Abstract
We report on the NMR-based solution structure of the 93del d(GGGGTGGGAGGAGGGT) aptamer, a potent nanomolar inhibitor of HIV-1 integrase. This guanine-rich DNA sequence adopts an unusually stable dimeric quadruplex architecture in K+ solution. Within each 16-nt monomer subunit, which contains one A·(G·G·G·G) pentad sandwiched between two G·G·G·G tetrads, all G-stretches are parallel, and all guanines are anti with the exception of G1, which is syn. Dimer formation is achieved through mutual pairing of G1 of one monomer, with G2, G6, and G13 of the other monomer, to complete G·G·G·G tetrad formation. There are three single-nucleotide double-chain-reversal loops within each monomer fold, such that the first (T5) and third (A12) loops bridge three G-tetrad layers, whereas the second (A9) loop bridges two G-tetrad layers and participates in A·(G·G·G·G) pentad formation. Results of NMR and of integrase inhibition assays on loop-modified sequences allowed us to propose a strategy toward the potential design of improved HIV-1 integrase inhibitors. Finally, we propose a model, based on molecular docking approaches, for positioning the 93del dimeric DNA quadruplex within a basic channel/canyon formed between subunits of a dimer of dimers of HIV-1 integrase.
Keywords: dimeric quadruplex, DNA aptamer
HIV-1 integrase catalyzes the integration of proviral DNA into the host-cell genome, a reaction critical for efficient viral replication. HIV-1 integrase is necessary and sufficient for the first two steps of DNA integration (1): (i) 3′-end processing, where two nucleotides are removed from each 3′-end of the viral DNA; (ii) strand transfer, where each 3′-processed viral DNA end is attached to the host-cell DNA. As a result, HIV-1 integrase constitutes an attractive target for drugs against AIDS (2, 3).
Guanine-rich oligonucleotides have been identified as potent inhibitors of HIV-1 integrase (4, 5). In particular, a recent study (6) has identified a 16-nt guanine-rich sequence d(GGGGTGGGAGGAGGGT), designated 93del, which inhibits both the processing and strand transfer functions of HIV-1 integrase at nanomolar concentrations.
Guanine-rich oligonucleotides can form a variety of G-quadruplex architectures involving stacked planar G·G·G·G tetrads. This polymorphism of G-quadruplex topologies reflects the range of alternate strand directionalities, loop connectivities, and syn/anti distribution of guanine bases around G-tetrads (7–16). Higher order pairing alignments involving G-tetrads have been recently identified, and these include pentads (17), hexads (18), and heptads (19) for sequences containing guanine stretches separated by adenine residues.
Here, we present the NMR-based solution structure of 93del. We establish that this 16-nt DNA sequence adopts an unusually stable dimeric quadruplex architecture in K+ solution. Each monomer subunit contains two G·G·G·G tetrads and one A·(G·G·G·G) pentad, where all of the G-stretches are parallel and linked by three single-nucleotide double-chain-reversal loops. In our attempts to design potent HIV-1 integrase inhibitors, we have modified individual residues within each of the three loops and probed changes in the folding topology and in the inhibitory activity against HIV-1 integrase. This work has allowed us to propose a strategy toward the potential design of improved HIV-1 integrase inhibitors. In addition, we could identify a potential complementary cavity for 93del binding on the surface of a recently reported crystal structure of tetrameric HIV-1 integrase containing the N-terminal and catalytic core domains (20). We have used molecular docking to propose a model for the structure of the complex between 93del and the tetramer of HIV-1 integrase.
Methods
DNA Sample Preparation. Unlabeled and site-specific 2%-15N, 13C-labeled DNA oligonucleotides were chemically synthesized and purified as described (9, 21). Samples were dialyzed successively against 50 mM KCl solution and against water. Unless otherwise stated, the strand concentration of the NMR samples was typically 1–9 mM; the samples contained 90 mM KCl; pH was adjusted to 7 with HCl and KOH.
Gel Filtration Chromatography. A Synchropack GPC100 column (Synchrom, Lafayette, IN) was used for stoichiometry determination by gel filtration chromatography at room temperature (22). The elution buffer contained 70 mM KCl and 20 mM potassium phosphate, pH 7.
CD Measurements. CD spectra of oligonucleotides were recorded on a JASCO J-710 spectropolarimeter. They were averages of at least five scans from 200 to 320 nm. Experimental conditions were as follows: strand concentration, 30 μM; temperature, 5°C; 100 mM KCl; 10 mM potassium phosphate, pH 7.
NMR Spectroscopy. NMR experiments were performed on 500- and 600-MHz Varian and Bruker spectrometers at 25°C, unless otherwise specified. Resonance assignments were obtained by using site-specific low-enrichment labeling (21) and through-bond correlations at natural abundance (22) (imino-H8 by JRHMBC; H8-H2 by HMBC; H8/6-H1′ by HSQC and sHMBC; H1′-H2′/2″/3′/4′/5′/5″ by COSY and TOCSY; H5′/5″/4′/3′-P by [1H-31P]-TOCSY and [1H-31P]-HSQC), and independently verified by using NOESY experiments. Interproton distances were measured by using NOESY experiments performed in H2O (50- and 200-ms mixing times) and in D2O (50-, 75-, 100-, 150-, 200-, 250-, and 300-ms mixing times).
