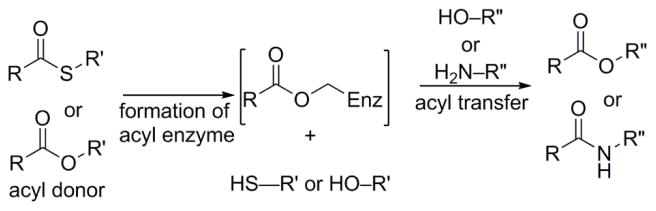
Abstract
Enzymes within a family often catalyze different reactions. In some cases, this variety stems from different catalytic machinery, but in other cases the machinery is identical; nevertheless, the enzymes catalyze different reactions. In this review, we examine the subset of α/β-hydrolase fold enzymes that contain the serine-histidine-aspartate catalytic triad. In spite of having the same protein fold and the same core catalytic machinery, these enzymes catalyze seventeen different reaction mechanisms. The most common reactions are hydrolysis of C–O, C–N and C–C bonds (Enzyme Classification (EC) group 3), but other enzymes are oxidoreductases (EC group 1), acyl transferases (EC group 2), lyases (EC group 4) or isomerases (EC group 5). Hydrolysis reactions often follow the canonical esterase mechanism, but eight variations occur where either the formation or cleavage of the acyl enzyme intermediate differs. The remaining eight mechanisms are lyase-type elimination reactions, which do not have an acyl enzyme intermediate and, in four cases, do not even require the catalytic serine. This diversity of mechanisms from the same catalytic triad stems from the ability of the enzymes to bind different substrates, from the requirements for different chemical steps imposed by these new substrates and, only in about half of the cases, from additional hydrogen bond partners or additional general acids/bases in the active site. This detailed analysis shows that binding differences and non-catalytic residues create new mechanisms and are essential for understanding and designing efficient enzymes.
Keywords: hydrolase, lyase, mechanism, divergent evolution, catalytic triad, oxyanion hole, X-ray structures
Graphical Abstract

1. Introduction
The ability of enzymes to catalyze reactions more efficiently than synthetic catalysts often draws admiration from chemists. There is an idealized notion that enzymes control reactions by restricting and focusing the rearrangements of electrons to just the desired reaction. In a similar mindset, Knowles called triosephosphate isomerase a perfect enzyme since it catalyzes its reactions as fast as substrates can diffuse to the active site.1 Indeed the rate acceleration by enzymes is dramatic and can reach factors of 1013.2
This admiration grows further as chemists and biochemists struggle to design and improve enzymes. The state of the art in computational enzyme design makes ~100 predictions; of these a handful are catalytically active, but the rate accelerations are still far from those of natural enzymes.3,4 Designing improvements in naturally occurring enzymes still requires creating and testing tens of thousands of variants.5 These difficulties make clear that there are many aspects of efficient enzyme catalysis that we do not yet understand.
Another puzzle about enzymes is their catalytic promiscuity. Enzymes can catalyze, besides their natural reaction, other reactions.6–8 For example, carbonic anhydrase catalyzes not only the hydration of carbon dioxide, but also efficiently catalyzes the hydrolysis of esters. Such sloppiness seems surprising for highly efficient and optimized catalysts.
A third puzzle is how enzymes with similar active sites catalyze different reactions. Divergent evolution creates families of enzymes with similar active sites.9,10 Some differ in their substrate specificity, but others differ in the reaction that they catalyze. The details that determine which reaction is catalyzed are critical to understanding why enzymes are efficient and how to improve them. This review focuses on identifying these differences for one group of enzymes: α/β-hydrolase fold enzymes with a Ser-His-Asp active site. We suggest that both catalytic promiscuity and tendency to catalyze different reactions with the same machinery come from 1) the ability to bind different substrates, 2) the requirements for different chemistry by these substrates and in about half of the cases, 3) changes in hydrogen bond partners or general acids/bases in the active sites that create new mechanistic steps.
The Ser-His-Asp catalytic triad occurs in several protein folds. For example, it occurs in the α/β-hydrolase fold, subtilisin fold, and chymotrypsin fold indicating that evolution converged on the same solution for catalysis several times.11 This review considers only examples within the α/β-hydrolase fold family. Focusing on one protein fold makes it easier to identify differences for each reaction type.
Two public databases focus on the α/β-hydrolase fold superfamily of proteins. The ESTHER database7,12,13 gathers and annotates gene and protein sequences as well as biochemical, pharmacological and structural data. The α/β-hydrolase fold enzyme family 3DM database14 is a structure-based multiple-sequence alignment of ~60,000 α/β-hydrolase fold enzymes with separate sequence alignments for subfamilies if a structure is available for that subfamily.
Within the α/β-hydrolase fold superfamily, 75% (~44,000) contain a Ser-His-Asp (Glu) catalytic triad. The diversity of catalytic activities in this groupis the focus of this review. This review compares catalytic activities within this subset of the α/β-hydrolase fold superfamily to provide a better understanding of the versatility of this catalytic core. Of these, 16,600 (37%) have unknown function and will not be considered further. The largest group with known function are esterases and lipases (12,300 sequences, 28%), followed by peptidases (8,600 sequences, 19%). The remaining 7,000 sequences (16%) include other hydrolases as well as the less common activities like oxidoreductase and lyase. Besides these activities, we also include unnatural activities in this review. Unnatural activities are promiscuous reactions catalyzed by these enzymes, but which have no natural role. Researchers previously reviewed the protein structure aspects of the α/β-hydrolase fold enzyme family15–18 and the breadth of different catalytic activities in this family,19–21 but we focus on the Ser-His-Asp subset of enzymes and the mechanistic diversity within it.
Although we include catalytically promiscuous reactions of the serine-histidine-aspartate enzymes, most of the review compares different, albeit similar, enzymes that use different catalytic mechanisms. To organize these differences, we start with the Enzyme Commission (EC) classification groups, but extend it as needed to discuss varying mechanisms. If the enzymes differ in any of the first three numbers of the four number classification, then they are different reactions. If they differ in only the fourth number, then they differ only in substrate specificity. Differences in substrate specificity will only be included when the reaction mechanism differs. Khersonsky & Tawfik22 used a similar classification to distinguish between catalytic and substrate promiscuity of enzymes; here we also use it to compare different enzymes. For reactions that have not yet been classified by the Enzyme Commission, we use the most likely classification groups.
2. Structure and substrate-binding site of α/β-hydrolases
Most α/β-hydrolase fold enzymes contain two domains: a catalytic domain and a cap or lid domain. The α/β-hydrolase fold refers to the catalytic domain. This domain contains the core catalytic machinery - the Ser-His-Asp catalytic triad and the oxyanion hole - and it contains part of the substrate binding site. The lid or cap domain forms the rest of the substrate-binding site, see below.
The common structure of the catalytic domain is a central β-sheet of eight strands interconnected by α-helices and with strand 2 running antiparallel to the rest, Figure 1. This structure also positions the catalytic triad residues and the oxyanion hole residues on loops connecting the β-strands and α-helices.15–18
Figure 1.

Schematic of the α/β-hydrolase fold showing sequence of α-helices (red rectangles) and β-sheets (blue arrows) and location of the catalytic triad residues and the oxyanion loop. The oxyanion loop positions one main chain N-H to donate a hydrogen bond to the oxyanion. The other N-H comes from the residue after the catalytic serine. These two residues form the oxyanion hole.
The catalytic domain varies in size and also in some details of the catalytic machinery, but none of these differences provide explanations for the different reactivities discussed later in the review. For example, instead of a Ser-His-Asp catalytic triad, some α/β-hydrolase-fold enzymes contain a Ser-His-Glu triad (~16% of the Asp + Glu total). In a few cases, the catalytic acid is located on a different loop.
The oxyanion hole part of the catalytic machinery can also show differences. In most cases, the oxyanion hole is formed by hydrogen bonds from two main chain N-H’s. These N-H’s bind the carbonyl oxygen and stabilize the oxyanion of the tetrahedral intermediates. One variation in the oxyanion hole is a third hydrogen bond donor. In lipase B from Candida antarctica, the side chain Oγ-H of one of the oxyanion hole residues, Thr40, also donates a hydrogen bond. Another variation in the oxyanion hole is replacing the main chain N-H from the oxyanion hole with a side chain interaction. For example, lipase A from C. antarctica uses the side chain of Asp9523 and dipeptidyl peptidase IV uses the side chain hydroxyl of Tyr547.24,25
The cap or lid domain forms the top part of the substrate binding site. The structure, amino acid sequence and location within the linear vary widely. Often the cap domain is ~100 amino acid residues with mostly helical secondary structure inserted between strands β6 and β7 of the catalytic domain. For example, the lid domain of lipase A from C. antarctica consists of 95 amino acids in six helices.23 However, the lid domain can be missing completely as in cutinase;26 it can be small like the 40 amino acids in MCP hydrolase27 or it can be large like the 355 amino acids in prolyl oligopeptidase.28 The lid domain sometimes occurs in a different location; for example, in 2,6-dihydroxy-pseudo-oxynicotine (DHPON) hydrolase, the lid domain is at the N-terminus. Besides a mainly helical secondary structure, it can also be mainly β-sheet secondary structure as in prolyl oligopeptidase.28 Since the lid domain adds to substrate binding, it also contributes to substrate specificity. For example, the large lid in prolyl oligopeptidase creates a tunnel that excludes structured peptides from the active site and accounts for the preference of this peptidase for short oligopeptides over larger, structured proteins. In another example, the lid domain of dipeptidyl peptidase IV contains a double glutamate motif that binds the terminal NH3+ group of a peptide.24,25 The location of this binding site relative to the catalytic serine allows only dipeptides to be cleaved. Although the lid domain does not contain any of the core catalytic machinery (Ser-His-Asp triad) or oxyanion hole, it can contain residues that are essential to catalysis. For example, the lid domain of DHPON hydrolase contains an arginine, which is essential for catalytic activity, see section 7 below. Creating an epoxide hydrolase from an esterase required exchanging portions of the lid.29
The active site serine divides the substrate-binding site into two parts: the acyl binding side and the alcohol (or nucleophile) binding site, Figure 2. This description assumes the substrate is an ester. The details of this binding site differ between the α/β-hydrolases. For example, in Candida rugosa lipase contains a wide open binding site that accommodates esters of tertiary alcohols, while lipase B from C. antarctica contains a much smaller binding site.30,31
Figure 2.
The substrate-binding site of α/β-hydrolases. A) Schematic showing how the active site accommodates different parts of a tetrahedral intermediate. The active site contains a region to stabilize the oxyanion as well as regions for the acyl and alcohol parts of an ester. B) Ribbon diagram of carboxylesterase Est30 from Geobacillus stearothermophilus (cap domain in gray, catalytic domain in green; pdb id: 1tqh) with tetrahedral intermediate for hydrolysis of propyl acetate and the catalytic serine and histidine in sticks representation. The active site lies between the two domains. The inset shows a close-up of the active site. The Ser-His-Asp triad (aspartate not shown) and the oxyanion hole residues are in the catalytic domain. The cap domain contributes substrate binding residues.
3. Canonical esterase mechanism
The canonical esterase mechanism uses the core Ser-His-Asp catalytic machinery, and is the reference point for other mechanisms discussed in this review. This mechanism is a ping-pong bi-bi reaction involving an acyl-serine enzyme intermediate, Scheme 1. Either the formation or release of the acyl enzymes may be rate limiting. The acyl enzyme forms via the first tetrahedral intermediate and is released via a second tetrahedral intermediate. To optimize these reactions, computations suggest that the catalytic triads of esterases adopt a rigid compromise geometry that minimizes reorganization during this multistep mechanism.32
Scheme 1.
Canonical esterase mechanism for hydrolysis of methyl acetate, a typical ester. The top left structure shows the free enzyme with the catalytic triad (Asp-His-Ser) and the oxyanion hole (two main chain amide N-H’s). The first step is binding the substrate, methyl acetate, in the active site. Attack of serine on the ester carbonyl carbon yields the first tetrahedral intermediate, Td1. The catalytic histidine acts as a base to deprotonate the serine in this step. Reformation of the carbon-oxygen double bond leads to the release of methanol and the formation of the acyl enzyme intermediate. Histidine acts as an acid in this step enabling the leaving group to be methanol instead of methoxide. Next, water binds to the acyl enzyme intermediate. The active site histidine again acts as a base to deprotonate the water so that it can attack the carbonyl carbon of the acyl enzyme to form the second tetrahedral intermediate, Td2. Lastly, reformation of the carbon-oxygen double bond releases acetic acid and restores the free enzyme state. To draw the mechanistic steps clearly, only selected lone pairs of electrons are shown.
When acylation is faster than deacylation, the acyl enzyme intermediate accumulates and can be characterized by spectroscopy, by mass spectrometry and even by X-ray crystallography. Typically, either mutagenesis of key residues or unusual reaction conditions slow release of the acyl enzyme. Crystal structures of the acyl enzyme intermediate have been solved for juvenile hormone esterase (pdb id: 2fj033) and palmitoyl protein thioesterase (pdb id:1eh534). Surprisingly, a carboxylesterase Est30 crystal structure showed a tetrahedral intermediate (pdb id: 1tqh35). The crystallization conditions presumably stabilized this intermediate allowing it to accumulate (Figure 2 above).
In other cases, crystallographers solved crystal structures of phosphonates or sulfonate covalently linked to the catalytic serine. Phosphonates mimic the first tetrahedral intermediate, while sulfonates mimic the second tetrahedral intermediate, Chart 1. Structures of phosphonates bound to the active site have been solved for many lipases; for example, Burkholderia cepacia lipase (pdb id: 1hqd,36 1ys137), two Candida lipases (pdb id: 1lbs,38 1lpm39), and human lysosomal phospholipase A2 (pdb id: 4×9540) A structure of sulfonate bound to an esterase (pdb id: 3ia241) has also been solved. These structures, combined with detailed kinetic studies, firmly established the canonical esterase mechanism.
Chart 1.

Phosphonates mimic the first tetrahedral intermediate in the hydrolysis of esters just before loss of the alcohol, while sulfonates mimic the second tetrahedral intermediate where water has added to the acyl enzyme intermediate.
