SUMMARY
Autism spectrum disorder (ASD) is a heterogeneous disease, but genetically defined models can provide an entry point to studying the molecular underpinnings of this disorder. We generated germline mutant mice with loss-of-function mutations in Chd8, a de novo mutation strongly associated with ASD, and demonstrate that these mice display hallmark ASD behaviors, macrocephaly, and craniofacial abnormalities similar to patient phenotypes. Chd8+/− mice display a broad, brain-region specific dysregulation of major regulatory and cellular processes, most notably histone and chromatin modification, mRNA and protein processing, Wnt signaling, and cell cycle regulation. We also find altered synaptic physiology in medium spiny neurons of the nucleus accumbens. Perturbation of Chd8 in adult mice recapitulates improved acquired motor learning behavior found in Chd8+/− animals, suggesting a role for CHD8 in adult striatal circuits. These results support a mechanism linking chromatin modification to striatal dysfunction and the molecular pathology of ASD.
Graphical abstract
INTRODUCTION
Autism spectrum disorder (ASD) remains a poorly understood disease despite major recent advances in identifying risk alleles and associated symptoms. Sequencing-based studies have identified over 800 risk alleles, highlighting the genetic complexity of ASD (Abrahams and Geschwind, 2008; Iossifov et al., 2012; O’Roak et al., 2011; O’Roak et al., 2012a; O’Roak et al., 2012b; Parikshak et al., 2013). One approach to dissecting this complexity is to create mouse models that carry mutations that mirror those in patients (Nestler and Hyman, 2010; Silverman et al., 2010), providing an entry point to studying the impact of risk alleles identified through genome sequencing.
One of the genes most strongly associated with ASD is chromodomain helicase DNA-binding protein 8 (CHD8), which encodes an ATP-dependent chromatin remodeler (Bernier et al., 2014; Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012a; O’Roak et al., 2012b; Sanders et al., 2012; Talkowski et al., 2012; Zahir et al., 2007). The first evidence for its role in ASD was the identification of disruptive CHD8 mutations in two unrelated children with cognitive impairment and developmental delay (Zahir et al., 2007). Further investigation into balanced chromosomal abnormalities (Talkowski et al., 2012) and de novo exome sequencing of ASD patients suggested an important role for CHD8 in the brain (Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012a; O’Roak et al., 2012b; Sanders et al., 2012). Functional analysis using knockdown in human cells in vitro indicated that CHD8 regulates many ASD risk genes involved in neurodevelopment and synaptic function (Cotney et al., 2015; De Rubeis et al., 2014; Sugathan et al., 2014; Wilkinson et al., 2015). Based on these lines of evidence, Bernier et al. performed targeted resequencing of 3,730 children with ASD or developmental delay and proposed a subtype of ASD patients with mutations in CHD8 and specific phenotypes (Bernier et al., 2014). To gain insight into the role of CHD8 in the brain, we generated mice carrying Chd8 heterozygous loss-of-function (LOF) mutations, the predominant form found in ASD patients. Chd8+/− mice present with macrocephaly, craniofacial abnormalities, and behavioral deficits. Analysis of genome-wide CHD8 binding sites and brain-wide gene expression profiles shows brain region-specific enrichments for other ASD-associated genes as well as histone and chromatin modification, mRNA processing, protein folding, and cell cycle. We find a nucleus accumbens (NAc)-specific upregulation in Wnt signaling, highlighting the importance of CHD8 regulation in this brain region. Based on this regulatory profile and the observed behavioral phenotypes, we investigated the electrophysiology of medium spiny neurons (MSNs) within the NAc and observed a decrease in local inhibitory signaling coinciding with an increase in spontaneous excitatory activity. Finally, in vivo perturbation of Chd8 in the NAc of wild-type adult animals recapitulates the acquired motor learning phenotype found in Chd8+/− mice, linking striatal circuits to the observed phenotypes. These data provide insight into the role of CHD8 in the brain as well as its contribution to ASD.
RESULTS
Generation of a Chd8 LOF mutant mouse via Cas9-mediated germline editing
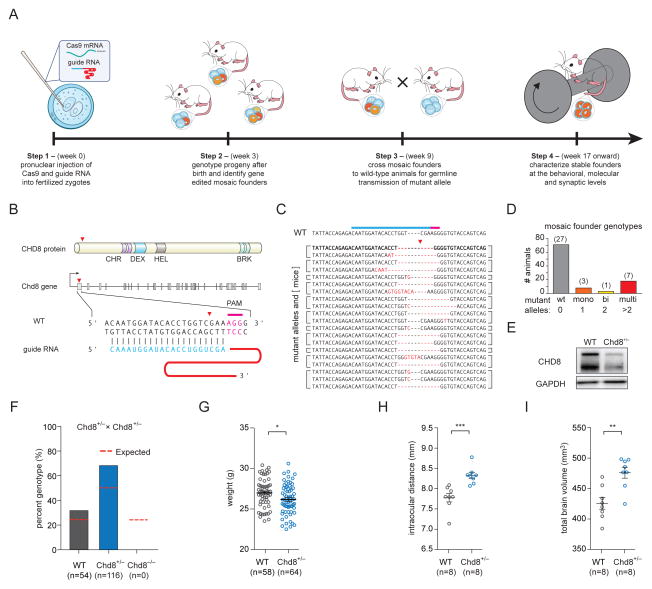
To study the role of CHD8 expression in the brain, we generated germline mutant mice using Cas9 (Figure 1A). We designed three single guide RNAs (sgRNAs) targeting the Chd8 gene and tested their efficiency by transient transfection in mouse N2A cells followed by insertion and deletion (indel) analysis (Figure 1B and Figure S1). The optimal sgRNA was identified, co-injected with Cas9 mRNA into the pronucleus of C57BL/6 single cell zygotes, and implanted into recipient mothers at the two-cell stage (Wang et al., 2013). Progeny were born with a variety of mutant alleles harboring indels at the target site (Figure 1C). Genotyping tail tissue revealed that individual animals had between 1 and 4 unique alleles, indicating that the first generation of gene edited progeny were mosaic. These animals likely resulted from unique editing events after division of the single cell zygote. To characterize the distribution of genotypes, animals were classified as having zero (wild-type, n = 27), one (monoallelic, n = 3), two (biallelic, n = 1), or more than two (multiallelic, n = 7) mutant allele(s) (Figure 1D and Figure S1).
Figure 1. Generation and morphological characterization of germline mutant Chd8+/− mice.
(A) Workflow for generating and characterizing germline edited mice.
(B) Chd8 targeting strategy and sgRNA design. Also see Figure S1.
(C) Sequences of Chd8 edited alleles in mosaic founders. Data represent all mutant alleles found within 38 animals from 6 recipient mothers. Bolded Chd8 allele with 7-nt deletion represents LOF mutation in established mouse line used for all phenotypic characterizations.
(D) Classification and quantification of Chd8-targeted mosaic founder genotypes. Mice were classified as having zero (WT, n = 27), one (monoallelic, n = 3), two (biallelic, n = 1), or >2 (multiallelic, n = 7) mutant allele(s). Also see Figure S1.
(E) CHD8 protein expression in whole brain lysates from wild-type and Chd8+/− embryonic day 18 mice, showing reduced expression [WT (n = 3) 100 ± 4 % SEM; Chd8+/− (n = 4) 71 ± 3 % SEM] in heterozygous mutant mice. Each lane was loaded with 5 μg of protein with GAPDH as loading control.
(F) Plot of expected (red dashed line) versus actual genotype ratios demonstrating homozygous null animals are embryonic lethal. Also see Figure S2.
(G) Weights of 10-week old male Chd8+/− mice compared to wild-type littermates [WT (n = 58) 26.9 ± 0.2 g SEM; Chd8+/− (n = 64) 26.1 ± 0.2 g SEM, two-tailed t-test p-value = 0.016].
(H) Intraocular distances of 10-week old male Chd8+/− mice compared to wild-type littermates [WT (n = 8) 7.8 ± 0.1 mm SEM; Chd8+/− (n = 8) 8.33 ± 0.08 mm SEM, two-tailed t-test p-value = 0.001].
(I) Total brain volume of 10-week old male Chd8+/− mice compared to wild-type littermates [WT (n = 8) 430 ± 10 mm3 SEM; Chd8+/− (n = 8) 476 ± 9 mm3 SEM, two-tailed t-test p-value = 0.002].
Mutations in CHD8 identified in patients are most often LOF, and therefore we reasoned that a Cas9-mediated indel causing a frameshift mutation within an early constitutive exon would be sufficient to disrupt protein expression. To establish a mouse line with a single, germline transmitted LOF mutation in Chd8, we crossed all of the first generation Chd8 mutant progeny (n = 11) to wild-type mice. Within each resulting litter at least one progeny harbored a mutant allele identified within the parent. We also identified new alleles not found in the tail snips of parents. Therefore, we refer to the first generation of germline edited progeny as ‘mosaic founders’ to distinguish them from true founders with germline transmission of a single unique allele. One founder with germline transmission of a Chd8 allele containing a 7-nucleotide deletion in exon 1 that causes a frameshift mutation (Figure 1C, bold sequence) was selected to establish the Chd8+/− strain, which was used for all further analyses. Heterozygous mice showed approximately half the expression of CHD8 as compared to wild-type littermates (Figure 1E). As expected, homozygous mutant animals (Chd8−/−) were not viable (Figure 1F) (Nishiyama et al., 2004). Chd8+/− mice were viable and fertile but had reduced body size [WT (n = 58) 26.9 ± 0.2 g SEM; Chd8+/− (n = 64) 26.1 ± 0.2 g SEM, two-tailed t-test p-value = 0.016](Figure 1G and Figure S2A). Taken together our results demonstrate that Cas9-mediated zygote editing results in mosaic founders with potential for germline transmission of Chd8 LOF alleles, and with additional crosses single alleles can be selected.