Structure Calculation. The structure of 93del was calculated on the basis of NMR restraints by using the x-plor program (23). A total of 100 initial structures were embedded and optimized by using the distance geometry simulated annealing protocol. The 10 best structures were selected and subjected to distance-restrained molecular dynamics calculations and then refined against the intensities of well resolved NOESY cross-peaks at various mixing times. During the intensity refinement, the NMR R-factor (23) improved from the initial value of 5.0% to 1.6%, together with a simultaneous improvement in structure convergence. The detailed procedure is described in Supporting Text, which is published as supporting information on the PNAS web site.
Molecular Docking. The docking between the DNA G-quadruplexes and the tetrameric HIV-1 IN1–212 was performed by using the gramm program (24, 25). Because the crystal structure of the enzyme has underdetermined regions (segments 46–56 and 139–149 for each subunit), the low-resolution generic docking with grid step 6.8 Å and grid size 64 Å was used. The DNA molecule was rotated with 20°-angle intervals. The energy score for repulsion was 6.5 a.u.
HIV-1 Integrase Inhibition Assays. The disintegration assays used the “dumbbell” substrate db-Y1 (26). The processing and strand transfer assays used oligonucleotides derived from the U5 end of the HIV-1 LTR as described (6). For inhibition assays, the inhibitors were incubated with HIV-1 integrase for 30 min at 37°C for the disintegration reaction (10 min at 37°C for the processing and strand transfer reactions) before adding substrate. The detailed procedure is described in Supporting Text.
Results
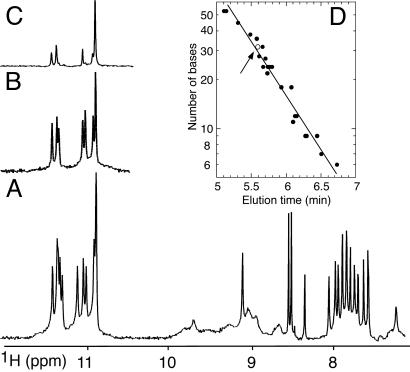
93del Forms Stable Dimeric Quadruplex. The NMR spectrum of 93del in K+ solution is of exceptional quality, with sharp and well resolved imino and nonexchangeable base protons (Fig. 1A). The number and intensity of peaks reflect formation of a single species. The sharp guanine imino protons at 10.5–11.5 ppm (Fig. 1 A) are characteristic of G-tetrad formation. A subset of these imino protons exchange slowly with water, to the extent that eight of them remain after 3 days in D2O at room temperature (Fig. 1B). The structure is unusually stable: several of these imino protons persist in D2O, even after heating the sample in a boiling water bath for 10 min (Fig. 1C).
Fig. 1.
NMR spectra of 93del. (A) The 600-MHz proton spectrum (7.0–12.0 ppm) of 93del in 90 mM K+ H2O solution at 25°C. (B and C) The corresponding imino proton spectra of 93del after dissolution in D2O for 3 days at room temperature (B) and 2 months at room temperature followed by 10 min at 100°C (C). (D) Stoichiometry determination of 93del by gel filtration chromatography. The known number of residues of several reference oligonucleotides is plotted (on a log scale) as a function of the elution time. The straight line is an interpolation through these data. The elution time of 93del (1 μMto 1 mM) is represented by an open circle and emphasized by an arrow.
The stoichiometry of the structure formed by the 16-nt 93del sequence was determined by gel filtration chromatography using a column calibrated with DNA and RNA oligonucleotides of known molecular weight. The gel filtration profile displayed a peak whose elution time corresponds to a molecular weight of 32 nucleotides, indicating a dimer (Fig. 1D). The NMR spectrum under the same experimental conditions corresponds to a single species (as mentioned above). Furthermore, the G-tetrad alignments (identified below) are consistent with a dimeric quadruplex architecture.
Folding Topology of 93del Dimeric Quadruplex. NMR resonances of 93del were unambiguously assigned to their positions in the sequence by the site-specific low-enrichment approach (21) using 2%-15N, 13C-labeled samples and through-bond correlations at natural abundance (22, 27) (data not shown and Fig. 6, which is published as supporting information on the PNAS web site).
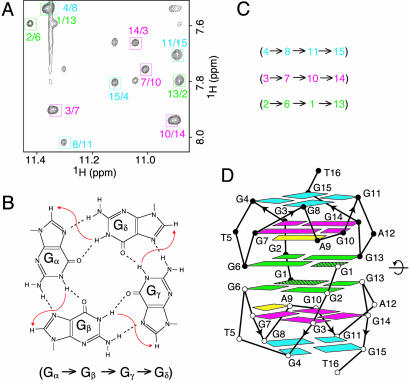
The G-tetrad alignments were identified from NOESY spectra (Fig. 2A), based on the characteristic guanine imino-H8 connectivity patterns around individual G-tetrads (Fig. 2B). The G-tetrad alignment information (Fig. 2C) formed the basis for generation of the 93del twofold symmetric dimeric quadruplex folding topology (Fig. 2D). Each strand in the dimeric quadruplex uses all 12 guanines to form G4·G8·G11·G15, G3·G7·G10·G14, and G2·G6·G1·G13 tetrads. Four G-stretches (2–3–4, 6–7–8, 10–11, and 13–14–15) are oriented in the same direction and represent four columns that support the three G-tetrad planes within each monomer.