4. Overview of differences in reaction mechanisms
Table 1 lists seventeen different reaction mechanisms catalyzed by α/β-hydrolase fold enzymes with a serine-histidine-aspartate catalytic triad and the reasons that new mechanistic steps arise. Table S1 lists example reactions along with summaries of the mechanistic details. The rest of this review examines each reaction in more detail. Researchers have identified the mechanistic details of most of these reactions with confidence; the discussion below notes when the steps are still speculative.
Table 1.
Possible reasons why the mechanisms of Ser-His-Asp enzymes in the α/β-hydrolase fold family change to create seventeen different reactions mechanisms.
| mechanisma | Ser/OxHb | substrate requires new mechanistic steps | different binding of substrate creates new mechanistic steps | new H-bonds or general acids/bases create new mechanistic stepsc |
|---|---|---|---|---|
| hydrolase-type | ||||
| perhydrolase(EC 1.11.1.18)promiscuous | yes/yes | no | H-bond acceptor speeds formation of acyl enzyme | |
| acyl transferase(EC 2.3.1) | yes/yes | no | Instead of water, the acceptor (alcohol or amine) binds to nucleophile site | H-bond donor deactivates water |
| canonical esterase(also lipase, thioesterase)(EC 3.1.1 & 3.1.2) | yes/yes | no | reversed binding of ester possible, which prevents histidine interaction with leaving alcohol. Not counted as a different mechanism. | |
| lactonase(EC 3.1.1.81) | yes/yes | lone pairs of leaving group oxygen are incorrectly oriented in tetrahedral intermediate to be protonated | general acid from lid domain replaces catalytic histidine to protonate leaving oxygen. | |
| phosphotriesterase (EC 3.1.8) | yes/yes | yes, too crowded for water to attack from the normal location | a new histidine acts as general base/acid for water attack from a different direction | |
| peptidase (EC 3.4 & 3.5) | yes/yes | tetrahedral intermediate forms in non-reactive orientation of leaving amide N–H | H-bond acceptor from lid domain or from substrate speeds inversion at nitrogen to move lone pair into reactive orientation | |
| MCP hydrolase(EC 3.7.1.8) | yes/yes | anionic substrate is unreactive to nucleophilic attack; protonation by serine starts reaction | substrate bound in less stable non-planar conformation to limit delocalization of negative charge | substrate acts as general base |
| DHPON hydrolase(EC 3.7.1.19) | yes/yes | tautomerization forms carbonyl needed for reaction | glutamate, oriented by an arginine from lid, catalyzes tautomerization | |
| cis-trans isomerase(EC 5.1.99)promiscuous | no, relactonization competes with hydrolysis of acyl enzyme | |||
| lyase-type | ||||
| direct epoxidation & conjugate addition(EC 1.11, 4.1.99, 4.2.99, 4.3.3, 4.4.1)promiscuous | no/yes | yes, hydrolysis not possibled | ||
| dioxygenase(EC 1.13) | yes/yes | yes, hydrolysis not possibled | dioxygen, not carbon group binds in oxyanion hole | |
| decarboxylase(EC 4.1.1) | no/no | yes, hydrolysis not possibled | blocked oxyanion hole requires carbonyl to bind in different location | catalytic histidine acts as general acid, not base |
| (S)-HNL(EC 4.1.2.47) | yes/no | yes, hydrolysis not possibled | blocked oxyanion hole requires carbonyl to bind in different location | lysine stabilizes leaving cyanide |
| (R)-HNL(EC 4.1.2.10)promiscuous | yes/yes | yes, hydrolysis not possibled | proposed nitrile binding in oxyanion hole | |
| aldolase(EC 4.1.2)promiscuous | no/yes | yes, hydrolysis not possibled | ||
| 1,4-elimination(EC 4.2.99.20) | yes/yes | yes, hydrolysis not possibled | ||
| racemasee(EC 5.1.99)promiscuous | no/yes | yes, hydrolysis not possibled | proposed nitrile binding in oxyanion hole | |
Promiscuous reactions are indicated. MCP = meta-cleavage product in aromatic compound degradation; DHPON = 2,6-dihydroxy-pseudo-oxynicotine; HNL = hydroxynitrile lyase.
Indicates whether catalytic serine is essential for the reaction and whether the oxyanion hole is used in the mechanism.
Hydrogen bond partners and general acids/bases come from the catalytic domain unless otherwise noted.
The substrate cannot hydrolyze, thereby opening up new mechanistic possibilities.
Proposed mechanism; limited information available.
Figure 3 groups these reactions according to differences in mechanism. Many hydrolases – esterases, thioesterases, and lipases – follow the canonical esterase mechanism, while the remaining sixteen reaction mechanisms differ. Eight of these sixteen are variations on the canonical mechanism. They involve an acyl enzyme intermediate and use the oxyanion hole to stabilize tetrahedral intermediates, but differ either in the formation or the cleavage of the acyl enzyme intermediate. The remaining eight mechanisms are lyase-type mechanisms without an acyl enzyme intermediate. The lyase-type mechanism uses only general acid-base catalysis together with binding to orient substrates for reaction. In four of the lyase-type mechanisms, the catalytic serine is not required. The serine does not prevent reaction, so we include it with serine-histidine-aspartate triad reactions.
Figure 3.

Venn diagrams of differences between the seventeen different reactions catalyzed by α/β-hydrolase fold enzymes with a Ser-His-Asp catalytic triad. Nine reactions follow a hydrolase-mechanism with an acyl enzyme intermediate. The first is the canonical esterase mechanism. Eight other mechanisms are similar, but differ in either the formation of the acyl enzyme (five examples) or the release of the acyl enzyme (three examples). Eight reactions follow a lyase-type mechanism where no acyl enzyme forms. Some of these reactions do not require active site serine, while the others do, but use it differently. The oxyanion hole is used by most reactions to bind the oxygen of a carbonyl, but it can also bind a nitrile nitrogen, a dioxygen molecule, or nothing at all. The decarboxylase mechanism does not use the catalytic serine, nor the oxyanion hole.
In the hydrolase mechanisms, the oxyanion hole always binds the carbonyl oxygen and stabilizes the oxyanion of the tetrahedral intermediates. For the lyase-type mechanisms, the role of the oxyanion hole is more varied. The oxyanion hole binds a carbonyl oxygen in four cases, but it stabilizes an enolate or similar species, not a tetrahedral intermediate. The oxyanion hole also binds a nitrile nitrogen in two cases, nothing in another two cases, and dioxygen in one case.
Half of the sixteen non-canonical mechanisms involve new hydrogen bonds (four examples) or new general acids/bases in the mechanisms (four examples, Table 1). Usually the new interactions come from the enzyme, but sometimes they come from within the substrate. In another case the the catalytic histidine makes a new interaction. Usually histidine acts as a base, but in the decarboxylase, it acts as an acid.
Although it is the enzyme doing the catalysis, the substrate can create new steps by preventing the usual steps from occurring. For example, in many lyase reactions the substrate is an aldehyde or ketone that cannot hydrolyze. The catalytic histidine can still deprotonate the serine Oγ, and this nucleophile may attack the carbonyl carbon to form a tetrahedral intermediate; however, this tetrahedral intermediate can only revert to the starting compounds. In these cases, the serine Oγ may act as a base to form an enolate and this enolate can lead to new reactions. Thus, the inability of the substrate to follow the canonical mechanistic steps allows new steps to occur.
X-ray crystal structures have also been solved for enzymes that follow variations of the canonical esterase mechanism. Structures of covalent acyl enzyme intermediates bound to two acyltransfersases (deacetylcephalosporin C acetyltransferase; pdb id: 2vax42 and homoserine transsuccinylase; pdb id: 2vjd43), a peptidase (tricorn-interacting factor F1 peptidase; pdb id: 1xqw44), and a variant of a C-C hydrolase (MCP hydrolase; pdb id: 3v1n45) were solved. The structure of a sulfonate bound to kynurenine formamidase (pdb id: 4e1446) was reported and tetrahedral intermediates bound to human dipeptidyl peptidase IV (pdb id: 1nu8,24 1r9n25) and lactonase (pdb id: 1hl747) were reported.
Evidence supporting the lyase type mechanism was obtained from crystal structures of hydroxynitrile lyases with substrate acetone cyanohydrin (pdb id: 1sc948) and inhibitor benzoic acid (pdb id: 1gxs49). Bound product in the active site of menaquinone synthase also supports the proposed mechanism (pdb id: 4mys50).
The variations in mechanism come from both natural and promiscuous reactions. Using the canonical mechanism as the reference point, the remaining sixteen mechanisms in Table 1 are variations. Among the eight variations on the hydrolase mechanism, only two are for promiscuous reactions. Among the eight lyase mechanisms, four are for promiscuous reactions. Overall, ten of the variations in mechanisms are natural reactions, while six are promiscuous reactions.
5. EC 1 oxidoreductases
EC 1.11 peroxide as donor
The oxidation of organic substrates with hydrogen peroxide catalyzed by lipases or esterases is a promiscuous, unnatural reaction. Two different mechanisms were discovered. One is a direct oxidation, while the other is an indirect oxidation where the enzyme catalyzes perhydrolysis to form a peroxy acid and this peroxy acid spontaneously oxidizes the organic substrate. The overall reaction is the same in both cases - incorporation of one oxygen from hydrogen peroxide into the substrate. The EC number is the same, but the mechanisms differ.
Direct epoxidation
Lipase B from C. antarctica catalyzes a promiscuous direct epoxidation of α,β-unsaturated aldehydes with hydrogen peroxide, Scheme 2A.51 The reaction occurs in the active site because no reaction occurs when a known active-site binding inhibitor blocks the active site. The reaction is similarly fast with either WT CAL-B or the mutant where alanine replaces the active site serine; thus, no acyl enzyme intermediate is possible. Also, there cannot be a peracid intermediate (see perhydrolysis below) because the reaction mixture did not contain carboxylic acids or esters. The apparent kcat was ~0.5 s−1 and the reaction showed no enantioselectivity.
Scheme 2.
Direct epoxidation of α.β-unsaturated aldehydes with hydrogen peroxide catalyzed by CAL-B or the CAL-B S105A variant. A) The epoxidation occurs in buffer or organic solvent and yields racemic epoxide. The active site serine is not needed for catalysis so no acyl enzyme intermediate can form. B) The proposed mechanism supported by calculations involves deprotonation of H2O2 by the active site histidine and stabilization of the oxyanion intermediate by the oxyanion hole.
Since no acyl enzyme forms, this is a lyase-type mechanism. Experiments and theoretical calculations suggest that catalysis involves 1) base activation of the hydrogen peroxide by the catalytic histidine, 2) stabilization of oxyanion intermediate by the oxyanion hole and 3) positioning the reactants for reaction, Scheme 2B. The reaction likely follows a two-step mechanism. First, the addition of hydrogen peroxide to the β-carbon to form an oxyanion intermediate and second, the collapse of this intermediate to form an epoxide and eliminate water. The catalytic histidine deprotonates the attacking oxygen of hydrogen peroxide in the first step and protonates the leaving water oxygen in the second step. The oxyanion hole of CAL-B contains three potential hydrogen bond donors: main chain N–H of Gln106, main chain N–H of Thr40 and γ-OH of Thr40. Calculations suggest that only the last two contribute to stabilization of the oxyanion intermediate in this case. There are no special hydrogen bonds or general acid/base interactions. In this review, we call this mechanism the conjugate addition mechanism. Several other reactions discussed in section 8 below follow this mechanism.
The mechanism cannot be a hydrolysis because the substrate – an α,β-unsaturated aldehyde – cannot hydrolyze. Nevertheless, similar mechanistic steps occur during the new reaction. The carbonyl group binds in the oxyanion hole which polarizes the carbonyl as in hydrolysis of an ester. Since the substrate contains an α,β-unsaturated double bond, this α,β-bond is also polarized for nucleophilic attack at the β-position. The catalytic histidine acts as a base and can deprotonate the catalytic serine, a water molecule or a hydrogen peroxide molecule. Any of these three can attack the carbonyl carbon, but these attacks form tetrahedral intermediates that can only return to starting materials. With some adjustments in structure, any of these three nucleophiles can attack the β-carbon. Only the attack of hydrogen peroxide creates a path to a stable product. Thus, the binding of a new substrate, which cannot undergo the natural reaction, leads to a new intermediate. The role of the catalytic histidine as general base is the same as its role in hydrolysis.
Perhydrolysis (bromoperoxidase)
Bromoperoxidases, for example bromoperoxidase A1 Streptomyces aureofaciens,52 catalyze the formation of hypobromite from bromide and hydrogen peroxide, Scheme 3A. The resulting hypobromite reacts spontaneously with organic compounds to introduce bromine. Most bromoperoxidases contain a heme or metal cofactor, but bromoperoxidases in the α/β-hydrolase fold family do not and are called cofactor-independent bromoperoxidases. Their peroxidase activity requires acetate or other carboxylic acids and is more precisely called perhydrolase activity.53,54 These cofactor-independent bromoperoxidases catalyze the conversion of acetate and hydrogen peroxide to peracetic acid. Next, the peracetic acid, which is more reactive than hydrogen peroxide, reacts with bromide to form hypobromite, Scheme 3B. This hypobromite reacts with organic compounds as before. Thus, the bromoperoxidase activity of α/β-hydrolases is an enzyme-catalyzed perhydrolysis of carboxylic acids. In this review, the bromoperoxidases are called perhydrolases to emphasize the chemical step that they catalyze.
Scheme 3.

Enzyme-catalyzed perhydrolysis accounts for the bromoperoxidase activity in the α/β-hydrolase family. A) Bromoperoxidases catalyze the formation of hypobromite from bromide and peroxides like hydrogen peroxide. In a second step, hypobromite reacts spontaneously with organic compounds like monochlorodimedone to replace a hydrogen with bromine. B) Some α/β-hydrolases show bromoperoxidase activity in acetate buffer. The α/β-hydrolase catalyze the perhydrolysis of acetic acid to form peracetic acid. Next, the peracetic acid spontaneously reacts with bromide to form hypobromite. This hypobromite can spontaneously brominate organic compounds as in panel A.
