Abstract
The NMR structures of the recombinant prion proteins from chicken (Gallus gallus; chPrP), the red-eared slider turtle (Trachemys scripta; tPrP), and the African clawed frog (Xenopus laevis; xlPrP) are presented. The amino acid sequences of these prion proteins show ≈30% identity with mammalian prion proteins. All three species form the same molecular architecture as mammalian PrPC, with a long, flexibly disordered tail attached to the N-terminal end of a globular domain. The globular domain in chPrP and tPrP contains three α-helices, one short 310-helix, and a short antiparallel β-sheet. In xlPrP, the globular domain includes three α-helices and a somewhat longer β-sheet than in the other species. The spatial arrangement of these regular secondary structures coincides closely with that of the globular domain in mammalian prion proteins. Based on the low sequence identity to mammalian PrPs, comparison of chPrP, tPrP, and xlPrP with mammalian PrPC structures is used to identify a set of essential amino acid positions for the preservation of the same PrPC fold in birds, reptiles, amphibians, and mammals. There are additional conserved residues without apparent structural roles, which are of interest for the ongoing search for physiological functions of PrPC in healthy organisms.
Keywords: nonmammalian species, transmissible spongiform encephalopathy
The prion protein (PrP) has attracted a lot of interest because of its relation to transmissible spongiform encephalopathies (TSEs), which are a group of invariably fatal neurological diseases (1). Healthy organisms that do not express a prion protein, such as suitably selected transgenic laboratory animals, cannot develop a TSE (2), and the “protein-only hypothesis” further attributes TSE-causing infectivity to an aggregated “scrapie form” of PrP (PrPSC) that has been isolated from brain tissue of diseased organisms (1). Although TSEs have only been documented for mammalian species, PrP has been identified in a wider range of higher organisms, which on an evolutionary scale extends at least down to amphibians (3–9). In apparent contrast to the high sequence conservation among mammalian PrPs, no physiological function has been reliably attributed to the “cellular form” of PrP (PrPC) found in healthy organisms.
In view of its critical role in TSEs, the prion protein has also attracted considerable interest by structural biologists. So far, atomic resolution structure determination was focused on recombinant mammalian prion proteins (10–18), which have recently been shown to represent the protein architecture of PrPC (19). As a group, the mammalian PrPs have ≈90% sequence identity (4). Here, we present the NMR structures of recombinant PrP from chicken, turtle, and frog (chPrP, tPrP, and xlPrP, respectively), each of which has ≈30% sequence identity with mammalian PrP. We then exploit this low homology in searches, based on comparison of the three-dimensional structures, for conserved amino acids with apparent roles in maintaining a common PrPC-fold, and for nonstructural conserved amino acids that might provide leads to the unknown physiological function of PrPC.
Materials and Methods
Protein Preparation. The chPrP gene was provided to us by D. A. Harris (Washington University School of Medicine, St. Louis), the tPrP gene was given to us by T. Simonic (Università di Milano, Milano, Italy), and the xlPrP gene was obtained from a cDNA library. All three genes were cloned into the vector pRSETA, and the proteins were expressed in Escherichia coli. For the purification of the recombinant proteins, we followed described procedures (20).
