Abstract
Certain bloodsucking insects deliver nitric oxide (NO) while feeding, to induce vasodilation and inhibit blood coagulation. We have expressed, characterized, and determined the crystal structure of the Cimex lectularius (bedbug) nitrophorin, the protein responsible for NO storage and delivery, to understand how the insect successfully handles this reactive molecule. Surprisingly, NO binds not only to the ferric nitrophorin heme, but it can also be stored as an S-nitroso (SNO) conjugate of the proximal heme cysteine (Cys-60) when present at higher concentrations. EPR- and UV-visible spectroscopies, and a crystallographic structure determination to 1.75-Å resolution, reveal SNO formation to proceed with reduction of the heme iron, yielding an Fe-NO complex. Stopped-flow kinetic measurements indicate that an ordered reaction mechanism takes place: initial NO binding occurs at the ferric heme and is followed by heme reduction, Cys-60 release from the heme iron, and SNO formation. Release of NO occurs through a reversal of these steps. These data provide, to our knowledge, the first view of reversible metal-assisted SNO formation in a protein and suggest a mechanism for its role in NO release from ferrous heme. This mechanism and Cimex nitrophorin structure are completely unlike those of the nitrophorins from Rhodnius prolixus, where NO protection is provided by a large conformational change that buries the heme nitrosyl complex, highlighting the remarkable evolution of proteins that assist insects in bloodfeeding.
Keywords: crystal structure, heme protein, nitrophorin, S-nitrosocysteine, S-nitroso
Nitric oxide (NO) is a reactive molecule produced at low concentrations in all higher animals for regulating activities such as blood pressure, wound healing, and memory formation, and at higher concentrations to kill infectious invaders (1). Many NO interactions of biological importance involve heme-containing proteins. NO is synthesized by NO synthase (NOS), a dimeric heme protein that converts l-arginine and oxygen to citrulline and NO (2). NO signaling often involves soluble guanylate cyclase, a heterodimeric heme protein that catalyzes cGMP formation after NO binding at the heme center (3). NO delivery by bloodfeeding insects is through heme-containing nitrophorins (4, 5).
Because NO is a free-radical molecule with a half-life of <1 sec when exposed to biological tissue, protection of the molecule during storage and transport is required. Reaction products of unprotected NO include higher nitrogen oxides, tyrosine nitrations, heme oxidation or reduction, and various S-nitrosations (S-nitroso, SNO). Formation of Cys-SNO from NO and free thiol in vivo has been demonstrated for many proteins (6); however, the functional significance of this attractive model for regulation remains uncertain (7). For example, formation of hemoglobin Cysβ93-SNO (Hb-SNO) has been postulated to be reversible and to be involved in blood pressure regulation; however, tight binding or reaction with oxygen at the hemoglobin ferrous center would seem to preclude this possibility (8).
Nitrophorins are bound to NO and protected against nucleophilic agents in the low pH saliva of bloodsucking insects. During a blood meal, saliva is injected into the victim, where dilution and higher pH lead to release of NO from nitrophorin, vasodilation, and improved feeding by the insect (9, 10). The nitrophorin heme is ferric (FeIII) rather than ferrous, as found in the globins, so that oxygen binding is eliminated and NO transport and release facilitated, because oxygen does not bind to ferric heme and NO binding is 6-7 orders of magnitude weaker to ferric heme than to ferrous heme (11, 12). The nitrophorins from Rhodnius prolixus (the kissing bug) accomplish storage and delivery in part through a ruffled ferric-stabilized heme and a pH-dependent conformational change that places the NO molecule in an unreactive hydrophobic environment (11, 13-16). The nitrophorin from Cimex lectularius (the bedbug) also stores and releases NO in a pH-dependent manner by using a ferric heme protein but does so with a protein evolutionarily unrelated to the Rhodnius nitrophorins (10). Here, we present the structure and mechanism for this class of nitrophorin.
Materials and Methods
Protein Expression and Purification. The Cimex lectularius nitrophorin (cNP) gene (10) was modified for insertion into the expression vector pET17b by PCR and placed into Escherichia coli, yielding expression strain BL21DE3-cNP. Apoprotein cNP was expressed in insoluble form, solubilized in 6 M guanidine hydrochloride/5 mM DTT/1 mM EDTA/30 mM Tris·HCl, pH 7.5, and the remaining insoluble material was separated by centrifugation at 100,000 × g. The solubilized apoprotein was refolded in 30 mM Tris·HCl (pH 7.5)/0.8 M NaCl/1 mM EDTA/10 mM DTT/15 μM PMSF, and dialyzed against 100 mM sodium phosphate, pH 7.0. Heme was incorporated by the dropwise addition of 10 mM hemin solution in DMSO. Incorporation was considered complete when the ratio of the absorbance at 389 nm (Soret) to 280 nm (protein) reached ≈1.0. Excess hemin was removed by centrifugation after adjusting the pH to 5.0 with acetic acid. The protein was purified on a Q-Sepharose column and then on a Sephacryl S-100 column. The fractions containing cNP were pooled, concentrated by ultra-filtration using Centricon YM-30 filters, and equilibrated with an appropriate 20 mM buffer.