Since water and hydrogen peroxide are similar, one might expect that all hydrolases would also catalyze perhydrolysis and that the mechanisms would be the same. Surprisingly, only some hydrolases catalyze perhydrolysis and those that do vary in their abilities. A good perhydrolase has a kcat > ~5 s−1. Below we identify a hydrogen bond acceptor interaction that improves the perhydrolysis activity, but there are likely other features, currently unknown, that promote or hinder perhydrolysis. Perhydrolysis is a promiscuous unnatural reaction, the natural function of these esterases that catalyze perhydrolysis of carboxylic acids may be lactone hydrolysis.55,56
Pseudomonas fluorescens esterase (PFE) catalyzes the slow perhydrolysis of acetic acid, kcat ~0.1 s−1, Scheme 4. Two variants of this esterase are much better catalysts for perhydrolysis. PFE-L29P catalyzes perhydrolyis 43-fold faster57 (kcat ~5 s−1) and PFE L29I catalyzes perhydrolysis 83-fold faster58 (kcat ~10 s−1).
Scheme 4.

Perhydrolysis of acetic acid yields peracetic acid, top equation. The reaction involves an acetyl enzyme intermediate, whose formation limits the reaction rate, bottom equation.
There are two ways that the reaction mechanism can change to increase the rate of perhydrolysis. Perhydrolysis of acetic acid involves formation of an acetyl serine intermediate followed by cleavage of the intermediate by hydrogen peroxide, Scheme 4. The rate determining step of perhydrolysis is the first step, formation of the acetyl enzyme intermediate. Increasing the rate of acetyl enzyme formation would increase the rate of perhydrolysis. Another possibility is to increase the selectivity of the cleavage step for hydrogen peroxide over water. Reaction with water regenerates the starting acetic acid, while reaction with hydrogen peroxide yields product.
Both PFE variants are better perhydrolases because they form the acyl enzyme faster; their selectivity for hydrogen peroxide over water is unchanged or lower as compared to wild type. For example, PFE-L29P catalyzed the isotope exchange between acetic acid and H218O 26 times faster than the wild-type, an increase that is similar to the 43-fold increase in kcat for perhydrolysis. The selectivity of L29P PFE for hydrogen peroxide over water was up to two fold lower than that for wild type.57
X-ray crystal structures suggest a molecular basis for the faster formation of acetyl enzyme, Chart 2. Thus, perhydrolysis follows the canonical hydrolase mechanism, but the formation of the acetyl enzyme is faster due to an extra hydrogen bond acceptor from the enzyme.
Chart 2.
Active site differences between esterase and variants with higher perhydrolase activity affect the rate of acetyl enzyme formation. A) In variant PFE-L29P, the main chain carbonyl of Trp28 accepts a hydrogen bond from the leaving water of the tetrahedral intermediate. This interaction speeds acetyl enzyme formation. B) In wild type PFE, this carbonyl group is farther from the active site. It can also accept a hydrogen bond via a water molecular bridge, but this the bridge can also donate a hydrogen bond, which would hinder water loss. C) In variant PFE-L29I, an acetate, held by the main chain amides of Ile29 and Leu30, accepts a hydrogen bond. A negatively charged acetate is a better hydrogen bond acceptor than water, which may account for the faster perhydrolysis catalyzed by PFE-L29I as compared to PFE-L29P. D) The general mechanism for a faster perhydrolase is to position a hydrogen bond acceptor for the leaving water in the tetrahedral intermediate.
Other perhydrolases in the α/β-hydrolase family catalyze perhydrolysis of esters.59 These perhydrolases are also serine hydrolases, but lack the proline in the oxyanion loop. The rate determining steps and competing reactions for these ester perhydrolases differ from the acid perhydrolases described above, but the mechanism is similar to the canonical esterase mechanism. Ester perhydrolases are not included in this review.
EC 1.13 dioxygenase
Dioxygenases catalyze the addition of both atoms of oxygen to a substrate, typically using metal or organic cofactors.60,61 One exception is an α/β-hydrolase fold family dioxygenase, which does not use metals or cofactors. The 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (HOD) from Arthrobacter nitroguajacolicus Rü61a converts 1H-3-hydroxy-4-oxoquinaldine (QND) to carbon monoxide and N-acetylanthranilate, Scheme 5.62,63 This dioxygenase cleaves two carbon-carbon bonds using oxygen as the oxidant. Direct reaction of organic compounds with oxygen is usually slow because electrons are paired in organic compounds, while oxygen has unpaired electrons. The key step of this dioxygenase is deprotonation of the substrate to create an anion that is intrinsically reactive toward molecular oxygen.64 The higher electron energy of an anion allows a single electron transfer from the anion to oxygen to form a [substrate radical – superoxide anion radical] pair.
Scheme 5.
Bacterial oxidation of quinaldine (R = CH3) to catechol. The α/β-hydrolase 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (HOD) converts 1H-3-hydroxy-4-oxoquinaldine (QND) to carbon monoxide and N-acetylanthranilate by oxidative cleavage of the C2–C3 and C3–C4 bonds.
Like other α/β-hydrolase enzymes, the active site is at the interface between the catalytic and cap domains. The binding site is flat and circular with a basin at the end containing the oxyanion hole residues. Steady-state kinetics suggest that 1H-3-hydroxy-4-oxoquinaldine (QND) binds first in the flat portion of the active site, Scheme 6.65 The catalytic serine oxygen donates a hydrogen bond to the C4 carbonyl oxygen of QND, the 3-OH group of QND donates a hydrogen bond to the catalytic histidine and the NH of QND donates a hydrogen bond to the main chain carbonyl oxygen of Trp36. Replacement of serine with alanine weakens substrate binding, but does not affect the reaction rate,65 leading to the proposed mechanism involving only the His-Asp dyad.
Scheme 6.
Proposed reaction mechanism proposed for co-factor free dioxygenase cleavage of QND with oxygen. The reaction does not involve an acyl enzyme intermediate and the Ser-His-Asp triad serves only as a general acid-base catalyst. The aspartate residue of the catalytic triad (Asp126) is not shown for clarity. Similarly only selected electrons are shown. Dioxygen is shown in green.
To start the reaction, the catalytic histidine deprotonates the 3-OH.66 The oxyanion hole binds molecular oxygen below the substrate’s ring.67 The overall positive electrostatic potential of the oxyanion hole region suggests a single electron transfer from the substrate to oxygen to form a [substrate radical – superoxide anion radical] pair.67 Coupling of these two radicals forms a peroxide intermediate, which can attack the carbonyl, forming the endoperoxide. Models of the endoperoxide intermediate in the active site suggest a close interaction between Trp160 and leaving CO that destabilizes this intermediate, favoring the release of CO. In the final step, the organic substrate removes a proton from His251 and leaves the active site.
Thus, the key features that enable this new chemistry are 1) binding of the quinaldine and oxygen substrates, 2) the inability of the substrate to undergo hydrolysis and 3) electrostatic environment that favors the electron transfer to form the [substrate radical – superoxide anion radical] pair.
6. EC 2 transferase
EC 2.3 acyltransferase
Acyltransferases (Enzyme Commission number 2.3) catalyze the transfer an acyl group from a donor, usually an ester or thioester, to an acceptor, usually an alcohol or amine thereby forming esters or amides. For example, the enzyme DAC-acetyltransferase catalyzes the transfer of an acetyl group from acetyl-CoA to the alcohol deacetylcephalosporin C (DAC) in the final step in the synthesis of the antibiotic cephalosporin C, Chart 3.42 Like hydrolases, acyltransferases form an acyl enzyme intermediate, but acyltransferases transfer this acyl group to an acceptor, avoiding hydrolysis.
Chart 3.
A) The final step of cephalosporin C biosynthesis involves acetyl transfer from acetyl-CoA to deacetyl cephalosporin C (DAC). B) An X-ray structure shows many hydrogen bonds and ion pair interactions that bind the alcohol DAC (blue lines) in the alcohol site of DAC-acetyltransferase. These interactions favor binding of this alcohol instead of water and therefore favor acetyl transfer over acetyl hydrolysis. The structure shows the catalytic serine 149 in the acetylated form.
The mechanistic difference between an acyl transferase and esterase occurs in the second stage – cleavage of the acyl enzyme intermediate, Scheme 7. Acyl transferases favor reaction with an amine or alcohol acceptor, while esterases favor reaction with water. This difference is not absolute: hydrolysis of the acyl donor is a side reaction of acyltransferases, and esterases in an organic solvent catalyze acyl transfer to acceptors other than water. Examples include resolution of alcohols by enantioselective acylation, regioselective acylation of sugars, and synthesis of polymers by ring-opening polymerization of lactones.68 This preference in acyltransferases comes from both features that favor the amine or alcohol acceptor over water and features that disfavor water.
Scheme 7.
Like hydrolases, acyl transferases form an acyl-serine enzyme intermediate, but unlike hydrolases, acyl transferase transfer this group to an acceptor (usually an alcohol or amine), not to water. The acyl donor is usually an ester or thioester.
In some cases, crystal structures show specific binding interactions between the alcohol acceptor and the alcohol-binding site of the acyltransferase. These interactions favor binding of the alcohol over water and thus, favor acyl transfer over hydrolysis. For example, the active site of acyltransferases Srf TE and Mycobacterium antigens are hydrophobic so favor binding of a hydrophobic acceptor. In a second example, the enzyme DAC-acetyltransferase also tightly binds the alcohol acceptor DAC (KM = 0.04 mM;69 0.3 mM70). An X-ray crystal structure of DAC bound to DAC-acetyltransferase (pdb id: 2vav42) shows multiple hydrogen bonds and ion pair interactions that bind and orient this alcohol for reaction, Chart 3. In a third example, homoserine transacetylase catalyzes the transfer of the acetyl group from acetyl-CoA to homoserine. A model of the tetrahedral intermediate for acetyl transfer to homoserine based on the X-ray structure of the enzyme shows ion pairs between the homoserine amino group and Asp338 and the homoserine carboxyl group and Arg212.71 Both of these residues are absolutely conserved in homoserine transacetylases. This binding positions the homoserine hydroxyl in the ideal location for acetyl transfer from Ser143.
Acyl transferases may also reduce the reactivity of water by a subtle difference in the conformation of the oxyanion loop in the active site. The conformation of the oxyanion loop differs in acyltransferases and esterases,41 Chart 4. In esterases, an amide carbonyl oxygen points toward the alcohol binding region, while in acyl transferases, an amide N–H points to this region. An X-ray structure of an esterase containing a bound sulfonate transition state analog shows a bridging water molecule that can act as a base to increase the nucleophilicity of the attacking water molecule. In contrast, the interaction in acyltransferases involves the acidic N–H, which decreases the reactivity of water. This activation or deactivation disappears for alcohol nucleophiles because the alcohol R group displaces the bridging water molecule.
Chart 4.
A different main chain conformation of the oxyanion loop in acyltransferases can lower the reactivity of water, but not alcohols. In esterases (top), the main chain carbonyl oxygen acts as a base via a bridging water molecule to activate the attacking water molecule. In acyltransferases, the N–H acts as an acid via a bridging water molecule to deactivate the attacking water molecule. For the acyl transfer reaction, the attacking nucleophile is an alcohol, which is larger than water. This alcohol displaces the bridging water molecule thereby eliminating any activating or deactivating effects. Acyltransferases may also favor binding of the incoming alcohol nucleophile.
Several lipases favor acyl transfer to alcohols59,72,73 or hydroxylamine74–76 ( over hydrolysis even in aqueous solution. For example, lipase from Candida parapsilosis converts ethyl oleate and hydroxylamine to the oleylhydroxamic acid in 40% yield.74 The preference for alcohols is likely due to a hydrophobic nucleophile site that favors binding an alcohol over water. Engineering a more hydrophobic nucleophile site increased the acyltransferase versus hydrolase activity of C. antarctica lipase A by ~50%.77 The preference for hydroxylamine is in part because hydroxylamine is a good nucleophile, but other factors must also contribute since enzymes differ in their preference for hydroxylamine over water.
Thus, acyl transferases follow the canonical esterase mechanism, but differ in the release of the acyl enzyme. Transfer to an acceptor is favored over water by binding site changes that favor binding of the acceptor and a hydrogen bond donation that deactivates water.
7. EC 3 hydrolases
EC 3.1.1 C–O carboxylic acid esterases
Esterase
Section 3 above described the canonical esterase mechanism. This is a natural reaction catalyzed by many hydrolases. For example, an esterase in tobacco catalyzes hydrolysis of methyl salicylate as part of defense signal.78
Here we describe an example of different substrate binding, which changed the mechanism of an ester hydrolysis by preventing the catalytic histidine from acting as a general acid. We do not include this variation as a new mechanism among the seventeen possible mechanisms.
Esterase 2 from Alicyclobacillus acidocaldarius catalyzes the hydrolysis of both n-hexanoate and n-dodecanoate p-nitrophenyl esters, but the rate limiting step differs for the two substrates.79 For the n-hexanoate, formation of the acyl enzyme is fast, as expected for an ester with a good leaving group like p-nitrophenol. Cleavage of the acyl enzyme is the rate limiting step. However for the n-dodecanoate, formation of the acyl enzyme is surprisingly slow, and limits the rate. Pre-steady state kinetics show a burst of p-nitrophenol release for the n-hexanoate consistent with fast acylation, but no burst for the n-dodecanoate.
A crystal structure of an enzyme variant containing n-hexadecanoylsulfonate covalently bound to the active site serine suggested an explanation - the two substrates bind differently. The n-hexanoyl substrate likely binds normally with the n-hexanoyl group in the acyl binding site and the p-nitrophenol in the alcohol binding site. The reaction mechanism follows the canonical esterase mechanism. In contrast, the n-hexadecanoyl inhibitor, and likely the n-dodecanoyl substrate, are too long for the acyl binding site and instead bind in a reversed orientation with the acyl group in the alcohol binding site and the p-nitrophenyl group in the acyl binding site. This reversal places the oxygen of the p-nitrophenol leaving group too far from the catalytic histidine to accept a hydrogen bond. This lack of a hydrogen bond slows the collapse of the first tetrahedral intermediate and accounts for the slower acylation for this substrate. The details of the mechanism for release of the acyl enzyme are unknown; one possibility is that the longer dodecanoate moves to bind to the acyl binding site and the mechanism proceeds as in the canonical esterase mechanism.
