Abstract
The Legionella pneumophila Dot/Icm system is a type IV secretion apparatus that transfers bacterial proteins into eukaryotic host cells. The RalF protein is a substrate engaged and translocated into host cells by the Dot/Icm system. In this study, the mechanism of Dot/Icm-mediated translocation of RalF has been investigated. It was determined that RalF translocation into host cells occurs before bacterial internalization. Sequences essential for RalF translocation were located at the C terminus of the RalF protein. A fusion protein consisting of a 20-aa C-terminal RalF peptide appended to the calmodulin-dependent adenylate cyclase domain of the Bordetella pertussis adenylate cyclase protein was translocated into host cells by the Dot/Icm system. A leucine (L372) residue at the -3 position in relation to the RalF C terminus was critical for translocation. Consistent with RalF L372 playing an important role in substrate recognition by the Dot/Icm system, most other Dot/Icm substrates were found to have amino acid residues with similar physical properties at their -3 or -4 C-terminal positions. These data demonstrate that the Dot/Icm system can transfer bacterial proteins that modulate host cellular functions before uptake and indicate that substrate recognition involves a C-terminal translocation signal. Thus, Legionella has the ability to engage synthesized substrate proteins and transfer them into host cells on contact, enabling Legionella to rapidly alter transport of the vacuole in which it resides.
Keywords: ADP ribosylation factor, type IV secretion systems, vacuole biogenesis
Found ubiquitously in fresh water environments, the bacterial pathogen Legionella pneumophila can replicate within protozoan host cells that feed on bacteria (1). When inhaled by humans, Legionella can replicate within alveolar macrophages and cause a severe pneumonia known as Legionnaires disease (2, 3). To replicate within eukaryotic cells, Legionella modulates transport of the vacuole in which it resides (4-6). Legionella-containing vacuoles evade fusion with lysosomes and are remodeled into an endoplasmic reticulum-derived organelle that supports bacterial replication (7-10). Modulation of vacuole transport requires the Legionella Dot/Icm system (11-14).
The Legionella Dot/Icm system is a protein secretion apparatus related to type IV secretion systems found in other bacteria (15, 16). As predicted for most type IV secretion systems, the Dot/Icm apparatus will engage proteins in the cytosol of the bacterial cell and translocate them into eukaryotic host cells (17-20). RalF is one of the proteins translocated into host cells by the Dot/Icm apparatus (20). RalF functions as an exchange factor for the eukaryotic ADP ribosylation factor (ARF) family of small GTPase proteins (20). ARF proteins are highly conserved and play an important role in regulating vesicular transport between the endoplasmic reticulum and the Golgi apparatus in all eukaryotic cells (21). By stimulating the exchange of GDP for GTP on ARF, the RalF protein is able to activate ARF and retain ARF on the membrane of Legionella-containing vacuoles. In addition to RalF, recent studies indicate that the Dot/Icm system transfers at least 10 other proteins into host cells during infection (17-20). Although multiple substrates of the Dot/Icm system have now been identified, the mechanisms by which the Dot/Icm system recognizes these proteins and translocates them into host cells is poorly understood.
To analyze the translocation of Yersinia enterocolitica Yop proteins into host cells by a type III secretion system, the Cornelis group (22) developed a reporter system that quantitatively measures the delivery of bacterial proteins into the cytosol of host cells by specialized secretion systems. In this approach, the adenylate cyclase (Cya) domain of the Bordetella pertussis Cya toxin is fused to putative substrate proteins transferred into host cells by a given secretion system. Because the Cya enzyme requires calmodulin as a cofactor and calmodulin is present only in the cytosol of eukaryotic cells, translocation of a Cya fusion protein into host cells can be measured by determining intracellular cAMP levels. This approach has been used recently to demonstrate translocation of two Legionella proteins (LepA and LepB) into mouse macrophages by the Dot/Icm system (18). Here, we use the Cya system to investigate the requirements for translocation of RalF into host cells by the Dot/Icm system.
Materials and Methods
Bacterial Strains, Plasmids, and Media. All Legionella were derivatives of L. pneumophila strain Lp01 (14). Strains defective in dot/icm genes and the ralF gene were described (20, 23-26). Details of plasmid construction are given in Supporting Text and Table 3, which are published as supporting information on the PNAS web site. Legionella were grown on charcoal yeast extract plates containing appropriate antibiotics (10 μg/ml chloramphenicol or kanamycin) as described (23, 27).
Cell Culture. CHO FcγRII cells were cultured at 37°C in 5% CO2 in α-MEM plus 10% FBS as described (8). Mouse bone marrow-derived macrophages were cultured from A/J mice as described (28). As indicated, gentamicin (100 μg/ml) was added to tissue culture medium to kill extracellular Legionella.