UV-Visible Spectroscopy. A saturated NO solution (2 mM NO) was used for titration of 2 ml of deoxygenated cNP (5 μM) in 100 mM sodium citrate (pH 5.6) or 100 mM potassium phosphate (pH 8.0) in a septum-closed quartz cuvette. Ferric cNP-NO formation was complete when the NO concentration reached ≈50 μM. For experiments with saturating NO, 20 μl of deoxygenated 0.5 mM cNP was added to a septum-closed quartz cuvette containing 2 ml of NO-saturated 100 mM sodium citrate or potassium phosphate buffer. Spectra were measured by using a Varian Cary 300 dual-beam spectrophotometer at room temperature, using identical solutions without protein for the blank in all cases. All NO gas was first passed through a 2M NaOH solution to remove other nitrogen oxides. Because such compounds accumulate in tanks of NO to levels that cannot be readily removed by NaOH, all NO solutions were spectroscopically monitored for nitrite (λmax = 354 nm, ref. 17). cNP is sensitive to nitrogen oxide contaminants, especially where NO gas is extensively blown over the protein solution, giving rise to a species with a Soret band at ≈420 nm, possibly reflecting a six-coordinate heme with two ligands from solution rather than linkage to Cys-60.
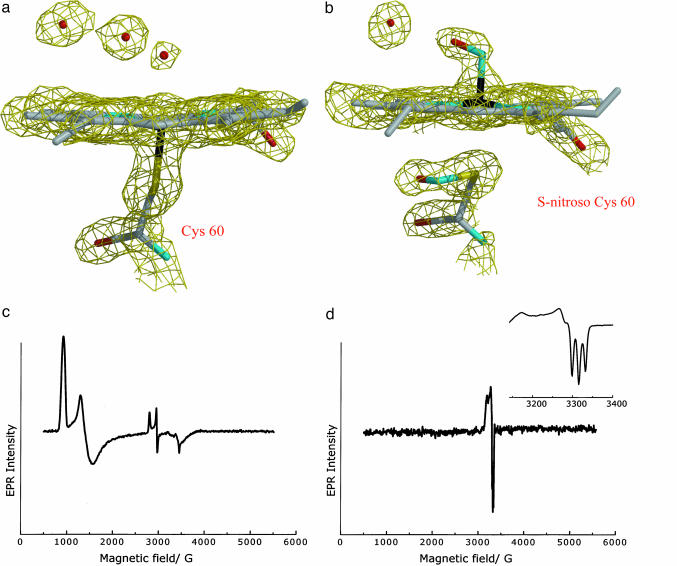
EPR Spectroscopy. Solutions (1 mM) of cNP for EPR experiments were buffered at pH 5.6 or 7.0 with 100 mM sodium citrate or 100 mM potassium phosphate, respectively. One hundred microliters of this solution was placed in a rubber septum closed EPR tube and argon was blown above the solution for 10 min. The NO complexes were prepared by brief (≈5 sec) or prolonged (≈15 min) blowing of NO gas anaerobically over the deoxygenated cNP solution in the EPR tube and freezing in liquid nitrogen. EPR spectra were recorded at 4.2 K on an X-band Bruker ESP-300E spectrometer equipped with a helium cryostat. The following settings were used. For cNP (pH 7.5, Fig. 2c): microwave frequency of 9.342 GHz, power at 0.2 mW, receiver gain of 1 × 105, modulation frequency of 100 kHz, and modulation amplitude of 4 G. For cNP-NO (pH 5.6, Fig. 2d): microwave frequency of 9.338 GHz, power at 2 μW, receiver gain of 1 × 105, modulation frequency of 100 kHz, and modulation amplitude of 4 G. Fig. 2d Inset: microwave frequency of 9.340 GHz, power at 2 μW, receiver gain of 1 × 105, modulation frequency of 100 kHz, and modulation amplitude of 4 G. For both spectra of Fig. 2d, the signal for excess NO (g = 1.98) has been subtracted (18). Spectra of cNP-NO, pH 5.6, and cNP-NO after first reducing the protein with sodium dithionite (pH 5.6), were prepared similarly to those at pH 7.5.
Fig. 2.
Electron density and EPR spectra. (a) Electron density for heme in the aqua complex (2 Fo - Fc). The view is flipped with respect to Fig. 1, and heme propionate A has been removed for clarity. Similar electron density is obtained if NO is first added and then removed from the crystal. (b) Electron density for cNP-NO, displaying both NO-heme and SNO. (c) EPR spectra of cNP at 4.2 K in frozen aqueous phosphate buffer (pH 7.5) showing the major high-spin EPR signal (g = 7.25, 4.75, and 1.90) and minor low-spin signal (g = 2.37, 2.25, and 1.94). The high-spin signal is characteristic of five-coordinate thiolate-bound ferriheme centers (44), and the low-spin signal is characteristic of low-spin ferriheme thiolate coordination with a sixth ligand such as water (45). (d) EPR spectrum of cNP in the presence of excess NO (pH 7.5). The wide-field sweep shown demonstrates that there is no other high- or low-spin FeIII signal remaining upon addition of the NO. (Inset) Expansion of the EPR spectrum of a second sample of cNP in the presence of excess NO. The spectrum is characteristic of five-coordinate FeII-NO (g = 2.105, 2.044, 2.013, nitrogen hyperfine splitting, AN = 16.8 G) (46), a {FeNO}7 center. Similar spectra are obtained at pH 5.6, or on addition of NO after chemical reduction of the protein with sodium dithionite.