Macrocephaly and abnormal craniofacial features in Chd8+/− mice
CHD8 mutant patients frequently exhibit macrocephaly and craniofacial abnormalities (Bernier et al., 2014). To determine whether Chd8+/− mice recapitulate similar features, we utilized ex vivo high-resolution brain magnetic resonance imaging (MRI) to assess intraocular distance and total brain volume in 10-week old males. We found an increase in both intraocular distance [WT (n = 8) 7.8 ± 0.1 mm SEM; Chd8+/− (n = 8) 8.33 ± 0.08 mm SEM, two-tailed t-test p-value = 0.001](Figure 1H) as well as total brain volume [WT (n = 8) 430 ± 10 mm3 SEM; Chd8+/− (n = 8) 476 ± 9 mm3 SEM, two-tailed t-test p-value = 0.002](Figure 1I) in Chd8+/− mice compared to wild-type littermates. Thus, Chd8+/− mice recapitulate these patient-like morphological phenotypes.
CHD8 is expressed in most cell types throughout the brain
To understand the function of CHD8, we first determined the developmental expression profile of CHD8 protein in wild-type C57BL/6 mouse brain by performing western blots on whole brain samples collected throughout embryonic development (E11.5-E19.5) as well as in neonates (P0) and adults (Figure S2B). We found that CHD8 protein was expressed strongly during embryonic development, but also remains observable in neonates and adults (Figure S2B–C). These results are consistent with results from human and macaque brains (Bernier et al., 2014).
To determine whether CHD8 expression was limited to a specific cell type, we prepared sections from adult (10-week old males) Chd8+/− mice and wild-type littermates. Immunofluorescence imaging shows that CHD8 expression is punctate and localized within the nucleus of almost every cell (CHD8 and DAPI positive) (Figure S2C–E). Specifically, we find that CHD8 is expressed in mature neurons (NeuN positive), interneurons (parvalbumin (PV) positive), oligodendrocytes (CNP1 positive), and astrocytes (GFAP positive) (Figure S2D–E). Interestingly, CHD8 is expressed in most, but not all DAPI and NeuN positive cells, suggesting a cellular population or a dynamic state in which CHD8 is not expressed.
CHD8 affects pathways involved in cell cycle as well as histone and chromatin modification
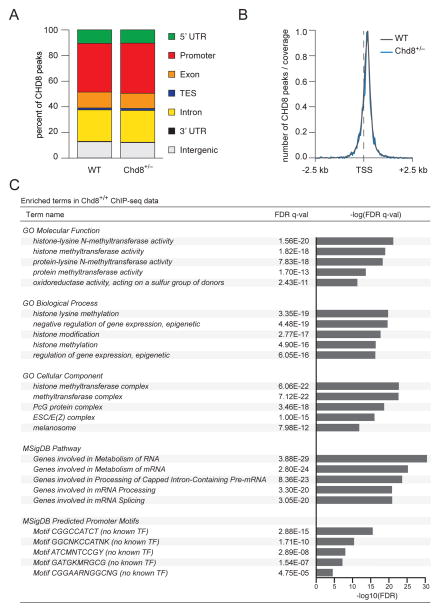
CHD8 is a chromatin modifier and transcription factor that has been shown to bind to ~2000 transcriptionally active genes in stable cell lines (Subtil-Rodriguez et al., 2014). It therefore may regulate downstream genes associated with ASD risk. In support of this possibility, transcriptional profiling of human neural progenitor cells in which CHD8 was knocked down identified several dysregulated ASD-associated genes as well as other genes associated with synapse and brain development (Cotney et al., 2015; Sugathan et al., 2014; Wilkinson et al., 2015). We investigated the genome-wide binding profile of CHD8 in the cortex of 10-week old male mice to identify likely target genes as well as dysregulated pathways. As a control both ChIP-seq inputs and IgG conditions on age and genotype tissue were utilized. Peaks were called for each genotype-control pair using MACS2 (FDR cutoff of 1%) and only peaks shared between input and IgG conditions were considered. We found that CHD8 binding sites are enriched in promoters (38% of all CHD8 peaks) (Figure 2A) with peaks centered on the transcriptional start site (TSS) (Figure 2B). These observations were consistent in both Chd8+/− mice and wild-type littermates. A Gene Ontology (GO) enrichment analysis of CHD8 binding sites in wild-type mice shows enrichment of numerous and diverse terms (Figure 2C). Most notably, terms with the lowest FDR consistently show enrichment for histone and chromatin modification as well as alterations in mRNA and protein processing.
Figure 2. ChIP-seq of adult Chd8+/− cortex shows peak enrichment near transcriptional start sites and in critical cellular pathways.
(A) Bar chart of CHD8 binding peaks as a percentage of total peaks showing CHD8 primarily binds in promoters (WT 38 %; Chd8+/− 39 %) for both genotypes. Replicate somatosensory cortices from wild-type (n = 2) and Chd8+/− mice (n = 2) were microdissected and used for ChIP-seq. ChIP-Seq controls were both input and IgG for each genotype. Peaks were called for each genotype and each control using MACS2 (FDR cutoff of 1%). Only peaks shared between input and IgG for a particular genotype were considered. Annotations were made using HOMER with the mouse mm10 genome assembly and annotation.
(B) Histogram of CHD8 peaks around the transcription start site (TSS). Distance from TSS was calculated using HOMER with the mouse mm10 genome assembly and annotation.
(C) Functional interpretation and gene ontology of CHD8 peaks in Chd8+/+ cortex using GREAT(McLean et al., 2010). Enriched terms for Molecular Function, Biological Process, Cellular Component, MSigDB Pathway, and MSigDB Predicted Promoter Motifs are shown. Also see Figure S3.
Cortical lamination, major cell types, and late stage cortical progenitor number in the somatosensory cortex do not vary between Chd8+/− and wild-type mice
CHD8 binding site enrichments suggest alterations in cell cycle and cortical development. CHD8 binds to other ASD-associated genes that are major orchestrators of cortical development, namely Ctnnb1 (beta-catenin), Ankrd11, Foxg1, and BAF complex members Arid1a and Bcl11b. The observed macrocephaly (Figure 1I) along with alterations in cell cycle and the dysregulation of master regulators of cortical development led us to test whether a reduction in CHD8 disrupts lamination and specification of cortical neuron subtypes. We investigated morphology and major cell types within the cortex by examining immunostained brain sections collected from 21-day old male mice. Morphological analysis using Nissl staining shows no overt phenotype present in the somatosensory cortex of Chd8+/− mice compared to wild-type littermates (Figure S3A–B). Upon immunostaining for Cux1, a marker for layer II/III/IV projection neurons, and Bcl11b, a marker for layer V/VI projection neurons, we observed no significant differences between Chd8+/− mice and wild-type littermates within the somatosensory cortex (Figure S3B). Similarly, immunostaining for parvalbumin (PV) positive interneurons and Olig2 positive oligodendrocytes showed no overt differences within the somatosensory cortex of Chd8+/− mice compared to wild-type littermates (Figure S3B).
We next tested whether a reduction in CHD8 resulted in defects within the cortical progenitor population. In particular, we examined both the number of cortical progenitors as well as the cell cycle length in embryonic day 15.5 (E15.5) embryos. We performed intraperitoneal injections of BrdU and Edu in pregnant dams 120 and 30 minutes prior to euthanasia (Figure S3C) (Mairet-Coello et al., 2012; Watanabe et al., 2015). Examination of brain sections showed no increase in the number of cortical progenitor cells as measured by BrdU incorporation within the somatosensory cortex of Chd8+/− mice compared to wild-type littermates [WT (n = 6) 240 ± 10 BrdU+ cells SEM; Chd8+/− (n = 6) 241 ± 9 BrdU+ cells SEM, two-tailed t-test p-value = 0.990](Figure S3D). Finally, we examined whether CHD8 was altering the cell cycle length, as would be predicted from previous studies as well as enrichment for cell cycle regulation genes enriched in CHD8 peaks. However, examination of brain sections showed no increase in either the total cell cycle length [WT (n = 6) 9.4 ± 0.5 hours SEM; Chd8+/− (n = 6) 10.3 ± 0.1 hours SEM, two-tailed t-test p-value = 0.107](Figure S3E) or the length of S-phase [WT (n = 6) 2.8 ± 0.1 hours SEM; Chd8+/− (n = 6) 3.0 ± 0.3 hours SEM, two-tailed t-test p-value = 0.430](Figure S3F) within the somatosensory cortex of Chd8+/− mice compared to wild-type littermates. Taken together, these data suggest that reduction in CHD8 in mice results in no gross defects in specification, migration or lamination of different subtypes in the neocortex.
Chd8+/− mice exhibit broad gene expression changes throughout the brain
Considering CHD8 is not only expressed in cortex but throughout the brain in most cell types we set out to characterize the brain-wide transcriptional changes resulting in a decrease of CHD8 in an unbiased way. We performed RNA sequencing on microdissected tissue from 10-week old males. We investigated brain regions previously implicated in ASD, namely the medial prefrontal cortex (mPFC), dorsal striatum (DS), NAc, ventral tegmental area (VTA), hippocampal formation (HPF), amygdala (AM), and lateral hypothalamus (LH).