Fig. 2.
G-tetrad alignments and global topology of 93del. (A) NOESY spectrum (150-ms mixing time) of 93del at 25°C. The guanine imino-H8 cross-peaks are framed and labeled with the imino proton assignment in the first position and that of the H8 proton in the second position. (B) Characteristic guanine imino-H8 NOE connectivity patterns around a Gα·Gβ·Gγ·Gδ tetrad as indicated with arrows (connectivity between Gδ and Gα is implied). (C) Characteristic guanine imino-H8 NOE connectivities observed for G4·G8·G11·G15 (cyan), G3·G7·G10·G14 (magenta), and G2·G6·G1·G13 (green) tetrads. (D) The proposed dimeric quadruplex topology of 93del based on the G-tetrad alignments outlined in C.
Three single-residue loops T5, A9, and A12 connect G4 to G6, G8 to G10, and G11 to G13, respectively. The identification of NOEs between A9(H8) and G10(H8) (peak a in Fig. 7, which is published as supporting information on the PNAS web site) and between A9(H8) and G7(NH2) (data not shown) established formation of G7·(A9-G10) triad (28) as part of the A9·(G3·G7·G10·G14) pentad, similar to results published previously, which identified markers for pentad formation (17).
The strong intensity of the intraresidue H8-H1′ NOE cross-peak for G1 (Fig. 7), reflects a syn glycosidic conformation for this residue. The intermediate intensities observed for the H8-H1′ NOE cross-peaks of G6 and G13 reflect interstrand contributions (see analysis of the structure outlined below). The weaker intensities observed for all remaining H6/8-H1′ NOE cross-peaks reflect anti glycosidic conformations for all other nucleotides. The anti conformations of all guanines other than G1 are consistent with the relative magnitude of J-couplings (3JC8-H1′ > 3JC4-H1′), manifested by the observation of C8-H1′ (not shown), but not C4-H1′ peaks in a multibond correlation experiment (29).
The anchoring of the two monomers to generate the dimer (Fig. 2D) is achieved with syn G1 of one monomer completing G-tetrad formation through pairing with anti G2, G6, and G13 of the other monomer. The alignment of monomers across the dimeric interface was defined by a number of observed interstrand connectivities, which included NOEs from G13(H8), G6(H8), and G10(H8) of one subunit to G13(H1′), G6(H1′), and G1(H5′/5″) of the other subunit, respectively.
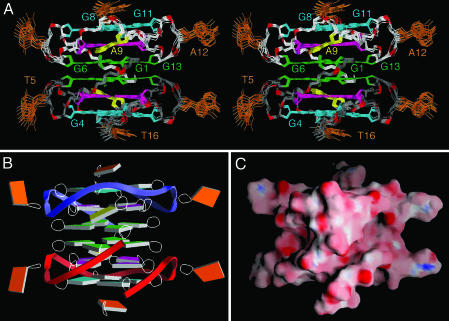
Solution Structure of 93del Dimeric Quadruplex. The structure of 93del was calculated on the basis of NMR restraints (Table 1, which is published as supporting information on the PNAS web site) by using the x-plor program (23). A stereoview of 10 superpositioned lowest energy intensity-refined structures of 93del is shown in Fig. 3A. There is excellent convergence for the stacked core of the dimeric quadruplex (rms deviation for heavy atoms, 0.57 ± 0.06 Å), with only the looped out bases T5 and A12 and 3′-terminal base T16 being less well defined amongst the refined structures. Slab and surface views of one representative refined structure of 93del are shown in Fig. 3 B and C, respectively.
Fig. 3.
Refined structures of the 93del dimeric quadruplex. (A) Stereoview of 10 superpositioned refined structures of 93del. The G4·G8·G11·G15 tetrad is colored cyan, the A9·(G3·G7·G10·G14) pentad is colored magenta (A5 is colored yellow), and the G2·G6·G1·G13 tetrad is colored green, with the remaining bases in orange. The backbone of the top and bottom monomer is colored white and gray, respectively. The phosphorus atoms are colored red, and the exocyclic backbone oxygens deleted in the interest of clarity. (B and C) Slab (B) and surface (C) views of a representative refined structure of 93del are shown. Positive and negative electrostatic potentials are depicted in blue and red, respectively.
The dimeric quadruplex structure of 93del is based on six continuously stacked G-tetrads, two of which are part of A·(G·G·G·G) pentads (Fig. 3A). Two central G-tetrads adopt anti·anti·syn·anti alignments and four others adopt anti·anti·anti·anti alignments (Fig. 2D). The stacking patterns between adjacent tetrad and pentad planes (Fig. 8 A–C, which is published as supporting information on the PNAS web site) show partial stacking between five-membered rings or between the five- and six-membered rings of the guanine bases.