NMR Measurements and Structure Calculations. NMR measurements were performed at 20°C on Bruker DRX500, DRX600, and DRX750 and Avance900 spectrometers. The protein samples used were uniformly 15N-labeled and 13C, 15N-labeled tPrP(23–225), tPrP(121–225), chPrP(23–225), chPrP(121–225), and xlPrP(90–222). In this notation, the numbers in parentheses identify the first and last residues in the individual constructs, and the numeration for human PrP is used as explained in the caption to Fig. 1. The NMR samples were in ether 95% H2O/5% D2O or 99.9% D2O, contained 5 mM sodium acetate buffer at pH 4.5 (pH 4.3 for chPrP), and the protein concentration was 0.6–1.0 mM. The programs prosa (21) and xeasy (22) were used for data processing and spectral analysis, respectively. Sequence-specific resonance assignments for the proteins were obtained by using standard triple-resonance NMR experiments (23). Distance constraints for the input for the structure determination were obtained from three-dimensional 13C-resolved [1H, 1H]-NOESY, three-dimensional 15N-resolved [1H, 1H]-NOESY, and two-dimensional [1H, 1H]-NOESY spectra recorded at a proton frequency of 750 or 900 MHz with mixing times of 40 or 50 ms. The automatic nuclear Overhauser enhancement (NOE) assignment module candid (24) implemented in the program dyana (25) was used for the structure calculations. dyana was also used to convert NOE intensities into upper distance constraints according to an inverse sixth power peak volume-to-distance relationship, to remove meaningless constraints, and to derive constraints for the backbone torsion angles ϕ and Ψ from the Cα chemical shift values (26, 27). The final round of structure calculation with dyana was started with 100 randomized conformers. The 20 conformers with the lowest final target function values were energy-minimized in a water shell with in the program opalp (28, 29) using the amber force field (30). The program molmol (31) was used to analyze the results of the structure calculations (Table 1) and to prepare the drawings of the structures.
Fig. 1.
Sequence alignment of hPrP, chPrP, tPrP, and xlPrP, where hPrP represents “mammalian-type PrP.” At the top is the residue numbering for hPrP, which is used throughout this manuscript, i.e., insertions and deletions in chPrP, tPrP, and xlPrP required for maximal coincidence with hPrP are not consecutively numbered. In this alignment, the amino acids shown in red are identical in all four PrPs, and the ones displayed in blue are identical in chPrP, tPrP, and hPrP. The black box identifies the segment containing polypeptide repeats (see text) for which no individual alignments were attempted. The GPI attachment site is identified with a green box, and the glycosylation sites (asparagine attachment site and nearest-next threonine/serine) is identified with light blue boxes. The orange boxes indicate segments with higher than average sequence conservation that do not have an apparent stabilizing role in the PrP fold, and might thus be conserved for functional reasons. These include the polypeptide segment 23–42 with the N-terminal signaling-peptide cleavage site, which has been suggested to be the signaling-peptide for reinternalization of PrP (36), a Src homology 3 (SH3)-binding motif of residues 100–110 (37, 38), the segment 113–128 representing a predicted transmembrane helix (39, 40), and the segment 146–155 in hPrP, chPrP, and tPrP that shows similarity to the laminin α2-chain (see text). The pink boxes indicate segments with high amino acid identity that appear to be needed for the stability of the “PrP-fold” (see text). At the bottom, the regular secondary structures in the globular domain of the four proteins are indicated. The sequence alignment was performed interactively so as to align a maximal number of identical residues. For the globular domain (121–230), the alignment was also based on visual inspection of the three-dimensional structures. In chPrP, the insertion at the end of helix α2 was divided into two segments to properly align the glycosylation site 197–199.
Table 1. Input for the structure calculation and characterization of the energy-minimized NMR solution structures of tPrP(121–225), chPrP(121–225), and xlPrP(90–222).
| Quantity* | chPrP(121–225) | tPrP(121–225) | xlPrP(90–222) |
|---|---|---|---|
| NOE upper distance limits | 1,889 | 1,522 | 2,283 |
| Dihedral angle constraints | 123 | 110 | – |
| Residual target function value, Å2 | 1.62 ± 0.20 | 1.75 ± 0.18 | 1.99 ± 0,19 |
| Residual distance constraint violations | |||
| Number > 0.1 Å | 29 ± 2 | 30 ± 4 | 24 ± 4 |
| Maximum, Å | 0.16 ± 0.03 | 0.14 ± 0.01 | 0.14 ± 0.01 |
| Residual dihedral angle constraint violations | |||
| Number > 2.0° | 1 ± 1 | 0 | – |
| Maximum, ° | 2.13 ± 0.80 | 1.21 ± 0.71 | – |
| Amber energies, kcal/mol | |||
| Total | –4,633 ± 98 | –4,694 ± 69 | –5,215 ± 60 |
| van der Waals | –285 ± 17 | –309 ± 16 | –244 ± 17 |
| Electrostatic | –5,285 ± 94 | –5,265 ± 74 | –6,032 ± 59 |
| rms deviation to the averaged coordinates (Å)† | |||
| Backbone (N, Cα, C′) | 0.72 ± 0.14 | 0.85 ± 0.13 | 0.75 ± 0.13 |
| (135–200, 211–237) | (125–224) | (127–166, 172–225) | |
| All heavy atoms | 1.17 ± 0.13 | 1.33 ± 0.15 | 1.19 ± 0.12 |
Except for the top two entries, the average for the 20 conformers with the lowest residual dyana target function values and the standard deviation among them are given. NOE, nuclear Overhauser enhancement
The numbers in parentheses identify the polypeptide segments for which the rms deviation was calculated
Database Searches. The SwissProt and TrEMBL databases (32) of proteins were searched by using the pattinprot tool at the Pole Bio-Informatique Lyonnaise (http://pbil.ibcp.fr). The databases were searched by using the query E-x (2)-[WY]-[WY]-x-E-N.
