genes are regulated through the ER/AP-1 pathway (9–11). The identification and characterization of model ER/AP-1-regulated genes, such as the ovalbumin, collagenase-1, IGF-1, and c-Myc genes, has aided in our understanding of this pathway (12–15).
Estrogen-dependent activation through the ER/ERE pathway involves a variety of coactivators that function with liganded ERs to modify histones (e.g., SRC-p300/CBP complexes), alter chromatin structure (e.g., Swi/Snf), and recruit RNA polymerase II (e.g., TRAP/DRIP/Mediator) (16). Many coactivators, such as the SRC proteins and the Med220 subunit of Mediator, bind directly to the AF-2 of agonist-bound ER through short α-helical “LXXLL” motifs called NR boxes (17). In general, antagonists fail to establish a proper AF-2 conformation and thus block receptor–coactivator interactions (17, 18). The mechanistic details of estrogen-dependent activation through the ER/AP-1 pathway are much less clear, although similar sets of coactivators are likely to be involved. In both pathways, the DNA-bound factors act as nucleation sites for subsequent coactivator recruitment (6).
Although animal- and cell-based assays have increased our understanding of the biology of the ER/AP-1 pathway, our understanding of the underlying molecular mechanisms of this pathway are limited. In some cases, the published literature has provided conflicting results regarding the domains of ER required for its activity in the ER/AP-1 pathway, as well as the protein–protein interactions required for activation in the ER/AP-1 pathway (7, 19, 20). Herein, we describe the development of a biochemical assay for examining ligand-regulated transcription through the ER/AP-1 pathway with chromatin templates. Our assay conditions recapitulate the altered pharmacology of SERMs that have been reported by using various cell-based assays. We have used this system to study the molecular mechanisms underlying ligand-regulated transcription in the ER/AP-1 pathway. A key conclusion from our studies is that different enhancer/activator combinations in the ERα and AP-1 pathways use coactivators in distinct ways.
Materials and Methods
Synthesis and Purification of Recombinant Proteins. FLAG-tagged hERα(1–595), hERαΔAB(180–595), hERαΔDBD(1–180/263– 595), hERα82G, hERαL540Q, and hERβ(1–530) were expressed in Sf9 cells by using a baculovirus vector and purified by anti-FLAG affinity chromatography as described (21, 22). His6-tagged hERαΔLBD(1–301) was expressed in Escherichia coli and purified by using standard nickel-nitrilotriacetic acid affinity chromatography. Full-length his6-tagged c-Fos/c-Jun heterodimers were expressed in E. coli and purified by using a denaturing/renaturing protocol as described (23). GST-Med220(RID), GST-SRC2(RID), and GST-SRC2(PID) were expressed in E. coli and purified by using glutathione affinity chromatography as described (24, 25).
Chromatin Assembly and in Vitro Transcription Assays. The plasmid templates p2AP1-E4 and p2ERE-E4 contain two copies of the human collagenase-1 AP1 and Xenopus vitellogenin ERE sequences, respectively, located upstream of the adenovirus E4 core promoter. The control template, pE4, is similar but lacks the AP-1 sites and EREs. The natural collagenase-1 promoter construct (-73 to +60) was kindly provided by Steve Nordeen. In vitro chromatin assembly and transcription reactions were carried out as described (21, 26). Briefly, wild-type or mutant ER proteins (40 nM) and ligands (400 nM) were added during chromatin assembly, whereas the GST-fused polypeptides (600 nM) and chemical inhibitors (0.25 to 5 μM of LysCoA or H3-CoA-20) were added after chromatin assembly was completed. In vitro transcription was performed by using HeLa cell nuclear extract as a source of c-Fos/c-Jun and the RNA polymerase II transcription machinery. For the inhibitor experiments shown in Fig. 5, recombinant c-Fos/c-Jun (5 nM) was added during chromatin assembly for the “Fos + Jun/AP1 sites” condition. Because of dilution during reaction setup, the final concentrations of factors and ligands in the transcription assays were 30% of the concentrations indicated for the chromatin assembly reactions. Single-round transcription assays (see Fig. 3) and mock chromatin assembly reactions (Fig. 6, which is published as supporting information on the PNAS web site) were performed as described (21). RNA products from the transcription reactions were analyzed by primer extension (26). The assays were quantified by PhosphorImager analysis with imagequant version 1.2 software (Molecular Dynamics). All transcription reactions were carried out in duplicate, and each experiment was performed three or more times to ensure reproducibility.