The different loop connectivities are plotted in Fig. 8 D–F. Two single-residue (A12 and T5) double-chain-reversal loops, which connect adjacent parallel strands and bridge three G-tetrad layers, adopt similar alignments, with the A12 and T5 bases pointing outwards from the quadruplex scaffold. This finding is consistent with the lack of sequential H6/8-H1′ NOE connectivity of these residues with the preceding and following residues (Fig. 7). The single-residue (A9) double-chain-reversal turn, which connects G8 to G10 and bridges two G-tetrad layers, participates in A9·(G3·G7·G10·G14) pentad formation. The A9 base (in yellow) is tilted relative to the plane of the G3·G7·G10·G14 tetrad (in magenta) (Fig. 3A). The G1 residue from one monomer stacks over the G7·(A9·G10) triad (Fig. 9A, which is published as supporting information on the PNAS web site) from the other monomer (Fig. 9B). This positioning of the G7·(A9·G10) triad is supported by several unusual proton chemical shifts (see legend of Fig. 9).
The sugar-phosphate backbones of individual monomers are continuously aligned across the dimeric interface of the 93del quadruplex (Fig. 3A). The alignment is interrupted between G1, G6, G10, and G13 of one subunit and G10, G6, G1, and G13 of the other subunit (Figs. 2D and 9 C and D). There are two wide and two narrow grooves, whose dimensions are highlighted in yellow in Fig. 9 E and F. Their shapes are quite different, as shown in an alternate surface view (Fig. 3C).
NMR and CD Spectra of 93del and Modified Sequences. The importance of individual residues to the maintenance of the dimeric quadruplex folding topology of 93del was systematically probed with either deletion of terminal residues or substitution of single loop residues (Table 2, which is published as supporting information on the PNAS web site).
The NMR spectrum of 93del lacking G1 (seq1) is very different from that of 93del, with the former exhibiting a doubling of resonances (Fig. 10, which is published as supporting information on the PNAS web site). By contrast, the NMR spectrum of 93del lacking T16 (seq2) is almost identical to that of 93del, except for the lack of the T16 peak (Fig. 10).
The NMR spectra of 93del after T5-to-A5 substitution in loop 1(seq3), A9-to-T9 substitution in loop 2 (seq10), and A12-to-T12 substitution in loop 3 (seq4), are very similar to the spectrum of unmodified 93del, except for the chemical shifts of the substituted residues (Fig. 10). Indeed, the NMR spectral characteristics of the 93del dimeric quadruplex were not perturbed when T5 in loop 1 was substituted by dinucleotides TT, TA, or AT or by trinucleotide TTA, when A9 in loop 2 was substituted by dinucleotides TT, AT, TA, or AA, when A12 in loop 3 was substituted by dinucleotide AA, or even when both A9 in loop 2 and A12 in loop 3 were simultaneously substituted by thymine residues (Table 2).
CD spectra of 93del and all modified sequences (Table 2) exhibit a positive peak at 260 nm (Fig. 11 A, which is published as supporting information on the PNAS web site), characteristic of parallel-stranded G-quadruplexes (30). A comparison of the CD spectra of 93del and seq1 is shown in Fig. 11B.
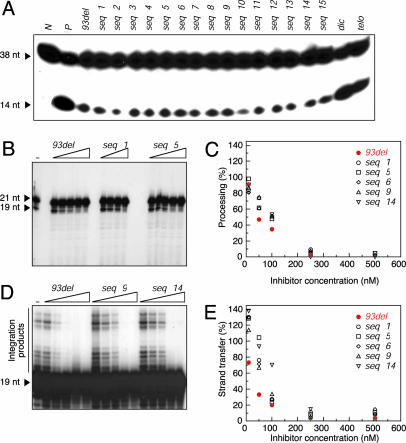
HIV-1 Integrase Inhibition Assays on 93del and Modified Sequences. To test the integration inhibitory activities of 93del and the modified sequences described above, we performed assays on the reverse “disintegration” reaction by HIV-1 integrase (31). Our results indicated that all modified 93del sequences inhibited the disintegration reaction, with an efficiency approximately comparable to that of 93del (Fig. 4A). This finding contrasts with very weak inhibition efficiency, if any, of a standard Watson–Crick duplex (lane dic) or a G-quadruplex topology formed by a fragment of the human telomeric G-rich strand (lane telo). It should also be noted that 93del lacking G1 (seq1), which cannot form the 93del dimeric quadruplex scaffold, was still as efficient as 93del in inhibiting HIV-1 integrase in this assay (lanes seq1 and 93del).
Fig. 4.
HIV-1 integrase inhibition assays of 93del and modified analogs. (A) Disintegration assays. Lanes N and P are controls without and with HIV-1 integrase alone, respectively. Other lanes are inhibition assays with added inhibitors as indicated, where telo stands for the human telomeric sequence d[AGGG(TTAGGG)3] and dic stands for the d(CGCGAATTCGCG) dodecamer. The concentration of all inhibitors was 300 nM, except for dic being 1 μM. The DNA substrate and disintegration product are monitored as bands of 38 and 14 nt, respectively. (B–E) The processing and strand transfer assays. (B and D) Examples of the processing (B) and strand transfer (D) reactions inhibited by various concentrations of 93del and modified sequences. Lanes marked “-” are controls with HIV-1 integrase alone. Other lanes are inhibition assays with added inhibitors as indicated. The concentrations of seq1 are 10, 50, 100, and 500 nM; the concentrations of 93del, seq5, seq9, and seq14 are 10, 50, 100, 250, and 500 nM. In the processing reaction, the substrate and product are monitored as bands of 21 and 19 nt, respectively. In the strand transfer reaction, the substrate is monitored as the 19-nt band and the integration products are monitored as bands >19 nt. (C and E) Plots of the processing (C) and strand transfer (E) reaction efficiency as a function of the concentration of inhibitors.
