Abstract
The NMR structures of the recombinant cellular form of the prion proteins (PrPC) of the cat (Felis catus), dog (Canis familiaris), and pig (Sus scrofa), and of two polymorphic forms of the prion protein from sheep (Ovis aries) are presented. In all of these species, PrPC consists of an N-terminal flexibly extended tail with ≈100 amino acid residues and a C-terminal globular domain of ≈100 residues with three α-helices and a short antiparallel β-sheet. Although this global architecture coincides with the previously reported murine, Syrian hamster, bovine, and human PrPC structures, there are local differences between the globular domains of the different species. Because the five newly determined PrPC structures originate from species with widely different transmissible spongiform encephalopathy records, the present data indicate previously uncharacterized possible correlations between local features in PrPC three-dimensional structures and susceptibility of different mammalian species to transmissible spongiform encephalopathies.
Keywords: mammalian species, feline transmissible spongiform encephalopathy, scrapie
The prion protein (PrP) in mammalian organisms has attracted keen interest because of its relation to a group of invariably fatal neurodegenerative diseases, the transmissible spongiform encephalopathies (TSEs) or “prion diseases,” which include bovine spongiform encephalopathy (BSE), Creutzfeldt–Jakob disease in humans, feline spongiform encephalopathy, and scrapie in sheep. It is well established that expression of the host-encoded PrP is essential for TSE propagation (1, 2). In transgenic mice lacking the gene that encodes PrP, TSEs could not be observed, and the susceptibility toward TSE of these mice could only be restored by reestablishing PrP expression (3). High sequence conservation of PrP in mammalian species (4) indicates that this protein is functionally important in the healthy organism (1, 2), but the search for this unknown function is still ongoing.
PrP was identified in the context of TSEs in an aggregated “scrapie” isoform of PrP (PrPSc) (5), which copurifies with the infective agent (6). This osbservation, the apparent stability of the infectious agent under DNA/RNA denaturing conditions (7), and the unusual progression of the disease (8) led to the “protein-only hypothesis.” This hypothesis proposes that the major component, if not the only one, of the infectious particle causing TSE is a protein, i.e., presumably PrPSc (1, 7–9).
An early observation in TSE infections has been the species barrier (10). Compared with transmission with infectious material from the same species, the incubation time for onset of TSEs is prolonged if a given species is challenged with infectious brain homogenate originating from another species. The incubation time may be reduced by consecutive passages within the new host, whereby the adaptation to the new host can take several generations for the disease to show clinical signs (11). In vivo and in vitro experiments indicated that the species barrier for infectious transmission of TSEs is somehow related to the extent of PrP sequence homology between the species involved (12, 13) (Fig. 1). Following the protein-only hypothesis, the compatibility of the PrPs from the originating species to the new host should actually be a decisive factor for the propagation of the disease because the covalent structure of PrP in the PrPSc form assumed to be identical to that in the cellular isoform of (PrPC) present in healthy organisms (1). Overall, however, inspection of the amino acid sequence of PrP has not been conclusive to even qualitatively assess either the species barrier for TSEs between different species or the susceptibility of a given species to TSE (4, 14–16). For example, the species barrier transmission of BSE to feline species seems to be readily overcome, as indicated by numerous cases of feline spongiform encephalopathy during the BSE crisis in the United Kingdom, whereas there have been no reports of TSE-infected dogs (17). These observations present an apparent contrast with the facts that the number of amino acid exchanges between bovines cats or dogs is almost equal, with 14 and 13, respectively (15, 18), and that the sequences of the dog PrP (cPrP) and cat PrP (fPrP) differ only in four positions within the fragment 121–230 (Fig. 1).
Fig. 1.
Amino acid sequence alignment of the polypeptide fragment 125–231 for the following prion proteins (numeration of hPrP by following ref. 15): cow, bPrP; sheep, ovPrP; dog, cPrP; cat, fPrP; pig, scPrP; mouse, mPrP; Syrian hamster, shPrP; and human, hPrP. At the top, the locations of the regular secondary structures in bPrP(121–231) are indicated, and the complete sequence of bPrP is given. For the other species, only the residue positions with amino acid exchanges with respect to bPrP are indicated (and a deletion at position 230 of hPrP is indicated by •).