Cya Assay. CHO FcγRII cells were replated into 48-well tissue culture plates 1 day before infection. Medium was exchanged at 30 min before infection with that containing rabbit anti-Legionella antiserum diluted at ratio of 1:1,000. Cytochalasin D (20 μM) or chloramphenicol (100 μg/ml) was added to the wells as indicated. Legionella were added to each well (1.2 × 106 bacteria per well) and centrifuged onto a confluent monolayer of host cells (4 × 104 cells per well) for 5 min at 180 × g. Plates were immediately warmed in a 37°C water bath for 5 min, then placed in a CO2 incubator for a total of 1 h. Cells were washed three times with ice-cold PBS and lysed in 100 μl of extraction solution (50 mN HCl/0.1% Triton X-100) on ice. After boiling for 5 min, extracts were neutralized with 6 μl of 0.5 M NaOH and cAMP was extracted with 2 vol of ethanol. Insoluble materials were pelleted by centrifugation, and the soluble materials containing cAMP were lyophilized. The cAMP levels were determined for each extract by using an ELISA kit (Amersham Biosciences, RPN-225).
Immunoblot Analysis. Proteins for Legionella cells in suspension were precipitated after the addition of 10% trichloroacetic acid, and protein pellets were washed with acetone. Sample preparation was carried out in parallel with the Cya assay. Total protein from an estimated 2 × 107 Legionella bacteria was separated by SDS/10% PAGE. Protein transfer to Immobilon P (Millipore) membranes, and immunoblot analysis to detect the M45 epitope by using the mAb45 Ab were carried out as described (23, 29).
Localization of ARF1-GFP in Infected Host Cells. ARF1-GFP localization in mouse macrophages and CHO FcγRII cells was assayed as described (8, 20). Briefly, cells were transfected with a retroviral vector carrying the gene for ARF1-GFP and plated onto cover glass in 24-well tissue culture plates. Legionella were added to host cells at a multiplicity of infection of 50, and the plates were centrifuged at 180 × g for 5 min. Plates were incubated for 10 min at 37°C, the extracellular bacteria were washed out, and then plates were further incubated for a total of 1 h. Immunofluorescent analysis of infected macrophages was carried out as described (8, 20).
Results
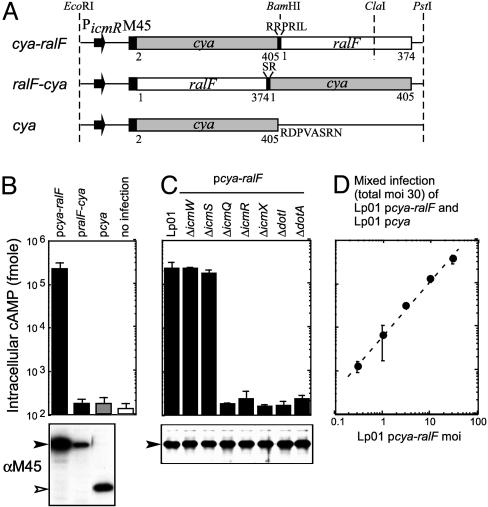
A Cya-Based Reporter System to Analyze Translocation of RalF into Host Cells. Fusion proteins were constructed to test whether RalF translocation could be assessed by using the Cya fusion approach. The Cya2-405 domain was fused both to the N and C termini of the full-length RalF protein. Genes encoding these RalF fusion proteins were inserted into the broad-host range vector pMMB207M45NT such that the M45 epitope tag in this vector was appended to the N terminus of each fusion protein. Transcription of the genes encoding these fusion proteins was driven by the promoter for the Legionella icmR gene (Fig. 1A). CHO cells producing the FcγRII protein were used as eukaryotic hosts. These data show that cAMP levels increased >1,000-fold in CHO FcγRII cells after infection by Legionella, producing the Cya-RalF fusion protein (Fig. 1B). By contrast, infection with Legionella producing the RalF-Cya fusion or Cya alone did not result in a dramatic increase in cAMP levels. Importantly, cAMP levels remained unchanged after infection of CHO FcγRII cells when the Cya-RalF protein was produced in Legionella defective in components of the Dot/Icm system (Fig. 1C). The only exception was that translocation of Cya-RalF was still observed for Legionella defective in the icmS or icmW genes, which encode accessory proteins that are not required for all Dot/Icm-dependent activities (23, 26, 30). Intracellular cAMP levels in CHO FcγRII cells showed a linear increase over a 2-log range in relation to the multiplicity of Legionella producing Cya-RalF (Fig. 1D). These results indicate that the Cya fusion assay is a remarkably sensitive and quantitative tool to analyze RalF translocation into host cells by the Dot/Icm system.
Fig. 1.