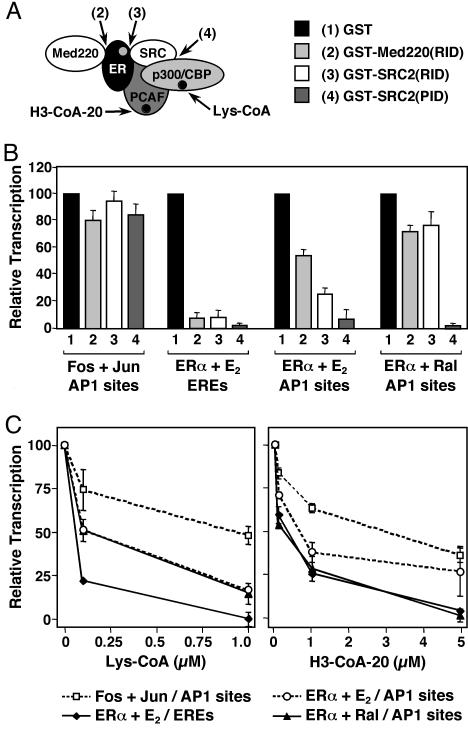
Fig. 5.
Distinct coactivator usage in the ERα, AP-1, and ERα/AP-1 pathways. Effects of polypeptide and chemical inhibitors on ERα-, AP-1-, and ERα/AP-1-dependent transcription. Templates containing two AP-1 sites or two EREs upstream of the adenovirus E4 promoter were assembled into chromatin and transcribed in the presence or absence of c-Fos/c-Jun heterodimers, ERα, and ligands, as indicated. Each bar or point represents the mean ± SEM for three or more determinations. (A) Schematic representation of protein–protein interactions blocked by the GST-fused polypeptide inhibitors and enzymatic activities blocked by the chemical inhibitors. (B) Distinct protein–protein interactions are differentially required by the ERα, AP-1, and ERα/AP-1 pathways. Shown is a summary of multiple experiments performed in the presence of the GST-fused polypeptide inhibitors, as indicated. (C) p300/CBP and PCAF acetyltransferase activities are differentially required by the ERα, AP-1, and ERα/AP-1 pathways. Shown is a summary of multiple experiments performed in the presence of the chemical inhibitors, as indicated.
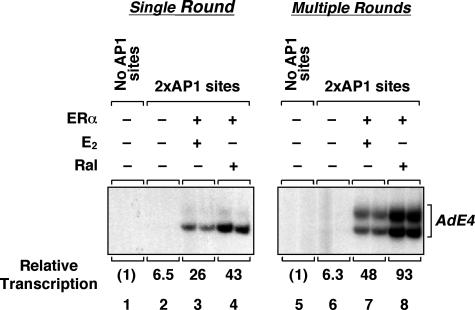
Fig. 3.
ERα enhances transcription through AP-1 by promoting transcription initiation. Templates containing no AP-1 sites or two AP-1 sites upstream of the adenovirus E4 promoter were assembled into chromatin and transcribed in the presence of ERα,E2, and Ral as indicated. Sarkosyl was added to 0.25% after the initiation of transcription by the addition of rNTPs to limit transcription to a single round. Note that transcription initiates primarily from the most 3′ start site of the AdE4 promoter in experiments in which Sarkosyl is added (21, 25).
Immobilized DNA Template Assays. A single end-biotinylated fragment of p2AP1-E4 containing the AP-1 sites and the adenovirus E4 promoter was immobilized on M280-streptavidin Dynabeads as recommended by the manufacturer (Dynal). A similar fragment from pE4, lacking the AP-1 sites, was used as a control. For each binding reaction, beads containing 300 ng of immobilized DNA template were blocked in binding buffer (20 mM Hepes, pH 7.6/100 mM KCl/0.2 mM EDTA/20% wt/vol glycerol/0.5 mM DTT) containing 0.5 mg/ml BSA and 0.5 mg/ml recombinant human insulin for 2 h at room temperature. The blocked beads were incubated with HeLa cell nuclear extract (200 μg per reaction) and nonspecific competitor DNA (1 μg per reaction) in the absence or presence of 1 nM of wild-type or mutant ERα and 100 nM of ligand for 1 h at room temperature. After washing three times in binding buffer containing 0.2% vol/vol Nonidet P-40 and 0.3 mg/ml insulin, the specifically bound proteins were released from the beads by boiling in SDS/PAGE loading solution and analyzed by Western blotting using antibodies to ERα or FLAG (for detection of ERα) or antibodies to c-Jun and c-Fos (for detection of AP-1).