Stopped-Flow Kinetics. Stopped-flow measurements were carried out by using an RSM 1000 rapid-scanning spectrophotometer (OLIS, Bogart, GA). Experiments at moderate NO concentration were performed at 25°C in either 20 mM sodium citrate (pH 5.6) or 100 mM potassium phosphate (pH 8.1), whereas those at 1 mM NO were performed at 10°C in either 20 mM sodium citrate (pH 5.6) or 20 mM potassium phosphate (pH 8.1). In both cases, data were collected at a rate of 1,000 scans per second with the monochromator wavelength centered at 460 nm. Rate constants were computed with the program sigmaplot (SPSS, Chicago).
Crystallization. The cNP crystals grew spontaneously as thin needles from solutions containing 22% polyethylene glycol (PEG) 4000, 200 mM Li2SO4, 100 mM Tris·HCl (pH 8.6) at room temperature, by using a hanging-drop method. A significant improvement in crystal morphology was achieved by seeding thin cNP needles into drops preequilibrated against the same solution as above, supplemented with ethanol to 12% vol/vol. A typical cNP crystal obtained in this way grew overnight to dimensions of 0.4 × 0.2 × 0.1 mm. The native data were obtained from a cNP crystal soaked in 45% PEG/Tris-HCl, pH 7.5, for 20 min to remove ethanol, and then frozen in liquid N2. The cNP-NO complex was obtained by first washing a cNP crystal anaerobically for 1 h in an argon-saturated solution (45% PEG 4000/100 mM sodium citrate, pH 5.6), moving the crystal to a similar solution saturated with NO for 20 min, and flash-freezing the crystal in liquid N2. To test for reversibility of the Cys-SNO conjugate, a cNP-NO crystal was prepared as above but was returned to an argon-saturated solution (pH 7.4) for 1 h and then flash-frozen.
Structure Determinations. The cNP crystals belong to space group P21 with cell constants a = 49.20, b = 42.10, c = 65.85 Å, β = 95.9° at 100 K. In-house data were measured with a Rigaku RAXIS IV++ detector by using Cu Kα radiation and Osmic (Rigaku/MSC, The Woodlands, TX) mirrors. Phasing information was acquired from three sources: multiwavelength anomalous dispersion (MAD) data at the iron absorption edge in a native cNP crystal, and multiple isomorphous replacement anomalous scattering data from two derivatives, xenon and mercury. The MAD data were collected at beamline 9-2 at the Stanford Synchrotron Radiation Laboratory (Stanford, CA) at two wavelengths, 1.7406 Å (edge) and 1.5306 Å (remote), by using the Area Detector Systems Corporation (Poway, CA) Quantum 4 detector. The xenon derivative was obtained from a crystal equilibrated for 1 min in 50% PEG 4000, saturated in a xenon chamber (Hampton, San Diego) with xenon gas for 10 min at 200 psi, flash-frozen in liquid N2, and examined in-house. The mercury derivative was obtained from a crystal soaked for 24 h in mother liquor supplemented with mercury acetate to a 10 mM concentration. The crystal was briefly moved to 50% PEG 4000 and flash-frozen. The data were acquired at the National Synchrotron Light Source (Brookhaven, NY) beamline X12C, by using 1.0099-Å x-rays and the Area Detector Systems Corporation Quantum 4 detector. All data were measured at 100 K.
Data were processed with d*trek (19) and scaleit (20). The edge MAD data were used as a tentative native data set during phasing. The dispersive difference Patterson map for the MAD data and difference Patterson maps for the derivatives revealed a single iron peak (10 σ), a single xenon peak (40 σ), and two low-occupancy mercury sites. Heavy-atom parameters were refined with mlphare and dm (20), resulting in phases to 3.0 Å (figure of merit = 0.79), and a partially interpretable electron density map. Multiple rounds of manual rebuilding with o (21), phase combination and extension with sigmaa (20), and refinement and simulated annealing with cns (22) led to a nearly complete model of the protein. Final refinement was with refmac 5 (23). Refinement statistics are in Table 1, and phasing statistics are in Table 2, which is published as supporting information on the PNAS web site.