To more quantitatively compare the integration inhibition efficiencies of 93del and several modified sequences (including seq1, which lacks G1, and seq5, seq6, seq9, and seq14, which contain a 2- or 3-nt loop, Table 2), we used the processing and the strand transfer reactions (1). The inhibition assays were performed with various inhibitor concentrations (Fig. 4 B–E). In both reactions, 93del was the best inhibitor among the tested analogs, with IC50 being ≈50 nM (vs. 100 nM for other sequences) in the processing (Fig. 4C) and ≈25 nM (vs. 70–130 nM for other sequences) in the strand transfer (Fig. 4E). Kinetic measurements on the processing and strand transfer reactions also indicated that 93del was slightly more potent than seq1 (Fig. 12, which is published as supporting information on the PNAS web site).
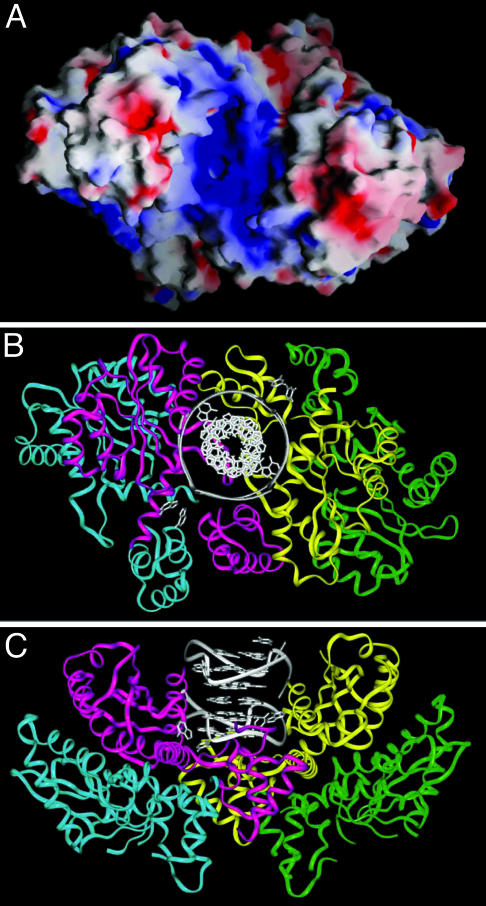
Docking DNA G-Quadruplexes onto Tetrameric HIV-1 Integrase. HIV-1 integrase (288 aa) is composed of three domains: N-terminal, catalytic core, and C-terminal domains (1). It has been shown to form both dimers and tetramers in solution (32). The individual domains have been determined by x-ray and NMR studies to form dimers (33–35). Dimer formation has also been observed in the crystal structures of IN52–288, which contains the catalytic core linked to the C-terminal domain (36), and of IN1–212, which contains the N-terminal domain linked to the catalytic core (20). The authors noted that an HIV-1 integrase tetramer, based on crystal lattice contacts in IN1–212, exhibits positively charged channels suitable for DNA binding (20), and also bears a resemblance to Tn5, a related bacterial transposase that adopts a tetrameric alignment of subunits (37).
Our starting point is the tetrameric (actually dimer of dimers) model of HIV-1 IN1–212 (20). There is a positively charged channel within this dimer of dimers (Fig. 5A), which appears to be complementary both in shape and charge to the 93del dimeric quadruplex fold (Fig. 3C). We computed the surface complementarity between the 93del quadruplex and the IN1–212 tetramer by using gramm, a molecular docking program (24, 25), and analyzed 100 structures of the complex exhibiting minimal energy. All complexes with high scores positioned the 93del quadruplex within the positively charged cavity of the IN1–212 tetramer in one of two alternate orientations (Fig. 13, which is published as supporting information on the PNAS web site). In the 10 best structures, the 93del quadruplex is oriented as shown in Fig. 5 B and C.
Fig. 5.
Models of 93del dimeric quadruplex bound to tetrameric HIV-1 integrase. (A) Molecular surface of the tetramer of IN1–212. Positive and negative electrostatic potentials are shown in blue and red, respectively. (B and C) Two views of the model of the complex between 93del dimeric quadruplex and tetrameric IN1–212 (see text). The 93del structure is colored in gray; four monomers of IN1–212 are colored in red, blue, yellow, and green, respectively.
In contrast, the docking calculation performed with the intramolecular parallel-stranded G-quadruplex formed by the human telomeric repeats (Protein Data Bank code 1KF1) (13) resulted in no complexes (among the 10 best-score complexes), where the quadruplex is positioned within the above described channel of the tetramer of HIV-1 integrase.