Results
The NMR structures of recombinant chPrP(23–225), tPrP(23–225), and xlPrP(99–222) (see Fig. 1 for the sequence numeration used) all contain a globular domain comprising a C-terminal segment of ≈100 residues. N-terminal to this structured domain there is a flexibly disordered tail of 110 residues for chPrP(23–225), 118 residues for tPrP(23–225), and 69 residues for mature xlPrP (Fig. 1). These global architectures coincide with those of mammalian PrPs(10–18), and they were clearly evidenced by different dispersion of the 1H chemical shifts and different values of the steady-state 15N{1H}-NOEs for the tail and the globular domain in each protein. Here, we focus on the NMR structure determination of the globular domain in the three prion proteins.
The NMR structures of the globular domains were determined in the constructs chPrP(121–225), tPrP(121–225), and xlPrP(90–222). Nearly complete resonance assignments were obtained, and chemical shift lists have been deposited in the BioMagResBank (33–35). The structure determination protocols are presented in Table 1. The three structures are shown in Fig. 2 B–D.
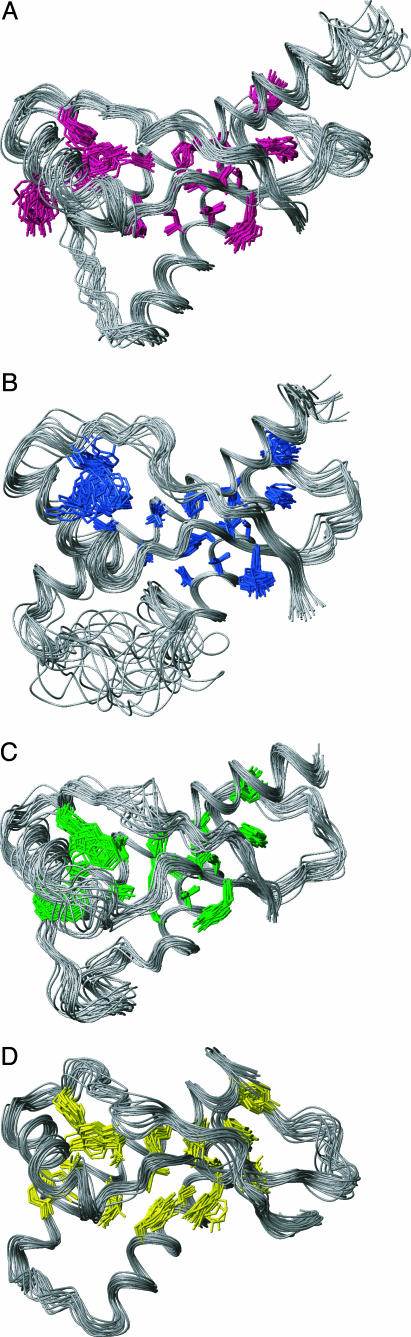
Fig. 2.