Thus, ester hydrolysis normally follows the canonical esterase mechanism, but the details may differ in individual cases.
Lipase
Lipases catalyze the hydrolysis of triglycerides and also follow the canonical esterase mechanism. For example, lipase from C. rugosa (pdb id: 1trh80) is a broad specificity enzyme with applications such as generation of flavors from milk fats. The key difference between lipase activity and esterase activity is that triglycerides are water-insoluble substrates. Lipases contain a hydrophobic region on their lid domains. This hydrophobic region binds them to the water-triglyceride interface where the triglyceride can partition into the active site.81
Lactonase
Although lactones are simply cyclic esters, their shapes differ due to the different conformation along the Ccarbonyl–Oalcohol bond, Chart 5. Esters have a strong preference (4–5 kcal/mol) to orient the alcohol carbon cis to the carbonyl oxygen, in part because this orientation cancels the dipoles along the carbonyl C=O and the Calcohol–Oalcohol bond. In lactone rings smaller than eight, this conformation is impossible, so the alcohol carbon orients trans to the carbonyl oxygen.
Chart 5.

The cyclic nature of a lactone creates a different shape in the alcohol part as compared to an ester. In esters, the alcohol carbon orients cis to the carbonyl oxygen to cancel the two dipoles shown. In lactones, rings smaller than eight force the alcohol carbon trans to the carbonyl oxygen. The different alcohol carbon location also changes the location of the lone pairs on the alcohol oxygen in esters versus lactones.
The different shape of lactones as compared to esters requires two differences in a lactonase as compared to an esterase. First, the binding site for a lactone should favor the cyclic form of a lactone over the extended form of an ester. The X-ray structure of N-acyl homoserine hydrolase from Ochrobactum sp. indeed shows such a binding site.82 This lactonase catalyzes the hydrolysis of the homoserine lactone ring, Scheme 8. To bind the substrate in the active site without reaction, the researchers used the inactive Ser102Gly variant. The structure reveals an active site similar to esterases, but with no room to accommodate an alcohol group in the extended conformation. This lack of space favors binding of lactones over esters. In another case, reducing the size of the alcohol binding site in an esterase increased its affinity for the lactone more than 50-fold.55
Scheme 8.
Reaction and key catalytic steps catalyzed by N-acyl homoserine lactone hydrolase. A) The lactonase catalyzes hydrolysis of the homoserine lactone ring. B) Proposed mechanism for formation of the acyl enzyme intermediate based on the X-ray structure with bound substrate (pdb id: 4g8b). The lone pairs on the lactone’s alcohol oxygen point away from the catalytic histidine and accept a hydrogen bond from Tyr160. We propose that this residue, which is essential for catalysis, acts as the proton donor in place of the catalytic histidine.
The second difference required by the different shape is an alternative proton donor. The lone pairs on the alcohol oxygen of a lactone point in the opposite direction as compared to esters. The lone pairs in lactones point away from the catalytic histidine; protonation requires a proton donor in a different orientation. The crystal structure of the inactive Ser102Gly variant of N-acyl homoserine lactonase with bound substrate shows cap residue Tyr160 positioned to donate a proton to the lactone, Scheme 8. Mutagenesis of Tyr160 to glycine eliminated lactonase activity showing that Tyr160 is essential for catalysis. We propose that this tyrosine acts as the proton donor in place of the catalytic histidine as shown below. Tyr160 lies opposite His248 in the active site in an ideal location to protonate a lactone substrate.
EC 3.1.2 thioesterases
Thioesterases catalyze hydrolysis of thioesters, often protein thioesters, Scheme 9. The two available structures of thioesterases in the α/β-hydrolase-fold family did not reveal features unique to a thioesterase, nor did these thioesterases have a clear preference for thioesters. The active site of palmitoyl protein thioesterase is similar to that of an esterase.34 Although this thioesterase did not catalyze hydrolysis of any of the oxoesters tested,83 the ester substrates were not exact mimics of the thioesters, so shape specificity may also have contributed to the lack of activity. Thioesters are also more reactive than oxoesters in general. The active site structure of myristoyl acyl carrier protein thioesterase from Vibrio harveyi, which also catalyzes hydrolysis of oxoesters, showed a missing N-H donor for the oxyanion hole, so the structure may not reveal the catalytically active conformation. The authors suggested that catalysis might require a structural rearrangement similar to that required in some lipases84 (Lawson et al.; 1994). Thus, thioesterase catalysis likely follows the canonical esterase mechanism.
Scheme 9.
Palmitoyl-protein thioesterase (EC 3.2.1.22) catalyzes hydrolysis of palmitoyl cysteine residues on the surface of H-Ras protein.83
EC 3.1.8 phosphate triester hydrolases
Organophosphates can irreversibly inhibit serine esterases by forming stable covalent adducts with the catalytic serine, see Chart 1 above. Organophosphorus compounds are used as insecticides, drugs and nerve agents. For example, the anti glaucoma drug echothiophate acts by forming a covalent adduct with cholinesterases, Scheme 10. Loss of the thiol leaving group leaves a stable phosphoryl enzyme intermediate. Water cannot attack this intermediate because 1) the water binding site is blocked by one of the O-alkyl groups of the phosphonate, 2) the active site histidine is still protonated and cannot act as a base and 3) the phosphorus center is more crowded than the carbon in an acyl enzyme intermediate.
Scheme 10.
Organophosphates inhibit esterases by forming stable phosphoryl enzyme intermediates, but a histidine in a new region of the active site can promote hydrolysis. A. Slow hydrolysis of the cholinesterase inhibitor echothiophate by a human butyrylcholinesterase variant. B. The Gly117His substitution allows positioning and activation of a water molecule to hydrolyze phosphorylserine intermediate.
Insects evolve resistance to insecticides by enabling new mechanistic steps that allow hydrolysis of the intermediate. The new steps, enabled by a single amino acid substitution, activate a water molecule in another part of the active site. For example, the Glu137Asp replacement in Lucilia cuprina (blowfly) carboxylesterase confers resistance to the organophosphate insecticide chlorfenvinphos.85 Glu137 originates from the catalytic domain. Mutation of the equivalent residue in butylcholinesterase, Gly117His, enabled it to catalyze hydrolysis of ecothiophate and the insecticide paraoxon, Scheme 10.86,87 The new histidine activates a water molecule in a region different from the normal water binding site and promotes attack at the phosphorus, Scheme 10B. Although this rate is slow, it corresponds to a 100,000-fold enhancement over the spontaneous rate. Thus, the new general base creates a new mechanism.
EC 3.4 C–N bond hydrolysis in peptides: peptidases
Peptidases or proteases catalyze the hydrolysis of C–N bonds in peptides. For example, prolyl oligopeptidase (EC 3.4.21.26) cleaves the amide bond after proline in neuropeptides such as oxytocin.88 For assays, researchers often use chromogenic substrates, Scheme 11.
Scheme 11.
Prolyl oligopeptidase catalyzed catalyzes the hydrolysis of amide links after proline. The natural substrates are various peptides; the substrate shown is a chromogenic substrate often used in assays.
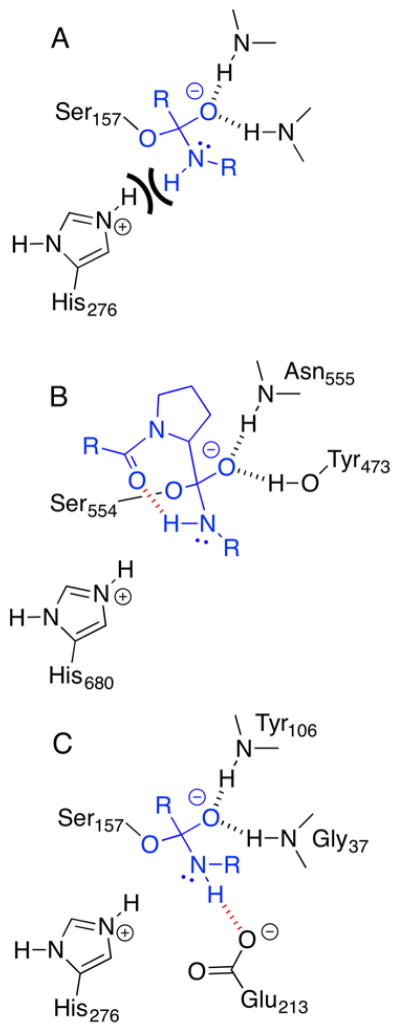
While esterases have similar active sites to peptidases, esterases do not catalyze peptide hydrolysis or catalyze hydrolysis very slowly. One hypothesis focuses on the oxygen atom in esters versus the N-H at the corresponding position in amides. Syrén and Hult89 proposed that the hydrogen atom on the amide nitrogen may disrupt formation of the key catalytic hydrogen bond between the catalytic histidine and the nitrogen atom of the amide substrate. The ester oxygen does not have an attached hydrogen atom, so a similar disruption cannot occur, Chart 6A.
Chart 6.
The N–H on an amide may interfere with catalysis, but peptidases avoid this interference with hydrogen bond acceptors that speed inversion at the nitrogen. A) Breakdown of the first tetrahedral intermediate in the hydrolysis of an amide requires protonation of the nitrogen. If the N-H of the amide, not the lone pair of the amide, points toward the catalytic histidine, it prevents protonation. B) Prolyl endopeptidases use an intramolecular hydrogen bond promote inversion of the nitrogen. The prolyl carbonyl oxygen of the prolyl peptide substrate accepts a hydrogen bond from the N-H of the leaving amide. This drawing is based on an X-ray structure of the catalytically inactive Ser554Ala variant complexed with an octapeptide91 (pdb id:1e8m). C) Glutamate 213 is a hydrogen bond acceptor in the active site of tricorn-interacting aminopeptidase F1, which may similarly promote inversion of the nitrogen.
During formation of the tetrahedral intermediate, stereoelectronic effects favor an orientation of the amide that is unreactive for the collapse of the tetrahedral intermediate. Stereoelectronic effects favor pointing the lone pair anti to the incoming serine, which orients the nitrogen lone pair away from the histidine.90 Moving it to a reactive orientation requires either rotation of the nitrogen or inversion of its configuration. Rotation is likely hindered in an active site, so inversion is the likely mechanism.
Peptidases catalyze the inversion of the nitrogen configuration using hydrogen bond acceptors. In some cases this hydrogen bond acceptor comes from the substrate, but in other cases it comes from the enzyme. The two mechanisms are considered to be one and the same. For the prolyl endopeptidase, the hydrogen bond acceptor is the substrate’s prolyl carbonyl oxygen, Chart 6B.91 A crystal structure of the catalytically inactive Ser554Ala variant of prolyl oligopeptidase complexed with an octapeptide shows a hydrogen bond between the NH of the leaving group and a carbonyl group of the prolyl residue in the peptide. Another observation consistent with this explanation is that lipases catalyze hydrolysis of amides that contain an internal hydrogen bond acceptor faster than similar substrates without the hydrogen bond acceptor.92,93 In other cases, such as tricorn-interacting aminopeptidase F1,94 the enzyme active site contains a hydrogen bond acceptor. The inactive S105A mutant of F1 was crystalized with a dipeptide that reveals a hydrogen bond between the NH leaving group and lid residue Glu213 (pdb id: 1xqw94), Chart 6C.
To test the importance of hydrogen bond acceptors in amide hydrolysis, two groups introduced a hydrogen bond acceptor in the active site of a lipase95,96 to increase the rate of amide hydrolysis. The Ile189Met or Ile189Glu variants of CAL-B catalyzed hydrolysis of 4-nitroacetanilide 17- to 24- times faster than the wild type enzyme, respectively. Ile189 originates from the lid. These variants catalyzed the hydrolysis of esters slightly slower (70–80% of wt rate), also consistent with the notion that the hydrogen bond only increases the rate of amide hydrolysis.
EC 3.5 C–N bond hydrolysis in substrates other than peptides
Kynurenine formamidase (EC 3.5.1.9) converts N-formyl-L-kynurenine to formic acid and kynurenine in the tryptophan degradation pathway, Scheme 12.97 This reaction is also an amide hydrolysis, but its EC number differs from peptidases (EC 3.4) because the amide is not a peptide amide. The mechanism is likely similar to that for amidases. Phenylmethylsulfonyl fluoride inactivates the enzyme indicating a serine nucleophile mechanism and the X-ray structure of the Drosophila enzyme shows a typical Ser-His-Asp catalytic triad. The mechanism likely involves the substrate-assisted catalysis like the one in peptidases where an internal hydrogen bond from the substrate orients the amide N-H away from the catalytic histidine. Substrate docking into the X-ray structure of the formamidase from Drosophila47 did not identify what distinguishes this amidase from esterases, but here we suggest that an internal hydrogen bond promotes nitrogen inversion as discussed above for the prolyl peptidases.
Scheme 12.
Kynurenine formamidase catalyzes hydrolysis of an amide during tryptophan degradation. An internal hydrogen bond, shown in red dashes, may contribute to catalysis by preventing the N-H from disrupting the catalytic histidine.
EC 3.7 hydrolysis of C–C bonds
meta-cleavage product hydrolase
Microbial oxidation of aromatic compounds usually proceeds via meta-cleavage products, Scheme 13. (The designation meta refers to cleavage between a hydroxylated carbon and an adjacent non-hydroxylated carbon.) These meta-cleavage products are vinylogous 1,5-diketones, drawn below as an enol tautomer, which extends the conjugation. Meta-cleavage product hydrolases (MCP-hydrolases) catalyze hydrolysis of C-C bonds in these vinylogous 1,5-diketones.98
Scheme 13.
Microbial oxidation of biphenyl proceeds via the biphenyl meta-cleavage product (MCP) 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA). The MCP is the dienol tautomer of a vinylogous 1,5-diketone. The MCP hydrolase catalyzes the hydrolysis of the C5–C6 bond yielding benzoic acid and 2-hydroxypenta-2,4-dienoic acid.