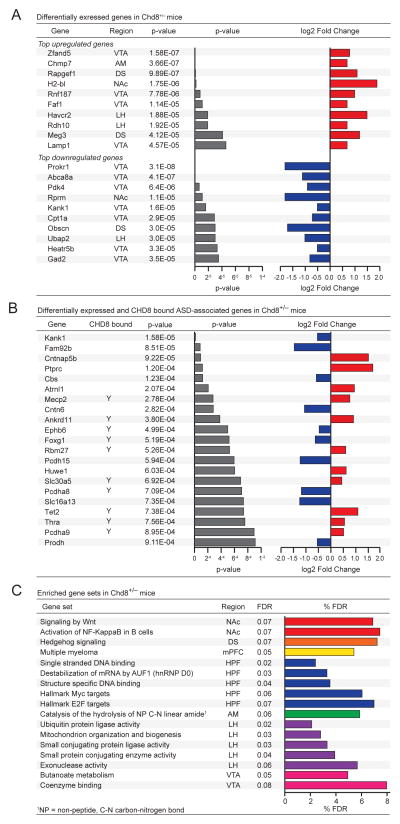
We performed differential expression analysis within each region and found brain region-specific dysregulation in Chd8+/− mice compared to wild-type littermates (Figure S4A). The top 10 up- and down-regulated genes irrespective of brain region are shown (Figure 3A). Dysregulated genes found within more than one region are Eif2b5 (DS and AM), Nbl1 (NAc and LH), Mgp (HPF and AM). Select differentially expressed genes within the NAc were validated by RT-qPCR (Figure S4B). Among the differentially expressed genes we find genes previously associated with ASD, some of which are well characterized causal mutations while others are uncharacterized (Figure 3B). In particular, we find transcription factors essential for the development of the brain (i.e., FoxG1), global regulators of the epigenome (i.e., Mecp2 and Tet2), as well as many neuronal and synaptic adhesion molecules, such as Kank1, Cntnap5b, Cntn6, Ankrd11, Pcdh15, Pcdha8, and Pcdha9. Many of these genes are also directly bound by CHD8 (Figure 3B). To identify entire pathways dysregulated in Chd8+/− mice, we performed gene set enrichment analysis (GSEA)(Figure 3C). Consistent with previous reports that CHD8 acts as a negative regulator of Wnt signaling, we identified a NAc-specific positive enrichment for Wnt signaling.
Figure 3. RNA-seq of adult Chd8+/− brain regions shows globally dysregulated genes and pathways.
(A) Table of the top 10 upregulated (top) and downregulated (bottom) differentially expressed genes. Differential expression analysis using DEseq2 was performed on a TPM expression matrix from RNA sequencing libraries generated from different brain regions when comparing 10-week old male Chd8+/− mice and wild-type littermates. Also see Figure S4.
(B) Table of differentially expressed ASD-associated genes in different brain regions when comparing 10-week old male Chd8+/− mice and wild-type littermates. Genes bound by CHD8 are denoted with Y.
(C) Enriched gene sets in Chd8+/− mice. Gene set enrichment analysis was performed on RNA sequencing libraries generated from different brain regions when comparing 10-week old male Chd8+/− mice and wild-type littermates. Table of enriched gene sets with FDR below 8%.
Chd8+/− mice exhibit synaptic dysfunction within MSNs in the NAc
The observations that Wnt signaling and synaptic adhesion molecules are dysregulated in the NAc prompted us to further characterize the role of CHD8 in this region. To study whether mutation of Chd8 results in altered synaptic transmission, we assayed several electrophysiological parameters of MSNs in the core region of the NAc by whole cell slice recording utilizing aged-matched littermates between 6~8 weeks old.
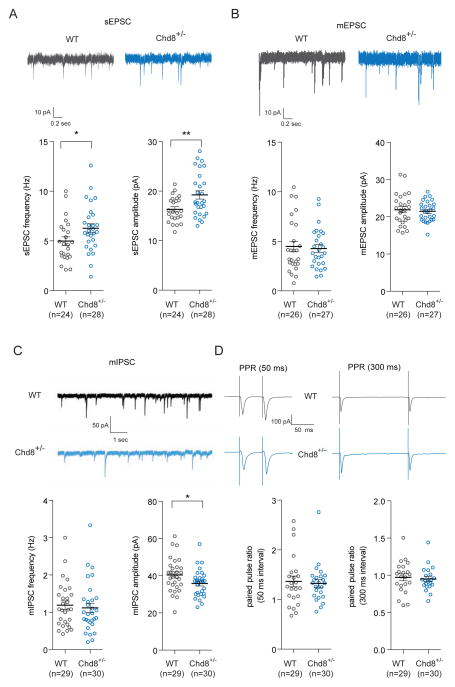
First, we recorded spontaneous excitatory postsynaptic current (sEPSC) and observed an increase in sEPSC frequency [WT (n = 24) 5.0 ± 0.4 Hz SEM; Chd8+/− (n = 28) 6.3 ± 0.5 Hz SEM, two-tailed t-test p-value = 0.048] and amplitude [WT (n = 24) 16.3 ± 0.5 pA SEM; Chd8+/− (n = 28) 19.2 ± 0.8 pA SEM, two-tailed t-test p-value = 0.006] in Chd8+/− mice compared to wild-type littermates (Figure 4A). These results suggest that excitatory inputs onto MSNs of NAc is enhanced. Then, we measured miniature excitatory postsynaptic current (mEPSC) and observed neither an increase in frequency [WT (n = 26) 4.5 ± 0.5 Hz SEM; Chd8+/− (n = 27) 4.3 ± 0.4 Hz SEM, two-tailed t-test p-value = 0.760] nor amplitude [WT (n = 26) 21.9 ± 0.8 pA SEM; Chd8+/− (n = 27) 21.5 ± 0.5 pA SEM, two-tailed t-test p-value = 0.674] in Chd8+/− mice compared to wild-type littermates (Figure 4B).
Figure 4. Chd8 mutation leads to striatal dysfunction.
(A) (Top) Representative sEPSC traces from MSNs in the core region of the NAc of Chd8+/− mice and wild-type littermates. Chd8+/− mice displayed both an increase in (Left) sEPSC frequency [WT (n = 24) 5.0 ± 0.4 Hz SEM; Chd8+/− (n = 28) 6.3 ± 0.5 Hz SEM, two-tailed t-test p-value = 0.048] as well as (Right) sEPSC amplitude [WT (n = 24) 16.3 ± 0.5 pA SEM; Chd8+/− (n = 28) 19.2 ± 0.8 pA SEM, two-tailed t-test p-value = 0.006] compared to wild-type littermates.
(B) (Top) Representative mEPSC traces from MSNs in the core region of the NAc of Chd8+/− mice and wild-type littermates. Chd8+/− mice had no difference in either (Left) mEPSC frequency [WT (n = 26) 4.5 ± 0.5 Hz SEM; Chd8+/− (n = 27) 4.3 ± 0.4 Hz SEM, two-tailed t-test p-value = 0.760] or (Right) mEPSC amplitude [WT (n = 26) 21.9 ± 0.8 pA SEM; Chd8+/− (n = 27) 21.5 ± 0.5 pA SEM, two-tailed t-test p-value = 0.674] compared to wild-type littermates.
(C) (Top) Representative mIPSC traces from MSNs in the core region of the NAc of Chd8+/− mice and wild-type littermates. Chd8+/− mice had no increase in (Left) mIPSC frequency [WT (n = 29) 1.2 ± 0.1 Hz SEM; Chd8+/− (n = 30) 1.1 ± 0.1 Hz SEM, two-tailed t-test p-value = 0.663] but did have a decrease in (Right) mIPSC amplitude [WT (n = 29) 40 ± 2 pA SEM; Chd8+/− (n = 30) 36 ± 1 pA SEM, two-tailed t-test p-value = 0.036] compared to wild-type littermates.
(D) (Top) Representative paired-pulse ratio traces. No difference was observed between PPR for intervals of (Left) 50 ms [WT (n = 23) 1.4 ± 0.1 PPR SEM; Chd8+/− (n = 25) 1.3 ± 0.1 PPR SEM, two-tailed t-test p-value = 0.755] or (Right) 300 ms [WT (n = 22) 0.97 ± 0.05 PPR SEM; Chd8+/− (n = 21) 0.95 ± 0.04 PPR SEM, two-tailed t-test p-value = 0.711] in Chd8+/− mice compared to wild-type littermates.
To further study the synaptic properties of MSNs, we measured inhibitory synaptic transmission onto MSNs of the NAc. We observed no difference in the frequency of miniature inhibitory postsynaptic current (mIPSC) in MSNs between genotypes [WT (n = 29) 1.2 ± 0.1 Hz SEM; Chd8+/− (n = 30) 1.1 ± 0.1 Hz SEM, two-tailed t-test p-value = 0.663]. However, we observed a decrease in the amplitude of mIPSC [WT (n = 29) 40 ± 2 pA SEM; Chd8+/− (n = 30) 36 ± 1 pA SEM, two-tailed t-test p-value = 0.036] in Chd8+/− mice compared to wild-type littermates (Figure 4C). To further investigate whether cortical inputs and presynaptic components contribute to the increased excitatory inputs onto MSNs in the NAc, we examined the paired pulse ratio (PPR) between EPSCs using parasagittal slice preparation as demonstrated previously (Rothwell et al., 2014). We observed no difference in PPR with an interval of 50 ms [WT (n = 23) 1.4 ± 0.1 PPR SEM; Chd8+/− (n = 25) 1.3 ± 0.1 PPR SEM, two-tailed t-test p-value = 0.755] or 300 ms [WT (n = 22) 0.97 ± 0.05 PPR SEM; Chd8+/− (n = 21) 0.95 ± 0.04 PPR SEM, two-tailed t-test p-value = 0.711] in Chd8+/− mice compared to wild-type littermates (Figure 4D). These results suggest that a local decrease of inhibitory transmission may contribute to the enhanced excitatory inputs onto MSNs in the NAc in Chd8+/− mice.