Table 1. Crystallographic data and refinement statistics.
| cNP-H2O* | cNP-NO | |
|---|---|---|
| Resolution, Å | 1.80 | 1.75 |
| Multiplicity | 3.6 | 3.4 |
| Completeness,†% | 98.5 (96.5) | 98.7 (98.4) |
| I/σ(I)† | 11.0 (2.4) | 20.0 (2.6) |
| Rmerge† | 0.064 (0.31) | 0.043 (0.24) |
| Rcryst/Rfree | 0.15/0.20 | 0.17/0.21 |
| rmsd bonds, Å/angles, ° | 0.022/2.0 | 0.023/1.8 |
Crystals were in space group P21 with cell dimensions a = 49.2 Å, b = 42.1 Å, c = 65.9 Å, β = 95.9°. Data were measured in-house (λ = 1.54 Å, T = 100 K)
Overall values. Outer-shell values are in parentheses
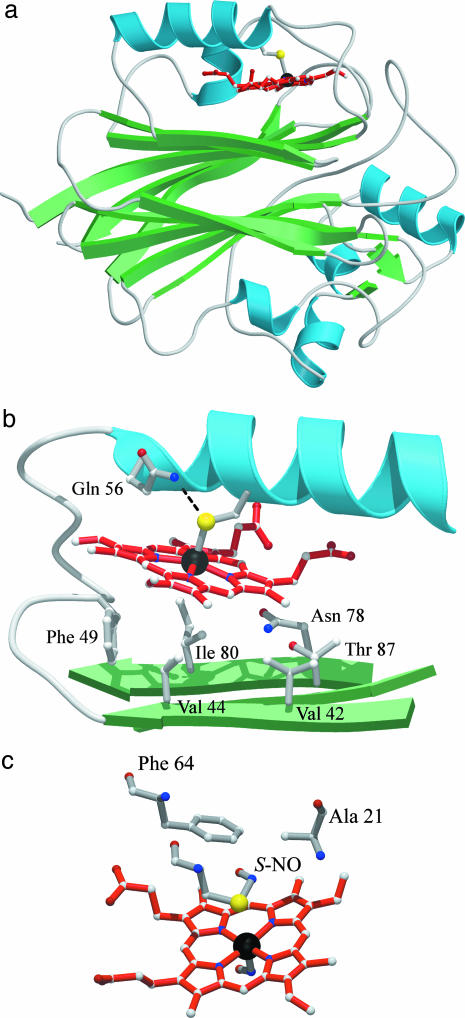
Final models for cNP and cNP-NO are composed of 280 of 282 residues in the protein sequence and have all residues in the allowed regions of a Ramachandran plot. Residues 51 and 55, linking the proximal helix to the rest of the molecule, and residues 93-103, connecting two β-strands, have several side chains with poor electron density. Residues 1-2 display no electron density and were not included in the final models. The structures were validated with procheck (24). Fig. 1 was prepared by using molscript (25). Fig. 2 was prepared by using bobscript (26) and raster3d (27).
Fig. 1.
cNP structure. (a) Ribbons representation of the cNP fold. (b) Closeup of the binding pocket. (c) S-nitroso Cys-60 in cNP-NO.
Results and Discussion
cNP Structure. We expressed cNP, a 32-kDa protein, in E. coli. Small amounts of protein could be obtained from soluble fractions, but the bulk of the expression was in insoluble inclusion bodies. The latter could be refolded and reconstituted with heme, yielding protein with identical spectral properties, both in the presence and absence of NO, to the soluble fraction and to protein derived from the insect (10). The cNP structure was determined to 1.8-Å resolution by using a combination of MAD and multiple isomorphous replacement anomalous scattering phasing (Fig. 1 and Table 1).
The cNP fold is unlike that of the nitrophorins from Rhodnius prolixus but similar to two functionally unrelated non-heme proteins, an exonuclease (28), and inositol polyphosphate 5-phosphatase (29), which display structural similarity Z scores of 15.1 and 30.2, respectively (30). The fold consists of an extensive β-sandwich motif, with heme inserted into a hydrophobic pocket above one face of the sandwich (Fig. 1). A cis peptide bond occurs between residues 224 and 225, near the edge of the β-sandwich. Heme is linked through its iron to the protein via Cys-60 (2.42 Å), which is part of a proximal helix. The proximal pocket is similar to that found in cytochrome P450 and NOS, despite no overall structural similarity to either protein. In this arrangement, the proximal cysteine is near the N terminus of the proximal helix, the dipole for which may help stabilize the thiolate anion, and is also hydrogen bonded to Gln-56, which may strengthen NO binding, as suggested previously for ligand binding by cytochrome P450 (31) and NOS (32). However, the orientation of the proximal cysteine differs among these proteins, in that the torsional angle described by atoms CA—CB—SG—Fe is 168° for cNP (Fig. 1b), whereas in cytochrome P450 and NOS, this angle is closer to 90°. This difference may facilitate thiolate release from the cNP heme iron when NO binds by preventing the sulfur atom from following the iron toward the distal pocket on binding NO (see below), whereas in cytochrome P450 and NOS, this angle is closer to 90°, allowing the sulfur atom to follow iron movements through rotation around the CA-CB bond. The cNP distal pocket is relatively small (35 Å3), hydrophobic, and contains three ordered solvent molecules, one of which appears weakly associated with heme iron (3.1 Å, Fig. 2a). The heme itself is moderately ruffled with the heme iron shifted 0.36 Å out of the heme plane, toward the proximal cysteine.