The meta-cleavage product in the oxidation of biphenyl is 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA). The 2-hydroxyl group readily deprotonates (pKa= 7.3) to form the dianion because this planar molecule can delocalize the negative charge over many atoms, Scheme 14. X-ray structures show that binding to the enzyme twists the molecule to a non-planar conformation.45 This twist and the binding of the carbonyl oxygen to the oxyanion hole localize the negative charge to C5 and the oxyanion.
Scheme 14.
The MCP-hydrolase substrate, HOPDA, deprotonates readily to the dianion where the negative charge delocalizes over many atoms; only two of the many possible resonance structures are shown. Binding to the active site twists this substrate to a non-planar conformation called the keto form due to keto resonance at C2. The twist and the H-bond donors from the oxyanion hole localize the charge to C5 and the oxyanion oxygen. The reaction starts from this keto form.
The structure of BphD S112A mutant was solved with the substrate bound in this nonplanar form, Figure 4. The active site of MCP-hydrolases contains five residues not found in esterases, Table 2. Four of these (Asn111, Phe175, Arg190, Trp260; numbering from the Burkholderia xenovorans enzyme) contact the dienoate part of the substrate,99 which corresponds to the alcohol binding region of an esterase. Replacement of any of these residues with alanine decreased kcat/KM by greater than 99%, which is consistent with an essential role in catalysis. The remaining conserved residue, Cys263, lies in the second sphere of residues. Replacement of this cysteine with alanine decreased the kcat/KM by only 30%, so this cysteine is not essential to catalysis.100 and its roles is unclear. MCP hydrolases also have a nonpolar site that binds the phenyl moiety and corresponds to the acyl binding site of an esterase.45 Esterases rarely accept benzoate esters, so this region must also differ in MCP hydrolases.
Figure 4.
Crystal structure of BphD S112A with HOPDA (blue) in the nonplanar keto form (pdb id: 2PUH). Catalytic triad is shown in grey carbons. The C-C hydrolases contain conserved substrate-binding residues shown in magenta carbons (side chain interacts with substrate) or green carbons (backbone interacts with substrate). Residues from the catalytic domain include Gly42, Asn111, Ser112, Met113, Asp237, His265 and Trp266. Phe175 and Arg190 belong to the cap domain. Another conserved residue, Cys263, lies outside the active site and is not shown.
Table 2.
Residues conserved in MCP-hydrolases (BphD numbering) as compared to the corresponding residues commonly found in esterases.100
| BphDa | Esterase |
|---|---|
| Asn111 | His, Phe |
| Phe175 | Asp |
| Arg190 | Ser |
| Cys263 | Phe, Ala, Leu |
| Trp266 | Gly |
BphD = MCP hydrolase from B. xenovorans LB400
The unique feature of the mechanism for C-C bond hydrolysis is that the substrate, not the catalytic histidine, is the base that deprotonates the active site serine, Scheme 15.45, 101 The negatively charged C5 acts as the base to deprotonate Oγ of Ser112. Upon deprotonation, the Oγ anion attacks the C6 carbonyl to form the first tetrahedral intermediate. The reason for the change in mechanism is that the substrate is unreactive as the dianion. A negative charge at C5 reduces any partial positive charge at the C6 carbonyl making it unreactive to nucleophilic attack. If His265 deprotonates Oγ of Ser112 nothing further occurs. Only when the substrate C5 deprotonates Oγ of Ser112 does it create a reactive substrate. Thus, the properties of the substrate change the mechanism of the reaction. Another way to look at this reaction is that the serine protonates the anionic substrate, which both creates the serine nucleophile and activates the substrate to nucleophilic attack. Other serine hydrolases do not catalyze hydrolysis of C-C bonds likely because they cannot bind the polar substrate or cannot bind it in a reactive conformation.
Scheme 15.
Proposed mechanism for MCP-hydrolases with residues numbered to match those in BphD from the biphenyl degradation pathway of B. xenovorans LB400. The dianion of HOPDA binds in a non-planar conformation, which localizes the negative charge to C5. This negative charge, not the histidine, deprotonates the serine to create the nucleophile. The rest of the reaction steps are similar to those in the canonical esterase mechanism.
The rest of the mechanism follows the canonical esterase mechanism. Collapse of the first tetrahedral intermediate breaks the carbon-carbon bonds to release 2-hydroxypenta-2, 4-dienoic acid (HPD) and form the benzoyl-enzyme intermediate. The formation of this benzoyl-enzyme intermediate does not require the catalytic histidine and the His265Gln variant of the enzyme accumulates the benzoyl enzyme intermediate, allowing for characterization by X-ray crystallography.45
Next, water binds, is deprotonated by the His265, and attacks the benzoyl-enzyme intermediate to generate the second tetrahedral intermediate. Collapse of the second tetrahedral intermediate releases benzoate.
Many MCP hydrolases also catalyze the hydrolysis of esters.76,102,103 Substitution of any of the five conserved residues (Asn111, Phe175, Arg190, Cys263, Trp266) in MhpC (similar experiments in E. coli MCP-hydrolase76 and other MCP hydrolases104) increased or left unchanged the rate of ester hydrolysis, but decreased HOPDA hydrolysis. This difference is consistent with the notion that these residues are important for binding HOPDA in a catalytically productive orientation. Substitution of the active site histidine with glutamine eliminates hydrolysis of esters, but for HOPDA only the second step is slowed.103 This result is consistent with the proposal that the HOPDA substrate deprotonates the serine for the MCP-hydrolase reaction, but cannot for the esterase reaction.
Thus, the change in mechanism comes from the ability of MCP hydrolases to bind the new, polar substrate in a reactive conformation and the new chemical steps created by this substrate.
2,6-dihydroxypseudooxynicotine hydrolase (DHPON hydrolase)
The bacterial pathway for nicotine metabolism contains a C-C bond hydrolase that hydrolyzes 2,6-dihydroxy-pseudo-oxynicotine to γ-N-methyl-aminobutyrate and 2,6-dihydroxy-pyridine, Scheme 16.105
Scheme 16.
Degradation of nicotine by the bacteria Arthrobacter nicotinovorans includes a hydrolysis of a C–C bond in DHPON catalyzed by DHPON hydrolase. If DHPON hydrolase is not present, DHPON irreversibly cyclizes to 2,6-dihydroxy-N-methyl-myosmine.
The 110-residue lid domain forms part of the substrate binding site and appears in X-ray structures in both an open and closed conformation.106 Researchers suggest that the substrate binds to the open conformation, which then closes for reaction. Most lid domains are inserted between β6 and β7, but the lid domain in DHPON hydrolase occurs as the N-terminus. This insertion location is not believed to affect the function of the lid domain.
The first step of the suggested mechanism is an enol to keto tautomerization similar to that in the MCP hydrolases, Scheme 17. This tautomerization breaks the conjugation between the carbonyl and aromatic ring to create a more electrophilic carbonyl for attack as well as a better leaving group. The closed conformation creates an ion pair between Glu148 and Arg18, a residue from the lid domain. Replacement of either residue with alanine decreases activity at least 50-fold. Glu148 deprotonates the phenol OH and protonates C3. The closed conformation also distorts the substrate from a planar geometry at C3 to bent at C3 to favor the enol tautomer, which has a tetrahedral configuration at C3.
Scheme 17.
Proposed mechanism for hydrolysis of the C–C bond in DHPON involves an enol to keto tautomerization followed by steps similar to those for ester hydrolysis. Glu148 catalyzes the enol to keto tautomerization by deprotonation of the phenolic OH at C2 followed by protonation at C3. An acyl enzyme intermediate forms at Ser217 in this mechanism.
The next steps are similar to the canonical esterase mechanism. The His-Asp pair deprotonate Ser217 Oγ-H, which then attacks the carbonyl carbon, forming the first tetrahedral intermediate. Collapse of this intermediate produces the first product, 2,6 -dihydroxypyridine. Water is then activated by the His-Asp dyad, and adds to the acyl-enzyme intermediate. Collapse of this second tetrahedral intermediate produces γ-N-methyl-aminobutyrate and regenerates the resting state of the enzyme. Replacement of Ser217 with alanine decreased activity >500-fold indicating that it is essential for catalysis.
The researchers also suggested another mechanism where water directly attacks the substrate and no acyl enzyme intermediate forms (not shown).106 This suggestion was based on a similar hypothesis for the MCP hydrolase. Subsequent research ruled out direct attack by water for the MCP hydrolase mechanism and confirmed an acyl enzyme intermediate, so the direct attack mechanism is also unlikely for DHPON hydrolase.
In summary, the large N-terminal lid domain helps to bind the substrate DHPON. Similar to MCP-hydrolases, DHPON hydrolase begins catalysis with an enol to keto tautomerization initiated by Glu148, followed by formation of the acyl enzyme intermediate. Unlike the MCP hydrolases, in DHPON hydrolases, the catalytic histidine deprotonates the serine Oγ.
8. EC 4 lyases
Enzymes in enzyme classification group 4 are lyases. Lyases cleave C-C, C-O, C-N bonds, not by hydrolysis or oxidation, but by elimination yielding a double bond. The mechanisms for the lyases in the α/β-hydrolase family do not involve an acyl enzyme intermediate, but use only general acid-base catalysis. In most cases the oxyanion hole is used, but the first two examples are cases where it is not used.
EC 4.1.1 C–C carboxy lyases (decarboxylases)
Tomato methyl ketone synthase 1 (MKS1), catalyzes the decarboxylation of β-ketoacids to form methylketones for plant defense, Scheme 18.107 The EC number for this enzyme is not yet assigned but it belongs with the other decarboxylases in E.C. 4.1.1 (carboxyl-lyases). Decarboxylation of β-ketoacids is facile in the carboxylic acid form, but slow in the carboxylate form.
Scheme 18.
Decarboxylation of β-ketomyristic acid catalyzed by methyl ketone synthase from tomato (MKS1). A) Tomato leaves convert β-ketomyristic acid, an intermediate in fatty acid biosynthesis, into 2-tridecanone for defense against insects. B) The active site contains a threonine that blocks the oxyanion hole and a catalytic triad of Ala-His-Asn, although the Ser-His-Asp variant is only 0.6-fold slower. The oxyanion hole and catalytic serine are not shown because there is no evidence that they contribute to catalysis. The key step is the protonation of the carboxylate by the catalytic histidine to the acid form, which decarboxylates yielding the enol and carbon dioxide. The enol later tautomerizes to the keto form (not shown).
The X-ray crystal structure shows an α/β-hydrolase fold and an unusual catalytic triad of Ala-His-Asn. The Ser-His-Asn, Ala-His-Asp and Ser-His-Asp variants have similar activity (1.2-fold, 0.8-fold and 0.6-fold, respectively). Thus, the catalytic serine does not help or hinder catalysis and the Asn variant is only slightly better than Asp. Many close homologs, which are likely similar decarboxylases, contain a serine as the nucleophile. For this reason, this example is included in this review with other Ser-His-Asp enzymes.
The active site contains a tunnel to bind the straight chain portion. This tunnel lies between the catalytic and cap domains similar to the acyl group binding site in some lipases. The oxyanion hole region is blocked by Thr18, which forces the carboxylate to find a different binding orientation in the active site. The activity of the Thr18Ala variant is 7-fold lower than wt MKS1.The threonine has the same role and corresponding location as the threonine in the active site of (S)-hydroxynitrile lyases in the next section.
Thus, the key catalytic step in the proposed mechanism is protonation of the substrate’s carboxylate by a protonated active site histidine.107 Blocking the oxyanion hole prevents the oxyanion from binding in this hole, where it cannot be protonated. Once the carboxylate has been protonated, decarboxylation proceeds rapidly. This is the only example where neither the active site serine, nor the oxyanion hole, are used.
EC 4.1.2 C-C bond cleavage, aldehyde lyases
(S)-Hydroxynitrile lyases
Hydroxynitrile lyases are plant enzymes that catalyze the release of cyanide from cyanogenic glycosides as a defense against insects, Scheme 19. The natural substrate of most HNLs is the achiral acetone cyanohydrin, but they also catalyze the S-enantioselective cleavage of mandelonitrile. HNL’s exist in several protein fold families; here we focus on those in the α/β-hydrolase fold family. The best characterized (S)-HNL is HNL from rubber tree, Hevea brasiliensis (HbHNL). Royal DSM uses HbHNL in the manufacture of an insecticide precursor to catalyze the reverse reaction, addition of hydrogen cyanide to aldehydes.108
Scheme 19.
Linamarin is a cyanogenic glucoside in plants. Hydrolysis of the glucoside by glycosidases yields acetone cyanohydrin. Hydroxynitrile lyase from Hevea brasiliensis catalyzes the elimination of hydrogen cyanide from this cyanohydrin.
Besides the Ser-Asp-His catalytic triad, (S)-HNL’s have threonine and lysine residues in the active site. X-ray crystal structures of HbHNL with substrate acetone cyanohydrin and product acetone48 show that the threonine residue blocks the oxyanion hole region causing the carbonyl of acetone to orient different from the carbonyl of esters in an esterase, Scheme 20. The positive charge of the lysine residue’s side chain stabilizes the negative charge on nitrile and may raise the pKa of the catalytic histidine.
Scheme 20.
Mechanism for the cleavage of acetone cyanohydrin by (S)-HNL from Hevea brasiliensis39,95. Blocking of the oxyanion hole by threonine forces the acetone carbonyl group into a different orientation from that for the ester carbonyl group in esterases. In HNL, the serine Oγ interacts with the oxygen of the carbonyl group, while in esterases it interacts with the carbon.
The accepted mechanism for HbHNL is general acid-base catalysis with a single transition state and no covalent intermediate,109 Scheme 20. The substrate acetone cyanohydrin binds in the active site by forming hydrogen bonds through its hydroxyl group with Oγ’s of Ser80 and Thr11 (numbering according to HbHNL). His235 removes a proton from Ser80, which in turn deprotonates the substrate’s hydroxyl group, which causes formation of a C-O double bond and release of cyanide. Next, His235 protonates the cyanide to complete the reaction.