Chd8+/− mice display ASD-like behavioral phenotypes
To examine whether Chd8+/− mice manifest phenotypic outcomes relevant to diagnostic symptoms found in patients with ASD, such as anxiety, repetitive behavior and impaired social interactions, we performed a panel of well-characterized behavioral assays with age-matched wild-type and Chd8+/− littermates.
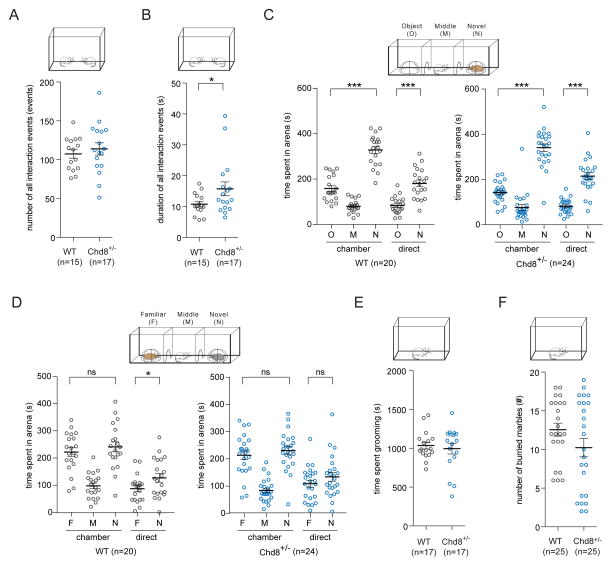
We first performed social behavioral tests of Chd8+/− mice at an early developmental stage using the juvenile social play paradigm. We utilized age-and gender-matched Chd8+/− and wild-type littermate pairs at postnatal day 23~25 as previous described (Bolivar et al., 2007; McFarlane et al., 2008; Yang et al., 2009). We found no difference in the total number of all interactive events between genotypes [WT (n = 15) 107 ± 6 events SEM; Chd8+/− (n = 17) 114 ± 8 events SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure 5A). However, we observed an increase in the total duration of all interactive events between in Chd8+/− mice compared to wild-type littermates [WT (n = 15) 71 ± 4 s SEM; Chd8+/− (n = 17) 90 ± 10 s SEM, one-way ANOVA with Bonferroni post hoc test p-value = 0.011](Figure 5B). By categorizing different patterns of reciprocal play behaviors, we found no difference in the total number or duration of any specific behavior between genotypes: nose-to-nose events [WT (n = 15) 28 ± 2 events SEM; Chd8+/− (n = 17) 27 ± 2 events SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5A), nose-to-anogenital sniffing events [WT (n = 15) 32 ± 2 events SEM; Chd8+/− (n = 17) 33 ± 3 events SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5B), following behavior events [WT (n = 15) 10 ± 2 events SEM; Chd8+/− (n = 17) 15 ± 4 events SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5C), direct interaction events [WT (n = 15) 37 ± 2 events SEM; Chd8+/− (n = 17) 37 ± 3 events SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5D), nose-to-nose duration [WT (n = 15) 10.8 ± 0.9 s SEM; Chd8+/− (n = 17) 16 ± 2 s SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5E), nose-to-anogenital sniffing duration [WT (n = 15) 18 ± 2 s SEM; Chd8+/− (n = 17) 21 ± 3 s SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5F), following behavior duration [WT (n = 15) 6.0 ± 0.8 s SEM; Chd8+/− (n = 17) 13 ± 4 s SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5G), direct interaction duration [WT (n = 15) 36 ± 2 s SEM; Chd8+/− (n = 17) 41 ± 5 s SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05](Figure S5H).
Figure 5. Chd8+/− mice display a mild defect in social behavior and normal repetitive behaviors.
(A) During juvenile social play we observed no difference in the total number of all interactive events between Chd8+/− and wild-type littermate mouse pairs [WT (n = 15) 107 ± 6 events SEM; Chd8+/− (n = 17) 114 ± 8 events SEM, one-way ANOVA with Bonferroni post hoc test p-value > 0.05]. Also see Figure S5.
(B) During juvenile social play we observed an increase in the total duration of all interactive events between Chd8+/− and wild-type littermate mouse pairs [WT (n = 15) 71 ± 4 s SEM; Chd8+/− (n = 17) 90 ± 10 s SEM, one-way ANOVA with Bonferroni post hoc test p-value = 0.011].
(C) (Top) Sociability test of the three-chambered social approach task showing both (Left) wild-type and (Right) Chd8+/− mice display significant preference for the novel mouse compared to the novel object [WT (n = 20) 160 ± 10 (novel object chamber, O) 80 ± 6 (middle chamber, M) 330 ± 20 (novel mouse chamber, N) 84 ± 8 (novel object direct interaction, O) 180 ± 10 (novel mouse direct interaction, N) s SEM. One-way ANOVA with Bonferroni post hoc test: novel object versus novel mouse chamber p-value <0.001, novel object versus novel mouse direct interaction p-value < 0.001; Chd8+/− (n = 24) 140 ± 9 (novel object chamber, O) 80 ± 10 (middle chamber, M) 340 ± 20 (novel mouse chamber, N) 82 ± 6 (novel object direct interaction, O) 220 ± 10 (novel mouse direct interaction, N) s SEM. One-way ANOVA with Bonferroni post hoc test: novel object versus novel mouse chamber p-value <0.001, novel object versus novel mouse direct interaction p-value < 0.001]. Also see Figure S5.
(D) (Top) Social novelty test of the three-chambered social approach task showing (Left) wild-type but not (Right) Chd8+/− mice display significant preference for the novel mouse compared to the familiar mouse [WT (n = 20) 220 ± 20 (familiar mouse chamber, F) 100 ± 10 (middle chamber, M) 240 ± 20 (novel mouse chamber, N) 90 ± 10 (familiar mouse direct interaction, F) 130 ± 20 (novel mouse direct interaction, N) s SEM. One-way ANOVA with Bonferroni post hoc test: familiar versus novel mouse chamber p-value > 0.05, familiar versus novel mouse direct interaction p-value = 0.040; Chd8+/− (n = 24) 210 ± 20 (familiar mouse chamber, F) 84 ± 8 (middle chamber, M) 230 ± 10 (novel mouse chamber, N) 110 ± 10 (familiar mouse direct interaction, F) 130 ± 20 (novel mouse direct interaction, N) s SEM. One-way ANOVA with Bonferroni post hoc test: familiar versus novel mouse chamber p-value = 0.988, familiar versus novel mouse direct interaction p-value = 0.292].
(E) Quantification of self-grooming events during a one-hour period showed no difference in grooming behavior between genotypes [WT (n = 17) 1040 ± 40 s SEM; Chd8+/− (n = 17) 1000 ± 70 s SEM, two-tailed t-test p-value = 0.637].
(F) In the marble burying test, Chd8+/− and wild-type littermates showed no difference in the number of marbles buried [WT (n = 23) 12.6 ± 0.8 marbles SEM; Chd8+/− (n = 25) 10 ± 1 marbles SEM, two-tailed t-test p-value = 0.125].
We then performed the three-chambered social approach task with age-matched Chd8+/− mice and wild-type. As previously described, the three-chambered assay involves habituation, a sociability test, and a social novelty (Chadman et al., 2008; Chao et al., 2010; McFarlane et al., 2008; Moy et al., 2004; Silverman et al., 2010; Yang et al., 2009). No preference for either side of the test apparatus was observed for mice in either group during the habituation phase (Figure S5I–J). In the sociability test of the three-chambered assay, we counterbalanced the object and social sides for each experimental mouse, and experimental mice were given free access to interact with a novel mouse or a novel object. During this phase, both Chd8+/− mice and wild-type littermates displayed significant preference for the novel mouse compared to the novel object [WT (n = 20) 160 ± 10 (novel object chamber) 80 ± 6 (middle chamber) 330 ± 20 (novel mouse chamber) 84 ± 8 (novel object direct interaction) 180 ± 10 (novel mouse direct interaction) s SEM. One-way ANOVA with Bonferroni post hoc test: novel object versus novel mouse chamber p-value <0.001, novel object versus novel mouse direct interaction p-value < 0.001; Chd8+/− (n = 24) 140 ± 9 (novel object chamber) 80 ± 10 (middle chamber) 340 ± 20 (novel mouse chamber) 82 ± 6 (novel object direct interaction) 220 ± 10 (novel mouse direct interaction) s SEM. One-way ANOVA with Bonferroni post hoc test: novel object versus novel mouse chamber p-value <0.001, novel object versus novel mouse direct interaction p-value < 0.001](Figure 5C). In the social novelty test phase, experimental mice were given free access to interact with a novel mouse or a familiar mouse. In this phase, wild-type littermates but not Chd8+/− mice displayed significant preference for the novel mouse compared to the familiar mouse [WT (n = 20) 220 ± 20 (familiar mouse chamber) 100 ± 10 (middle chamber) 240 ± 20 (novel mouse chamber) 90 ± 10 (familiar mouse direct interaction) 130 ± 20 (novel mouse direct interaction) s SEM. One-way ANOVA with Bonferroni post hoc test: familiar versus novel mouse chamber p-value > 0.05, familiar versus novel mouse direct interaction p-value = 0.040; Chd8+/− (n = 24) 210 ± 20 (familiar mouse chamber) 84 ± 8 (middle chamber) 230 ± 10 (novel mouse chamber) 110 ± 10 (familiar mouse direct interaction) 130 ± 20 (novel mouse direct interaction) s SEM. One-way ANOVA with Bonferroni post hoc test: familiar versus novel mouse chamber p-value = 0.988, familiar versus novel mouse direct interaction p-value = 0.292](Figure 5D). We observed no difference in locomotion, as measured by the total number of entries into the side chambers, between wild-type littermates and Chd8+/− mice during both the sociability test [WT (n = 20) 41 ± 4 entries SEM; Chd8+/− (n = 24) 40 ± 4 entries SEM, two-tailed t-test p-value = 0.889](Figure S5K–M) and the social novelty test [WT (n = 20) 52 ± 6 entries SEM; Chd8+/− (n = 24) 42 ± 4 s SEM, two-tailed t-test p-value = 0.131](Figure S5N–P). These results indicate that adult Chd8+/− mice show a mild deficit in social interaction behavior in the social novelty but not the sociability test of the three-chambered social approach task.