Results
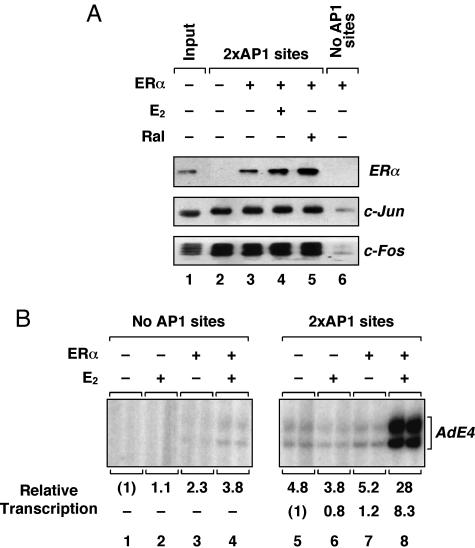
ERα Is Recruited to DNA and Activates Transcription Through AP-1. To begin our analysis of ER-dependent transcription through AP-1, we examined the recruitment of purified recombinant ERα to immobilized DNA templates lacking or containing two AP-1 sites (Fig. 1A). These assays were performed in the presence of HeLa cell nuclear extract, which was used as a source of native c-Fos and c-Jun (Fig. 7, which is published as supporting information on the PNAS web site). Specifically bound factors were analyzed by Western blotting. As expected, c-Fos and c-Jun bound to the immobilized template dependent on the presence of AP-1 sites, but independent of ERα (Fig. 1 A). ERα, with or without estradiol (E2) or raloxifene (Ral), also bound to the template dependent on the presence of AP-1 sites (Fig. 1 A) and on the presence of HeLa cell nuclear extract (data not shown). Although the binding of ERα to the template appeared to be enhanced in the presence of ligands in some experiments, these effects were not consistently observed (see, for example, Fig. 4B). These results indicate that ERα can be recruited to a promoter that lacks EREs by associating with DNA-bound factors, such as c-Fos/c-Jun heterodimers, at AP-1 sites.
Fig. 1.
ERα is recruited to DNA and activates transcription through AP-1. (A) Native DNA-bound Fos/Jun heterodimers recruit recombinant ERα to an immobilized DNA template containing AP-1 sites. Associated proteins were detected by Western blotting with antibodies ERα, c-Jun, and c-Fos. E2, 17β-estradiol; Ral, raloxifene. (B) Liganded ERα activates transcription through AP-1 sites with chromatin templates in vitro. Templates containing no (Left)or two (Right) AP-1 sites upstream of the adenovirus E4 promoter were assembled into chromatin and transcribed in the presence of ERα and E2 as indicated.
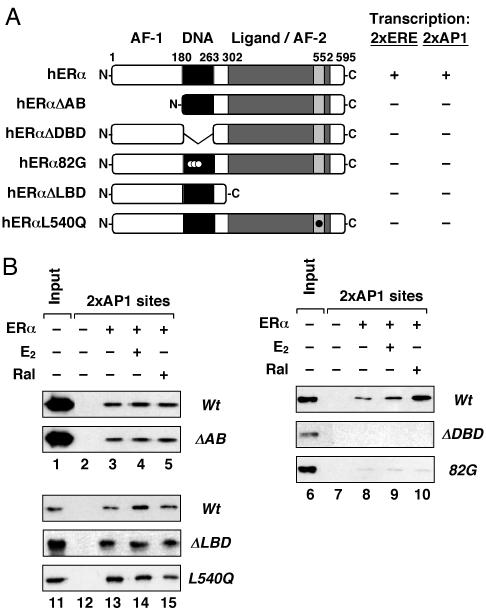
Fig. 4.
Distinct domains of ERα are required for recruitment and transcription activation through AP-1. (A) Multiple domains of ERα are necessary for efficient ERE- and AP-1-dependent transcription. (Left) Schematic representation of the ERα mutants used in these studies. Templates containing two EREs or two AP-1 sites upstream of the adenovirus E4 promoter were assembled into chromatin and transcribed in the presence of wild-type or mutant ERα± E2 (and Ral, for the AP-1 template). The results of transcription assays are summarized (Right), with “+” indicating 100% activity and “-” indicating <5% of transcriptional activity of wild-type ERα. (B) The DBD of ERα is required for recruitment by native DNA-bound Fos/Jun heterodimers to an immobilized DNA template containing AP1 sites. Bound ERα was detected by Western blotting using an antibody to ERα (or to FLAG, for the ERαΔAB experiment).