Quantitative analysis of RalF translocation into host cells. (A) Schematic representation of the Cya fusion constructs used to measure RalF translocation. Cya with RalF appended C-terminally (cya-ralF) and N-terminally (ralF-cya) are shown, as is the construct used as a negative control that produces only Cya (cya). The promoter and the translation initiation region of the Legionella icmR gene initiates transcription and translation of the fusion in each construct. All constructs have an N-terminal M45 epitope for immunological detection of the gene products. Regions derived from the Cya toxin of the Bordetella cya gene and the Legionella ralF gene are designated with amino acid residue numbers. Additional amino acid residues located at the fusion junctions and at the end of the Cya protein that resulted from the cloning procedures used are also indicated. These genes were ligated into pMMB207 and the resulting plasmids were designated as pcya-ralF,pralF-cya, and pcya, respectively. (B) CHO FcγRII cells were infected with wild-type Legionella harboring the indicated plasmids The y axis has cAMP levels detected for host cells infected for 1 h plotted on a logarithmic scale. Results are the average ± SD from an experiment performed in triplicate. Immunoblots shown below correspond to Cya protein levels inside of the Legionella cells used for the infection. (C) Requirements for Dot and Icm components in Cya-RalF translocation were examined. Isogenic Legionella mutants defective in the gene indicated at the top were derived from the wild-type strain Lp01. Immunoblots shown below correspond to Cya protein levels inside of the Legionella cells used for the infection. (D) The dynamic range of the Cya translocation assay was examined by adding Legionella at a multiplicity of infection (moi) of 30 to CHO FcγRII cells. The inoculum used was comprised of Legionella producing either Cya-RalF (pcya-ralF) or Cya alone (pcya) combined at different ratios. Plotted on the x axis is the number (moi) of Legionella producing Cya-RalF. The linear relationship between cAMP levels and the amount of Cya-RalF producing bacteria added indicate a dynamic range for this assay of at least 2 orders of magnitude.
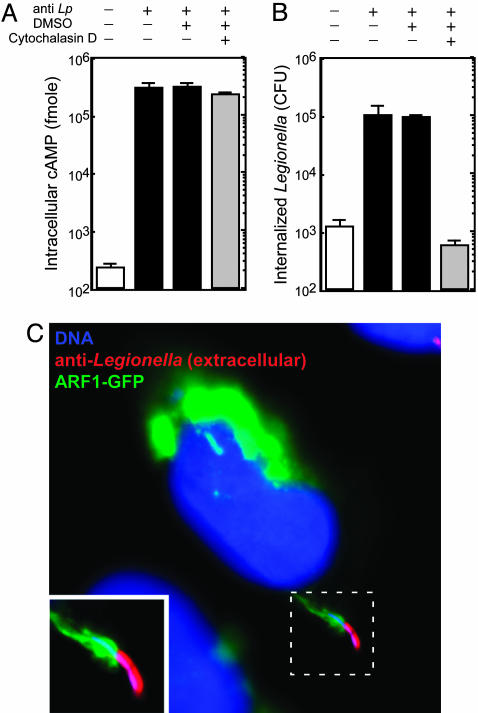
RalF Translocation Occurs Before Legionella Are Internalized. Previous studies (11) show that Legionella entry into CHO FcγRII cells depends on IgG-opsonization of bacteria. This finding was confirmed by showing that most nonopsonized Legionella centrifuged onto CHO FcγRII cells were killed upon addition of gentamicin to the culture medium, which indicates that they were internalized inefficiently (Fig. 2B, white bar). Translocation of Cya-RalF in this system also depended on IgG-opsonization of bacteria (Fig. 2A, white bar). Because Legionella adhere poorly to CHO FcγRII cells, these data suggested that intimate contact between host cells and/or uptake of Legionella is important for RalF translocation. To test whether adherence is sufficient for RalF translocation, CHO FcγRII cells were treated with Cytochalasin D to prevent the internalization of adherent IgG-opsonized Legionella. These data show that the Cytochalasin D treatment blocked Legionella internalization (Fig. 2B, gray bar) but did not prevent translocation of Cya-RalF into CHO FcγRII cells (Fig. 2A, gray bar). These data suggest that RalF is transferred into host cells by Legionella before bacteria are internalized. Because RalF translocation mediates the recruitment of ARF to vacuoles containing Legionella, we also examined translocation of the native RalF protein by fluorescence microscopy by using CHO FcγRII cells, producing ARF1-GFP. These CHO FcγRII cells were fixed 5 min after infection, and extracellular bacteria were identified by staining with anti-Legionella antibodies before permeabilization of the host cell plasma membrane. These experiments revealed dramatic ARF1-GFP recruitment to nascent Legionella-containing vacuoles (Fig. 2C). These data indicate that RalF translocation requires intimate contact with host cells but does not require bacterial internalization.
Fig. 2.