With the aim to obtain more detailed insight into possible correlations between PrPC structure and PrPC function in health and disease, we started years ago an investigation of the three-dimensional structures of recombinant PrPs (19). The relevancy of this approach has recently been substantiated by the demonstration that recombinant PrP has the same fold as PrPC (20, 21). The lack of the posttranslational modifications in PrP expressed in Escherichia coli thus has at most very limited local effects the protein molecular architecture (21). So far, three-dimensional structures in solution have been reported for combinant PrPC of the widely used laboratory animals mouse PrP (19) and Syrian hamster PrP (22), and cattle PrP (bPrP) (23) and human PrP (hPrP) (24). A crystal structure is available the globular domain of a sheep PrP (ovPrP) (25). This paper now presents the prion protein NMR structures of the pig (scPrP), dog cPrP, the cat fPrP, and two variant ovPrPs. This selection three-dimensional PrPC structures includes the prion protein from a species that has so far been resistant to the challenge with BSE-infected food in the natural environment, i.e., cPrP (26). For scPrP, observation of neurologic disorder after challenge with BSE-infected brain homogenate (27) has so far not been followed up with the standard procedures that would qualify the disease as a TSE (1).
Materials and Methods
Cloning, Expression, and Purification of the Prion Proteins. The genes for various ovPrP polymorphisms were provided to us by Dr. A. Bossers (Central Institute for Animal Disease Control, Lelystad, The Netherlands), and the constructs for cPrP(residues 121–231), cPrP(23–231), fPrP(121–231), fPrP(23–231), scPrP(121–231), and scPrP(23–231) were cloned from total DNA. All genes were cloned into the vector pRSETA, and the proteins were expressed in E. coli. For the purification of the recombinant proteins, we followed procedures described in refs. 28 and 29.
NMR Measurements and Structure Calculations. NMR measurements were performed at 20°C on Bruker DRX500, DRX600, DRX750, and Avance900 spectrometers. The protein samples used were uniformly 15N-labeled and 13C, 15N-labeled scPrP(121–231), cPrP(121–231), fPrP(121–231), ovPrP with histidine at position 168 {ovPrP[H168](121–231)}, and ovPrP with arginine at position 168 {ovPrP[R168](121–231)} and 15N-labeled scPrP(23–231), cPrP(23–231), fPrP(23–231), and ovPrP[H168](23–231). The proteins were dissolved at concentrations of 0.5–1.0 mM either in 95% H2O/5% 2H2O or 99.9% 2H2O containing 5 mM sodium acetate at pH 4.5. The programs prosa (30) and xeasy (31) were used for data processing and spectral analysis, respectively. Sequence-specific resonance assignments for the proteins were obtained by using standard triple-resonance NMR experiments (32).
Steady-state 15N{1H}-nuclear Overhauser enhancements (NOEs) of 15N-labeled scPrP(23–231), cPrP(23–231), fPrP(23–231), and ovPrP[H168](121–231) were measured with recovery delays and proton saturation periods of 4 sec (33).
Distance constraints for the structure calculations were obtained from three-dimensional 13C-resolved [1H, 1H]-NOESY and three-dimensional 15N-resolved [1H, 1H]-NOESY spectra recorded at a proton frequency of 750 or 900 MHz with mixing times of 40 or 50 ms. For scPrP(121–231) and ovPrP[H168](121–231), the automatic NOE assignment module candid (34), implemented in the program dyana (35), was used for the structure calculation. For cPrP(121–231), fPrP(121–231), and ovPrP[R168](121–231), automatic NOE identification was added by using the program suite atnos (36)/candid (34)/dyana (35) for the structure calculation. The program dyana (35) was also used to convert NOE intensities into upper-limit distance constraints according to a sixth power peak volume-to-distance relationship, to remove meaningless constraints, and to derive constraints for the backbone torsion angles ϕ and Ψ from Cα chemical shift values (37, 38). The final round of structure calculation was started by using 100 randomized conformers. The 20 conformers with the lowest residual dyana target function values were energy-minimized in a water shell with the program opalp (39, 40) by using the amber force field (41). The program molmol (42) was used to analyze the results of the protein structure calculations (Table 1) and to prepare the drawings of the structures (Figs. 2 and 3).