To assess the repetitive behavior of Chd8+/− mice, we utilized two common behavioral paradigms investigating grooming and burying behavior. To assess self-grooming behavior, experimental animals were placed inside the test chamber for a one-hour period and we did not observe a difference in the total grooming time between genotypes [WT (n = 17) 1040 ± 40 s SEM; Chd8+/− (n = 17) 1000 ± 70 s SEM, two-tailed t-test p-value = 0.637](Figure 5E). Moreover, no skin lesions were found on any Chd8+/− mouse or wild-type littermate for the duration of our study. In the marble burying assay, which takes place in a novel testing cage with 24 marbles introduced, we did not observe a difference in the number of marbles buried between genotypes [WT (n = 23) 12.6 ± 0.8 marbles SEM; Chd8+/− (n = 25) 10 ± 1 marbles SEM, two-tailed t-test p-value = 0.125](Figure 5F). Together, these results suggest that Chd8+/− mice do not display repetitive grooming or burying behaviors.
To assess the memory performance of Chd8+/− mice, we conducted a memory test by subjecting mice to either a contextual or toned fear conditioning task. During the training phase, Chd8+/− mice exhibited a similar percentage of freezing behavior compared to wild-type littermates [repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 for each time point](Figure S6A). Percentages of freezing time per 30 s bin across the training were compared and no difference was detected between genotypes. After being conditioned to aversive electrical shocks, mice were placed into the test apparatus with identical context 24 hours later. During the contextual fear conditioning we observed no difference in freezing time between genotypes [WT (n = 16) 34 ± 3 % SEM; Chd8+/− (n = 21) 35 ± 3 % SEM, two-tailed t-test p-value = 0.788](Figure S6B). Similarly, during the tone fear conditioning we observed no difference in freezing time between genotypes [WT (n = 16) 31 ± 2 % SEM; Chd8+/− (n = 21) 34 ± 2 % SEM, two-tailed t-test p-value = 0.231](Figure S6C). We did observe a significant difference in freezing time between the baseline and tone conditions for both genotypes [WT (n=16) baseline 6 ± 1 % SEM; tone 30 ± 2 % SEM, two-tailed t-test p-value < 0.001; Chd8+/− (n=21) baseline 4.9 ± 0.9 % SEM; tone 34 ± 2 % SEM, two-tailed t-test p-value < 0.001](Figure S6C). These data suggest that both contextual fear conditioning and tone based fear conditioning are intact in Chd8+/− mice.
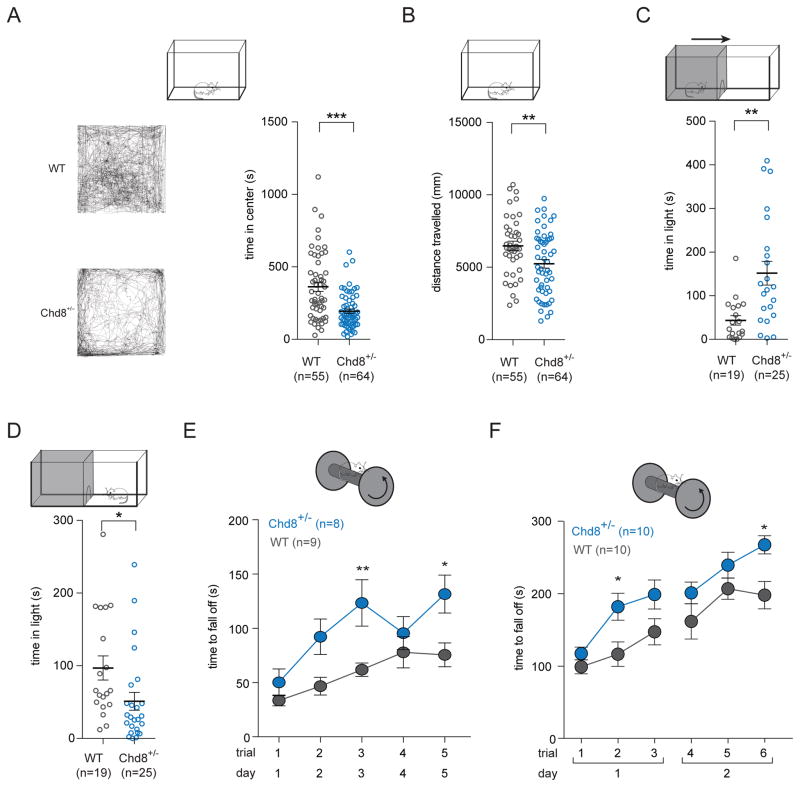
Many individuals with ASD experience various degrees of anxiety, including a population of ASD patients with CHD8 mutations (Bernier et al., 2014; Merner et al., 2016; White et al., 2009). Increased anxiety-like behaviors were also frequently detected in many mouse models of ASD (Kazdoba et al., 2014; McGill et al., 2006; Zhou et al., 2016). To more generally assess the behaviors of the Chd8+/− animals we conducted the open field test. We found that Chd8+/− mice spent less time in the center [WT (n = 55) 360 ± 30 s SEM; Chd8+/− (n = 64) 190 ± 20 s SEM, two-tailed t-test p-value < 0.001](Figure 6A) of the arena and also exhibit reduced total distance moved [WT (n = 41) 6.5 ± 0.3 m SEM; Chd8+/− (n = 55) 5.2 ± 0.3 m SEM, two-tailed t-test p-value = 0.062](Figure 6B) compared to wild-type littermates. Less time spent in the center of the arena is often interpreted as an anxiety-like phenotype (Bailey and Crawley, 2009). We further probed this phenotype utilizing the dark-light emergence test. In the dark-light emergence test, which takes place in a two-chamber arena with differential lighting intensity, we observed an increase in the latency to enter the light side of the arena from the dark side [WT (n = 19) 40 ± 10 s SEM; Chd8+/− (n = 22) 150 ± 30 s SEM, two-tailed t-test p-value = 0.001](Figure 6C) as well as the total time spent in the light side of the arena [WT (n = 19) 100 ± 20 s SEM; Chd8+/− (n = 25) 50 ± 20 s SEM, two-tailed t-test p-value = 0.029](Figure 6D) in Chd8+/− mice compared to wild-type littermates. Together, these results suggest Chd8+/− mice display anxiety-like behaviors in the open field and dark-light emergence tests.
Figure 6. Chd8+/− mice display anxiety-like behavior and increased acquired motor learning.
(A) (Left) Open field traces for animals with median center time values. (Right) In the open field test, Chd8+/− mice spent less time in the center compared to wild-type littermates [WT (n = 55) 360 ± 30 s SEM; Chd8+/− (n = 64) 190 ± 20 s SEM, two-tailed t-test p-value < 0.001]. Also see Figure S6.
(B) In the open field test, Chd8+/− mice showed reduced locomotion compared to wild-type littermates [WT (n = 41) 6.5 ± 0.3 m SEM; Chd8+/− (n = 55) 5.2 ± 0.3 m SEM, two-tailed t-test p-value = 0.062].
(C) In the dark-light emergence test, Chd8+/− mice spent more time in the dark side of the arena before crossing over to the light side of the arena compared to wild-type littermates [WT (n = 19) 40 ± 10 s SEM; Chd8+/− (n = 22) 150 ± 30 s SEM, two-tailed t-test p-value = 0.001].
(D) In the dark-light emergence test, Chd8+/− mice spent less time in the light side of the arena compared to wild-type littermates [WT (n = 19) 100 ± 20 s SEM; Chd8+/− (n = 25) 50 ± 20 s SEM, two-tailed t-test p-value = 0.029].
(E) In the rotarod performance test, Chd8+/− mice (n = 10) spent more time on the rotating rod before falling off compared to wild-type littermates (n = 10). One trial was performed per day for five days [repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) 0.088 (Trial 2) 0.008 (Trial 3) > 0.05 (Trial 4) 0.020 (Trial 5)].
(F). In the rotarod performance test, Chd8+/− mice (n = 8) spent more time on the rotating rod before falling off compared to wild-type littermates (n = 9). Three trials were performed per day for two days [repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) 0.042 (Trial 2) 0.201 (Trial 3) 0.604 (Trial 4) > 0.05 (Trial 5) 0.025 (Trial 6)].