To examine the transcriptional activity of ERα at AP-1 sites, we used an in vitro chromatin assembly and transcription system. Plasmid templates lacking or containing two AP-1 sites upstream of the adenovirus E4 promoter (pE4 and p2AP1-E4) were assembled into chromatin in the presence of ERα and E2 by using an extract prepared from Drosophila embryos (S190). The assembled templates were transcribed by using HeLa cell nuclear extract as a source of both c-Fos/c-Jun and the RNA polymerase II transcription machinery. As shown in Fig. 1B, a modest (≈5-fold) AP-1 site-dependent activation of transcription was observed in the absence of ERα, likely because of the native c-Fos/c-Jun in the HeLa extract (compare lanes 1 and 5). In contrast, a robust AP-1 site-dependent activation of transcription was observed in the presence of ERα and E2 (≈30-fold versus no ERα/no AP-1 sites; compare lanes 1 and 8). The modest activation by ERα and E2 in the absence of the AP-1 sites (compare lanes 1 and 4) is likely due to a cryptic AP-1 site in the vector backbone located upstream of the promoter (ref. 27 and data not shown). Similar results were observed with ERβ (Fig. 2 and data not shown). Taken together, our biochemical assays provide a clear indication that ERα can be recruited to and activate transcription from a promoter template that lacks EREs, but contains AP-1-binding sites.
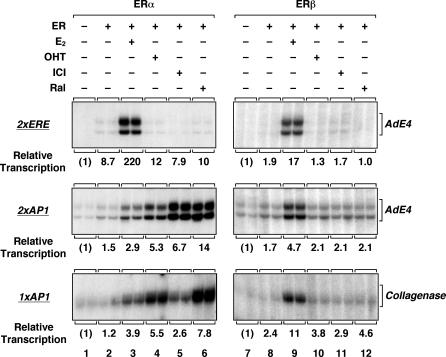
Fig. 2.
ERα and ERβ support ERE- and AP-1 site-dependent transcription with synthetic and natural promoters. Templates containing two EREs (Top) or two AP-1 sites (Middle) upstream of the adenovirus E4 promoter, or a fragment of the collagenase-1 promoter with the consensus AP-1 site at -73/-60 (Bottom), were assembled into chromatin and transcribed in the presence of ERα or ERβ and ligands, as indicated. E2, 17β-estradiol; OHT, 4-hydroxytamoxifen; ICI, ICI164384; Ral, raloxifene.
ERα, but Not ERβ, Supports SERM-Activated Transcription Through AP-1. Previous studies have shown that SERMs, including Ral, hydroxytamoxifen (OHT), and ICI164384 (ICI), can stimulate ER-dependent transcription with AP-1 site-containing promoters, but typically not with ERE-containing promoters (7, 8). We examined this altered (“nonclassical”) pharmacology by using our in vitro chromatin assembly and transcription system (Fig. 2). Templates containing synthetic or natural promoters with EREs or AP-1 sites were assembled into chromatin and transcribed in the presence of ERα and various ligands. For comparison, we also examined the activity of ERβ under similar conditions. As expected, E2, but not Ral, OHT, or ICI, functioned as an agonist with ERα and ERβ at a synthetic ERE-containing promoter, although ERα was a more potent activator than ERβ (Fig. 2 Top), as we have shown (22). E2 also functioned as an agonist with ERα and ERβ at AP-1 site-containing promoters, although in this case ERβ was a more potent activator than ERα (Fig. 2 Middle). Interestingly, Ral, OHT, and ICI functioned as agonists with ERα, but not ERβ (i.e., <2-fold effect), at synthetic and natural AP-1 site-containing promoters, with Ral having the most potent activity (7- to 10-fold enhancement vs. no ligand) (Fig. 2 Middle and Bottom). The E2- and SERM-activated transcription by ERα with AP-1 site-containing promoters was only observed with chromatin templates (Fig. 6), suggesting a role for chromatin in the transcription process. Collectively, these results indicate that altered pharmacology of SERMs with ERα at an AP-1 site-containing promoter can be recapitulated with chromatin templates assembled in vitro.
Liganded ERα Promotes the Efficient Assembly of Transcription Preinitiation Complexes at an AP-1 Site-Containing Promoter. We have shown that ERα has a dual role in transcription at ERE-containing promoters, increasing both the efficiency of transcription initiation and the number of subsequent rounds of reinitiation (21). To examine how ERα might affect transcription at an AP-1 site-containing promoter, we performed single-round transcription experiments with chromatin templates. Limiting transcription to a single round allows examination of transcription preinitiation complex assembly leading to productive transcription initiation (28). To limit transcription to a single round, we added the detergent Sarkosyl after the initiation of transcription (i.e., 10 s after the addition of rNTPs). Under these conditions, Sarkosyl inhibits transcription reinitiation, but not elongation of transcriptionally engaged RNA pol II and, thus, a single round of transcription is achieved (28). Both E2- and Ral-bound ERα efficiently activated transcription in a single round (Fig. 3), indicating an enhancement of preinitiation complex formation at the promoter. However, the effects of E2- and Ral-bound ERα on transcription reinitiation, determined by comparison to corresponding multiple round transcription experiments, were modest (i.e., 2- to 3-fold; Fig. 3 and data not shown). These results suggest that the primary transcription initiation event may be the major target for ligand-dependent regulation in the ERα/AP-1 pathway.