Table 1. Input for the structure calculation and characterization of the energy-minimized NMR structures of scPrP(121–231), fPrP(121–231), cPrP(121–231), ovPrP[H168](121–231), and ovPrP[R168](121–231).
| scPrP | fPrP | cPrP | ovPrP[H168] | ovPrP[R168] | |
|---|---|---|---|---|---|
| NOE upper distance limits | 1,922 | 1,454 | 1,479 | 2,064 | 1,622 |
| Dihedral angle constraints | 110 | 114 | 122 | 114 | 94 |
| Residual target function value, Å2 | 0.99 ± 0.19 | 1.77 ± 0.28 | 1.93 ± 0.21 | 0.98 ± 0.21 | 1.61 ± 0.23 |
| Residual distance constraint violations | |||||
| Number >0.1 Å | 19 ± 3 | 27 ± 4 | 25 ± 5 | 31 ± 3 | 20 ± 5 |
| Maximum, Å | 0.13 ± 0.01 | 0.16 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 | 0.22 ± 0.00 |
| Residual dihedral angle constraint violations | |||||
| Number >2.0° | 1 ± 1 | 1 ± 1 | 1 ± 1 | 1 ± 1 | 0 ± 1 |
| Maximum, ° | 1.8 ± 0.8 | 1.94 ± 0.45 | 2.90 ± 0.85 | 2.3 ± 0.8 | 1.45 ± 0.78 |
| amber energies (kcal/mol) | |||||
| Total | –4,628 ± 63 | –4,797 ± 105 | –4,657 ± 99 | –4,960 ± 73 | –4,651 ± 68 |
| Van der Waals | –300 ± 12 | –280 ± 15 | –283 ± 16 | –341 ± 14 | –123 ± 14 |
| Electrostatic | –5,236 ± 56 | –5,497 ± 85 | –5,313 ± 99 | –5,542 ± 68 | –5,448 ± 64 |
| rms deviation to the averaged coordinates,* Å | |||||
| bb (N, Cα, C′) | 0.78 ± 0.13 | 0.74 ± 0.14 | 0.70 ± 0.12 | 0.76 ± 0.10 | 0.94 ± 0.18 |
| (125–228) | (125–166, 172–225) | (125–166, 172–225) | (125–228) | (127–166, 173–225) | |
| All heavy atoms | 1.20 ± 0.17 | 1.23 ± 0.13 | 1.16 ± 0.11 | 1.24 ± 0.14 | 1.46 ± 0.24 |
Except for the top two entries, the average for the 20 conformers with the lowest residual dyana target function values and the standard deviation among them are given.
bb, backbone. The numbers in parentheses indicate the residues for which the rms deviation values were calculated
Fig. 2.
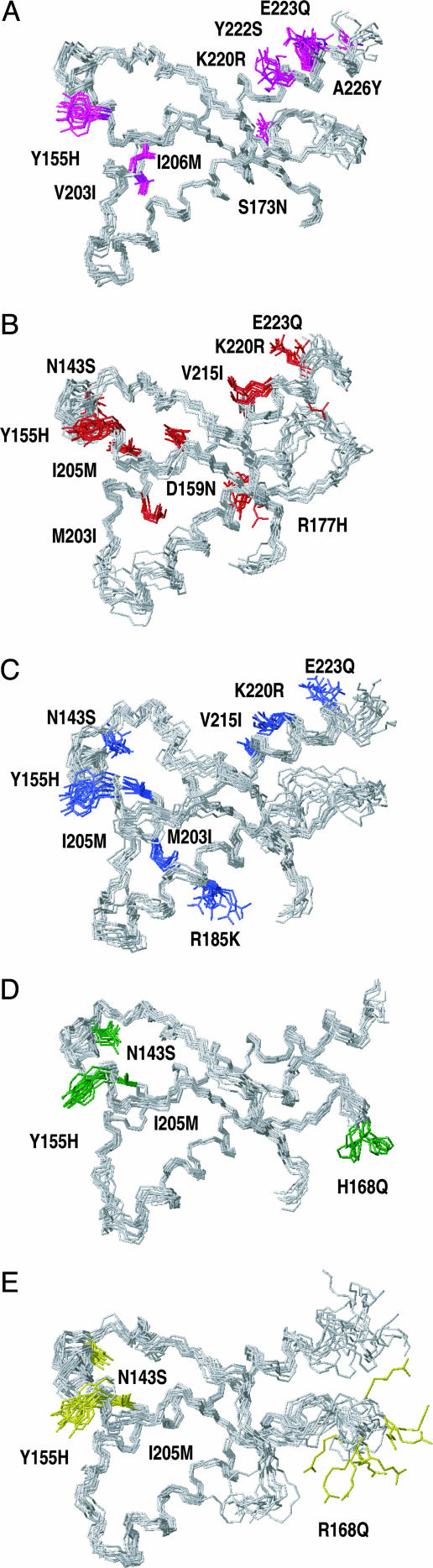
NMR structures of the globular domains in the five prion proteins studied in this paper. Each structure is shown as a bundle of 20 energy-minimized conformers, with gray coloring of the backbone and species-specific coloring of the amino acid side chains that are different from bPrP. For each species, the amino acid replacements relative to bPrP are identified by indication with the one-letter code of the amino acid in the species considered, the sequence position, and the amino acid in bPrP. The conformers were aligned for best fit of the backbone heavy atoms of the residues 128–166 and 172–220, and displayed are the residues 125–227. (A) scPrP(121–231) (side chains pink); (B) cPrP(121–231) (side chains red); (C) fPrP(121–231) (side chains blue); (D) ovPrP[H168](121–231) (side chains green); (E) ovPrP[R168](121–231) (side chains yellow).