The RalF protein is translocated into host cells by extracellular Legionella. (A) CHO FcγRII cells were infected with Legionella producing Cya-RalF. Indicated above the graph is the addition (+) of opsonizing Ab (anti-Lp), DMSO, or Cytochalasin D dissolved in DMSO. Cya-RalF translocation levels were determined by measuring cAMP levels in host cells 1 h after infection. All values are the average ± SD for an experiment performed in triplicate. (B) In a parallel assay to that shown in A, CHO FcγRII cells were infected with Legionella producing Cya-RalF for 15 min and gentamicin was then added to kill extracellular bacteria. Internalized Legionella were determined 1 h after infection by plating bacteria that survived the gentamicin treatment on agar plates and counting colony-forming units. All values are the average ± SD for an experiment performed in triplicate. (C) CHO FcγRII cells producing ARF1-GFP (green) were infected for 5 min with opsonized Legionella (Lp01). Before permeabilization of the host cell plasma membrane, extracellular Legionella were stained with an anti-Legionella Ab (red). Host and bacterial DNA was stained with DAPI (blue) after permeabilization. This fluorescent micrograph shows an extracellular Legionella bacterium recruiting ARF1-GFP to the site of internalization.
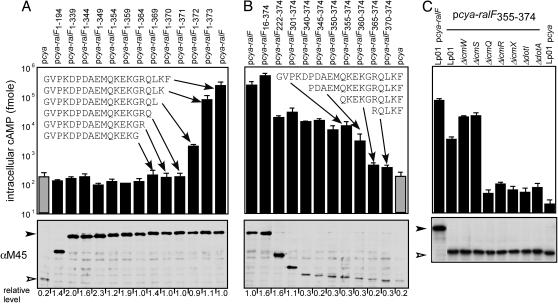
The C Terminus of RalF Contains an Amino Acid Sequence That Is Necessary and Sufficient for Dot/Icm-Mediated Protein Translocation. Domains in RalF required for protein translocation into host cells were determined by deletion analysis. These data show that deletion of RalF residues 372-374, a C-terminal deletion of only 3 aa, abolished RalF translocation into host cells (Fig. 3A, pcya-ralF1-371). Immunoblot analysis shows that the Cya-RalF fusion proteins were produced at similar levels in Legionella (Fig. 3A Lower), which means that the defect in cAMP production is directly related to the efficiency of protein translocation. These data indicate that truncation of three or more C-terminal residues from Cya-RalF eliminate translocation of this fusion protein into host cells by the Dot/Icm system. Consistent with the C-terminal domain of RalF being important for translocation, modifications of the RalF C terminus adversely affected translocation, as determined by recruitment of ARF1-GFP to Legionella-containing vacuoles (Table 1). These modifications included adding an epitope tag to the C terminus of RalF (ralF-M45) and a short C-terminal truncation (ralF1-339) of the native RalF protein. Taken together, these data indicate that the C-terminal region of RalF is necessary for substrate transfer by the Dot/Icm system.
Fig. 3.
Identification of a RalF translocation signal by deletion analysis. (A and B) CHO FcγRII cells were infected with Legionella producing either C-terminal truncations of Cya-RalF (A) or N-terminal RalF truncations fused to Cya (B). Protein translocation was measured by determining the intracellular cAMP levels for CHO FcγRII cells infected with Legionella (Lp01) harboring the plasmid indicated above the graph. Immunoblots shown below each graph correspond to Cya protein levels inside of the Legionella cells used for the infection. Scanning densitometry was used to determine the amount of each fusion protein produced relative to the amount of the Cya-RalF protein and these values are indicated below the immunoblot. Inserted in each graph are the C-terminal amino acid sequences for the indicated Cya fusion proteins. (C) Bacterial products required for translocation of Cya-RalF355-374 into CHO FcγRII cells were investigated by using the Legionella dot and icm mutants indicated above the graph. Immunoblots shown below each graph correspond to the steady-state levels of each Cya protein inside of the Legionella cells used for the infection. All cAMP values are the average ± SD for an experiment performed in triplicate.
Table 1. Modification of the RalF C terminus disrupts ARF1-GFP localization to the Legionella-containing vacuole.
| ARF1-GFP localization,* % of vacuoles positive
|
|||
|---|---|---|---|
| Strain | Plasmids | Average | SD |
| Lp01 | None | 23.8 | 6.3 |
| Lp01 ΔralF | Vector | 1.3 | 1.2 |
| Lp01 ΔralF | pralF | 42.1 | 6.4 |
| Lp01 ΔralF | pralF-M45 | 1.3 | 1.1 |
| Lp01 ΔralF | pralF1-339 | 0.0 | 0.0 |
| Lp01 ΔralF ΔdotA | pralF | 0.0 | 0.0 |
Mouse bone marrow-derived macrophages producing ARF1-GFP were infected with the indicated Legionella strains for 1 h, and GFP-ARF1 localization was assayed by fluorescence microscopy. Recruitment of ARF1-GFP to the Legionella-containing vacuoles was scored, and values are presented as the percent of vacuoles that stained positive for ARF1-GFP. Indicated are the average and SD for an experiment performed in triplicate in which a minimum of 50 vacuoles were scored for each sample.