Given the observation of reduced locomotion in the open field test (Figure 6A), one may expect that Chd8+/− mice develop impaired motor skills in addition to elevated anxiety-like behaviors. To gain a deeper understanding of phenotypes related to decreased locomotion, we performed the rotarod test. We first used a rotarod performance test paradigm where animals were tested once a day for five days and found that Chd8+/− mice outperformed weight-matched wild-type littermates [repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) 0.088 (Trial 2) 0.008 (Trial 3) > 0.05 (Trial 4) 0.020 (Trial 5)](Figure 6E). Experimental conditions for rotarod vary widely across laboratories so to confirm our result we reproduced the phenotype in a later generation of mice using a second paradigm with different experimental conditions, namely three trials per day for two days in weight-matched animals [repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) 0.042 (Trial 2) 0.201 (Trial 3) 0.604 (Trial 4) > 0.05 (Trial 5) 0.025 (Trial 6)](Figure 6F). Together, these data suggest that Chd8+/− mice show reduced locomotion in the open field test and an increase in acquired motor learning in the rotarod performance text.
Perturbation of Chd8 in wild-type adult mice recapitulates behavioral phenotypes
Increased acquired motor learning is a feature shared among several other ASD mouse models, including PTEN knockout (Kwon et al., 2006), NLGN3 R451C missense mutations (Chadman et al., 2008), and NLGN3 knockout (Rothwell et al., 2014) among others (Etherton et al., 2009; Nakatani et al., 2009; Penagarikano et al., 2011). Interestingly, MSN and NAc-specific mutation of NLGN3 (R451C or LOF) results in an increase in acquired motor learning in the rotarod performance test (Rothwell et al., 2014).
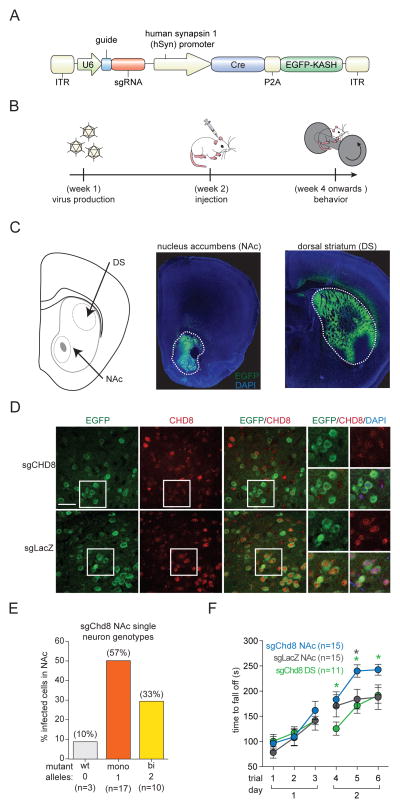
To test whether CHD8 expression in the NAc of adults was required for the acquired motor learning phenotype we first validated that Chd8 was expressed in the NAc of adult animals (Figure S7A). We then stereotactically injected adeno-associated virus (AAV) vectors containing Chd8-targeting sgRNA (Figure 7A–B) into either the NAc or the dorsal striatumof Cas9 knockin mice (Platt et al., 2014) (Figure 7C). Control mice were injected with an AAV carrying a non-targeting sgRNA (sgLacZ) into the NAc. Six weeks after injection we performed immunohistochemistry (Figure 7D), microdissected the injected region, and genotyped individual FACS sorted EGFP-KASH-tagged fluorescent nuclei by Illumina sequencing. AAV-mediated delivery of Chd8-targeting sgRNA mediated robust mutagenesis of the Chd8 allele. We observed cells with zero (wild-type, 10%), one (monoallelic, 57%), or two (biallelic, 33%) mutant alleles (Figure 7E). We then performed the rotarod performance test on injected animals and found that perturbation of Chd8 in the NAc (sgChd8-NAc) but not the dorsal striatum (sgChd8-DS) improved acquired motor learning in the rotarod performance test compared to control injected animals (sgLacZ-NAc) [sgChd8-NAc versus sgLacZ-NAc: repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value = 0.840 (Trial 1) > 0.05 (Trial 2) 0.732 (Trial 3) > 0.05 (Trial 4) > 0.026 (Trial 5) 0.031 (Trial 6); sgChd8-DS versus sgLacZ-DS: repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) > 0.05 (Trial 2) > 0.05 (Trial 3) > 0.05 (Trial 4) > 0.05 (Trial 5) > 0.05 (Trial 6); sgChd8-NAc versus sgChd8-DS: repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) > 0.05 (Trial 2) 0.815 (Trial 3) 0.036 (Trial 4) 0.011 (Trial 5) 0.074 (Trial 6)](Figure 7F). By contrast, in the open field test, we observed no differences between genotypes (Figure S7B–C). Taken together these results demonstrate that CHD8 expression in adults is not required for the increased anxiety-like or decreased locomotor behavior but is required for acquired motor learning in the rotarod performance test.
Figure 7. In vivo perturbation of Chd8 in adult mice recapitulates increased acquired motor learning phenotype.
(A) Diagram of AAV vector for sgRNA expression, Cas9 induction, and nuclei labeling in neurons of the Cre-dependent Cas9 mice.
(B) Workflow for generating and characterizing somatically edited Cre-dependent Cas9 mice.
(C) (Left) Schematic representation of a brain slice with the nucleus accumbens (NAc) and dorsal striatum (DS) target regions indicated. (Right) Representative immunofluorescence images four weeks post-injection showing AAV infected, EGFP-expressing neurons within the NAc and DS. Enclosed regions outline the targeted region. Also see Figure S7.
(D) Representative immunofluorescence images of AAV injected nucleus accumbens. Scale bar represents 50 μm.
(E) Indel analysis on Illumina sequencing reads from FACS sorted neuronal nuclei showing single cells with zero (wild-type, 10%), one (monoallelic, 57%), or two (biallelic, 33%) mutant alleles.
(F) In the rotarod performance test, sgChd8-NAc AAV injected mice (n = 15) spent more time on the rotating rod before falling off compared to sgChd8-DS (n = 11) and sgLacZ-NAc (n = 15) AAV injected control animals [sgChd8-NAc versus sgLacZ-NAc: repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value = 0.840 (Trial 1) > 0.05 (Trial 2) 0.732 (Trial 3) > 0.05 (Trial 4) > 0.026 (Trial 5) 0.031 (Trial 6); sgChd8-DS versus sgLacZ-DS: repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) > 0.05 (Trial 2) > 0.05 (Trial 3) > 0.05 (Trial 4) > 0.05 (Trial 5) > 0.05 (Trial 6); sgChd8-NAc versus sgChd8-DS: repeated-measures two-way ANOVA with Bonferroni post hoc test, adjusted p-value > 0.05 (Trial 1) > 0.05 (Trial 2) 0.815 (Trial 3) 0.036 (Trial 4) 0.011 (Trial 5) 0.074 (Trial 6)]. Also see Figure S7.
DISCUSSION
Our findings demonstrate an important role for CHD8 in neurodevelopment, physiology, and behavior. We showed that Chd8+/− mice exhibit a reduction in CHD8 expression, macrocephaly and craniofacial abnormalities as well as altered behavior, including anxiety-like behavior, a mild social behavior defect, and increased acquired motor learning, but we did not observe a change in repetitive behavior. Together, we find that Chd8+/− mice display some but not all of the phenotypic outcomes relevant to the diagnostic symptoms found in human patients.
ChIP- and RNA-seq revealed a broad role for CHD8 in genome regulation, including control of cell cycle, histone and chromatin modifications, and mRNA and protein processing. These observations were brain region-specific, suggesting differential effects across cells and circuits in the developing and adult brain. We find alterations in expected pathways based on previous studies (i.e., Wnt and p53 signaling) as well as novel effectors connecting chromatin modification (i.e., Hdac4 and Chd7) and histone methylation (i.e., Mecp2 and Tet2) to development (i.e., Ctnnb1 (beta-catenin), FoxG1, Arid1b, and Bcl11a) and the synapse (i.e., Ankr11, and Shank1-3, and Pcdha8-9). These data provide support that CHD8 influences the expression of many genes and pathways some of which likely result in the observed neuropathology.
CHD8 LOF mutations are strongly associated with ASD, and evidence suggesting that CHD8 plays a causal role in neurodevelopment and ASD neuropathology is mounting (Bernier et al., 2014; Iossifov et al., 2014; O’Roak et al., 2012a; O’Roak et al., 2012b). Early insights into the function of CHD8 revealed a modulatory role in Wnt signaling, which may lead to altered neurogenesis and cortical development. We confirmed that CHD8 is a negative regulator of Wnt signaling. In keeping with this, we observed morphological phenotypes, namely macrocephaly and craniofacial abnormalities, but not an overt phenotype in the major cortical cell types, lamination of the cortex, or number and cell cycle length of mid-stage cortical progenitors. These findings suggest that cell type specification and radial lamination of the cortex, which are are mid- to late-stage cortical developmental processes, are largely intact in Chd8+/− mice.
Chd8 mutation results in striatal dysfunction and an increased acquired motor learning behavioral phenotype mediated by synaptic transmission within MSNs in the NAc. These findings combined with previous reports implicating the striatum (Di Martino et al., 2011; Hollander et al., 2005; Peca et al., 2011) and in particular the NAc (Dolen et al., 2013; Grueter et al., 2013; Gunaydin et al., 2014; Rothwell et al., 2014), strongly suggest the NAc is an important node for social behavior and ASD pathology. Circuit mapping studies will be valuable for further understanding the links between MSNs in the NAc, striatal circuits, and ASD risk alleles in the context of ASD pathology.