NMR structures of the globular domains of hPrP, chPrP, tPrP, and xlPrP. The polypeptide backbone fold for the residues 126–230 (see Fig. 1 for the sequence numbering used) and the core side chains with <20% solvent accessibility are shown for each species, as a superposition of the 20 conformers used to represent the NMR structure. The following side chains are included (see Fig. 1 for the sequence information): 141, 149 (only for chPrP and xlPrP), 150 (only for hPrP and xlPrP), 161, 162, 164 (only for xlPrP), 175, 176, 179, 180 (only for hPrP, chPrP, and tPrP), 183, 184, 205, 206, 209, 210, 213, 214, and 218. (A) hPrP (side chains shown in pink). (B) chPrP (side chains shown in blue). (C) tPrP (side chains shown in green). (D) xlPrP (side chains shown in yellow).
In the orientation of the proteins in Fig. 2, the N terminus is on the right, pointing to the lower right corner, and the C terminus is in the upper right corner. The regular secondary structures (Fig. 1) are arranged in the three-dimensional structure as follows: The strand β1 runs from the lower right to the upper left immediately after the N terminus, helix α1 is in the upper left corner and propagates toward the center of the structure, where the polypeptide chain then forms the β2-strand of the antiparallel β-sheet. A loop of residues 166–175 on the extreme right leads to the start of the helix α2, which ends at the lowest point of the structure. Loops of variable length in the different proteins (Fig. 1) lead from the end of α2 to the start of helix α3 on the left of the structure, from where α3 propagates to the C terminus in the upper right corner. In chPrP, insertion of 11 residues in the loop linking the helices α2 and α3 (Fig. 1) is accommodated in a disordered spatial arrangement of this polypeptide segment (Fig. 2B). Each structure has a well ordered core of hydrophobic side chains, which are conserved among the four species (Figs. 1 and 2).
Discussion
In this section, we conduct searches for possible correlations between amino acid sequence (Fig. 1), three-dimensional structure of the PrPC form of the proteins in Fig. 2, and functional properties of prion proteins. For the unstructured N-terminal tail, these considerations are based primarily on the primary structures (Fig. 1) and on literature data on the biochemistry and cell biology of PrPC. For the globular domain, most of the conclusions result from detailed comparison of the available mammalian and nonmammalian PrPC structures (Fig. 2).
Sequence Conservation in the N-terminal Flexible Tail and PrPC Trafficking. The prion protein sequences from mammals (represented here by human PrP, hPrP), chicken, turtle, and frog show several regions of above-average identity (Fig. 1). In the N-terminal flexibly disordered part of the protein, this includes a glycine-rich and positively charged stretch of 15–20 residues at the chain end, which triggers the cleavage of the N-terminal signal sequence 1–22, and has been implicated in subcellular trafficking of PrPC (36). A highly conserved region of residues 100–110 forms a Src homology 3 (SH3)-binding motif, which in mammals binds the C-terminal SH3 domain of Grb2 (37, 38). Another highly conserved segment of residues 113–128 represents a putative transmembrane helix (39, 40); part of this segment is deleted in xlPrP (Fig. 1). There is also the region made up of polypeptide repeats, with octarepeats in mammalian PrP, hexarepeats in bird and reptile PrP, and no readily apparent repeat pattern in xlPrP (Fig. 1). The tentative functional assignments for these highly conserved sequence elements are further validated by the three-dimensional structures to the extent that in all of the different species they are located in the flexibly disordered, highly solvent-accessible “tail.” The most clear-cut implication results with regard to trafficking of PrPC, because the structural elements known to be important for cellular trafficking are highly conserved in the PrP sequences (Fig. 1) and the PrPC three-dimensional structures. These are the N-terminal signaling sequence (not shown in Fig. 1) that directs PrP to the endoplasmatic reticulum, the C-terminal signaling sequence that attaches to the glycosylphosphatidylinositol (GPI) anchor and directs PrPC to lipid rafts within the membrane (41), and sequence 23–42, which directs the prion protein to clathrin-coated pits or caveolae like domains, and therefore controls the endocytosis of the prion protein (42).