The key features of the (S)-HNL mechanism is no use of the oxyanion hole and no acyl-enzyme intermediate. Serine is not a nucleophile but a proton shuttle between substrate and catalytic histidine. A lysine in the leaving group site stabilizes the negative charge on the leaving cyanide.
(R)-Hydroxynitrile lyase (AtEST5)
Initial annotation of the EST5 gene in Arabidopsis thaliana classified it as an esterase, and later characterization confirmed that it catalyzes ester hydrolysis.110 The EST5 enzyme also catalyzes cleavage of mandelonitrile (42 s−1),111 but not of the natural substrate of other HNLs, acetone cyanohydrin. Further, A.thaliana does not contain cyanogenic glucosides. AtEST5 is most likely an esterase with a promiscuous hydroxynitrile lyase activity.
AtEST5 differs from HbHNL in its enantiopreference toward mandelonitrile, in its catalytic residues, and likely in its reaction mechanism. AtEST5 is highly enantioselective for (R)-mandelonitrile, but HbHNL and other HNL’s in the α/β-hydrolase family favor (S)-mandelonitrile. AtEST5 lacks the key catalytic lysine found in HbHNL and has a methionine at the corresponding location. AtEST5 also lacks the threonine found in HbHNL and has an asparagine at the corresponding location.
Andexer et al.112 docked the substrate in the active site and proposed a possible mechanism for AtEST5, Scheme 21. The key feature is binding the nitrile substituent in the oxyanion hole. Although no X-ray structures show nitrile bound to the oxyanion hole in α/β-hydrolase fold enzymes, Zhu et al.113 reported a structure of nitrile bound to an oxyanion hole in a dioxygenase, which has a different fold. Thus, the proposed binding of nitrile in the oxyanion hole is a reasonable hypothesis.
Scheme 21.
Proposed mechanism for cleavage of mandelonitrile by (R)-HNL from A. thaliana based on docking calculations.112 AtEST5 binds the substrate in an orientation that differs from both esterases (carbonyl is not in oxyanion hole) and from HbHNL (nitrile is in oxyananion hole). X-ray structures have not confirmed this proposed binding. For clarity, the catalytic aspartate is not shown.
The substrate hydroxyl group donates a hydrogen bond to His236 of the catalytic triad, while accepting a hydrogen bond from the δ-amide of Asn12.112 The reaction begins with proton abstraction from the hydroxyl group of mandelonitrile by His236. Next, elimination of cyanide forms the carbonyl group. The cyanide, stabilized by the oxyanion hole is protonated by Ser81, which then abstracts the proton from His236 to regenerate the starting state. In parallel with their opposite enantioselectivities, AtEST5 and HbHNL have opposite methods of proton abstraction and replacement. In AtEST5, the substrate hydroxyl proton is abstracted by the histidine and the cyanide is protonated by the serine. In HbHNL, the opposite happens: the hydroxyl proton is abstracted by the serine, and cyanide is protonated by histidine.
In summary, the active site differences between HbHNL and AtEST5 require the mechanisms to differ. Neither mechanism involves an acyl enzyme intermediate. The proposed mechanism for AtEST5 uses an oxyanion hole to stabilize the leaving cyanide group. Histidine directly deprotonates the substrate OH, while serine serves as proton shuttle to protonate the leaving cyanide. In contrast, in the HbHNL mechanism, the histidine deprotonated the substrate via a serine proton shuttle, and protonated the cyanide directly.
Aldolases
Lipase B from C. antarctica and several other lipases catalyze the aldol addition of an aldehyde to another carbonyl compound to form a β-hydroxy aldehyde,114–116 Scheme 22A. An active-site-targeted inhibitor stops the reaction indicating that the reaction occurs in the active site. The Ser105Ala variant catalyzes the reaction approximately two-fold faster than wild type indicating that the catalytic serine does not participate in the mechanism.
Scheme 22.
Lipase-catalyzed aldol addition involves general acid-base catalysis, but no acyl enzyme intermediate. A) Lipase B from C.antarctica catalyzes the addition of hexanal to a second molecule of hexanal to form a β-hydroxy aldehyde. This addition is not enantioselective, but lipase from pig pancreas shows low enantioselectivity (E ~2.5) in a similar reaction.116 B) Theoretical calculations indicate that lipase catalyzes both the enolate formation step and the enolate addition step. The active site serine is not shown because it is not essential for the reaction, while the active site aspartate is omitted for clarity.
Computer modeling suggests that CAL-B catalyzes both steps of the reaction: formation of the enolate and addition of the enolate to the carbonyl compound, Scheme 22B.114,115 Catalysis involves only general acid-base catalysis and no acyl enzyme intermediate. The oxyanion hole binds and polarizes the carbonyl; next, the catalytic histidine removes the α-proton to form enolate. In the enolate addition step, the protonated histidine acts as an acid to protonate the carbonyl compound. The reaction is slow – 65 d−1 or 0.0008 s−1. Molecular dynamics simulations suggest that reason for the slow reaction is that the substrates bind in non-reactive orientations most of the time.
A related promiscuous reaction is the Mannich reaction, where an enolate adds to an imine, instead of adding to an aldehyde in the aldol addition. Several lipases catalyzed the Mannich reaction between an imine of benzaldehyde and acetone; lipase from Mucor miehei gave the highest yield (89%).117 The reaction mechanism is likely similar to the aldol addition, but few mechanistic details are known.
In summary, CAL-B catalyzes an aldol addition using general acid-base catalysis. The substrates lack a leaving goup, so they cannot form an acyl enzyme. Indeed, replacement of the serine with alanine increased the reaction rate. The oxyanion hole polarizes the carbonyl to aid formation of the enolate intermediate.
Conjugate additions: C–C (4.1.99), C–O (4.2.99), C–N (4.3.3) and C–S (4.4.1) lyases
Conjugate additions (or 1,4-additions) are the addition of a nucleophile to an α,β-unsaturated carbonyl compound, Scheme 23. Depending on the nucleophile, these reactions belong to EC 4.1.99 (other carbon-carbon lyases), EC 4.2.99 (other carbon-oxygen lyases), EC 4.3.3 (amine lyases) or EC 4.4.1 (carbon-sulfur lyases). These reactions are discussed together because the mechanisms appears to be the same.
Scheme 23.
Lipases catalyze the addition of nucleophiles to α,β-unsaturated carbonyl compounds. These additions can form carbon-carbon, carbon-oxygen, carbon-nitrogen or carbon-sulfur bonds. v = measured rate at one substrate concentration.
Several lipases catalyze these conjugate addition reactions. For example, the Ser105Ala variant of lipase B from C. antarctica catalyzes the addition of acetyl acetone to acrolein at an impressive rate 4000 s−1, which corresponds to a rate acceleration >108 over the uncatalyzed reaction.118 Reaction mixtures of pure substrate with enzyme can warm to a boil due to the exothermic reaction. The addition reactions show little to no enantioselectivity.118–120 Others reported similar additions of an oxygen nucleophile,121,122 nitrogen nucleophile,123–126 or sulfur nucleophile.125
The reaction mechanism is similar to that for the direct epoxidation reaction in section 4 above. The oxyanion hole polarizes the α,β-unsaturated carbonyl compound while the catalytic histidine deprotonates the nucleophile. No acyl enzyme intermediate forms. The Ser105Ala variant of CAL-B, where formation of an acyl enzyme intermediate is impossible, is up to 100-fold faster that wild type CAL-B.118,127 After addition of the nucleophile, the protonated histidine protonates the α-carbon. In the direct epoxidation mechanism, this step differs. The epoxide forms and the protonated histidine protonates the leaving water from the hydroperoxide intermediate.
Lipases also catalyze the Morita-Baylis-Hillman reaction128 (conjugate addition followed by additional steps) and the addition of thiols to the double bond of vinyl esters.129,130 The reaction mechanisms of these reactions are unknown, but may be similar to those for conjugate additions.
In summary, binding the carbonyl oxygen of an α,β-unsaturated carbonyl compound in the oxyanion hole polarizes it for attack by a nucleophile. Histidine activates the nucleophile by deprotonation, and then protonates the α-carbon.
Lipase B from C. antarctica (CAL-B) catalyzes three different reaction mechanisms: canonical esterase mechanism, aldolase, and conjugate addition, which includes direct epoxidation. In addition, a variant of CAL-B catalyzes a fourth mechanism - peptide hydrolysis. This catalytic promiscuity of CAL-B is likely due to its stability, and abilities to bind a broad range of substrates and maintain activity in the organic solvents needed for some of these reactions. In addition, since it is often used by organic chemists for synthesis, it is often the first enzyme tested for new types of reactivity.
EC 4.2 C-O bond cleaving lyases
Menaquinone synthase, MenH (EC 4.2.99.20), catalyzes a 1,4-elimination reaction of pyruvate from a cyclohexene, Scheme 24A. It converts the 3-cyclohexene below to the corresponding 2,4-diene by elimination of pyruvate from C5 and a proton from C2. This bacterial enzyme is part of the vitamin K synthesis pathway.131 Researchers have solved the X-ray crystal structures of enzymes from both Escherichia coli131–133 and Staphylococcus aureus.134
Scheme 24.
Menaquinone synthase, MenH, catalyzes a 1,4-elimination of pyruvate from 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate. A) The ring numbering makes this reaction a 2,5-elimination for this substrate. The kcat value refers to the enzyme from E. coli.131 B) The proposed mechanism133 starts with the formation of an enolate, whose oxyanion is stabilized by the oxyanion hole. Elimination of pyruvate from this enolate is aided by protonation of the leaving group by the catalytic histidine. The amino acid numbers correspond to the E. coli enzyme.
X-ray crystallography (pdb id: 4mys50) revealed how product binds to the active site and lead to a proposed mechanism, Scheme 24B. The succinyl group binds in the region corresponding to the acyl binding site of esterases with the carbonyl group in the oxyanion hole. The cycloxenyl and enol pyruvyl groups bind in the alcohol binding site of esterases. Several conserved arginines within the active site (Arg90, Arg124, and Arg168; not shown in Scheme 24B) bind the three carboxylates of this polar substrate and position it for reaction.
The proposed mechanism involves general acid-base catalysis and no acyl enzyme intermediate. The reaction starts similarly to that in esterases. The substrate lies with the carbonyl in the oxyanion hole. The catalytic histidine, using Oγ of the catalytic serine as a proton shuttle, removes the proton at C2 to make an enolate. The oxyanion hole can stabilize the negative charge at the enolate oxygen. Elimination of pyruvate from this enolate, aided by the protonation of pyruvate by the catalytic histidine, yield the product cyclohexadiene.
9. EC 5 isomerases
EC 5.1.99 racemase
Lipase from B. cepacia (E.C. 3.1.1.3) catalyzes the slow racemization of N-methyl α-aminonitriles, Scheme 25A.135 This promiscuous unnatural reaction has not yet been assigned an Enzyme Classification number, but it will most likely fit into EC 5.1.99, which contains racemases and epimerases that act on other compounds.
Scheme 25.
A) Lipase from B. cepacia (BCL) catalyzes the racemization of N-methyl α-aminonitriles. B) The proposed mechanism is the reversible elimination of cyanide to form an imine intermediate. The catalytic histidine serves as the base, while the oxyanion hole stabilizes the released cyanide.
The suggested mechanism is the reversible elimination of cyanide to form an imine intermediate, Scheme 25B. There is no acyl enzyme intermediate; catalysis involves only acid-base reactions. This suggested mechanism was not tested experimentally.
EC 5.2 cis-trans isomerase
Dienelactone hydrolase (DLH) catalyzes the hydrolysis of dienelactone to maleylacetate, Scheme 26A, as part of the β-keto adipate pathway for bacterial degradation of aromatic compounds.136 The DLH catalytic triad consists of a nucleophilic cysteine (Cys123), a histidine (His202), and an aspartic acid (Asp171).137 Substitution of the catalytic cysteine with serine abolishes hydrolase activity, but enables isomerase activity, Scheme 26B.138 HPLC, proton NMR and thin layer chromatography confirmed the interconversion between the E- and Z-lactones. This isomerase activity is discussed here.
Scheme 26.
Hydrolysis catalyzed by wt DLH and isomerization catalyzed by DLH-Cys123Ser. A) Loss of the best leaving group, thiol, from the second tetrahedral intermediate leads to hydrolysis of the dienelactone by wt DLH. Steps for formation of the acyl enzyme and second tetrahedral intermediate are omitted for clarity. B) DLH-Cys123Ser catalyzes not hydrolysis, but the isomerization of the (Z)- and (E)-lactones. C. Possible mechanism for isomerization beginning with second tetrahedral intermediate. Hydrolysis of the acyl enzyme is slow because the serine Oγ is a poorer leaving group than the cysteine Sγ thus favoring reformation of the acyl enzyme. Rotation about the C2–C3 bond followed by tautomerization to enol form allows the acyl enzyme to recyclize to form the isomeric lactone.
The reason for the change in reaction is the the increase in the lifetime of acyl enzyme intermediate and the propensity of the acyl enzyme to relactonize.138 In wt DLH, attack of water on the acyl enzyme leads to the second tetrahedral intermediate shown in Scheme 26A. Loss of thiol leads to the hydrolysis product, maleylacetate. In contrast, the corresponding second tetrahedral intermediate in the DLH-Cys123Ser variant lacks good leaving group and is more likely to return to the acyl enzyme intermediate, Scheme 26C. This intermediate can isomerize the side chain by rotation about the C2–C3 bond followed by tautomerization to the enol form. Recyclization of the acyl enzyme to the lactone yields the isomeric lactone. Thus, the slow cleavage of the acyl enzyme intermediate allows isomerization of the lactone side chain and the propensity to reform a ring regenerates the lactone.
10. Concluding remarks
The Ser-His-Asp catalytic triad with oxyanion hole catalyzes a remarkable diversity of reactions using widely varying mechanisms. The range includes five different reaction classes and two general mechanisms: the hydrolase-type mechanism and the lyase-type mechanism. The rates of the reactions vary from a few turnovers per hour to thousands per second. The exact grouping of these mechanisms into seventeen different types is somewhat arbitrary, especially since the mechanistic details of some reactions are unknown.