NO Complex. The cNP-NO complex was obtained by first soaking a crystal in an anaerobic solution saturated with NO (pH 5.6) and then flash-freezing it. The diffraction data obtained from this crystal revealed a structure with NO bound to the heme in the distal pocket, as expected, but also with a modification to the proximal cysteine consistent with formation of a SNO conjugate (Fig. 2b). Refinement of this structure led to a slightly more saddled heme with the heme iron shifted 0.21 Å into the distal pocket. Sulfur of Cys-60 is 3.1 Å away from the heme iron in this model and possibly no longer attached. The Fe—N—O conjugate displays a highly bent geometry (119°) that is consistent with a ferrous-nitrosyl complex (33), and a Fe—N bond length (1.86 Å) that is slightly long for a five-coordinate heme-NO, suggesting that Cys-60 may still be weakly coordinated to the iron. The Cys-SNO conjugate refines with S—N bond length (1.7 Å), S—N—O bond angle (114°) and C—S—N—O dihedral angle (0°, syn geometry) that are typical of model compounds (34, 35). Remarkably, both NO additions are completely reversible: examination of a crystal first soaked with NO and then back soaked in an NO-free solution led to a crystal structure without modification of heme or Cys-60 (Fig. 5, which is published as supporting information on the PNAS web site).
The Cys-SNO conjugate occupies a small hydrophobic pocket bounded by the heme, Phe-64 and Ala-21 (Fig. 1c). In the absence of the Cys-SNO, Cys-60, Phe-64, and Ala-21 are all in van der Waals contact. In the presence of the SNO, these residues move apart, become slightly less well ordered, particularly Ala-21, and the hydrogen bond between Gln-56 and Cys-60 is lost. The SNO displays relatively high mobility (B ≈ 50 Å2) and lies in the only possible position in the proximal pocket.
EPR Spectroscopy. To further characterize the nitrosylation events, we undertook EPR spectroscopic measurements that detect transitions of electrons with unpaired spins. The unliganded protein displays an EPR signal consistent with a predominantly five-coordinate high-spin ferric system (five unpaired d electrons, Fig. 2c). Upon adding a moderate concentration of NO (≈50 μM, pH 5.6), the system becomes EPR-silent, which is consistent with a low-spin ferric (FeIII) NO complex [six paired electrons, five from iron, and one from NO, referred to as {FeNO}6 (36)]. However, upon adding a high concentration of NO (≈2 mM, pH 5.6), a resonance appears that is consistent with a five-coordinate, low-spin ferrous (FeII) nitrosyl complex ({FeNO}7, Fig. 2d). Thus, the EPR data are consistent with the NO inducing axial bond cleavage and heme reduction.
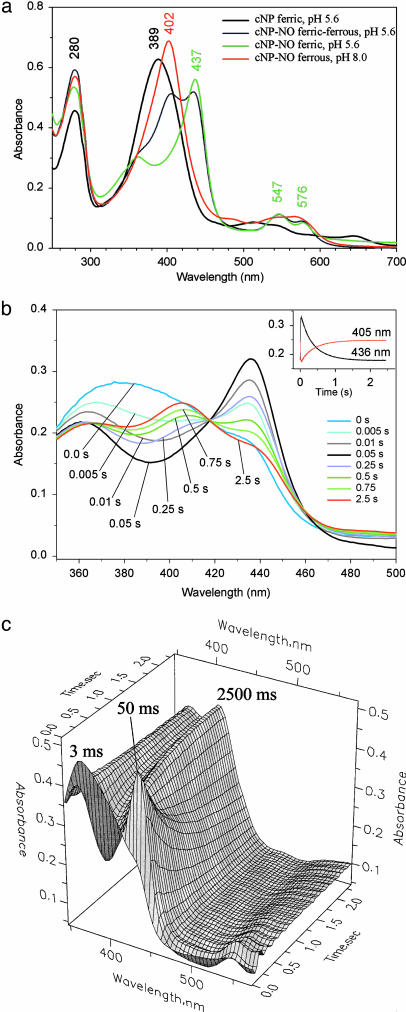
UV-Visible Spectroscopy. To investigate the order and mechanism of these events, we turned to static and stopped-flow electronic absorption spectroscopies. The absorption spectrum of the unliganded protein at pH 5.6 displays a prominent Soret band at 389 nm (Fig. 3a). Addition of moderate concentrations of NO (50 μM) led to a sharpened Soret band shifted to 437 nm, and α/β bands at 576 and 547 nm, respectively, values that are typical of thiolate-coordinated ferric heme-NO complexes. Higher concentrations of NO (from 200 μM to 2 mM) led to a decrease in the 437-nm band intensity, and the appearance of a new band at 405 nm that is consistent with formation of a ferrous cNP-NO complex. Reversal of NO binding could be achieved by blowing argon gas over the sample and was greatly facilitated by subsequently raising the pH, which leads to reduced affinity of the protein for NO (10). In contrast, adding saturating NO to cNP at pH 8 led to complete and irreversible formation of the ferrous heme-NO complex, suggesting that a competing reaction takes place, such as hydroxyl-dependent autoreduction of the nitrosylated heme (37).
Fig. 3.