Comparison of the Globular Domains of chPrP, tPrP, and xlPrP with hPrP. The three-dimensional structures of the globular domains determined in hPrP(121–230) (13), chPrP(121–225), tPrP(121–225), and xlPrP(90–222) clearly show extensive similarities. These similarities include the sequence locations of regular secondary structures (Fig. 1), the three-dimensional arrangement of the secondary structure elements (Fig. 2), and the presence of a hydrophobic core of amino acid side chains with low solvent accessibility (Fig. 2). In Fig. 3, this visual impression of structural similarity is substantiated with a more quantitative and detailed comparison.
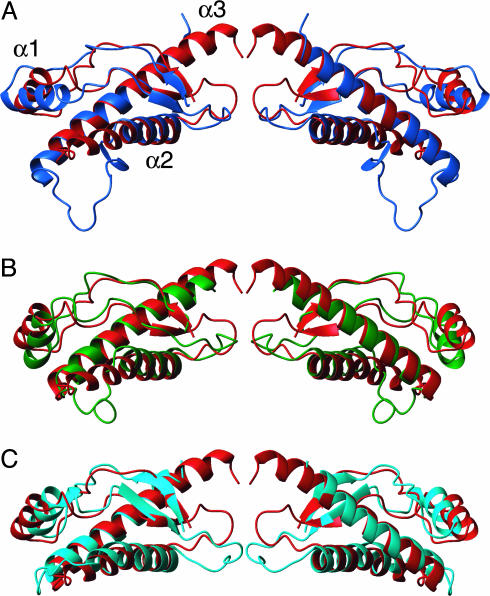
Fig. 3.
Comparison of the three nonmammalian PrPs investigated here with hPrP, representing the “mammalian-type” PrPC fold. Pairwise superpositions of the polypeptide backbone folds are shown. (A) chPrP(121–225) (blue) and hPrP(121–230) (red). (B) tPrP(121–225) (green) and hPrP(121–230) (red). (C) xlPrP(90–222) (cyan) and hPrP(121–230) (red). The protein fragments of residues 128–222 are displayed. The proteins were aligned for best fit of segments 128–131, 161–164, 173–187, and 201–222. The views on the right relate to those on the left by a 180° rotation about a vertical axis.
A tight superposition of the regular secondary structures in the four proteins shown in Fig. 2 is obtained if the helix α1 is not included in the fitting process. The backbone heavy atoms of the helices α2 and α3 and the β-sheet are then superimposable for any pair of the four PrPs with rms deviation values of 1.1 Å or less (Fig. 3). The ≈30% identity and 50% conservation of the sequence (Fig. 1) is then sufficient to ensure identical hydrophobic core packing (Fig. 2). When using this approach for structure superposition, the surface loops linking the regular secondary structures and the orientation of helix α1 show significant variation in the different PrPs (Fig. 3). This variation coincides with low amino acid conservation in the loops of the globular domain (Fig. 1). Some of these variations could be regarded as “structural signatures” for PrPs from the different evolutionary subgroups. A first example is loop 166–173, which shows dynamic disorder in hPrP (13), whereas it is more precisely defined in the other three proteins of Fig. 2. This loop appears to be stabilized by a long-range hydrogen bond between V171 and Y222 in tPrP, insertion of a proline in chPrP, and a 2-aa insertion in xlPrP. A second example is the C-terminal end of helix α2, which consists of a tetrathreonine segment in hPrP that clearly differs from the capped helices of chPrP and tPrP, and the kinked helix of xlPrP (Figs. 1 and 3). An additional “structural signature” for chPrP is provided by an insertion between the helices α2 and α3 (Figs. 1 and 2), which forms a flexibly disordered loop and an N-terminal elongation of helix α3 in chPrP(121–225) (Fig. 3). This insertion is conserved in all known avian PrP sequences (4).