Fig. 3.
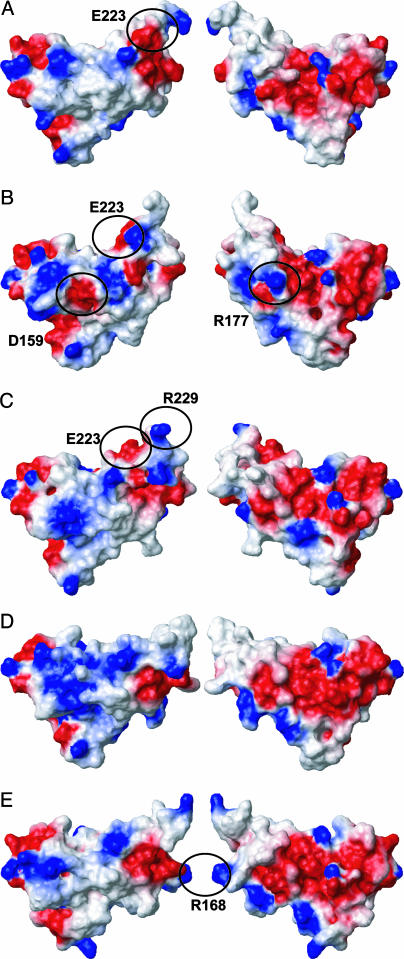
Surface views of the globular domains of the five proteins of Fig. 2. Shown are the residues 125–229. The presentation in Right relates to the one in Left through a 180° rotation around a vertical axis. The electrostatic surface potential is indicated in red (negative charge), white (uncharged), and blue (positive charge). The circles indicate charge differences relative to bPrP that are discussed in the text, and the amino acid residues are identified from which the charge differences originate. (A) scPrP(121–231), (B) cPrP(121–231), (C) fPrP(121–231), (D) ovPrP[H168](121–231), and (E) ovPrP[R168](121–231).
Results
For each of the four animal species for which the prion protein, or in the case of the sheep two different polymorphisms of the prion protein, were studied (Table 1), the mature full-length polypeptide chain with residues 23–231 (the residue numeration for human PrP (15) is used throughout this paper) and the stable partial sequence 121–231 (19) were cloned and expressed in E. coli. All nine recombinant proteins (ovPrP[R168](23–231) was not studied) were prepared with uniform 15N-labeling, and the five 121–231 fragments were also obtained with uniform 13C, 15N-labeling. The methods used for protein preparation and purification are described in Materials and Methods.
Following up on the approach used previously for the global characterization of other mammalian PrPs (23, 24, 43), hetero-nuclear 15N{1H}-NOEs were measured at 20°C for 15N-labeled scPrP(23–231), fPrP(23–231), cPrP(23–231), and ovPrP[H168](23–231). Each of the four proteins was thus shown to contain a structured region extending approximately from residues 125–226, with positive values for the 15N{1H}-NOEs, as expected for a globular protein with the size of PrP (44). At both ends of the globular domain, there are flexible peptide segments, as evidenced by negative values of the steady-state 15N{1H}-NOEs (data not shown). The C-terminal pentapeptide segment corresponds to a flexible linker between the structured domain of PrPC and the glycosylphosphatidylinositol anchor in the cell surface membrane (1, 2). The N-terminal polypeptide segment 23–124 forms an outstandingly long flexible tail, as evidenced by the observation in all four species that the residues 23–121 all show negative values of the 15N{1H}-NOEs for the backbone amide groups. This result coincides with corresponding data on all of the mammalian PrPs described in refs. 23, 24, 43, and 45. Because of different insertions relative to the human prion protein sequence (hPrP), this tail includes 103 residues for fPrP, 100 residues for cPrP and scPrP, and 101 residues for ovPrP (15).