UV-visible and stopped-flow spectra. (a) Representative absorption spectra for cNP. The cNP-NO (pH 8) spectrum is in the presence of excess (≈2 mM) NO (fully ferrous) and is identical to cNP-NO obtained from sodium dithionite-reduced protein and moderate NO. The cNP-NO (pH 5.6) spectra are in the presence of moderate (≈50 μM) NO (ferric) or excess NO (ferric/ferrous mixture). The peak maxima shift slightly between pH values of 5.6 and 8.0. The mixture of 402 nm (ferrous NO) and 437 nm (ferric NO) bands gives rise to peaks at 405 and 436 nm because of overlap. All peak assignments to ferric and ferrous states were confirmed by EPR spectroscopy. (b) Stopped-flow spectroscopy for cNP-NO formation. Representative time points are shown, emphasizing first the appearance and then the disappearance of the peak at 436 nm. (Inset) Change in absorption in the Soret bands for ferric and ferrous NO complexes. The apparent first-order rate constants for the loss of ferric-NO (k = 2.767 ± 0.005 s-1) and the gain of ferrous-NO (k = 2.626 ± 0.006 s-1) were essentially the same, which is consistent with linked processes. (c) Three-dimensional plot of the stopped-flow data. The ferric-NO peak is maximal at 50 ms and then recedes.
We used a rapid-scanning spectrophotometer attached to a stopped-flow device to monitor the kinetics of complex formation. Mixing cNP with 2 mM NO (pH 5.6) revealed a sequential order of events (Fig. 3b). Appearance of a Soret band at 436 nm, representing the ferric heme-NO complex, was completed by 50 ms and coincided with loss of the band at 389 nm (Fig. 3c). Lagging behind this was the appearance of a band at 405 nm, representing the ferrous heme-NO complex, concomitant with loss of intensity at 436 nm. Upon dilution of the pH 5.6 Fe-NO complex with an argon-saturated buffer solution, the ferrous band intensity diminished, and the ferric band increased, which is consistent with a reversible process.
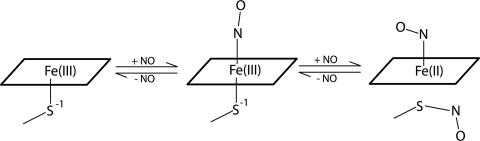
Model for cNP Function. Taken together, these data suggest a mechanism for reversible binding of NO to heme in cNP that incorporates a metal-assisted S-nitrosation (Fig. 4). Cys-SNO formation from S- and NO requires a one-electron oxidation, which is readily facilitated by metal centers (38). In cNP, this is accomplished through reversible reduction of the heme iron. In this mechanism, NO first binds to the ferric heme, giving rise to a complex that remains vulnerable to undesirable side reactions. Although not yet observed crystallographically, the loss of EPR signal and absorption spectral changes confirm the existence of the FeIII-NO complex. Subsequently, if excess NO is available, another NO molecule can react with Cys-60, resulting in electron abstraction by iron and leading to a neutral SNO conjugate. NO release, in response to lowered NO concentration, involves a simple reversal of these steps: decomposition of SNO is induced by FeII (38), leading to an FeIII-NO center, and release of NO from the now-ferric heme readily takes place, more so at higher pH. The detailed mechanism by which SNO formation and decomposition take place is not yet known, but could involve a concerted reaction, with SNO formation/decomposition occurring concomitantly with heme iron reduction/oxidation. Alternatively, a radical-based reaction may occur, in which homolytic cleavage of the Cys-FeIII bond leads to formation of a thiyl radical intermediate (RS·) that can react with NO to form SNO. Decomposition would proceed in reverse: homolytic cleavage of the SNO bond would again generate a thiyl radical, leading to reaction with FeII and formation of Cys-FeIII-NO.
Fig. 4.
Proposed mechanism for reversible heme nitrosylation and Cys-60 nitrosation in cNP.
A key component of our proposed mechanism is the coupling of heme reduction to SNO formation. In this regard, it is significant that the only other cysteine in cNP, Cys-90, located in a hydrophobic pocket near the surface of the protein, is unmodified by NO in the cNP-NO structure, as expected in the absence of oxygen and contaminating free metals. Further evidence in support of our proposed mechanism is the structure of ferrous cNP complexed with NO, prepared by reducing the heme with dithionite and transferring the ferrous cNP crystal to NO-saturated buffer (pH 5.6). In this structure, the Fe-NO is similar to the present structure; however Cys-60 is detached and unmodified (A.W., H. Badgandi, and W.R.M., unpublished results).
Conclusions. These data provide an observation of reversible SNO conjugation in a protein, and was accomplished by using dissolved NO gas rather than NO-donating compounds. The occurrence of SNO moieties in proteins for regulatory or signaling purposes has been widely proposed but difficult to confirm, as recently described for crystallographic studies of hemoglobin (39). The difficulty in characterizing these systems relates to the reactivity of the NO, the poor stability of SNO conjugates (40), and the one-electron oxidation/reduction required for forming/releasing the NO conjugate. Trace metals or oxygen can mediate the redox chemistry; however, these are often difficult to control and quantitate in vivo. With cNP, the metal center needed for SNO exchange is incorporated next to the key sulfur and is intimately coupled with NO binding at the metal. In this way, side reactions that would lead to loss of NO or reduction of heme are discouraged, which may well serve to protect the cNP-NO complex during storage in the insect's saliva.