Increased acquired motor learning has been reported for other mouse models of ASD (Michalon et al., 2012; Osterweil et al., 2013; Rothwell et al., 2014; Tian et al., 2015). For example, Rothwell et al. directly mapped the role of NLGN3 and NLGN3[R451C] in MSNs of the NAc to acquired motor learning. While we observed increased acquired motor learning by rotarod in Chd8+/− mice, it did not definitively implicate CHD8 expression in the NAc of adult mice. Therefore, we utilized CRISPR-Cas9 knockin mice to perturb Chd8 directly in vivo to map the relationship. In NAc-specific Chd8 perturbed mice, we were able to recapitulate the acquired motor learning phenotype observed in germline mutant animals. Together these data demonstrate a functional role of CHD8 in the adult brain in the NAc, which we directly link to behavioral phenotypes.
Recently, two independent groups reported the characterization of Chd8 loss of function mutations in mice (Durak et al., 2016; Katayama et al., 2016). These additional studies confirm that Chd8+/− mice show abnormal social and anxiety-like behavior as well as a macrocephaly phenotype, but no unifying mechanism for CHD8 function has emerged from those works. Together with these previous reports, our findings support the hypothesis that CHD8 acts by globally regulating many genes, highlighting several specific pathways that may be linked to animal behaviors relevant to ASD pathology. These studies provide a first look at the molecular and physiological consequences of LOF mutations in CHD8 on the developing and adult brain, elucidating critical defects as well as providing mechanistic insights. Our demonstration that Chd8+/− mice display hallmark features similar to those found in some ASD patients and characterization of the circuits underlying these behaviors open up broad avenues for future work. These results causally implicate CHD8 in ASD pathogenesis and provide a link between chromatin modification affecting the synapse and broader circuits connecting through the nucleus accumbens.
EXPERIMENTAL PROCEDURES
Germline editing of Chd8 with Cas9
sgRNAs were designed using the CRISPRtool (crispr.mit.edu) and tested by SURVEYOR assay (Transgenomic) according to the manufacturer’s protocol. The sequences of which are listed in supplemental Table S1 along with genomic primers. Human codon optimized Cas9 (from Streptococcus pyogenes) capped and polyadenylated mRNA and sgRNA RNA were co-injected by pronuclear injection at concentration of 200 ng/μl Cas9 mRNA and 50 ng/μl, and 200 ng/μl, respectively.
Immunofluorescence on adult brain
Adult male mice were transcardially perfused and brains were sectioned at 40 μm on a vibrating microtome and stained as previously described (Platt et al., 2014). Primary antibodies and dilutions used were as follows: anti-CHD8 (1:1000, Cell Signaling Technology, P58438), anti-NeuN (1:800, Cell Signaling Technology, 12943), anti-GFP (1:1600, Nacalai Tesque, GF090R), anti-parvalbumin (1:1000, Sigma Aldrich, P3088), anti-GFAP (1:1000, Aves Labs, GFAP), anti-S100b (1:1000, Abcam, ab4066) and anti-CNP1 (1:1000, Synaptic Systems, 255004). Secondary antibodies and dilutions used were as follows: AlexaFluor 405, 488, 568 and/or 647 secondary antibody (1:400, Life Technologies).
Immunofluorescence on embryonic and juvenile brain
E15.5 Embryos were dissected and fixed in 4% PFA overnight. P21 pups were transcardially perfused with PBS, then with 4% PFA, dissected, and postfixed in 4% PFA overnight. Brains were sectioned at 40 μm on a vibrating microtome (Leica VT1000S) and stained as previously described (Lodato et al., 2014). Primary antibodies and dilutions used were as follows: rat anti-CTIP2 antibody, 1:100 (Abcam ab18465); mouse anti-SATB2, 1:50 (Abcam ab51502); mouse anti-Pvalb, 1:1000 (Millipore MAB); Rabbit anti Olig2 (IBL-18953); Mouse anti Brdu (Millipore MAB); Rabbit anti Ki67(Abcam ab15580) and rabbit anti-CUX1, 1:100 (Santa Cruz CDP M-222). Secondary antibodies and dilutions used were as follows: AlexaFluor 405, 488, 568 and/or 647 secondary antibody (1:750, Life Technologies).
Quantification of BrdU incorporation and cell cycle length
To estimate the cell cycle length, we conducted a dual-pulse-labeling of DNA synthesis using 5-bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich) and 5-ethynyl-2′-deoxyuridine (EdU; Molecular Probes) as previously described (Mairet-Coello et al., 2012; Watanabe et al., 2015). Timed pregnant C57BL/6 females, crossed with Chd8+/− males, received one intraperitoneal injection of BrdU (50 mg/kg) and Edu (12.5 mg/kg) 120 and 30 minutes before sacrifice, respectively, and the ratios of cells that incorporated either or both BrdU and EdU were analyzed to estimate the cell cycle length. The detection of EdU-labeled cells was performed based on a fluorogenic click reaction (Salic and Mitchison, 2008). For Detection of BrdU, antigen retrieval was done by incubating sections in 2 N HCL for 20 minutes. For counting and colocalization analyses of BrdU, Edu and Ki67, confocal images of a 300 μm square ROI spanning the entire ventricular zone was imaged using Zeiss LSM 700 and analyzed by an independent investigator who was blinded to genotype and experimental conditions. At least 1000 cells were counted per section, 3–4 mid-cortex sections per genotype were counted. For cell cycle estimation was performed as previously described (Mairet-Coello et al., 2012; Watanabe et al., 2015). Briefly, all Ki67 cells present in the ROI were counted and the existence of co-staining with BrdU and/or Edu was noted. For BrdU incorporation, all BrdU positive cells present in the ROI were counted.
ChIP-seq
ChIP was performed as previously described (Hathaway et al., 2012). Somatosensory cortices were dissected from 10–11 week old adult males and homogenized. Tissues were fixed in 1% formaldehyde for 10 min at 25°C and then quenched in 0.125 M glycine on ice. Cross-linked cells were sonicated to produce chromatin fragments of 200 to 700 bp in length. Chromatin fragments were then incubated overnight at 4°C with anti-CHD8 antibody (Novus Biologicals, NB100-60417). DNA was extracted with phenol chloroform, followed by ethanol precipitation. ChIP-seq libraries were prepared according to the NEBNext protocol and sequenced using Illumina NextSeq. The reads were uniquely mapped to the mm10 genome utilizing Bowtie2 version 2.2.1(Langmead and Salzberg, 2012) and duplicated reads were removed with Samtools version 1.3 (Li et al., 2009) peaks were called using MACS2 version 2.1.1 (Zhang et al., 2008) with FDR cutoff of 5%. Binding site annotation and distance to TSS was performed using HOMER version 4.8 (Heinz et al., 2010). Functional enrichment and ontology was performed using GREAT version 3.0.0 (McLean et al., 2010).
RNA sequencing
Relevant brain regions were microdissected from 10–12 week old male mice and rapidly frozen on dry ice. RNA was purified by RNAeasy Plus Micro Kit (Qiagen) according to the manufacturer’s protocol. mRNA sequencing libraries were prepared using SMART-Seq2 (Picelli et al., 2013) and sequenced on the NextSeq system (Illumina). Transcripts were mapped and quantified using RSEM (Li and Dewey, 2011). Using a log2 transcripts per million expression matrix, differential expression analysis was performed using DEseq2 (Love et al., 2014) and nominal p-values are reported. Gene set enrichment analysis was performed using GenePattern (open source from the Broad Institute). Sample distances and hierarchical clustering were performed using GENE-E (open source from the Broad Institute) using average linkages and Pearson’s correlation. Heatmaps were also created using GENE-E using mean subtracted and standard deviation row normalized values.
Western blot
Protein lysates were prepared, quantified, and equally loaded on 4%–20% Tris-HCL Criterion Gel (Bio-Rad). Proteins were transferred onto PVDF membrane (Bio-Rad), blocked, and stained overnight using the following primary antibodies: anti-CHD8 (1:1000, Bethyl, A301-2214A), HRP conjugated anti-GAPDH (1:5000, Cell Signaling Technology, 3683), and HRP conjugated anti-ACTB (1:1000, Cell Signaling Technologies, 5125). Membranes were washed and stained with secondary antibodies: HRP-conjugated secondary antibodies (1:10,000, Cell Signaling Technology, 7074 and 7076). Membranes were washed, developed using SuperSignal West Femto Substrate (Pierce), and imaged on a gel imager (Bio-Rad).
Mouse behavior
For all behavioral experiments, unless otherwise noted, we used 10–14-week old group housed males weaned in groups with littermates of similar genotype. In all behavior experiments animals were randomized and experimenters were blinded to animal genotype during behavioral tests and data analysis. See SUPPLEMENTARY DATA for detailed description of each behavioral test.
Patch clamp slice electrophysiology
Slice preparation, data acquisition, and analysis were performed with experimenter blinded to mice genotype as described in previous reports (Peca et al., 2011; Rothwell et al., 2014; Zhou et al., 2016). See SUPPLEMENTARY DATA for detailed description of each measurement.
RNA-FISH
C57BL/6J male mice (Jackson Laboratories) adult (>6 weeks old) and day 17 (E17) embryonic brains were dissected and placed directly into either 10% neutral buffered formalin (NBF) or 4% paraformaldehyde (PFA) + 0.5% acetic acid and fixed for 1, 3, or 12 hours. After fixation, brains were embedded in paraffin and sliced at 5 μm thickness and mounted on glass slides without coverslips. These slides were processed using QuantiGene ViewRNA ISH Tissue (Affymetrix). FISH probes targeting the 5th to the 12th exon of Chd8 transcripts were ordered from Affymetrix. Slides were processed according to the Quantigene ViewRNA ISH protocol. Slides were imaged on a Zeiss Axio microscope under 20x or 40x objectives and fluorescent images were taken using a Cy3/TRITC filter for Fast Red and a DAPI filter for DAPI.