To determine whether RalF contains a C-terminal signal sequence sufficient for Dot/Icm-mediated protein translocation, N-terminal deletions of RalF were fused to Cya, and translocation of these fusion proteins was measured. Although some of the N-terminal deletions in RalF resulted in reduced levels of protein translocation, significant levels of protein translocation were detected for most of the Cya-RalF N-terminal deletion proteins (Fig. 3B). Importantly, translocation was detected when the last 20 or 15 aa of RalF were appended to Cya (Fig. 3B). Fusion proteins containing the last 10 or 5 aa of RalF were insufficient for Cya translocation, despite being produced at levels similar to the Cya-RalF360-374 protein (Fig. 3B). By using the Cya-RalF355-374 construct, it was determined that the 20-aa C-terminal region of RalF was sufficient to direct Dot/Icm-dependent translocation of the Cya reporter into host cells (Fig. 3C). Thus, the last 20 amino acid residues of RalF contain a translocation signal that is necessary and sufficient for the delivery of bacterial proteins into host cells by the Dot/Icm system.
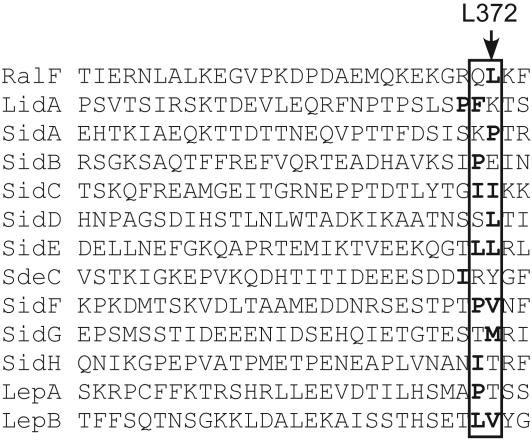
A Hydrophobic Residue Near the C Terminus Is Critical for RalF Translocation. Our data suggest a C-terminal amino acid sequence is important for translocation of substrate proteins by the Dot/Icm system. Attempts to align the C-terminal domains of all known Dot/Icm substrate proteins failed to identify a sequence or motif that is common to all substrates. However, a simple positional alignment of the C-terminal domains revealed that most of the substrate proteins had hydrophobic residues or a proline residue at the -3 or -4 positions in relation to the C-terminal amino acid (Fig. 4, boxed area). Additionally, a hydrophobic phenylalanine residue is located at the C-terminal -3 position of the MobA protein, which is a substrate of the RSF1010 DNA conjugation system that is also recognized as a substrate of the Dot/Icm system (19). RalF has a hydrophobic leucine residue at the -3 position in relation to the C terminus (Fig. 4, L372), and deletion analysis suggested that L372 is critical for RalF translocation (Fig. 3A, pcya-ralF1-371). To address whether L372 in RalF plays an important role in protein translocation by the Dot/Icm system, translocation efficiencies were compared for Cya-RalF fusion proteins with amino acid substitutions in RalF position 372. Substitutions to phenylalanine or proline (Table 2, L372F and L372P) had little effect on Cya-RalF translocation. Substitutions to small hydrophobic residues (Table 2, L372V and L372A) resulted in ≈5-fold reduction in Cya-RalF translocation. By contrast, substitutions to hydrophilic residues (Table 2, L372S and L372T) resulted in severe defects in Cya-RalF translocation. Because a 2-aa C-terminal truncation resulted in a significant decrease in Cya-RalF translocation, substitutions were made in the K373 amino acid at position -2 in RalF to determine whether a positive charge at this site is important for translocation. Unlike changes at L372, these data show that neither an alanine substitution nor charge alteration at the -2 position in RalF have a dramatic effect on the efficiency of Cya-RalF translocation (Table 2, K373A and K373E). These results indicate that RalF translocation by the Dot/Icm system requires a hydrophobic residue at the C-terminal -3 position and that the positive charge contributed by the K373 residue is not important.
Fig. 4.
A conserved location for C-terminal hydrophobic residues in Dot/Icm substrate proteins. C-terminal amino acid sequences of Dot/Icm substrate proteins are shown. Boxed are the -3 and -4 positions in relation to each protein's C terminus. Hydrophobic residues and proline residues within or adjacent to the boxed regions are bold. RalF L372 is depicted by an arrow.