Does SNO formation occur in the insect saliva? The answer to this is not known, but it seems likely to be the case. Previously reported EPR studies of whole-gland homogenates did not detect ferrous-NO protein in the salivary gland (41), but these measurements were made after dilution of the glands into a pH 7 buffer that favors loss of SNO by cNP. Formation of cNP-NO requires a relatively high NO concentration, higher than that normally found in the cytosol of oxygenated cells, but such concentrations may well be achieved in the Cimex salivary gland. Nitrophorin protein has been estimated to be as high as 40% of the total soluble protein in the salivary gland (10), and saturation of this protein would require an equally high concentration of NO, opening the way for SNO formation in a protective or storage capacity.
The cNP mechanism is in stark contrast to NO storage and transport by the Rhodnius nitrophorins. The latter belong to the lipocalin family and display an eight-stranded β-barrel structure, with ferric heme ligated through histidine rather than cysteine (5, 42). Binding of NO, especially at lower pH, leads to the collapse of two loops into the binding pocket, the expulsion of solvent, and the packing of hydrophobic groups against the NO moiety (13). Protection of NO in this arrangement is apparently through a physical barrier to reactive molecules. For cNP, protection is through the controlled and reversible chemical reaction involving SNO, an evolutionarily distinct mechanism achieving the same result.
Our cNP structure and proposed mechanism suggest Cys-SNO formation at metal centers may be common. For example, prolonged exposure of neuronal NOS (nNOS, ref. 43) to NO, or exposure of the W409F or W409Y mutant forms of nNOS to NO (32), leads to formation of a five coordinate FeII-NO complex with spectra similar to those of cNP. Whether SNO formation also occurs in these cases, or in any of the many proteins with cysteines linked to redox active metal centers, is not known, but this answer would be difficult to discover without freeze-trapping in the crystal, because the typically used spectroscopic signals do not readily detect sulfur adducts.
Supplementary Material
Acknowledgments
We thank Donald Shepley, Jacquie Brailey, and Robert E. Berry for protein expression and purification, Abreeza Zegeer for crystal preparations, and Arnold Raitsimring for assistance with EPR measurements. Portions of this work were performed at the National Synchrotron Light Source (Brookhaven, NY), beamline X12C, the Stanford Synchrotron Radiation Laboratory, beamline 9-2, and the Advanced Photon Source (Argonne, IL), beamline 14BMC. This work was supported in part by National Institutes of Health Grants HL62969 (to W.R.M.) and HL54826 (to F.A.W.) and an American Heart Association Fellowship (to E.M.M.). This paper was written while F.A.W. was on sabbatical leave in the Physics Institute at the University of Lübeck (Lübeck, Germany) with support from the Alexander von Humboldt Senior Award in Science.
Author contributions: A.W., E.M.M., F.A.W., and W.R.M. designed research; A.W., E.M.M., J.F.A., J.G.V., T.K.S., and W.R.M. performed research; A.W., E.M.M., F.A.W., and W.R.M. analyzed data; and A.W., F.A.W., and W.R.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: cNP, Cimex lectularius nitrophorin; NOS, NO synthase; SNO, S-nitroso; PEG, polyethylene glycol; MAD, multiwavelength anomalous dispersion.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1NTF and 1Y21).
References
- 1.Ignarro, L. J. (2000) Nitric Oxide Biology and Pathobiology (Academic, San Diego).
- 2.Alderton, W. K., Cooper, C. E. & Knowles, R. G. (2001) Biochem. J. 357, 593-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas, K. A., Pitari, G. M., Kazerounian, S., Ruiz-Stewart, I., Park, J., Schulz, S., Chepenik, K. P. & Waldman, S. A. (2000) Pharmacol. Rev. 52, 375-413. [PubMed] [Google Scholar]
- 4.Walker, F. A. & Montfort, W. R. (2000) in Advances in Inorganic Chemistry, eds. Sykes, A. G. & Mauk, G. (Academic, San Diego), Vol. 51, pp. 295-358. [Google Scholar]
- 5.Montfort, W. R., Weichsel, A. & Andersen, J. F. (2000) Biochim. Biophys. Acta 1482, 110-118. [DOI] [PubMed] [Google Scholar]
- 6.Stamler, J. S., Lamas, S. & Fang, F. C. (2001) Cell 106, 675-683. [DOI] [PubMed] [Google Scholar]
- 7.Forman, H. J., Torres, M. & Fukuto, J. (2002) Mol. Cell. Biochem. 234-235, 49-62. [PubMed] [Google Scholar]