In the remainder of this section and in Discussion, we focus primarily on the NMR structure determination of the constructs with residues 121–231 of the five aforementioned prion proteins and on an analysis of the resulting structures for the globular domains.
Resonance Assignments. For scPrP(121–231), complete resonance assignments were obtained for the entire polypeptide backbone. For cPrP(121–231), fPrP(121–231), ovPrP[H168](121–231), and ovPrP[R168](121–231), nearly complete assignments were obtained for the polypeptide backbone, the exceptions being the amide protons and amide nitrogens of Gln-168 (fPrP), Tyr-169 (cPrP and fPrP), Ser-170 (cPrP, ovPrP[H168], and ovPrP[R168]), Asn-171 (fPrP, cPrP, ovPrP[H168], and ovPrP[R168]), and Phe-175 (fPrP, cPrP, ovPrP[H168], and ovPrP[R168]). The amino acid side chain assignments are nearly complete, including all tyrosine, phenylalanine, and histidine ring resonances with the sole exception of Phe-198ζCH. The chemical shift lists of the five proteins have been deposited in the BioMagResBank (www.bmrb.wisc.edu) with the following entry codes: scPrP(121–231), 6380; cPrP(121–231), 6378; fPrP(121–231), 6377; ovPrP[H168](121–231), 6381; and ovPrP[R168](121–231), 6403.
Collection of Conformational Constraints and Structure Calculation. For scPrP(23–231) and ovPrP[H168](121–231), which were studied earlier than the other proteins, peak picking of the three-dimensional 15N-resolved and three-dimensional 13C-resolved [1H, 1H]-NOESY spectra was pursued interactively. The resulting lists of NOESY cross peaks together with chemical shift lists from the resonance assignments were used as input for automatic NOE assignment and structure calculation by using the standard protocol with the program package candid (34)/dyana (35).
For cPrP(121–231), fPrP(121–231), and ovPrP[R168](121–231), the automation of the structure determination process included the peak picking of the NOESY spectra by using the program package atnos/candid/dyana with a standard protocol (34–36). The input of NOE upper distance limits obtained for the individual proteins (Table 1) shows that the interactive peak picking resulted in an ≈25% higher total number of constraints and in improved convergence of the structure calculation, as evidenced by the lower residual dyana target function values.
The NMR Structures of the Globular Domains of scPrP, cPrP, fPrP, ovPrP[H168], and ovPrP[R168]. Table 1 shows that four of the five PrP structures were determined with comparable precision, as documented by backbone rms deviation values of 0.70–0.78 Å. The somewhat lower precision achieved for ovPrP[R168](121–231) is due to the fact that the NOESY data sets had to be recorded at 0.5 mM protein concentration as compared with ≈1.0 mM concentration for the other proteins.
In Fig. 2, the five structures are shown as bundles of 20 conformers (Table 1). The location of regular secondary structures coincides nearly identically with bPrP (Fig. 1). In Fig. 2, the amino acid exchanges relative to bPrP are indicated, which also serves as a guide to follow the polypeptide fold. The drawings start with residue 125 in the lower right, from where the polypeptide goes through the first β-strand 128–131 to the start of helix α1, which is at residue 143 in the upper left corner of the molecule. Following helix α1 from residues 144–154, the chain winds through the β-strand 161–164, which combines with residues 128–131 to form an antiparallel β-sheet, to the extreme right. A loop of residues 166–173 connects to helix α2 with residues 174–194, which leads to the lower left corner of the structure. A five-residue loop then leads to helix α3 with residues 200–226, which ends in the top right corner of the structure.
Some local features in the structures of Fig. 2 can all be directly related with the absence or very low intensity of the NMR signals for the backbone 15N-1H moieties of individual residues. First, the loop of residues 166–173 is disordered; a complete set of 15N-1H NMR signals could be observed only in scPrP(121–231) (Fig. 2A). Second, the start of helix α2 is poorly defined because the amide proton NMR signal of Phe-175 could not be detected, the sole exception being scPrP(121–231). Third, in all five proteins the helix α3 is somewhat nonregular near residue 220, which correlates with the observation that the 15N-1H NMR signals for one or several residues in the segment 218–222 have very low intensity. Local superposition of the residues 222–226 reveals the presence of two turns of well defined α-helix, also in ovPrP[R168](121–231) (Fig. 2E). Previously it was observed that the distortion of the helix α3 is particularly pronounced in murine PrP (19, 46).