Distinct Domains of ERα Are Required for AP-1-Dependent Recruitment of ERα and Transcription Activation. ERα has a number of distinct functional domains that could contribute independently to its recruitment to AP-1 site-containing promoters and the subsequent activation of transcription at those promoters. To examine this in detail, we used a set of ERα deletion and point mutants targeting the N-terminal A/B region containing AF-1, the DBD, and the C-terminal ligand binding domain/AF-2 (Fig. 4A Left and Fig. 8, which is published as supporting information on the PNAS web site). Individual deletion or mutation of any one of the three domains blocked ligand-dependent transcription (i.e., <5% of wild type) with both ERE- and AP-1 site-containing promoters in an in vitro chromatin assembly and transcription assay (Fig. 4A Right). In contrast, only the DBD was required for the recruitment of ERα to an AP-1 site-containing promoter in an immobilized template assay (Fig. 4B; compare the A/B and LBD mutants at Left with the DBD mutants at Right). Taken together, these results indicate that distinct domains of ERα are required for recruitment (i.e., the DBD) and activation (i.e., AF-1 and AF-2) at AP-1 site-containing promoters.
Distinct Coactivator Usage in the ERα, AP-1, and ERα/AP-1 Transcription Pathways. The transcriptional activities of ERα at EREs and Fos/Jun heterodimers at AP-1 sites require a variety of coactivators, including members of the steroid receptor coactivator family (SRCs or p160s), and the closely related acetyltransferases p300 and CBP (24, 29, 30). Thus, one might expect the same coactivators to be involved and possibly required in the ERα/AP-1 pathway, although the coactivators may be used in mechanistically distinct ways. To determine whether coactivator activities contribute in distinct ways to different ERα and AP-1 transcription pathways, we used a set of previously characterized GST-fused polypeptide inhibitors that block specific receptor–coactivator or coactivator–coactivator interactions, as illustrated in Fig. 5A (24, 25). They included the receptor interaction domains (RIDs) of SRC2 and Med220, and the p300/CBP interaction domain (PID) of SRC2. The inhibitors were used to compare the importance of specific protein–protein interactions in four distinct transcriptional pathways: (i) Fos/Jun heterodimers at AP-1 sites, (ii) ERα plus E2 at EREs, (iii) ERα plus E2 at AP-1 sites, and (iv) ERα plus Ral at AP-1 sites, as illustrated in Fig. 9, which is published as supporting information on the PNAS web site.
The effects of the inhibitors on in vitro transcription with chromatin templates are summarized in Fig. 5B. Note that in these experiments, the coactivators are provided by the HeLa cell nuclear extract used for the transcription assays. All three inhibitors had little effect on the activity of Fos/Jun heterodimers at AP-1 sites. This result was expected, given that SRCs and Med220 do not use the RIDs to interact with Fos and Jun and that SRC and p300/CBP may be recruited independently of each other by Fos and Jun, possibly abrogating the need for direct SRC-p300/CBP interactions. In contrast, all three inhibitors blocked the activity of ERα plus E2 at EREs, as we have shown, indicating a strong requirement for ERα-Med220, ERα-SRC, and SRC-p300/CBP interactions in that pathway (24, 25). The inhibitors had variable effects on the two ERα/AP-1 pathways (i.e., +E2 and +Ral). More specifically, Med220(RID) and SRC2(RID), which block ERα-Med220 and ERα-SRC interactions, respectively, had modest inhibitory effects compared to the ERα/E2/ERE pathway. However, the inhibitory effects of SRC(PID), which blocks SRC-p300/CBP interactions, were just as strong. Note that Ral blocks ERα-SRC interactions, so one would expect those interactions to be less important in the ERα/Ral/AP-1 pathway, as observed in Fig. 5B. Presumably, under those conditions, there is an alternate mode for p300/CBP recruitment (i.e., one that does not require ERα–SRC interactions, but does require SRC-p300/CBP interactions). Taken together, the results from these assays indicate that distinct ERα and AP-1 pathways require distinct sets of protein–protein interactions.