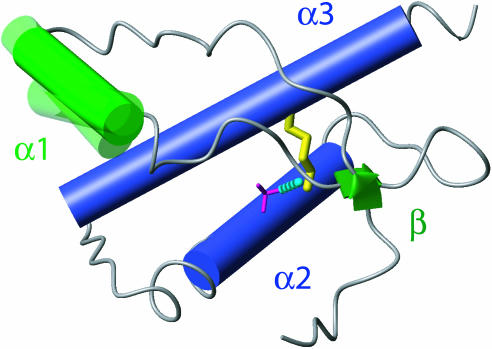
Minimal Scaffold Maintaining the PrPC Architecture in Prion Proteins. The structure comparisons in Fig. 3 indicate that the following structure elements are preserved in all of the proteins of Fig. 2, and form a scaffold that stabilizes the PrPC-type three-dimensional fold: The arrangement of the two long helices α2 and α3, their connection by a disulphide bond, the packing of the β-sheet against the helices α2 and α3 reinforced by a hydrogen bond between T(S)183 Oγ and Y162 HN, and the anchoring of the helix α1 against the protein core by two tyrosine or tryptophane residues in positions 149 and 150. Fig. 4 affords a visualization of this preserved scaffold of PrPC, which illustrates the variable orientation of helix α1 and indicates that variation of the C-terminal extension of helix α2 by about two turns (Fig. 1) can be accommodated in a “PrPC-fold.”
Fig. 4.
Structural features proposed to be essential for a stable PrPC fold. Helices are shown as cylinders, with α1 shown in green and α2 and α3 shown in dark blue. The β-sheet is represented by green arrows. The single disulfide bond is shown in yellow, and a hydrogen bond between Oγ of T183 and the backbone amide proton of Y162 is shown as a dashed cyan line. The backbone segments linking these structural features are shown as a gray spline function through the Cα positions. If the other elements shown are superimposed for best fit, the helix α1 adopts somewhat different orientations in the four proteins of Fig. 1.
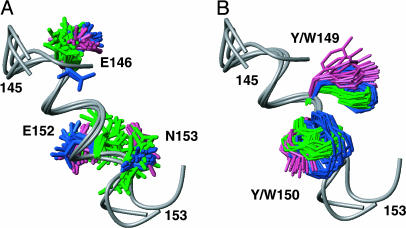
Indication for Function-Related Sequence Conservation in the Globular Domain of PrPC. Most of the highly conserved amino acids in the prion proteins shown in Fig. 1 have readily apparent structural roles in the PrPC fold (see legend to Fig. 2), but there are highly conserved residues in the peptide segment 146–153 of the helix α1 (Fig. 1) that could, because of their peripheral location, hardly be considered as structure-stabilizing elements (Fig. 5). The structural motif of residues 146–153 was therefore used to search a database of known protein sequences for functional sites. A search of three-dimensional structures gave several hits, which were exclusively helix–helix contacts within single domains. A more extensive search that included sequences for which no three-dimensional structures are available gave ≈100 hits, which eventually led to the indication of a significant relationship between PrP segment 146–153 and region 351–358 of the human laminin α2 chain, which has the sequence EECYYDEN. Because the laminin receptor precursor has been repeatedly suggested as a possible protein partner for PrPC (43–45), this indication of functional relations between the laminin α2 chain and part of the helix α1 in PrPC might be worth further study.
Fig. 5.
Conserved structural motif on the surface of the globular domains of chPrP, tPrP, and hPrP. There is no apparent role in stabilization of the protein fold, indicating that this motif might be conserved for reasons of the protein function. hPrP, chPrP, and tPrP have been superimposed for local best fit of the backbone atoms of residues 146–153, which form the helix α1 (the backbone is shown as a gray spline function). (A) With the side chains of the residues E146, E152, and N153. (B) With the side chains of the residues 149 and 150 (W or Y in the different species; see Fig. 1). hPrP side chains are shown in pink, chPrP side chains are shown in blue, and tPrP side chains are shown in green.
Acknowledgments
We thank Dr. T. Simonic for providing the turtle PrP plasmid, Dr. D. Harris for providing the chicken PrP plasmid, and Dr. J. Gurdon (University of Cambridge, Cambridge, U.K.) for the Xenopus laevis cDNA library. Financial support was obtained from the Schweizerischer Nationalfonds and the Eidgenössische Technische Hochschule (Zürich) through the National Center of Competence in Research Structural Biology.
Author contributions: K.W. designed research; L.C., D.A.L., D.R.P., and P.G. performed research; and L.C., D.A.L., D.R.P., and K.W. wrote the paper.