Table 2. RalF amino acid L372 is important for Cya-RalF translocation into host cells.
| cAMP level,† fmol
|
|||
|---|---|---|---|
| RalF substitution* | Average | SD | Translocation efficiency,‡ % |
| Wild-type | 1.6 × 105 | 1.4 × 104 | 100 |
| L372F | 1.5 × 105 | 2.8 × 104 | 99 |
| L372P | 1.4 × 105 | 2.4 × 104 | 89 |
| L372V | 3.2 × 104 | 5.2 × 103 | 20 |
| L372A | 2.9 × 104 | 3.4 × 103 | 18 |
| L372S | 4.5 × 103 | 1.1 × 103 | 2.7 |
| L372T | 7.0 × 102 | 1.5 × 102 | 0.3 |
| K373A | 1.3 × 105 | 1.7 × 104 | 81 |
| K373E | 1.5 × 105 | 1.1 × 104 | 93 |
| K373R | 2.0 × 105 | 1.5 × 104 | 125 |
Levels of expression of these Cya-RalF fusion proteins in Legionella were not affected by these mutations (data not shown).
Average levels of cAMP and SD for an assay performed in triplicate is shown.
Translocation efficiency was calculated as relative amounts of intracellular cAMP detected for the indicated RalF protein compared levels detected for with the wild-type RalF protein.
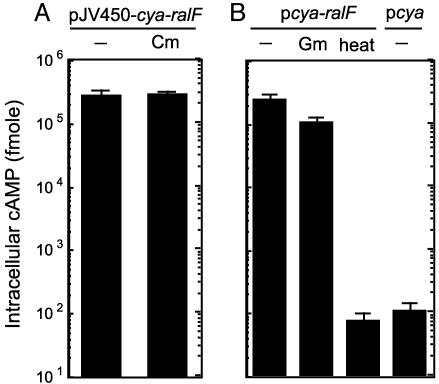
RalF Translocation Is Not Affected by Inhibitors of Protein Translation. The presence of a C-terminal translocation signal predicts that injection of the RalF protein by the Dot/Icm system would proceed only after translation of the protein has been completed. To determine whether RalF translocation requires ongoing bacterial protein synthesis, Legionella producing Cya-RalF were treated with chloramphenicol for 30 min before infection, and RalF translocation efficiencies were compared with untreated bacteria. These data show that chloramphenicol treatment had no effect on Cya-RalF translocation (Fig. 5A). Translocation efficiencies were also determined after Legionella producing Cya-RalF were treated with gentamicin, which irreversibly binds to ribosomes and inhibits translation. Treating Legionella with gentamicin (100 μg/ml) for 30 min resulted in an ≈1,400-fold reduction in bacterial viability, indicating that Legionella were highly susceptible to this concentration of gentamicin. Remarkably, when these gentamicin-treated Legionella were added to host cells, Cya-RalF translocation efficiencies were equivalent to those measured for untreated bacteria (Fig. 5B). As expected, heat-killed Legionella were unable to translocate Cya-RalF. These results are consistent with the Dot/Icm system, engaging and translocating RalF from cellular pools of completely synthesized protein, rather than translocation being linked to translation.
Fig. 5.
RalF translocation is not linked to protein translation. (A) Translocation of Cya-RalF (pJV450-cya-ralF) by wild-type Legionella (Lp01) was measured for untreated bacteria (-) and bacteria treated with chloramphenicol (Cm). Cya-RalF translocation was determined by measuring cAMP levels in host cells 1 h after infection. (B) Cya-RalF translocation was measured for Legionella that were untreated (-) or pretreated with gentamicin (Gm) or heat-inactivated at 80°C for 30 min (heat). Untreated Legionella producing Cya alone were used as a negative control to assess baseline levels of cAMP. All cAMP values are the average ± SD for an experiment performed in triplicate.
Discussion
Type IV secretion systems are used by many bacterial pathogens, presumably to translocate proteins that modulate host cell functions (31). The mechanisms by which substrate proteins are recognized and transported by type IV secretion systems are poorly understood. Here, we have examined translocation of the RalF protein into host cells by the Legionella Dot/Icm system. These data reveal several important features of the Dot/Icm translocation process, including requirements for host cell interaction and sequences within the RalF protein that are required for translocation.
It was determined that Legionella translocates Dot/Icm substrate proteins into host cells during the internalization process. Preventing Legionella uptake had no effect on delivery of the Cya-RalF protein into host cells. These data indicate that there must be a trigger that stimulates protein transfer by the Dot/Icm system that is encountered extracellularly. By initiating protein translocation before internalization, Legionella would be able to modulate host cell functions that are important for transporting plasma membrane-derived vacuoles along an endocytic pathway that results in fusion with lysosomes. Consistent with this idea, Dot/Icm-mediated modulation of host cell function before vacuole formation was apparent in fluorescent micrographs showing RalF-dependent recruitment of ARF1 to the sites of Legionella uptake. Thus, demonstration that Dot/Icm substrates are transferred during the internalization process sheds light on how Legionella are able to avoid endocytic fusion events that normally occur within minutes of bacterial internalization (12, 13).