Because SRC-p300/CBP interactions were required in both the ERα/E2/ERE and ERα/AP-1 pathways, we determined the requirement for p300/CBP acetyltransferase activity in these pathways. To test this requirement, we assayed the effects of a highly specific chemical inhibitor of p300/CBP acetyltransferase activity, Lys-CoA (31), by using in vitro transcription assays with chromatin templates. As shown in Fig. 5C, the addition of Lys-CoA inhibited transcription in all four pathways tested, but each pathway exhibited different sensitivities to the inhibitor. For example, Lys-CoA potently inhibited the activity of ERα plus E2 at EREs, but had a more modest effect on the activity of Fos/Jun heterodimers at AP-1 sites. In contrast, the effects of Lys-CoA on the ERα/AP-1 pathways were intermediate. Thus, although all four pathways require p300/CBP acetyltransferase activity for maximal transcription, each pathway has a distinct overall requirement. These results are consistent with the results from the SRC(PID) polypeptide inhibitor experiments shown in Fig. 5B. Similar results were obtained when we used an inhibitor of PCAF acetyltransferase activity, H3-CoA-20 (31), suggesting similar relative requirements for p300/CBP and PCAF acetyltransferase activities in a given pathway. Taken together, the results from the polypeptide and chemical inhibitor experiments reveal distinct coactivator usage by the AP-1, ERα/ERE, and ERα/AP-1 transcription pathways.
Discussion
A Biochemical Assay for Examining Ligand-Regulated Transcription in the ER/AP-1 Pathway. Here, we have described a biochemical assay for examining the mechanisms of ligand-regulated transcription in the ER/AP-1 pathway with chromatin templates. We found that (i) ERα/AP-1 complexes play a critical role in promoting the formation of stable RNA polymerase II preinitiation complexes leading to transcription initiation (Fig. 3), (ii) chromatin is a key determinant of estrogen and SERM signaling in the ERα/AP-1 pathway (Fig. 6), (iii) distinct domains of ERα are required for recruitment to DNA-bound Fos/Jun heterodimers and transcriptional activation at AP-1 sites (Fig. 4), and (iv) different enhancer/activator combinations in the ERα and AP-1 pathways use coactivators in distinct ways (Fig. 5). The latter two findings are discussed in more detail below.
Notably, our biochemical assay recapitulates the altered pharmacology of SERMs that have been reported by using various cell-based assays (7, 8). However, we observed some differences in the specific signaling outcomes compared to the previous studies, despite the fact that we used a similar reporter gene (e.g., collagenase-1) and cells (HeLa). Specifically, in contrast to Paech et al. (8), we found that Ral and Tam were more potent agonists than E2 with ERα/AP1 (Fig. 2) and E2, but not SERMs, activated through ERβ/AP1 (Fig. 2). These differences are likely due to experimental differences, including cell growth conditions (untransfected suspension cultures vs. transfected adherent cultures), chromatin status of the reporter template (chromatin-assembled vs. transiently transfected), endpoint assays (RNA transcribed in a 30-min reaction vs. luciferase activity produced in a 48-h transfection), and ER levels (titrated known amounts vs. overexpressed). The differences in the results of the two studies should serve as a caution that certain experimental parameters can affect the observed ligand responses in the ER/AP-1 pathway. Nonetheless, this assay system will greatly facilitate our understanding of the molecular and biochemical details of signal-regulated transcription in the ER/AP-1 pathway.
Distinct Domains of ERα Are Required for Recruitment and Activation in the ERα/AP-1 Pathway. The literature examining the domain(s) of ERα required for recruitment of the receptor to DNA-bound Fos/Jun heterodimers at AP-1 sites presents some conflicting results (7, 19, 20). Several independent studies using solution interaction assays with immobilized GST-ERα and recombinant c-Jun have variously found that the N-terminal A/B region, the DBD, or the hinge region of ERα could be responsible for interactions with c-Jun (7, 19, 20). These disparate results have been difficult to reconcile. We used an immobilized DNA assay with DNA bound native Fos/Jun heterodimers to show that the ERα DBD, but not the A/B region or the LBD, is required for the recruitment of the receptor to AP-1 sites (Fig. 4B). Interestingly, our results make a clear distinction between the ERα domain required for recruitment (i.e., the DBD) and the domains required for activation (i.e., the A/B region and the LBD). The use of DNA-bound Fos/Jun heterodimers in our assays provides a more accurate representation of the conditions under which ER and AP-1 interact.
Distinct Coactivator Usage by Different Enhancer/Activator Combinations in the ERα and AP-1 Pathways. We used our biochemical assay to compare the coactivator usage of four different transcription pathways involving ERα and AP-1: (i) Fos/Jun heterodimers at AP-1 sites, (ii) ERα plus E2 at EREs, (iii) ERα plus E2 at AP-1 sites, and (iv) ERα plus Ral at AP-1 sites, as illustrated in Fig. 9. Our results clearly show that the different enhancer/activator combinations use coactivators in distinct ways, even when comparing the two ERα/AP-1 pathways (i.e., +E2 and +Ral), which differ only in the ligand bound to ERα (Fig. 5). These results are consistent with recent studies showing that SERMs may promote the differential use of coactivators by ERα acting at EREs and AP-1 sites (32, 33). The use of inhibitory polypeptides, as shown in Fig. 5B, is instructive because they provide information about the protein–protein interactions required for ligand-regulated ERα activity. The fact that a given inhibitory polypeptide had different effects in the various pathways indicates that distinct protein–protein interactions and, hence, distinct protein surfaces, are required for each pathway. Collectively, these results indicate that the type of enhancer (i.e., ERE vs. AP-1 site) can affect the assembly of ERα-dependent transcription complexes. Our studies are reminiscent of studies with glucocorticoid receptor, which showed that the type of enhancer can play a major role in determining the protein–protein interactions required for the assembly of transcription complexes at target promoters (34, 35).