The most common reason for a new mechanism is that the substrate itself requires new steps. The changing the substrate from an ester to substrates that cannot hydrolyze (cyanohydrins, aldehydes & ketones, and others) requires a new reaction. These substrates follow a lyase-type reaction mechanism. Even for substrates that can hydrolyze, unique features of the substrate may require new mechanistic steps. For example, hydrolysis of a phosphotriester cannot follow the canonical mechanism because the phosphoryl enzyme intermediate blocks the region where water normally binds. In another example, lactones and peptides form tetrahedral intermediates with a lone pair orientation unsuitable for the next protonation step. A new mechanistic step is required for the reaction to continue. In three cases it is the enzyme, not the substrate, that determine the favored reaction. The substrates for perhydrolases, acyl transferases and lactone cis-trans isomerase can either hydrolyze or undergo a new reaction. The enzyme favors the new reaction and hydrolysis is a side reaction.
Although the substrate may require new steps, the enzyme does not need to provide these steps. One possibility is no reaction. The observation that the α/β-hydrolase fold enzymes catalyze this wide range of mechanisms is indeed remarkable.
None of the individual enzymes catalyze all seventeen reaction mechanisms. The most promiscuous enzyme is lipase B from C. antarctica and its variants, which catalyze four different mechanisms. One reason that a single enzyme cannot catalyze all seventeen mechanisms is that the substrates vary widely. Some are hydrophobic, some hydrophilic. Their shapes differ. To react, the substrate must bind to the active site in a catalytically productive orientation. It is unlikely that one active site could accommodate the wide range of substrate, especially since some of these orientations may be incompatible with one another. For example, some require the substrate to bind in the oxyanion hole, while others for the substrate to bind outside this region.
Much in the same way that a general acid can catalyze a wide range of reactions, so too a serine hydrolase can catalyze a wide range of reactions. Acid catalyzes both hydrolysis and elimination reactions; serine hydrolases also catalyze both reactions. In many cases, the substrate itself determines which reaction can occur because some substrates can undergo hydrolysis, while others cannot. In cases where the substrate can undergo several reactions, the serine hydrolase, unlike a general acid, can favor one reaction.
Supplementary Material
Acknowledgments
We thank the US National Institutes of Health (1R01GM102205-01) and the US National Science Foundation (CHE-1152804) for support of this research. We thank Prof. Dr. Uwe Bornscheuer and Dr. Alberto Nobili (U. of Greifswald) for searching the α/β-hydrolase 3DM database.
Footnotes
Table of twenty-one example reactions.
References
- 1.Albery W, Knowles J. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 2.Wolfenden R. Chem Rev. 2006;106:3379–3396. doi: 10.1021/cr050311y. [DOI] [PubMed] [Google Scholar]
- 3.Siegel JB, Gallaher JL, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiss G, Çelebi-Ölçüm N, Moretti R, Baker D, Houk KN. Angew Chem Int Ed. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 5.For example, Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Science. 2010;329:305–309. doi: 10.1126/science.1188934.
- 6.O’Brien PJ, Herschlag D. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 7.Bornscheuer UT, Kazlauskas RJ. Angew Chem Int Ed. 2004;43:6032–6040. doi: 10.1002/anie.200460416. [DOI] [PubMed] [Google Scholar]
- 8.Hult K, Berglund P. Trends Biotechnol. 2007;25:231–238. doi: 10.1016/j.tibtech.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Ohno S. In: Evolution by gene duplication. Soukup SW, editor. Springer-Verlag; New York: 1970. [Google Scholar]
- 10.Bergthorsson U, Andersson DI, Roth JR. Proc Natl Acad Sci USA. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodson G, Wlodawer A. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 12.Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A. Nucleic Acids Res. 2004;32:D145–D147. doi: 10.1093/nar/gkh141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A. Nucleic Acids Res. 2013;41:D423–D429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kourist R, Jochens H, Bartsch S, Kuipers R, Padhi SK, Gall MG, Böttcher D, Joosten HJ, Bornscheuer UT. ChemBioChem. 2010;11:1635–1643. doi: 10.1002/cbic.201000213. [DOI] [PubMed] [Google Scholar]
- 15.Ollis D, Cheah E, Cygler M, Dijkstra BW, Frolow F, Franken SM, Harel M, Remington S, Silman I, Schrag JD. Protein Eng Des Sel. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 16.Heikinheimo P, Goldman A, Jeffries C, Ollis DL. Structure. 1999;7:141–146. doi: 10.1016/s0969-2126(99)80079-3. [DOI] [PubMed] [Google Scholar]
- 17.Carr PD, Ollis DL. α/β hydrolase fold: an update. Protein Pept Lett. 2009;16:1137–1148. doi: 10.2174/092986609789071298. [DOI] [PubMed] [Google Scholar]
- 18.Nardini M, Dijkstra BW. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 19.Holmquist M. Curr Prot Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 20.Bugg TDH. Bioorg Chem. 2004;32:367–375. doi: 10.1016/j.bioorg.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Padhi SK, Fujii R, Legatt GA, Fossum SL, Berchtold R, Kazlauskas RJ. Chem Biol. 2010;17:863–871. doi: 10.1016/j.chembiol.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Khersonsky O, Tawfik DS. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 23.Ericsson DJ, Kasrayan A, Johansson P, Bergfors T, Sandström AG, Bäckvall JE, Mowbray SL. J Mol Biol. 2008;376:109–119. doi: 10.1016/j.jmb.2007.10.079. [DOI] [PubMed] [Google Scholar]
- 24.Thoma R, Löffler B, Stihle M, Huber W, Ruf A, Hennig M. Structure. 2003;11:947–959. doi: 10.1016/s0969-2126(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 25.Aertgeerts K, Ye S, Tennant MG, Kraus ML, Rogers J, Sang BC, Skene RJ, Webb DR, Prasad GS. Protein Sci. 2004;13:412–421. doi: 10.1110/ps.03460604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez C, De Geus P, Lauwereys M, Matthyssens G, Cambillau C. Nature. 1992;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- 27.Dunn G, Mohammed F, Coker A, Cooper JB, Robertson T, Garcia J-L, Bugg TDH, Wood SP. J Mol Biol. 2005;346:253–265. doi: 10.1016/j.jmb.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 28.Fülöp V, Böcskei Z, Polgár L. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 29.Jochens H, Stiba K, Savile C, Fujii R, Yu JG, Gerassenkov T, Kazlauskas RJ, Bornscheuer UT. Angew Chem Int Ed. 2009;48:3532–3535. doi: 10.1002/anie.200806276. [DOI] [PubMed] [Google Scholar]
- 30.Henke E, Bornscheuer UT, Schmid RD, Pleiss J. ChemBioChem. 2003;4:485–493. doi: 10.1002/cbic.200200518. [DOI] [PubMed] [Google Scholar]
- 31.Henke E, Pleiss J, Bornscheuer UT. Angew Chem Int Ed. 2008;41:3211–3213. doi: 10.1002/1521-3773(20020902)41:17<3211::AID-ANIE3211>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Smith AJT, Müller R, Toscano MD, Kast P, Hellinga HW, Hilvert D, Houk KN. J Am Chem Soc. 2008;130:15361–15373. doi: 10.1021/ja803213p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wogulis M, Wheelock CE, Kamita SG, Hinton AC, Whetstone PA, Hammock BD, Wilson DK. 2006;45:4045–4056. doi: 10.1021/bi0521644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellizzi JJ, III, Widom J, Kemp C, Lu JY, Das AK, Hofmann SL, Clardy J. Proc Natl Acad Sci USA. 2000;97:4573–4578. doi: 10.1073/pnas.080508097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P, Wang YF, Ewis HE, Abdelal AT, Lu C-D, Harrison RW, Weber IT. J Mol Biol. 2004;342:551–561. doi: 10.1016/j.jmb.2004.06.069. [DOI] [PubMed] [Google Scholar]
- 36.Liuc M, Tomic S, Lescic I, Ljubovic E, Sepac D, Sunjic V, Vitale L, Saenger W, Kojic-Prodic B. Eur J Biochem. 2001;268:3964–3973. doi: 10.1046/j.1432-1327.2001.02303.x. [DOI] [PubMed] [Google Scholar]
- 37.Mezzetti A, Schrag JD, Cheong CS, Kazlauskas RJ. Chem Biol. 2005;12:427–437. doi: 10.1016/j.chembiol.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Uppenberg J, Öhrner N, Norin M, Kleywegt GJ, Patkar S, Waagan V, Anthonsen T, Jones TA. Biochemistry. 1995;34:16838–16851. doi: 10.1021/bi00051a035. [DOI] [PubMed] [Google Scholar]
- 39.Cygler M, Grochulski P, Kazlauskas RJ, Schrag JD, Bouthillier F, Rubin B, Serreqi AN, Gupta AK. J Am Chem Soc. 1994;116:3180–3186. [Google Scholar]
- 40.Glukhova A, Hinkovska-Galcheva V, Kelly R, Abe A, Shayman JA, Tesme JJG. Nat Commun. 2015;6:6250. doi: 10.1038/ncomms7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y, Morley KL, Schrag JD, Kazlauskas RJ. ChemBioChem. 2011;12:768–776. doi: 10.1002/cbic.201000693. [DOI] [PubMed] [Google Scholar]
- 42.Lejon S, Ellis J, Valegard K. J Mol Biol. 2008;377:935–944. doi: 10.1016/j.jmb.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 43.Zubieta C, Arkus KAJ, Cahoon RE, Jex JM. J Biol Chem. 2008;283:7561–7567. doi: 10.1074/jbc.M709283200. [DOI] [PubMed] [Google Scholar]
- 44.Goettig P, Brandstetter H, Groll M, Göhring W, Konarev PV, Svergn DI, Huber R, Kim JS. J Biol Chem. 2005;280:33387–33396. doi: 10.1074/jbc.M505030200. [DOI] [PubMed] [Google Scholar]
- 45.Ruzzini AC, Ghosh S, Horsman GP, Foster LJ, Bolin JT, Eltis LD. J Am Chem Soc. 2012;134:4615–4624. doi: 10.1021/ja208544g. [DOI] [PubMed] [Google Scholar]
- 46.Han Q, Robinson H, Li J. Biochem J. 2012;446:253–260. doi: 10.1042/BJ20120416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Line K, Isupov MN, Littlechild JA. J Mol Biol. 2004;338:519–532. doi: 10.1016/j.jmb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Gruber K, Gartler G, Krammer B, Schwab H, Kratky C. J Biol Chem. 2004;279:20501–20510. doi: 10.1074/jbc.M401575200. [DOI] [PubMed] [Google Scholar]
- 49.Lauble H, Miehlich B, Förster S, Wajant H, Effenberger F. Biochemistry. 2002;41:12043–12050. doi: 10.1021/bi020300o. [DOI] [PubMed] [Google Scholar]
- 50.Sun Y, Yin S, Feng Y, Li J, Zhou J, Liu C, Zhu G, Guo Z. J Biol Chem. 2014;289:15867–15879. doi: 10.1074/jbc.M113.535641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svedendahl M, Carlqvist P, Branneby C, Allnér O, Frise A, Hult K, Berglund P, Brinck T. ChemBioChem. 2008;9:2443–2451. doi: 10.1002/cbic.200800318. [DOI] [PubMed] [Google Scholar]
- 52.Weng M, Pfeifer O, Krauss S, Lingens F, van Pée KH. J Gen Microbiol. 1991;137:2539–2546. doi: 10.1099/00221287-137-11-2539. [DOI] [PubMed] [Google Scholar]
- 53.Kirk O, Christensen MW, Damhus T, Godtfredsen SE. Biocatal Biotransfor. 1994;11:65–77. [Google Scholar]
- 54.Picard M, Gross J, Lübbert E, Tölzer S, Krauss S, van Pée KH, Berkessel A. Angew Chem Intl Ed. 1997;36:1196–1199. [Google Scholar]
- 55.Yin DLT, Bernhardt P, Morley KL, Jiang Y, Cheeseman JD, Purpero VM, Schrag JD, Kazlauskas RJ. Biochemistry. 2010;49:1931–1942. doi: 10.1021/bi9021268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honda K, Kataoka M, Sakuradani E, Shimizu S. Eur J Biochem. 2003;270:486–494. doi: 10.1046/j.1432-1033.2003.03407.x. [DOI] [PubMed] [Google Scholar]
- 57.Yin DLT, Kazlauskas RJ. Chem Eur J. 2012;18:8130–8139. doi: 10.1002/chem.201200052. [DOI] [PubMed] [Google Scholar]
- 58.Yin DLT, Purpero VM, Fujii R, Jing Q, Kazlauskas RJ. Chem Eur J. 2013;19:3037–3046. doi: 10.1002/chem.201202027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathews I, Soltis M, Saldajeno M, Ganshaw G, Sala R, Weyler W, Cervin MA, Whited G, Bott R. Biochemistry. 2007;46:8969–8979. doi: 10.1021/bi7002444. [DOI] [PubMed] [Google Scholar]
- 60.Bugg TDH. Curr Opin Chem Biol. 2001;5:550–555. doi: 10.1016/s1367-5931(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 61.Massey V. J Biol Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 62.Bauer I, Max N, Fetzner S, Lingens F. Eur J Biochem. 1996;240:576–583. doi: 10.1111/j.1432-1033.1996.0576h.x. [DOI] [PubMed] [Google Scholar]
- 63.Fischer F, Kunne S, Fetzner S. J Bacteriol. 1999;181:5725–5733. doi: 10.1128/jb.181.18.5725-5733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thierbach S, Bui N, Zapp J, Chhabra SR, Kappl R, Fetzner S. Chem Biol. 2014;21:217–225. doi: 10.1016/j.chembiol.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 65.Steiner RA, Janssen HJ, Roversi P, Oakley AJ, Fetzner S. Proc Natl Acad Sci U S A. 2010;107:657–662. doi: 10.1073/pnas.0909033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernandez-Ortega A, Quesne MG, Bui S, Heuts DPHM, Steiner RA, Heyes DJ, de Visser SP, Scrutton NS. J Biol Chem. 2014;289:8620–8632. doi: 10.1074/jbc.M113.543033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frerichs-Deeken U, Ranguelova K, Kappl R, Hüttermann J, Fetzner S. Biochemistry. 2004;43:14485–14499. doi: 10.1021/bi048735u. [DOI] [PubMed] [Google Scholar]
- 68.Bornscheuer UT, Kazlauskas RJ. Hydrolases in Organic Synthesis: Regio- and Stereoselective Biotransformations. 2. Wiley-VCH; Weinheim: 2005. [Google Scholar]
- 69.Scheidegger A, Gutzwiller A, Küenzi MT, Fiechter A, Niiesch J. J Biotechnol. 1985;3:109–117. [Google Scholar]
- 70.Matsuyama K, Matsumoto H, Matsuda A, Sugiura H, Komatsu K, Ichikawa S. Biosci Biotechnol Biochem. 1992;56:1410–1412. doi: 10.1271/bbb.56.1410. [DOI] [PubMed] [Google Scholar]
- 71.Mirza IA, Nazi I, Korczynska M, Wright GD, Berghuis AM. Biochemistry. 2005;44:15768–15773. doi: 10.1021/bi051951y. [DOI] [PubMed] [Google Scholar]
- 72.Neugnot V, Moulin G, Dubreucq E, Bigey F. Eur J Biochem. 2002;269:1734–1745. doi: 10.1046/j.1432-1327.2002.02828.x. [DOI] [PubMed] [Google Scholar]
- 73.Müller J, Sowa MA, Dörr M, Bornscheuer UT. Eur J Lipid Sci Technol. 2015 early view http://doi.org/10.1002/ejlt.201500292.