- 8.Gladwin, M. T., Lancaster, J. R., Jr., Freeman, B. A. & Schechter, A. N. (2003) Nat. Med. 9, 496-500. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro, J. M., Hazzard, J. M., Nussenzveig, R. H., Champagne, D. E. & Walker, F. A. (1993) Science 260, 539-541. [DOI] [PubMed] [Google Scholar]
- 10.Valenzuela, J. G. & Ribeiro, J. M. (1998) J. Exp. Biol. 201, 2659-2664. [DOI] [PubMed] [Google Scholar]
- 11.Andersen, J. F., Ding, X. D., Balfour, C., Shokhireva, T. K., Champagne, D. E., Walker, F. A. & Montfort, W. R. (2000) Biochemistry 39, 10118-10131. [DOI] [PubMed] [Google Scholar]
- 12.Ding, X. D., Weichsel, A., Andersen, J. F., Shokhireva, T. K., Balfour, C., Pierik, A. J., Averill, B. A., Montfort, W. R. & Walker, F. A. (1999) J. Am. Chem. Soc. 121, 128-138. [Google Scholar]
- 13.Weichsel, A., Andersen, J. F., Roberts, S. A. & Montfort, W. R. (2000) Nat. Struct. Biol. 7, 551-554. [DOI] [PubMed] [Google Scholar]
- 14.Maes, E. M., Weichsel, A., Andersen, J. F., Shepley, D. & Montfort, W. R. (2004) Biochemistry 43, 6679-6690. [DOI] [PubMed] [Google Scholar]
- 15.Roberts, S. A., Weichsel, A., Qiu, Y., Shelnutt, J. A., Walker, F. A. & Montfort, W. R. (2001) Biochemistry 40, 11327-11337. [DOI] [PubMed] [Google Scholar]
- 16.Shokhireva, T., Berry, R. E., Uno, E., Balfour, C. A., Zhang, H. & Walker, F. A. (2003) Proc. Natl. Acad. Sci. USA 100, 3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettache, N., Carter, T., Corrie, J. E., Ogden, D. & Trentham, D. R. (1996) Methods Enzymol. 268, 266-281. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich, M. P., Upadhyay, A. K., Riga, J., Arciero, D. M. & Hooper, A. B. (2002) Biochemistry 41, 4603-4611. [DOI] [PubMed] [Google Scholar]
- 19.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718-1725. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 21.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjelgard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thorton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 25.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 26.Esnouf, R. M. (1997) J. Mol. Graph. Model 15, 132-134, 112-113. [DOI] [PubMed] [Google Scholar]
- 27.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 28.Mol, C. D., Kuo, C.-F., Thayer, M. M., Cunningham, R. P. & Tainer, J. A. (1995) Nature 374, 381-386. [DOI] [PubMed] [Google Scholar]
- 29.Tsujishita, Y., Guo, S., Stolz, L. E., York, J. D. & Hurley, J. H. (2001) Cell 105, 379-389. [DOI] [PubMed] [Google Scholar]
- 30.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 31.Poulos, T. L. (1996) J. Biol. Inorg. Chem. 1, 356-359. [Google Scholar]
- 32.Couture, M., Adak, S., Stuehr, D. J. & Rousseau, D. L. (2001) J. Biol. Chem. 276, 38280-38288. [DOI] [PubMed] [Google Scholar]
- 33.Scheidt, W. R. & Ellison, M. K. (1999) Acc. Chem. Res. 32, 350-359. [Google Scholar]
- 34.Bartberger, M. D., Houk, K. N., Powell, S. C., Mannion, J. D., Lo, K. Y., Stamler, J. S. & Toone, E. J. (2000) J. Am. Chem. Soc. 122, 5889-5890. [DOI] [PubMed] [Google Scholar]
- 35.Arulsamy, N., Bohle, D. S., Butt, J. A., Irvine, G. J., Jordan, P. A. & Sagan, E. (1999) J. Am. Chem. Soc. 121, 7115-7123. [Google Scholar]
- 36.Enemark, J. H. & Feltham, R. D. (1974) Coord. Chem. Rev. 13, 339-406. [Google Scholar]
- 37.Hoshino, M., Maeda, M., Konishi, R., Seki, H. & Ford, P. C. (1996) J. Am. Chem. Soc. 118, 5702-5707. [Google Scholar]
- 38.Vanin, A. F., Malenkova, I. V. & Serezhenkov, V. A. (1997) Nitric Oxide 1, 191-203. [DOI] [PubMed] [Google Scholar]
- 39.Chan, N. L., Kavanaugh, J. S., Rogers, P. H. & Arnone, A. (2004) Biochemistry 43, 118-132. [DOI] [PubMed] [Google Scholar]
- 40.Williams, D. L. (1996) Methods Enzymol. 268, 299-308. [DOI] [PubMed] [Google Scholar]
- 41.Valenzuela, J. G., Walker, F. A. & Ribeiro, J. M. (1995) J. Exp. Biol. 198, 1519-1526. [DOI] [PubMed] [Google Scholar]
- 42.Weichsel, A., Andersen, J. F., Champagne, D. E., Walker, F. A. & Montfort, W. R. (1998) Nat. Struct. Biol. 5, 304-309. [DOI] [PubMed] [Google Scholar]
- 43.Migita, C. T., Salerno, J. C., Masters, B. S., Martasek, P., McMillan, K. & Ikeda-Saito, M. (1997) Biochemistry 36, 10987-10992. [DOI] [PubMed] [Google Scholar]
- 44.Peisach, J., Blumberg, W. E., Ogawa, S., Rachmilewitz, E. A. & Oltzik, R. (1971) J. Biol. Chem. 246, 3342-3355. [PubMed] [Google Scholar]
- 45.Chevion, M., Peisach, J. & Blumberg, W. E. (1977) J. Biol. Chem. 252, 3637-3645. [PubMed] [Google Scholar]
- 46.Wayland, B. B. & Olson, L. W. (1974) J. Am. Chem. Soc. 96, 6037-6041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.