Discussion
93del Adopts a Unique Compact Dimeric Quadruplex Topology. We have established that the 93del d(GGGGTGGGAGGAGGGT) oligonucleotide in K+ solution folds into a twofold symmetric dimeric quadruplex (Fig. 3A). Of the 16 bases in each monomer subunit, 13 (all 12 guanines and adenine A9) are paired, 2 (T5 and A12) are looped out, and 1 (T16) is a 3′-terminal stacked residue. Within each subunit, G-stretches are parallel, and loops that connect adjacent parallel strands are double-chain-reversal. Thus, the structure belongs to the family of all-parallel-stranded intramolecular quadruplexes (13–16, 18, 19). Importantly, the structure of the 93del dimeric quadruplex establishes that a single nucleotide, namely T5 of the first loop and A12 of the third loop (Fig. 3A), is sufficient to span three G-tetrad layers. Although a single-nucleotide segment may be too short to form an edgewise or diagonal loop in a G-quadruplex, recent data (16) suggested that it forms the most stable double-chain-reversal loop bridging three G-tetrad layers. The compact interlocking of symmetry-related subunits through (1 + 3) G2·G6·G1·G13 tetrad formation across the dimeric interface (Fig. 2D) constitutes an interesting feature of the 93del architecture and highlights a principle for robust dimeric quadruplex folding (see also ref. 17). The striking continuity of the backbone alignment of each of the four strands from individual subunits across the dimeric interface exhibits characteristics of nicked DNA architectures, which could constitute potential substrate sites for HIV-1 integrase.
Stability and Slow Unfolding Kinetics. The 93del dimeric quadruplex is very stable and/or exhibits extremely slow unfolding kinetics. The persistence of imino protons from the central G2·G6·G1·G13 tetrads in D2O solution at 100°C (Fig. 1C) demonstrates that the dimeric scaffold does not completely unfold even under extremely high temperature conditions. This has permitted observation of the 93del dimeric quadruplex at micromolar concentrations by NMR and even at a much lower concentration by denaturing gel electrophoresis (data not shown), despite its dimeric stoichiometry.
This finding suggests that the dimeric quadruplex scaffold of 93del, once formed, could persist in vivo over long time periods, and its compact structure could potentially inhibit nuclease-based degradation. This scaffold could also have favorable pharmacological properties because of its ability to retain its robust topology at very low concentrations.
Integrase Inhibitory Activity of Loop-Modified 93del Analogs. The NMR and CD spectra of loop-modified 93del sequences (Table 2) are consistent with an unchanged folding topology. All of these sequences and 93del exhibit comparable (within small variations) inhibitory activities against HIV-1 integrase (Fig. 4A), although slightly lower inhibitory activities were observed for sequences containing a 2- or 3-nt loop (Fig. 4 C and E). This finding establishes that the 93del quadruplex scaffold is the key component responsible for HIV-1 integrase inhibition. These results are supportive of earlier contributions on the targeting of HIV-1 integrase by guanine-rich sequences, where recognition specificity was attributed to the particular backbone topology of the folded G-quadruplex (4, 5). It should be noted that not all DNA quadruplexes inhibit HIV-1 integrase. Thus, the human telomeric sequence, which adopts distinct monovalent cation-dependent G-quadruplexes (9, 13), has almost no inhibitory activity (lane telo, Fig. 4A).
Integrase Inhibitory Activity of Truncated 93del Analogs. We next turn to two truncated analogs of 93del. One of these involves deletion of the 3′-terminal T residue (seq2, Table 2). Its NMR spectrum is very similar to that of 93del, indicating that the dimeric quadruplex topology is maintained. Furthermore, the inhibitory effects of this analog and of 93del are similar in the disintegration assay (Fig. 4A). Thus, 3′-T residue, which does not participate in tetrad formation, may be deleted with little effect on the 93del dimeric quadruplex topology or inhibitory function.
The second truncated analog of 93del involves deletion of the 5′-terminal G residue (seq1, Table 2). Our structural studies have established that G1 plays a critical role in completing formation of the central (1 + 3) G2·G6·G1·G13 tetrads (Fig. 2D), thereby anchoring the two monomers within the 93del dimeric quadruplex. Indeed, the NMR spectrum of seq1, which contains a doubling of sharp resonances, is very different from that of 93del (Fig. 10). In contrast, the CD spectra of both 93del and seq1 exhibit a positive peak at 260 nm (Fig. 11B), characteristic of parallel-stranded G-quadruplexes (30). It is conceivable that seq1 forms an alternate parallel-stranded quadruplex, whose topology and oligomeric status are yet unknown. The doubling of the resonances could reflect the presence of two interconverting conformers or alternately, formation of an asymmetric dimeric quadruplex. It is interesting that the inhibitory activity of seq1 against HIV-1 integrase is not much lower than that of 93del. This finding suggests that parallel-stranded quadruplex topologies, other than the 93del dimeric quadruplex, can also inhibit HIV-1 integrase.
Folding Topologies of Related G-Quadruplexes That Inhibit HIV-1 Integrase. The HIV-1 integrase inhibitor T30923 d(GGGTGGGTGGGTGGGT) oligonucleotide and its homologs T30177 and T30695 have been assigned to intramolecular quadruplex folds where adjacent strands are aligned antiparallel to each other around the quadruplex (5, 38, 39). This conclusion is actually at variance with the reported CD spectra in K+ solution (40), which showed a positive peak at 260 nm, characteristic of parallel-stranded G-quadruplexes. This (together with the sequence similarities between these oligonucleotides and 93del) raises some concern regarding the previously published topologies (38, 39). In our view, additional experimentation is warranted to rigorously differentiate between parallel and antiparallel alignment of strands, as well as between monomeric and dimeric stoichiometries, for the quadruplexes formed by the guanine-rich T30923, T30177, and T30695 sequences in K+ solution.