Abbreviations: PrP, prion protein; TSE, transmissible spongiform encephalopathy; PrPC, cellular PrP; chPrP, chicken PrP; tPrP, turtle PrP; xlPrP, frog PrP.
Data deposition: The sequences reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1U3M for chPrP(121–225), 1U5L for tPrP(121–225), and 1XU0 for xlPrP(90–222)].
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Büeler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339-1347. [DOI] [PubMed] [Google Scholar]
- 3.Oesch, B., Westaway, D., Walchli, M., McKinley, M. P., Kent, S. B., Aebersold, R., Barry, R. A., Tempst, P., Teplow, D. B., Hood, L. E., et al. (1985) Cell 40, 735-746. [DOI] [PubMed] [Google Scholar]
- 4.Wopfner, F., Weidenhofer, G., Schneider, R., von Brunn, A., Gilch, S., Schwarz, T. F., Werner, T. & Schätzl, H. M. (1999) J. Mol. Biol. 289, 1163-1178. [DOI] [PubMed] [Google Scholar]
- 5.Van Rheede, T., Smolenaars, M. M., Madsen, O. & De Jong, W. W. (2003) Mol. Biol. Evol. 20, 111-121. [DOI] [PubMed] [Google Scholar]
- 6.Harris, D. A., Falls, D. L., Johnson, F. A. & Fischbach, G. D. (1991) Proc. Natl. Acad. Sci. USA 88, 7664-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabriel, J. M., Oesch, B., Kretzschmar, H., Scott, M. & Prusiner, S. B. (1992) Proc. Natl. Acad. Sci. USA 89, 9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonic, T., Duga, S., Strumbo, B., Asselta, R., Ceciliani, F. & Ronchi, S. (2000) FEBS Lett. 469, 33-38. [DOI] [PubMed] [Google Scholar]
- 9.Strumbo, B., Ronchi, S., Bolis, L. C. & Simonic, T. (2001) FEBS Lett. 508, 170-174. [DOI] [PubMed] [Google Scholar]
- 10.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R. & Wüthrich, K. (1996) Nature 382, 180-182. [DOI] [PubMed] [Google Scholar]
- 11.James, T. L., Liu, H., Ulyanov, N. B., Farr-Jones, S., Zhang, H., Donne, D. G., Kaneko, K., Groth, D., Mehlhorn, I., Prusiner, S. B. & Cohen, F. E. (1997) Proc. Natl. Acad. Sci. USA 94, 10086-10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riek, R., Hornemann, S., Wider, G., Glockshuber, R. & Wüthrich, K. (1997) FEBS Lett. 413, 282-288. [DOI] [PubMed] [Google Scholar]
- 13.Zahn, R., Liu, A., Lührs, T., Riek, R., von Schroetter, C., Lopez Garcia, F., Billeter, M., Calzolai, L., Wider, G. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez Garcia, F., Zahn, R., Riek, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8334-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calzolai, L., Lysek, D. A., Güntert, P., von Schroetter, C., Riek, R., Zahn, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8340-8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haire, L. F., Whyte, S. M., Vasisht, N., Gill, A. C., Verma, C., Dodson, E. J., Dodson, G. G. & Bayley, P. M. (2004) J. Mol. Biol. 336, 1175-1183. [DOI] [PubMed] [Google Scholar]
- 17.Gossert, A. D., Bonjour, S., Lysek, D. A., Fiorito, F. & Wüthrich, K. (2005) Proc. Natl. Acad. Sci. USA 102, 646-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lysek, D. A., Schorn, C., Nivon, L. G., Esteve-Moya, V., Christen, B., Calzolai, L., von Schroetter, C., Fiorito, F., Herrmann, T., Güntert, P. & Wüthrich, K. (2005) Proc. Natl. Acad. Sci. USA 102, 640-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornemann, S., Schorn, C. & Wüthrich, K. (2004) EMBO Rep. 5, 1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahn, R., von Schroetter, C. & Wüthrich, K. (1997) FEBS Lett. 417, 400-404. [DOI] [PubMed] [Google Scholar]