A general theme that is emerging for type IV secretion system substrates is the presence of a C-terminal translocation signal (32-34). Here, we show that a 20-aa C-terminal region of the RalF protein is necessary and sufficient for translocation of proteins into host cell by the Legionella Dot/Icm system. Vergunst et al. (35) show that a C-terminal 20-aa region is sufficient for substrate translocation by the Agrobacterium tumefaciens VirB secretion system. Interestingly, a Cre fusion protein containing the RalF C terminus was not translocated into plant cells by A. tumefaciens (35), indicating that the RalF translocation domain is not recognized by the VirB system. Thus, it is likely that the C-terminal translocation domains of type IV substrate proteins have special features that allow specific recognition by their cognate secretion systems. Although the RalF C-terminal domain is clearly necessary and sufficient for substrate transfer by the Dot/Icm system, translocation of a Cya fusion protein containing only the last 20 aa of RalF was less efficient than translocation of the full-length RalF protein. This finding suggests that other domains in RalF may facilitate substrate recognition by the Dot/Icm system, which would increase the efficiency of substrate transfer. A possible candidate is the predicted integral membrane protein DotF, which was recently shown by two-hybrid analysis to interact with RalF (19).
Mutagenesis of the VirF translocation signal revealed arginine residues that are important for delivery of VirF into plant cells by the A. tumefaciens VirB system (35). One of the critical arginine residues in the VirF translocation signal is located at the -3 position in relation to the C-terminal amino acid. Unlike VirB substrates, we were unable to find a conserved arginine motif in Dot/Icm substrates by aligning the C-terminal domains of these proteins. However, similar to results from VirF deletion studies, our data indicate that the C-terminal -3 position in RalF is critical for type IV-mediated substrate translocation. Whereas a positively charged residue at the -3 position is important for VirF translocation, a hydrophobic residue at the -3 position was found to be important for RalF translocation. Although deletion analysis suggested a possible role for a lysine residue at position -2 in RalF, K373 substitutions to either a negatively charged amino acid or alanine did not affect Cya-RalF translocation dramatically. These data suggest that an electrostatic interaction mediated by the arginine at the -3 position in VirF is important for substrate recognition by the VirB system, whereas a hydrophobic interaction mediated by a leucine residue at the -3 position of RalF is important for substrate recognition by the Dot/Icm system. Our data indicate that RalF translocation by the Dot/Icm system is not coupled to protein translation, which is consistent with a model that predicts type IV secretion systems engage substrate proteins by recognizing a C-terminal translocation signal. Although chaperones may play an important role in substrate recognition, there is currently no evidence to suggest that type IV substrates are maintained in the bacterial cell in a partially unfolded state by interacting with chaperones. In the absence of chaperones, the prediction would be that these C-terminal translocation domains should be exposed and available for binding to components of the type IV secretion system. The RalF crystal structure has recently been solved (36), and, consistent with this prediction, these data show that the RalF C-terminal domain is located at the end of a long alpha helix comprised of amino acid residues 331-348 (Fig. 6, which is published as supporting information on the PNAS web site). This α-helix projects the C-terminal domain away from the globular domains of the folded RalF protein. Although disordered in the crystal structure, the RalF translocation signal consisting of amino acid residues 354-374 is exposed to solvent and accessible for interactions with other protein. From these data, we conclude that C-terminal domain of RalF is a translocation signal that is necessary for recognition by the Dot/Icm system. Future studies aimed at identifying proteins that interact with the C-terminal region of RalF should reveal key components of the Dot/Icm system that function in substrate recognition and initiate protein translocation into host cells.
Supplementary Material
Acknowledgments
We thank Dr. Guy Cornelis (Biozentrum, Universitat Basel, Basel) for providing pMS107, Dr. Annette Vergunst for communication of data before publication, and Dr. Shira Ninio for helpful discussions. This work was supported by National Institutes of Health Grants R01-AI41699 (to C.R.R.) and R01-GM61268 (to R.A.K.).
Author contributions: H.N., J.C.K., and C.R.R. designed research; H.N., E.D.C., and J.C.K. performed research; J.C.A. and R.A.K. contributed new reagents/analytic tools; H.N., J.C.K., and C.R.R. analyzed data; and H.N. and C.R.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office. Abbreviations: ARF, ADP ribosylation factor; Cya, adenylate cyclase.