Discussion
Comparative NMR studies with natural bovine PrPC isolated from calf brains showed that the three-dimensional structure of recombinant PrP prepared with the methods used in this paper corresponds to the polypeptide structure in natural PrPC containing all of the posttranslational modifications (21). In the following discussion, we therefore refer to the structures of Fig. 2 as the PrPC form of the prion protein.
All mammalian species studied so far (Fig. 2 and refs. 23, 24, 43, and 45) contain PrPC molecules with a flexibly extended N-terminal tail of length ≈100 residues and a C-terminal globular domain with ≈100 residues. The architecture of the globular domain is highly conserved in the different species, as was expected from the high degree of sequence identity (Fig. 1). Considering that the presently studied group (Fig. 2) includes species with widely different susceptibilities toward TSEs, we shall now search the preserved scaffold of the globular domain for local structure variations that might relate to different susceptibilities for developing TSE and, in particular, to variable stringency of the species barrier against infection with BSE.
Two areas of the globular domain of PrPC have been suggested to be important for the development of TSEs. First, the helix α1 has been implicated as a primary interaction site with the TSE-associated isoform PrPSc (1). Second, an epitope comprising the loop 166–172 and the C-terminal end of helix α3 has been suggested to be recognized by a conversion chaperone, i.e., “protein X” (47). Inspection of Fig. 2 then shows that species variations of the amino acid sequence are predominately located in or near these two molecular regions.
With regard to a possible role of helix α1 in TSE susceptibility, the five previously uncharacterized structures of Fig. 2 do not indicate any conclusive correlation. The helix has identical length, location, and orientation in all of the structures. Furthermore, species with or without a record of TSEs (cat and dog) and sheep polymorphisms with high and low TSE susceptibility all have identical sequences from residues 143–158 (Fig. 1).
Inspection of the amino acid substitutions in Fig. 2 indicates that there should be surface charge variations at or near the presumed protein X epitope (Fig. 3). A first intriguing observation results for the different sheep PrPs. In vivo and in vitro experiments that link BSE or scrapie susceptibility to the amino acid sequence of ovPrP (48, 49), showed that sheep carrying ovPrP[R168] are highly resistant to transmission of TSEs, whereas ovPrP[Q168] has been linked with high susceptibility and ovPrP[H168] with medium-high susceptibility to BSE or scrapie transmission (50). Position 168 is surface-exposed in the loop 166–173 and, therefore, has a dominant effect on the surface charges distribution in this region (Fig. 3). The positive charge of R168 in ovPrPC thus appears to protect healthy sheep when challenged with BSE infectivity or scrapie infectivity.
All four amino acid substitutions between the globular domains of cPrPC and fPrPC involve charged residues (Figs. 1 and 3). The presence of Asp-159 and Arg-177 in dogs causes unique charge distribution patterns on the front as well as the back side of cPrPC (Fig. 3), which might, from the presently available evidence, correlate with protection of dogs against challenge with BSE. fPrPC, in turn, shares the presence of a positive charge near the C terminus (Fig. 3C) with other TSE-susceptible species (Fig. 1).
Relative to bPrP, scPrP has a single charge-effective amino acid substitution in position 223 (Fig. 3A), which it shares, however, with both the dog and cat (Figs. 1 and 3), and which, therefore, would not appear to be critical with regard to TSE susceptibility. A charge-neutral amino acid replacement from Asn-173 in bPrP to Ser-173 in scPrP (Fig. 1) stabilizes the loop 166–173 in scPrPC to the extent that complete NMR assignments could be obtained. This amino acid substitution might thus affect the presumed protein X epitope (1, 47).
In conclusion, the seminal observation by the Weissmann group that expression of host-encoded PrP is a necessary condition for the development of a TSE (3) implies that each organism producing PrPC might be susceptible to spontaneous or transmitted TSE. Thus, although PrPSc has an increasingly prominent role in research on TSE diagnostics, it would appear that independent of the nature of the TSE-causing agent, PrPC will be a prime target for TSE prevention in healthy organisms and TSE treatment in disease. Detailed knowledge of PrPC three-dimensional structures will be an important part of the platform for such future endeavors.
Acknowledgments
We thank the Tierspital of the University of Zürich for the donation of cat, dog, and pig blood; Dr. A. Bossers for providing the different ovPrP genes; and Dr. G. Pioda for help with the structural determination of ovPrP[H168](121–231). This work was supported by the Schweizerischer Nationalfonds and the ETH Zürich through the National Center of Competence in Research “Structural Biology” and a Fannie and John Hertz Foundation fellowship (to L.G.N.).