93del Dimeric Quadruplex as a Template for Inhibitor Design. The unusual robustness of the 93del dimeric quadruplex makes it an ideal scaffold for systematic modifications aimed toward modulating inhibition specificity and affinity against HIV-1 integrase. One can alter the length or nature of the G-quadruplex by either adding or deleting G-tetrads, or alternately replacing G·G·G·G tetrads by their mixed G·C·G·C (11) or A·T·A·T (13) tetrad counterparts. One can also design sequences toward the goal of capping G-quadruplexes by base triads (28), given that base triads have been shown to be stabilized after stacking over G-tetrads (41).
Our studies suggest that all three loops within the 93del dimeric quadruplex can accommodate between one and three nucleotides (Table 2) without impacting on the core folding topology, while retaining HIV-1 integrase inhibitory activity. They also indicate loop composition (length, sequence)-dependent variations in the inhibition of HIV-1 integrase, and these could be expanded to modulate inhibitor activity.
Model of 93del–Integrase Complex. We have outlined a docking-based model of the 93del-integrase complex, where the dimeric quadruplex of 93del is positioned within a large basic channel/canyon (Fig. 5A) generated by a dimer of dimers of HIV-1 IN1–212 (Fig. 5 B and C). Insertion of 93del into this channel blocks access to amino acids D64, D116, and E152, which are catalytic residues essential for integrase function. The model does not include loop segments 46–56 and 139–149, which are disordered in the crystal structures, but could be important in complex formation. The loops of the DNA quadruplex may make contacts with the protein via these underdetermined segments (Fig. 13A) or the bottom of the channel (Fig. 13B). This finding would explain a slightly lower HIV-1 integrase inhibitory activity of modified sequences containing a 2- or 3-nt loop. It should be noted that a G-quadruplex structure with three 3-nt loops, such as that of the human telomeric repeats, would be too large to fit in the channel, consistent with almost no HIV-1 integrase inhibitory activity observed.
It should be kept in mind that the proposed model of the 93del–integrase complex requires substantiation and has been put forward in the spirit of stimulating further experimentation. The most important feature of our model is the concept that one macromolecule, a dimeric DNA quadruplex, can be positioned within an intersubunit channel/canyon of another macromolecule, a dimer of dimers of HIV-1 integrase. Furthermore, our contribution highlights the ability of multistranded DNA scaffolds, and the groups that project from them, to target channels/canyons at intersubunit interfaces of multimeric proteins.
Supplementary Material
Acknowledgments
We thank Dr. Maurice Guéron and Dr. Simon Litvak for critical reading of the manuscript. This research was supported by National Institutes of Health Grant GM34504. D.J.P. is a member of the New York Structural Biology Center supported by National Institutes of Health Grant GM66354. The work performed in M.-L.A.'s laboratory was supported by Sidaction and the Agence Nationale de Recherches contre le SIDA.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The coordinates for the structure of 93del have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1Y8D). The chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 6430).
References
- 1.Craigie, R. (2001) J. Biol. Chem. 276, 23213-23216. [DOI] [PubMed] [Google Scholar]
- 2.Neamati, N., Marchand, C. & Pommier, Y. (2000) Adv. Pharmacol. 49, 147-165. [DOI] [PubMed] [Google Scholar]
- 3.Tarrago-Litvak, L., Andréola, M. L., Fournier, M., Nevinsky, G. A., Parissi, V., de Soultrait, V. R. & Litvak, S. (2002) Curr. Pharm. Des. 8, 595-614. [DOI] [PubMed] [Google Scholar]
- 4.Mazumder, A., Neamati, N., Ojwang, J. O., Sunder, S., Rando, R. F. & Pommier, Y. (1996) Biochemistry 35, 13762-13771. [DOI] [PubMed] [Google Scholar]
- 5.Jing, N., Marchand, C., Liu, J., Mitra, R., Hogan, M. E. & Pommier, Y. (2000) J. Biol. Chem. 275, 21460-21467. [DOI] [PubMed] [Google Scholar]
- 6.de Soultrait, V. R., Lozach, P. Y., Altmeyer, R., Tarrago-Litvak, L., Litvak, S. & Andréola, M. L. (2002) J. Mol. Biol. 324, 195-203. [DOI] [PubMed] [Google Scholar]
- 7.Smith, F. W. & Feigon, J. (1992) Nature 356, 164-168. [DOI] [PubMed] [Google Scholar]