Our results also demonstrate that the type of ligand can play a role in determining the protein interaction surfaces used by ERα to recruit coactivators. Note, for example, the different requirements for ERα-SRC interactions in the ERα/AP-1 pathway with E2 and Ral (Fig. 5B). Specifically, we find that NR box-dependent ERα-SRC interactions are relatively unimportant in the presence of Ral (which is likely to block those interactions anyway), but SRCs are nonetheless still required to help recruit p300/CBP. These results suggest that NR box-independent interactions between AP-1 and SRCs, or ERα and SRCs, perhaps involving the N-terminal A/B region of ERα, may help to recruit SRCs in the ERα/AP-1 pathway. Interestingly, these “altered” interactions are ligand-dependent. Although the SRC interactions vary in the ERα/ERE and ERα/AP-1 pathways in the presence of different ligands (Fig. 5B), our results suggest that SRC-p300/CBP interactions are conserved. The differential expression and use of coactivators, like SRCs and p300/CBP, in different cells types may contribute to the cell-type specificity of SERM action in the ERα/ERE and ERα/AP-1 pathways. The biochemical assay that we have described herein will be useful for dissecting the underlying mechanisms of these pathways in more detail.
Supplementary Material
Acknowledgments
We thank Gary Isaacs, Kristine Frizzell, Mi Young Kim, and Steve Mauro for critical reading of this manuscript. We are grateful to Mi Young Kim (Cornell University) for generously providing mutant ERα expression constructs and Gary Isaacs (Cornell University) for recombinant c-Fos and c-Jun. This work was supported by National Institutes of Health Grants DK58110 (to W.L.K.) and GM62437 (to P.A.C.) and a postdoctoral fellowship from the Susan G. Komen Breast Cancer Foundation (to E.C.).
Author contributions: E.C., M.L.A., and W.L.K. designed research; E.C. and M.L.A. performed research; E.C., M.L.A., P.A.C., and W.L.K. analyzed data; and E.C., M.L.A., and W.L.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, estrogen receptor; SERM, selective estrogen receptor modulator; AF, activation function; ERE, estrogen response element; DBD, DNA-binding domain; LBD, ligand-binding domain; RID, receptor interaction domain; PID, p300/CBP interaction domain; OHT, hydroxytamoxifen; Ral, raloxifene; AP-1, activating protein-1.
References
- 1.Couse, J. F. & Korach, K. S. (1999) Endocr. Rev. 20, 358-417. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson, S., Makela, S., Treuter, E., Tujague, M., Thomsen, J., Andersson, G., Enmark, E., Pettersson, K., Warner, M. & Gustafsson, J. A. (2001) Physiol. Rev. 81, 1535-1565. [DOI] [PubMed] [Google Scholar]
- 3.Pettersson, K. & Gustafsson, J. A. (2001) Annu. Rev. Physiol. 63, 165-192. [DOI] [PubMed] [Google Scholar]
- 4.McDonnell, D. P. (2003) Endocrinology 144, 4237-4240. [DOI] [PubMed] [Google Scholar]
- 5.Jordan, V. C. (2004) Cancer Cell 5, 207-213. [DOI] [PubMed] [Google Scholar]
- 6.Kushner, P. J., Agard, D. A., Greene, G. L., Scanlan, T. S., Shiau, A. K., Uht, R. M. & Webb, P. (2000) J. Steroid Biochem. Mol. Biol. 74, 311-317. [DOI] [PubMed] [Google Scholar]
- 7.Webb, P., Lopez, G. N., Uht, R. M. & Kushner, P. J. (1995) Mol. Endocrinol. 9, 443-456. [DOI] [PubMed] [Google Scholar]
- 8.Paech, K., Webb, P., Kuiper, G. G., Nilsson, S., Gustafsson, J., Kushner, P. J. & Scanlan, T. S. (1997) Science 277, 1508-1510. [DOI] [PubMed] [Google Scholar]
- 9.Frasor, J., Danes, J. M., Komm, B., Chang, K. C., Lyttle, C. R. & Katzenellenbogen, B. S. (2003) Endocrinology 144, 4562-4574. [DOI] [PubMed] [Google Scholar]