References
- 1.Fields, B. S. (1996) Trends Microbiol. 4, 286-290. [DOI] [PubMed] [Google Scholar]
- 2.Fraser, D. W., Tsai, T. R., Orenstin, W., Parken, W. E., Beechan, H. J., Sharrar, R. G., Harris, J., Mallison, G. F., Martin, S. M., McDade, J. E., et al. (1977) N. Engl. J. Med. 297, 1189-1197. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz, M. A. & Silverstein, S. C. (1980) J. Clin. Invest. 66, 441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwitz, M. A. (1983) J. Exp. Med. 158, 2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horwitz, M. A. (1983) J. Exp. Med. 158, 1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwitz, M. A. & Maxfield, F. R. (1984) J. Cell Biol. 99, 1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson, M. S. & Isberg, R. R. (1995) Infect. Immun. 63, 3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagan, J. C. & Roy, C. R. (2002) Nat. Cell Biol. 4, 945-954. [DOI] [PubMed] [Google Scholar]
- 9.Coers, J., Monahan, C. & Roy, C. R. (1999) Nat. Cell Biol. 1, 451-453. [DOI] [PubMed] [Google Scholar]
- 10.Tilney, L. G., Harb, O. S., Connelly, P. S., Robinson, C. G. & Roy, C. R. (2001) J. Cell Sci. 114, 4637-4650. [DOI] [PubMed] [Google Scholar]
- 11.Kagan, J. C., Stein, M. P., Pypaert, M. & Roy, C. R. (2004) J. Exp. Med. 199, 1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiater, L. A., Dunn, K., Maxfield, F. R. & Shuman, H. A. (1998) Infect. Immun. 66, 4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy, C. R., Berger, K. & Isberg, R. R. (1998) Mol. Microbiol. 28, 663-674. [DOI] [PubMed] [Google Scholar]
- 14.Berger, K. H. & Isberg, R. R. (1993) Mol. Microbiol. 7, 7-19. [DOI] [PubMed] [Google Scholar]
- 15.Segal, G., Purcell, M. & Shuman, H. A. (1998) Proc. Natl. Acad. Sci. USA 95, 1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. (1998) Science 279, 873-876. [DOI] [PubMed] [Google Scholar]
- 17.Conover, G. M., Derre, I., Vogel, J. P. & Isberg, R. R. (2003) Mol. Microbiol. 48, 305-321. [DOI] [PubMed] [Google Scholar]
- 18.Chen, J., de Felipe, K. S., Clarke, M., Lu, H., Anderson, O. R., Segal, G. & Shuman, H. A. (2004) Science 303, 1358-1361. [DOI] [PubMed] [Google Scholar]
- 19.Luo, Z. Q. & Isberg, R. R. (2004) Proc. Natl. Acad. Sci. USA 101, 841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. (2002) Science 295, 679-682. [DOI] [PubMed] [Google Scholar]
- 21.Lippincott-Schwartz, J., Cole, N. B. & Donaldson, J. G. (1998) Histochem. Cell Biol. 109, 449-462. [DOI] [PubMed] [Google Scholar]
- 22.Sory, M. P. & Cornelis, G. R. (1994) Mol. Microbiol. 14, 583-594. [DOI] [PubMed] [Google Scholar]
- 23.Zuckman, D. M., Hung, J. B. & Roy, C. R. (1999) Mol. Microbiol. 32, 990-1001. [DOI] [PubMed] [Google Scholar]
- 24.Matthews, M. & Roy, C. R. (2000) Infect. Immun. 68, 3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews, H. L., Vogel, J. P. & Isberg, R. R. (1998) Infect. Immun. 66, 950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coers, J., Kagan, J. C., Matthews, M., Nagai, H., Zuckman, D. M. & Roy, C. R. (2000) Mol. Microbiol. 38, 719-736. [DOI] [PubMed] [Google Scholar]
- 27.Feeley, J. C., Gibson, R. J., Gorman, G. W., Langford, N. C., Rasheed, J. K., Mackel, D. C. & Blaine, W. B. (1979) J. Clin. Microbiol. 10, 437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Celada, A., Gray, P. W., Rinderknecht, E. & Schreiber, R. D. (1984) J. Exp. Med. 160, 55-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obert, S. O., Connor, R. J., Schmid, S. & Hearing, P. (1994) Mol. Cell. Biol. 14, 1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai, H. & Roy, C. R. (2001) EMBO J. 20, 5962-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cascales, E. & Christie, P. J. (2003) Nat. Rev. Microbiol. 1, 137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergunst, A. C., van Lier, M. C., den Dulk-Ras, A. & Hooykaas, P. J. (2003) Plant Physiol. 133, 978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vergunst, A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuink, T. J. & Hooykaas, P. J. (2000) Science 290, 979-982. [DOI] [PubMed] [Google Scholar]
- 34.Simone, M., McCullen, C. A., Stahl, L. E. & Binns, A. N. (2001) Mol. Microbiol. 41, 1283-1293. [DOI] [PubMed] [Google Scholar]
- 35.Vergunst, A. C., van Lier, M. C. M., den Dulk-Ras, A., Grosse Stüve, T. A., Ouwehand, A. & Hooykaas, P. J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amor, J. C., Swails, J., Roy, C. R., Nagai, H., Igmundson, A., Cheng, X. & Kahn, R. A. (November 1, 2004) J. Biol. Chem., 10.1074/jbc.M410820200.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.