- 8.Haider, S., Parkinson, G. N. & Neidle, S. (2002) J. Mol. Biol. 320, 189-200. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Y. & Patel, D. J. (1993) Structure (London) 1, 263-282. [DOI] [PubMed] [Google Scholar]
- 10.Wang, Y. & Patel, D. J. (1994) Structure (London) 2, 1141-1156. [DOI] [PubMed] [Google Scholar]
- 11.Kettani, A., Kumar, R. A. & Patel, D. J. (1995) J. Mol. Biol. 254, 638-656. [DOI] [PubMed] [Google Scholar]
- 12.Kuryavyi, V., Majumdar, A., Shallop, A., Chernichenko, N., Skripkin, E., Jones, R. & Patel, D. J. (2001) J. Mol. Biol. 310, 181-194. [DOI] [PubMed] [Google Scholar]
- 13.Parkinson, G. N., Lee, M. P. H. & Neidle, S. (2002) Nature 417, 876-880. [DOI] [PubMed] [Google Scholar]
- 14.Phan, A. T. & Patel, D. J. (2003) J. Am. Chem. Soc. 125, 15021-15027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seenisamy, J., Rezler, E. M., Powell, T. J., Tye, D., Gokhale, V., Joshi, C. S., Siddiqui-Jain, A. & Hurley, L. H. (2004) J. Am. Chem. Soc. 126, 8702-8709. [DOI] [PubMed] [Google Scholar]
- 16.Phan, A. T., Modi, Y. S. & Patel, D. J. (2004) J. Am. Chem. Soc. 126, 8710-8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, N., Gorin, A., Majumdar, A., Kettani, A., Chernichenko, N., Skripkin, E. & Patel, D. J. (2001) J. Mol. Biol. 311, 1063-1079. [DOI] [PubMed] [Google Scholar]
- 18.Kettani, A., Gorin, A., Majumdar, A., Hermann, T., Skripkin, E., Zhao, H., Jones, R. & Patel, D. J. (2000) J. Mol. Biol. 297, 627-644. [DOI] [PubMed] [Google Scholar]
- 19.Matsugami, A., Ouhashi, K., Kanagawa, M., Liu, H., Kanagawa, S., Uesugi, S. & Katahira, M. (2001) J. Mol. Biol. 313, 255-269. [DOI] [PubMed] [Google Scholar]
- 20.Wang, J. Y., Ling, H., Yang, W. & Craigie, R. (2001) EMBO J. 20, 7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan, A. T. & Patel, D. J. (2002) J. Am. Chem. Soc. 124, 1160-1161. [DOI] [PubMed] [Google Scholar]
- 22.Phan, A. T., Guéron, M. & Leroy, J. L. (2001) Methods Enzymol. 338, 341-371. [DOI] [PubMed] [Google Scholar]
- 23.Brünger, A. T. (1992) x-plor: A System for X-Ray Crystallography and NMR (Yale Univ. Press, New Haven, CT).
- 24.Katchalski-Katzir, E., Shariv, I., Eisenstein, M., Friesem, A. A., Aflalo, C. & Vakser, I. A. (1992) Proc. Natl. Acad. Sci. USA 89, 2195-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakser, I. A. (1996) Biopolymers 39, 455-464. [DOI] [PubMed] [Google Scholar]
- 26.Chow, S. A. (1997) Methods 12, 306-317. [DOI] [PubMed] [Google Scholar]
- 27.Phan, A. T. (2000) J. Biomol. NMR 16, 175-178. [DOI] [PubMed] [Google Scholar]
- 28.Kuryavyi, V. V. & Jovin, T. M. (1996) in Proceedings of the IX Conversation in Biomolecular Structure and Dynamics, eds. Sarma, R. H. & Sarma, M. H. (Adenine, Guilderland, NY), pp. 81-103.
- 29.Phan, A. T. (2001) J. Magn. Reson. 153, 223-226. [DOI] [PubMed] [Google Scholar]
- 30.Balagurumoorthy, P., Brahmachari, S. K., Mohanty, D., Bansal, M. & Sasisekharan, V. (1992) Nucleic Acids Res. 20, 4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow, S. A., Vincent, K. A., Ellison, V. & Brown, P. O. (1992) Science 255, 723-726. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, T. M., Engelman, A., Ghirlando, R. & Craigie, R. (1996) J. Biol. Chem. 271, 7712-7718. [DOI] [PubMed] [Google Scholar]
- 33.Eijkelenboom, A. P., Lutzke, R. A., Boelens, R., Plasterk, R. H., Kaptein, R. & Hard, K. (1995) Nat. Struct. Biol. 2, 807-810. [DOI] [PubMed] [Google Scholar]
- 34.Cai, M., Zheng, R., Caffrey, M., Craigie, R., Clore, G. M. & Gronenborn, A. M. (1997) Nat. Struct. Biol. 4, 567-577. [DOI] [PubMed] [Google Scholar]
- 35.Goldgur, Y., Dyda, F., Hickman, A. B., Jenkins, T. M., Craigie, R. & Davies, D. R. (1998) Proc. Natl. Acad. Sci. USA 95, 9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, J. C., Krucinski, J., Miercke, L. J., Finer-Moore, J. S., Tang, A. H., Leavitt, A. D. & Stroud, R. M. (2000) Proc. Natl. Acad. Sci. USA 97, 8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, P. & Mizzuchi, K. (1995) Cell 82, 209-220. [DOI] [PubMed] [Google Scholar]
- 38.Jing, N., Gao, X., Rando, R. F. & Hogan, M. E. (1997) J. Biomol. Struct. Dyn. 15, 573-585. [DOI] [PubMed] [Google Scholar]
- 39.Jing, N. & Hogan, M. E. (1998) J. Biol. Chem. 273, 34992-34999. [DOI] [PubMed] [Google Scholar]
- 40.Dapic, V., Abdomerovic, V., Marrington, R., Peberdy, J., Rodger, A., Trent, J. O. & Bates, P. J. (2003) Nucleic Acids Res. 31, 2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kettani, A., Basu, G., Gorin, A., Majumdar, A., Skripkin, E. & Patel, D. J. (2000) J. Mol. Biol. 301, 129-146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.