- 10.Kian Tee, M., Rogatsky, I., Tzagarakis-Foster, C., Cvoro, A., An, J., Christy, R. J., Yamamoto, K. R. & Leitman, D. C. (2004) Mol. Biol. Cell 15, 1262-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stossi, F., Barnett, D. H., Frasor, J., Komm, B., Lyttle, C. R. & Katzenellenbogen, B. S. (2004) Endocrinology 145, 3473-3486. [DOI] [PubMed] [Google Scholar]
- 12.Tora, L., Gaub, M. P., Mader, S., Dierich, A., Bellard, M. & Chambon, P. (1988) EMBO J. 7, 3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angel, P., Baumann, I., Stein, B., Delius, H., Rahmsdorf, H. J. & Herrlich, P. (1987) Mol. Cell. Biol. 7, 2256-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umayahara, Y., Kawamori, R., Watada, H., Imano, E., Iwama, N., Morishima, T., Yamasaki, Y., Kajimoto, Y. & Kamada, T. (1994) J. Biol. Chem. 269, 16433-16442. [PubMed] [Google Scholar]
- 15.Dubik, D. & Shiu, R. P. (1992) Oncogene 7, 1587-1594. [PubMed] [Google Scholar]
- 16.Acevedo, M. L. & Kraus, W. L. (2004) Essays Biochem. 40, 73-88. [DOI] [PubMed] [Google Scholar]
- 17.Klinge, C. M. (2000) Steroids 65, 227-251. [DOI] [PubMed] [Google Scholar]
- 18.Lonard, D. M. & Smith, C. L. (2002) Steroids 67, 15-24. [DOI] [PubMed] [Google Scholar]
- 19.Sabbah, M., Courilleau, D., Mester, J. & Redeuilh, G. (1999) Proc. Natl. Acad. Sci. USA 96, 11217-11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teyssier, C., Belguise, K., Galtier, F. & Chalbos, D. (2001) J. Biol. Chem. 276, 36361-36369. [DOI] [PubMed] [Google Scholar]
- 21.Kraus, W. L. & Kadonaga, J. T. (1998) Genes Dev. 12, 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung, E., Schwabish, M. A. & Kraus, W. L. (2003) EMBO J. 22, 600-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson, H. A. & Goodrich, J. A. (2001) Nucleic Acids Res. 29, E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, M. Y., Hsiao, S. J. & Kraus, W. L. (2001) EMBO J. 20, 6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acevedo, M. L. & Kraus, W. L. (2003) Mol. Cell. Biol. 23, 335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus, W. L. & Kadonaga, J. T. (1999) in Steroid/Nuclear Receptor Superfamily: A Practical Approach, ed. Picard, D. (Oxford Univ. Press, Oxford), pp. 167-189.
- 27.Grimm, S. L. & Nordeen, S. K. (1999) BioTechniques 27, 220-222. [DOI] [PubMed] [Google Scholar]
- 28.Hawley, D. K. & Roeder, R. G. (1987) J. Biol. Chem. 262, 3452-3461. [PubMed] [Google Scholar]
- 29.Lee, S. K., Kim, H. J., Na, S. Y., Kim, T. S., Choi, H. S., Im, S. Y. & Lee, J. W. (1998) J. Biol. Chem. 273, 16651-16654. [DOI] [PubMed] [Google Scholar]
- 30.Van Orden, K., Yan, J. P., Ulloa, A. & Nyborg, J. K. (1999) Oncogene 18, 3766-3772. [DOI] [PubMed] [Google Scholar]
- 31.Lau, O. D., Kundu, T. K., Soccio, R. E., Ait-Si-Ali, S., Khalil, E. M., Vassilev, A., Wolffe, A. P., Nakatani, Y., Roeder, R. G. & Cole, P. A. (2000) Mol. Cell 5, 589-595. [DOI] [PubMed] [Google Scholar]
- 32.Shang, Y. & Brown, M. (2002) Science 295, 2465-2468. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, H., Yi, X., Sun, X., Yin, N., Shi, B., Wu, H., Wang, D., Wu, G. & Shang, Y. (2004) Genes Dev. 18, 1753-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogatsky, I., Luecke, H. F., Leitman, D. C. & Yamamoto, K. R. (2002) Proc. Natl. Acad. Sci. USA 99, 16701-16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogatsky, I., Wang, J. C., Derynck, M. K., Nonaka, D. F., Khodabakhsh, D. B., Haqq, C. M., Darimont, B. D., Garabedian, M. J. & Yamamoto, K. R. (2003) Proc. Natl. Acad. Sci. USA 100, 13845-13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.