Author contributions: K.W. designed research; D.A.L., C.S., L.G.N., V.E.-M., B.C., L.C., C.v.S., F.F., T.H., and P.G. performed research; and D.A.L., C.S., L.C., and K.W. wrote the paper.
Abbreviations: PrP, prion protein; bPrP, bovine PrP; BSE, bovine spongiform encephalopathy; cPrP, dog PrP; fPrP, cat PrP; hPrP, human PrP; NOE, nuclear Overhauser enhancement; ovPrP, sheep PrP; ovPrP[H168], recombinant ovPrP with histidine at position 168; ovPrP[R168], recombinant ovPrP with arginine at position 168; PrPC, cellular isoform of PrP; PrPSc, scrapie isoform of PrP; scPrP, pig PrP; TSE, transmissible spongiform encephalopathy.
Data deposition: The atomic coordinates for a bundle of 20 conformers of each of the five NMR structures have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1XYQ for scPrP(121–231), 1XYJ for fPrP(121–231), 1XYK for cPrP(121–231), 1XYU for ovPrP[H168](121–231), and 1Y2S for ovPrP[R168](121–231)].
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weissmann, C., Enari, M., Klohn, P.-C., Rossi, D. & Flechsig, E. (2002) Proc. Natl. Acad. Sci. USA 99, 16378-16783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Büeler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339-1347. [DOI] [PubMed] [Google Scholar]
- 4.Schätzl, H. M., Da Costa, M., Taylor, L., Cohen, F. E. & Prusiner, S. B. (1995) J. Mol. Biol. 245, 362-374. [DOI] [PubMed] [Google Scholar]
- 5.Oesch, B., Westaway, D., Walchli, M., McKinley, M. P., Kent, S. B., Aebersold, R., Barry, R. A., Tempst, P., Teplow, D. B., Hood, L. E. & et al. (1985) Cell 40, 735-746. [DOI] [PubMed] [Google Scholar]
- 6.McKinley, M. P., Bolton, D. C. & Prusiner, S. B. (1983) Cell 35, 57-62. [DOI] [PubMed] [Google Scholar]
- 7.Alper, T., Cramp, W. A., Haig, D. A. & Clarke, M. C. (1967) Nature 214, 764-766. [DOI] [PubMed] [Google Scholar]
- 8.Griffith, J. S. (1967) Nature 215, 1043-1044. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner, S. B. (1982) Science 216, 136-144. [DOI] [PubMed] [Google Scholar]
- 10.Gajdusek, D. C. (1977) Science 197, 943-960. [DOI] [PubMed] [Google Scholar]
- 11.Hill, A. F. & Collinge, J. (2003) Trends Microbiol. 11, 578-584. [DOI] [PubMed] [Google Scholar]
- 12.Chen, S. G. & Gambetti, P. (2002) Neuron 34, 854-856. [DOI] [PubMed] [Google Scholar]
- 13.Kocisko, D. A., Priola, S. A., Raymond, G. J., Chesebro, B., Lansbury, P. T., Jr., & Caughey, B. (1995) Proc. Natl. Acad. Sci. USA 92, 3923-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Billeter, M., Riek, R., Wider, G., Hornemann, S., Glockshuber, R. & Wüthrich, K. (1997) Proc. Natl. Acad. Sci. USA 94, 7281-7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wopfner, F., Weidenhofer, G., Schneider, R., von Brunn, A., Gilch, S., Schwarz, T. F., Werner, T. & Schätzl, H. M. (1999) J. Mol. Biol. 289, 1163-1178. [DOI] [PubMed] [Google Scholar]
- 16.Van Rheede, T., Smolenaars, M. M., Madsen, O. & De Jong, W. W. (2003) Mol. Biol. Evol. 20, 111-121. [DOI] [PubMed] [Google Scholar]
- 17.Bartz, J. C., McKenzie, D. I., Bessen, R. A., Marsh, R. F. & Aiken, J. M. (1994) J. Gen. Virol. 75, 2947-2953. [DOI] [PubMed] [Google Scholar]
- 18.Lysek, D. A., Nivon, L. G. & Wüthrich, K. (2004) Gene 341, 249-253. [DOI] [PubMed] [Google Scholar]
- 19.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R. & Wüthrich, K. (1996) Nature 382, 180-182. [DOI] [PubMed] [Google Scholar]
- 20.Eberl, H., Tittmann, P. & Glockshuber, R. (2004) J. Biol. Chem. 279, 25058-25065. [DOI] [PubMed] [Google Scholar]