- 74.Fournand D, Vaysse L, Dubreucq E, Annaud A, Galzy P. J Mol Catal B Enzym. 1998;5:207–211. [Google Scholar]
- 75.Hacking MAPJ, van Rantwijk F, Sheldon RA. J Mol Catal B Enzym. 2001;11:315–321. [Google Scholar]
- 76.Li C, Hassler M, Bugg TDH. ChemBioChem. 2008;9:71–76. doi: 10.1002/cbic.200700428. [DOI] [PubMed] [Google Scholar]
- 77.Müller J, Sowa MA, Fredrich B, Brundiek H, Bornscheuer UT. ChemBioChem. 2015;16:1791–1796. doi: 10.1002/cbic.201500187. [DOI] [PubMed] [Google Scholar]
- 78.Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, Klessig DF, Tong L. Proc Natl Acad Sci U S A. 2005;102:1773–1778. doi: 10.1073/pnas.0409227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Simone G, Mandrich L, Menchise V, Giordano V, Febbraio F, Rossi M, Pedone C, Manco G. J Biol Chem. 2004;279:6815–6823. doi: 10.1074/jbc.M307738200. [DOI] [PubMed] [Google Scholar]
- 80.Grochulski P, Li Y, Schrag JD, Cygler M. Protein Sci. 1994;3:82–91. doi: 10.1002/pro.5560030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubin B. Nat Struct Biol. 1994;1:568–572. doi: 10.1038/nsb0994-568. [DOI] [PubMed] [Google Scholar]
- 82.Gao A, Mei G, Liu S, Wang P, Tang Q, Liu Y, Wen H, An X, Zhang L, Yan X, Liang D. Acta Crystallogr Sect D Biol Crystallogr. 2013;69:82–91. doi: 10.1107/S0907444912042369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Camp LA, Hofmann SL. J Biol Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 84.Lawson DM, Derewenda U, Serre L, Ferri S, Szittner R, Wei Y, Meighen EA, Derewenda ZS. Biochemistry. 1994;33:9382–9388. doi: 10.1021/bi00198a003. [DOI] [PubMed] [Google Scholar]
- 85.Newcomb RD, Campbell PM, Ollis DL, Cheah E, Russell RJ, Oakeshott JG. Proc Natl Acad Sci U S A. 1997;94:7464–7468. doi: 10.1073/pnas.94.14.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lockridge O, Blong RM, Masson P, Froment MT, Millard CB, Broomfield CA. Biochemistry. 1997;36:786–795. doi: 10.1021/bi961412g. [DOI] [PubMed] [Google Scholar]
- 87.Nachon F, Carletti E, Wandhammer M, Nicolet Y, Schopfer LM, Masson P, Lockridge O. Biochem J. 2011;434:73–82. doi: 10.1042/BJ20101648. [DOI] [PubMed] [Google Scholar]
- 88.Wilk S. Life Sci. 1983;33:2149–2157. doi: 10.1016/0024-3205(83)90285-0. [DOI] [PubMed] [Google Scholar]
- 89.Syrén PO, Hult K. ChemCatChem. 2011;3:853–860. [Google Scholar]
- 90.Syrén PO. FEBS J. 2013;280:3069–3083. doi: 10.1111/febs.12241. [DOI] [PubMed] [Google Scholar]
- 91.Fülöp V, Szeltner Z, Renner V, Polgár L. J Biol Chem. 2001;276:1262–1266. doi: 10.1074/jbc.M007003200. [DOI] [PubMed] [Google Scholar]
- 92.Balkenhohl F, Ditrich K, Hauer B, Ladner W. J Prakt Chem Chemik Zeit. 1997;339:381–384. [Google Scholar]
- 93.Cammenberg M, Hult K, Park S. ChemBioChem. 2006;7:1745–1749. doi: 10.1002/cbic.200600245. [DOI] [PubMed] [Google Scholar]
- 94.Goettig P, Groll M, Kim JS, Huber R, Brandstetter H. EMBO J. 2002;21:5343–5352. doi: 10.1093/emboj/cdf552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Syrén PO, Hendil-Forssell P, Aumailley L, Besenmatter W, Gounine F, Svendsen A, Martinelle M, Hult K. ChemBioChem. 2012;13:645–648. doi: 10.1002/cbic.201100779. [DOI] [PubMed] [Google Scholar]
- 96.Jung S, Kim J, Park S. RSC Adv. 2013;3:2590–2594. [Google Scholar]
- 97.Moore GP, Sullivan DT. Biochim Biophys Acta. 1975;397:468–477. doi: 10.1016/0005-2744(75)90137-0. [DOI] [PubMed] [Google Scholar]
- 98.Seah SYK, Labbé G, Nerdinger S, Johnson MR, Snieckus V, Eltis LD. J Biol Chem. 2000;275:15701–15708. doi: 10.1074/jbc.275.21.15701. [DOI] [PubMed] [Google Scholar]
- 99.Horsman GP, Bhowmik S, Seah SYK, Kumar P, Bolin JT, Eltis LD. J Biol Chem. 2007;282:19894–19904. doi: 10.1074/jbc.M702237200. [DOI] [PubMed] [Google Scholar]
- 100.Li C, Li J, Montgomery MG, Wood SP, Bugg TDH. Biochemistry. 2006;45:12470–12479. doi: 10.1021/bi061253t. [DOI] [PubMed] [Google Scholar]
- 101.Ruzzini AC, Bhowmik S, Ghosh S, Yam KC, Bolin JT, Eltis LD. Biochemistry. 2013;52:7428–7438. doi: 10.1021/bi401156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou H, Qu Y, Kong C, Wu Y, Zhu K, Yang J, Zhou J. Biotechnol Lett. 2012;34:1107–1113. doi: 10.1007/s10529-012-0880-0. [DOI] [PubMed] [Google Scholar]
- 103.Ruzzini AC, Horsman GP, Eltis LD. Biochemistry. 2012;51:5831–5840. doi: 10.1021/bi300663r. [DOI] [PubMed] [Google Scholar]
- 104.Alcaide M, Tornés J, Stogios PJ, Xu X, Gertler C, di Leo R, Bargiela R, Lafraya A, Guazzaroni M, López-Cortéz N, Chernikova T, Golyshina OV, Nechitaylo TY, Plumeier I, Pieper DH, Yakimov MM, Savchenko A, Golyshin PN, Ferrer M. Biochem J. 2013;454:157–166. doi: 10.1042/BJ20130552. [DOI] [PubMed] [Google Scholar]
- 105.Sachelaru P, Schiltz E, Igloi GL, Brandsch R. J Bacteriol. 2005;187:8516–8519. doi: 10.1128/JB.187.24.8516-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schleberger C, Sachelaru P, Brandsch R, Schulz G. J Mol Biol. 2007;367:409–418. doi: 10.1016/j.jmb.2006.12.068. [DOI] [PubMed] [Google Scholar]
- 107.Auldridge ME, Guo Y, Austin MB, Ramsey J, Fridman E, Pichersky E, Noel JP. Plant Cell. 2012;24:1596–1607. doi: 10.1105/tpc.111.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Poechlauer P, Skranc W, Wubbolts M. In: Asymmetric Catalysis on Industrial Scale. Schmidt E, Blaser H-U, editors. Wiley-VCH; Weinheim: 2004. pp. 149–164. [Google Scholar]
- 109.Schmidt A, Kratky C, Gruber K, Lamzin VS. J Biol Chem. 2008;283:21827–21836. doi: 10.1074/jbc.M801056200. [DOI] [PubMed] [Google Scholar]
- 110.Koo YJ, Yoon ES, Seo JS, Kim JK, Choi YD. J Korean Soc Appl Biol Chem. 2013;56:27–33. [Google Scholar]
- 111.Andexer JN, Langermann von J, Mell A, Bocola M, Kragl U, Eggert T, Pohl M. Angew Chem Int Ed Engl. 2007;46:8679–8681. doi: 10.1002/anie.200701455. [DOI] [PubMed] [Google Scholar]
- 112.Andexer JN, Staunig N, Eggert T, Kratky C, Pohl M, Gruber K. ChemBioChem. 2012;13:1932–1939. doi: 10.1002/cbic.201200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu X, van Pée KH, Naismith JH. J Biol Chem. 2010;285:21126–21133. doi: 10.1074/jbc.M110.120485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Branneby C, Carlqvist P, Magnusson AO, Hult K, Brinck T, Berglund P. J Am Chem Soc. 2003;125:874–875. doi: 10.1021/ja028056b. [DOI] [PubMed] [Google Scholar]
- 115.Branneby C, Carlqvist P, Hult K, Brinck T, Berglund P. J Mol Catal B Enzym. 2004;31:123–128. [Google Scholar]
- 116.Li C, Feng XW, Wang N, Zhou YJ, Yu XQ. Green Chem. 2008;10:616–618. [Google Scholar]
- 117.Li K, He T, Li C, Feng XW, Wang N, Yu XQ. Green Chem. 2009;11:777–779. [Google Scholar]
- 118.Svedendahl M, Hult K, Berglund P. J Am Chem Soc. 2005;127:17988–17989. doi: 10.1021/ja056660r. [DOI] [PubMed] [Google Scholar]
- 119.Strohmeier GA, SoviC T, Steinkellner G, Hartner FS, Andryushkova A, Purkarthofer T, Glieder A, Gruber K, Griengl H. Tetrahedron. 2009;65:5663–5668. [Google Scholar]
- 120.Xu JM, Zhang F, Wu Q, Zhang QY, Lin XF. J Mol Catal B Enzym. 2007;49:50–54. [Google Scholar]
- 121.Kitazume T, Ikeya T, Murata K. J Chem Soc, Chem Commun. 1986:1331–1333. [Google Scholar]
- 122.Kitazume T, Murata K, Kokusho Y, Iwasaki S. J Fluorine Chem. 1988;39:75–86. [Google Scholar]
- 123.Torre O, Alfonso I, Gotor V. Chem Commun. 2004:1724–1725. doi: 10.1039/b402244k. [DOI] [PubMed] [Google Scholar]
- 124.Torre O, Gotor-Fernández V, Alfonso I, García-Alles LF, Gotor V. Adv Synth Catal. 2005;347:1007–1014. [Google Scholar]
- 125.Carlqvist P, Svedendahl M, Branneby C, Hult K, Brinck T, Berglund P. ChemBioChem. 2005;6:331–336. doi: 10.1002/cbic.200400213. [DOI] [PubMed] [Google Scholar]
- 126.Cai Y, Wu Q, Xiao YM, Lv DS, Lin XF. J Biotechnol. 2006;121:330–337. doi: 10.1016/j.jbiotec.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 127.Svedendahl M, Jovanović B, Fransson L, Berglund P. ChemCatChem. 2009;1:252–258. [Google Scholar]
- 128.Reetz MT, Mondière RJG, Carballeira JD. Tetrahedron Lett. 2007;48:1679–1681. [Google Scholar]
- 129.Wu Q, Lou F, Liu BK, Lv DS, Lin XF. Adv Synth Catal. 2008;350:1959–1962. [Google Scholar]
- 130.Lou F, Liu BK, Wang JL, Pan Q, Lin XF. J Mol Catal B Enzym. 2009;60:64–68. [Google Scholar]
- 131.Jiang M, Cao Y, Guo ZF, Chen M, Guo Z. Biochemistry. 2007;46:10979–10989. doi: 10.1021/bi700810x. [DOI] [PubMed] [Google Scholar]
- 132.Jiang M, Chen X, Wu XH, Chen M, Wu YD, Guo Z. Biochemistry. 2009;48:6921–6931. doi: 10.1021/bi900897h. [DOI] [PubMed] [Google Scholar]
- 133.Johnston JM, Jiang M, Guo Z, Baker EN. PLoS ONE. 2013;8:e61325. doi: 10.1371/journal.pone.0061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dawson A, Fyfe PK, Gillet F, Hunter WN. BMC Struct Biol. 2011;11:19. doi: 10.1186/1472-6807-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vongvilai P, Linder M, Sakulsombat M, Humble MS, Berglund P, Brinck T, Ramström O. Angew Chem Int Ed Engl. 2011;50:6592–6595. doi: 10.1002/anie.201007373. [DOI] [PubMed] [Google Scholar]
- 136.Schmidt E, Knackmuss HJ. Biochem J. 1980;192:339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pathak D, Ashley G, Ollis D. Proteins Struct Funct Genet. 1991;9:267–279. doi: 10.1002/prot.340090405. [DOI] [PubMed] [Google Scholar]
- 138.Walker I, Hennessy JE, Ollis DL, Easton CJ. ChemBioChem. 2012;13:1645–1651. doi: 10.1002/cbic.201200232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.