- 21.Güntert, P., Dötsch, V., Wider, G. & Wüthrich, K. (1992) J. Biomol. NMR 2, 619-629. [Google Scholar]
- 22.Bartels, C., Xia, T. H., Billeter, M., Güntert, P. & Wüthrich, K. (1995) J. Biomol. NMR 6, 1-10. [DOI] [PubMed] [Google Scholar]
- 23.Bax, A. & Grzesiek, S. (1993) Acc. Chem. Res. 26, 131-138. [Google Scholar]
- 24.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Mol. Biol. 319, 209-227. [DOI] [PubMed] [Google Scholar]
- 25.Güntert, P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273, 283-298. [DOI] [PubMed] [Google Scholar]
- 26.Spera, S. & Bax, A. (1991) J. Am. Chem. Soc. 113, 5490-5492. [Google Scholar]
- 27.Luginbühl, P., Szyperski, T. & Wüthrich, K. (1995) J. Magn. Reson. B 109, 229-233. [Google Scholar]
- 28.Luginbühl, P., Güntert, P., Billeter, M. & Wüthrich, K. (1996) J. Biomol. NMR 8, 136-146. [DOI] [PubMed] [Google Scholar]
- 29.Koradi, R., Billeter, M. & Güntert, P. (2000) Comput. Phys. Commun. 124, 139-147. [Google Scholar]
- 30.Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M., Jr., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179-5197. [Google Scholar]
- 31.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-57. [DOI] [PubMed] [Google Scholar]
- 32.Bairoch, A. & Apweiler, R. (2000) Nucleic Acids Res. 28, 45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lysek, D. A., Calzolai, L. & Wüthrich, K. (2004) J Biomol NMR 30, 97. [DOI] [PubMed] [Google Scholar]
- 34.Calzolai, L., Lysek, D. A. & Wüthrich, K. (2004) J. Biomol. NMR 30, 97. [DOI] [PubMed] [Google Scholar]
- 35.Perez, D. & Wüthrich, K., J. Biomol. NMR, in press.
- 36.Nunziante, M., Gilch, S. & Schätzl, H. M. (2003) J. Biol. Chem. 278, 3726-3734. [DOI] [PubMed] [Google Scholar]
- 37.Spielhaupter, C. & Schätzl, H. M. (2001) J. Biol. Chem. 276, 44604-44612. [DOI] [PubMed] [Google Scholar]
- 38.Lysek, D. A. & Wüthrich, K. (2004) Biochemistry 43, 10393-10399. [DOI] [PubMed] [Google Scholar]
- 39.Hegde, R. S., Mastrianni, J. A., Scott, M. R., DeFea, K. A., Tremblay, P., Torchia, M., DeArmond, S. J., Prusiner, S. B. & Lingappa, V. R. (1998) Science 279, 827-834. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, R. S. & Harris, D. A. (2001) J. Biol. Chem. 276, 2212-2220. [DOI] [PubMed] [Google Scholar]
- 41.Harris, D. (1999) Clin. Microbiol. Rev. 12, 429-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilch, S., Nunziante, M., Ertmer, A., Wopfner, F., Laszlo, L. & Schätzl, H. M. (2004) Traffic 5, 300-313. [DOI] [PubMed] [Google Scholar]
- 43.Graner, E., Mercadante, A. F., Zanata, S. M., Forlenza, O. V., Cabral, A. L., Veiga, S. S., Juliano, M. A., Roesler, R., Walz, R., Minetti, A., et al. (2000) Brain Res. Mol. Brain Res. 76, 85-92. [DOI] [PubMed] [Google Scholar]
- 44.Gauczynski, S., Peyrin, J. M., Haik, S., Leucht, C., Hundt, C., Rieger, R., Krasemann, S., Deslys, J. P., Dormont, D., Lasmezas, C. I. & Weiss, S. (2001) EMBO J. 20, 5863-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rieger, R., Edenhofer, F., Lasmezas, C. I. & Weiss, S. (1997) Nat. Med. 3, 1383-1388. [DOI] [PubMed] [Google Scholar]