- 21.Hornemann, S., Schorn, C. & Wüthrich, K. (2004) EMBO Reports 5, 1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, T. L., Liu, H., Ulyanov, N. B., Farr-Jones, S., Zhang, H., Donne, D. G., Kaneko, K., Groth, D., Mehlhorn, I., Prusiner, S. B. & Cohen, F. E. (1997) Proc. Natl. Acad. Sci. USA 94, 10086-10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López García, F., Zahn, R., Riek, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8334-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahn, R., Liu, A., Lührs, T., Riek, R., von Schroetter, C., López García, F., Billeter, M., Calzolai, L., Wider, G. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haire, L. F., Whyte, S. M., Vasisht, N., Gill, A. C., Verma, C., Dodson, E. J., Dodson, G. G. & Bayley, P. M. (2004) J. Mol. Biol. 336, 1175-1183. [DOI] [PubMed] [Google Scholar]
- 26.Kirkwood, J. K. & Cunningham, A. A. (1994) Vet. Rec. 135, 296-303. [DOI] [PubMed] [Google Scholar]
- 27.Ryder, S. J., Hawkins, S. A., Dawson, M. & Wells, G. A. (2000) J. Comp. Pathol. 122, 131-143. [DOI] [PubMed] [Google Scholar]
- 28.Zahn, R., von Schroetter, C. & Wüthrich, K. (1997) FEBS Lett. 417, 400-404. [DOI] [PubMed] [Google Scholar]
- 29.Lysek, D. A. & Wüthrich, K. (2004) Biochemistry 43, 10393-10399. [DOI] [PubMed] [Google Scholar]
- 30.Güntert, P., Dötsch, V., Wider, G. & Wüthrich, K. (1992) J. Biomol. NMR 2, 619-629. [Google Scholar]
- 31.Bartels, C., Xia, T. H., Billeter, M., Güntert, P. & Wüthrich, K. (1995) J. Biomol. NMR 6, 1-10. [DOI] [PubMed] [Google Scholar]
- 32.Bax, A. & Grzesiek, S. (1993) Acc. Chem. Res. 26, 131-138. [Google Scholar]
- 33.Dayie, K. T. & Wagner, G. (1994) J. Magn. Reson. Ser. A . 111, 121-126. [Google Scholar]
- 34.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Mol. Biol. 319, 209-227. [DOI] [PubMed] [Google Scholar]
- 35.Güntert, P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273, 283-298. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Biomol. NMR 24, 171-189. [DOI] [PubMed] [Google Scholar]
- 37.Spera, S. & Bax, A. (1991) J. Am. Chem. Soc. 113, 5490-5492. [Google Scholar]
- 38.Luginbühl, P., Szyperski, T. & Wüthrich, K. (1995) J. Magn. Reson. Ser. B 109, 229-233. [Google Scholar]
- 39.Luginbühl, P., Güntert, P., Billeter, M. & Wüthrich, K. (1996) J. Biomol. NMR 8, 136-146. [DOI] [PubMed] [Google Scholar]
- 40.Koradi, R., Billeter, M. & Güntert, P. (2000) Comput. Phys. Commun. 124, 139-147. [Google Scholar]
- 41.Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M., Jr., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179-5197. [Google Scholar]
- 42.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-57. [DOI] [PubMed] [Google Scholar]
- 43.Riek, R., Hornemann, S., Wider, G., Glockshuber, R. & Wüthrich, K. (1997) FEBS Lett. 413, 282-288. [DOI] [PubMed] [Google Scholar]
- 44.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 45.Donne, D. G., Viles, J. H., Groth, D., Mehlhorn, I., James, T. L., Cohen, F. E., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13452-13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calzolai, L., Lysek, D. A., Güntert, P., von Schroetter, C., Riek, R., Zahn, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8340-8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaneko, K., Zulianello, L., Scott, M., Cooper, C. M., Wallace, A. C., James, T. L., Cohen, F. E. & Prusiner, S. B. (1997) Proc. Natl. Acad. Sci. USA 94, 10069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bossers, A., de Vries, R. & Smits, M. A. (2000) J. Virol. 74, 1407-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster, J. D., Parnham, D. W., Hunter, N. & Bruce, M. (2001) J. Gen. Virol. 82, 2319-2326. [DOI] [PubMed] [Google Scholar]
- 50.Belt, P. B., Muileman, I. H., Schreuder, B. E., Bos-de Ruijter, J., Gielkens, A. L. & Smits, M. A. (1995) J. Gen. Virol. 76, 509-517. [DOI] [PubMed] [Google Scholar]