AAV1/2 production
AAV1/2 was produced as previously described (McClure et al., 2011; Platt et al., 2014). Briefly, low passage HEK293FT cells (Life Technologies) were transfected with the plasmid of interest, pAAV1 plasmid, pAAV2 plasmid, helper plasmid pDF6, and PEI Max (Polysciences, Inc. 24765- 2). AAV particles were purified using HiTrap heparin columns (GE Biosciences 17-0406-01).
Stereotactic injection
Stereotactic injection was performed as previously described (Platt et al., 2014). Briefly, anesthesized male mice at least 6 weeks of age were injected with 1.25 μl of AAV virus bilaterally in the nucleus accumbens (AP +1.50, ML ± 1.10, DV −4.40) or the dorsal striatum (AP +0.70, ML ± 2.50, DV −2.40) at 100 nl/min. After injection the needle was left in place for 5 min before slowly being retracted.
Single nuclei preparation by FACS
Single nuclei experiments were performed as previously described (Platt et al., 2014). Briefly, 6 weeks post-injection the infected (EGFP+) regions were dissected, flash frozen on dry ice, and stored at −80°C until use. The tissue was gently homogenized in nuclei were isolated followed by gradient centrifugation. Nuclei labeled with Vybrant DyeCycle Ruby Stain (Life Technologies) and sorted using a BD FACSAria III. EGFP+ nuclei were sorted into individual wells of a 96 well-plate and used as an input for single cell indel analysis.
Animal work statement
All animal work was performed under the guidelines of Division of Comparative Medicine (DCM), with protocols (0414-024-17, 0414-027-17, and 0513-044-16) approved by Massachusetts Institute of Technology Committee for Animal Care (CAC), and were consistent with the Guide for Care and Use of Laboratory Animals, National Research Council, 1996 (institutional animal welfare assurance no. A-3125-01). Information regarding the gender and age/developmental stage is listed under specific experimental procedure descriptions.
Accession
Sequencing data are deposited in NCBI Sequence Read Archive under accession number PRJNA379430.
Supplementary Material
Acknowledgments
We thank the entire Zhang laboratory and Feng laboratory for thoughtful discussions and support. We thank P. Arlotta for support on analysis of development of neocortex and for mentoring A.S.S. We thank S. Hyman, E. Scolnick, and R. Macrae for helpful discussions. We thank M. Heidenreich, N. Habib, and M. Yim for technical assistance. We thank the Swanson Biotechnology Center for their support (ES Cell and Transgenics in particular). We thank L. Dennis for analyzing the behavioral experiments. We thank V. Natu, D. Wagh, and J. Coller at the Stanford Functional Genomics Facility for their help with sequencing. R.J.P. was supported by a National Science Foundation Graduate Research Fellowship under grant number 1122374, ETH Zurich internal funding, and NCCR-MSE internal funding. I.S. and Y.Z. are supported by the Simons Center for the Social Brain at MIT, postdoctoral fellowship. S.S. is supported by the NIMH (1F31MH111157). G.F. is supported by the McGovern Internal Funding Poitras Gift 1631119, the Stanley Center, the SFARI/Simons Foundation 6927482, and the Nancy Lurie Marks Family Foundation 6928117. F.Z. is supported by the NIH through NIMH (5DP1-MH100706 and 1R01-MH110049); NSF; the New York Stem Cell Foundation; the Howard Hughes Medical Institute; the Simons, Paul G. Allen Family, and Vallee Foundations; the Skoltech-MIT Next Generation Program; James and Patricia Poitras; Robert Metcalfe; and David Cheng. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. CRISPR reagents are available to the academic community through Addgene, and associated protocols, support forum, and computational tools are available via the Zhang lab website (http://www.genome-engineering.org).
Footnotes
AUTHOR CONTRIBUTIONS
RJP and FZ conceived of the study and oversaw experiments. RJP, YZ, IMS, ASS, NRW, JAK, JS, MD, SS, HRK performed experiments and analyzed results. GF and GRC oversaw experiments. RJP, IS, and FZ wrote the manuscript with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KR, Crawley JN. Anxiety-Related Behaviors in Mice. In: Buccafusco JJ, editor. Methods of Behavior Analysis in Neuroscience. Boca Raton (FL): 2009. [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar VJ, Walters SR, Phoenix JL. Assessing autism-like behavior in mice: variations in social interactions among inbred strains. Behav Brain Res. 2007;176:21–26. doi: 10.1016/j.bbr.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadman KK, Gong S, Scattoni ML, Boltuck SE, Gandhy SU, Heintz N, Crawley JN. Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 2008;1:147–158. doi: 10.1002/aur.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Kelly C, Grzadzinski R, Zuo XN, Mennes M, Mairena MA, Lord C, Castellanos FX, Milham MP. Aberrant striatal functional connectivity in children with autism. Biol Psychiatry. 2011;69:847–856. doi: 10.1016/j.biopsych.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolen G, Darvishzadeh A, Huang KW, Malenka RC. Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature. 2013;501:179–184. doi: 10.1038/nature12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak O, Gao F, Kaeser-Woo YJ, Rueda R, Martorell AJ, Nott A, Liu CY, Watson LA, Tsai LH. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci. 2016;19:1477–1488. doi: 10.1038/nn.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter BA, Robison AJ, Neve RL, Nestler EJ, Malenka RC. FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc Natl Acad Sci U S A. 2013;110:1923–1928. doi: 10.1073/pnas.1221742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander E, Anagnostou E, Chaplin W, Esposito K, Haznedar MM, Licalzi E, Wasserman S, Soorya L, Buchsbaum M. Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol Psychiatry. 2005;58:226–232. doi: 10.1016/j.biopsych.2005.03.040. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, Suyama M, Takumi T, Miyakawa T, Nakayama KI. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- Kazdoba TM, Leach PT, Silverman JL, Crawley JN. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res. 2014;3:118–133. doi: 10.5582/irdr.2014.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Molyneaux BJ, Zuccaro E, Goff LA, Chen HH, Yuan W, Meleski A, Takahashi E, Mahony S, Rinn JL, et al. Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat Neurosci. 2014;17:1046–1054. doi: 10.1038/nn.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G, Tury A, Van Buskirk E, Robinson K, Genestine M, DiCicco-Bloom E. p57(KIP2) regulates radial glia and intermediate precursor cell cycle dynamics and lower layer neurogenesis in developing cerebral cortex. Development. 2012;139:475–487. doi: 10.1242/dev.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure C, Cole KL, Wulff P, Klugmann M, Murray AJ. Production and titering of recombinant adeno-associated viral vectors. J Vis Exp. 2011:e3348. doi: 10.3791/3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2006;103:18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merner N, Forgeot d’Arc B, Bell SC, Maussion G, Peng H, Gauthier J, Crapper L, Hamdan FF, Michaud JL, Mottron L, et al. A de novo frameshift mutation in chromodomain helicase DNA-binding domain 8 (CHD8): A case report and literature review. Am J Med Genet A. 2016;170A:1225–1235. doi: 10.1002/ajmg.a.37566. [DOI] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, Jaeschke G, Bear MF, Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Nakayama K, Tsunematsu R, Tsukiyama T, Kikuchi A, Nakayama KI. Early embryonic death in mice lacking the beta-catenin-binding protein Duplin. Mol Cell Biol. 2004;24:8386–8394. doi: 10.1128/MCB.24.19.8386-8394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012a;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil EK, Chuang SC, Chubykin AA, Sidorov M, Bianchi R, Wong RK, Bear MF. Lovastatin corrects excess protein synthesis and prevents epileptogenesis in a mouse model of fragile X syndrome. Neuron. 2013;77:243–250. doi: 10.1016/j.neuron.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Sudhof TC. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil-Rodriguez A, Vazquez-Chavez E, Ceballos-Chavez M, Rodriguez-Paredes M, Martin-Subero JI, Esteller M, Reyes JC. The chromatin remodeller CHD8 is required for E2F-dependent transcription activation of S-phase genes. Nucleic Acids Res. 2014;42:2185–2196. doi: 10.1093/nar/gkt1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, Miles J, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A. 2014;111:E4468–4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Stoppel LJ, Heynen AJ, Lindemann L, Jaeschke G, Mills AA, Bear MF. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat Neurosci. 2015;18:182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Kageyama R, Ohtsuka T. Hbp1 regulates the timing of neuronal differentiation during cortical development by controlling cell cycle progression. Development. 2015;142:2278–2290. doi: 10.1242/dev.120477. [DOI] [PubMed] [Google Scholar]
- White SW, Oswald D, Ollendick T, Scahill L. Anxiety in children and adolescents with autism spectrum disorders. Clin Psychol Rev. 2009;29:216–229. doi: 10.1016/j.cpr.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B, Grepo N, Thompson BL, Kim J, Wang K, Evgrafov OV, Lu W, Knowles JA, Campbell DB. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry. 2015;5:e568. doi: 10.1038/tp.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Clarke AM, Crawley JN. Postnatal lesion evidence against a primary role for the corpus callosum in mouse sociability. Eur J Neurosci. 2009;29:1663–1677. doi: 10.1111/j.1460-9568.2009.06714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, Langlois S, Martin H, Willatt L, Marra MA, et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kaiser T, Monteiro P, Zhang X, Van der Goes MS, Wang D, Barak B, Zeng M, Li C, Lu C, et al. Mice with Shank3 Mutations Associated with ASD and Schizophrenia Display Both Shared and Distinct Defects. Neuron. 2016;89:147–162. doi: 10.1016/j.neuron.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.