Abstract
Gills of euryhaline teleosts are excellent models for studying osmotic-stress adaptation because they directly contact the aquatic environment and are an important effector tissue during osmotic stress. We acclimated tilapia (Oreochromis mossambicus) from fresh water (FW) to seawater (SW); performed suppression subtractive hybridization of gill mRNAs; and identified two transcription factors, osmotic stress transcription factor 1 (OSTF1) and the tilapia homolog of transcription factor II B (TFIIB), that are rapidly and transiently induced during hyperosmotic stress. mRNA levels increase 6-fold for OSTF1 and 4-fold for TFIIB, and they reach maxima 2 h after SW transfer. Protein levels increase 7.5-fold for OSTF1 and 9-fold for TFIIB, and they reach maxima 4 h after SW transfer. Induction of OSTF1 and TFIIB increases gradually with increasing salinity. Induction of OSTF1 and TFIIB is specific for osmotic stress and absent during oxidative stress (1 mM H2O2) or heat shock (+10°C). Bioinformatic analysis of OSTF1 reveals that it is a transcription factor of the TGF-β-stimulated clone 22/GILZ family. Because some mammalian homologs are strongly induced by glucocorticoids, OSTF1 may represent the molecular link between the SW hormone cortisol and transcriptional regulation of ion transport and cell differentiation in teleost gills. Coinduction of OSTF1 and TFIIB may serve to recruit TFIIB preferentially to OSTF1 target genes during hyperosmotic stress and compensate for reduced rates of transcription resulting from salt-induced chromatin compaction. We conclude that OSTF1 and TFIIB are critical elements of osmosensory signal transduction in euryhaline teleosts that mediate osmotic adaptation by means of transcriptional regulation.
Keywords: osmoregulation, osmotic stress
Euryhaline teleosts are osmoregulators that maintain plasma osmotic homeostasis largely by extrarenal NaCl transport. In seawater (SW) they actively secrete salt (NaCl), whereas in fresh water (FW) they actively absorb salt across gill epithelium. Accordingly, cellular organization of gill epithelium is different in FW compared with SW fish (1). During SW acclimation of euryhaline fish gill epithelium is extensively remodeled to account for altered requirements of ion transport and permeability. Remodeling includes changes in turnover of gill cells, altered gill cell differentiation, and modulation of expression and activity of ATPases, secondary active ion transporters, and structural proteins (2). Many adaptive responses to salinity change are based on transcriptional regulation. Recent work shows that mRNA levels of many ion and water transporters in gill epithelium are regulated by salinity. Examples include subunits of the Na+/K+-ATPase (3), the Na+/K+/2Cl- cotransporter (3), urea transporter (4), and aquaporin 3 (5). Also, other mRNAs such as 14-3-3 and C-type lectin are regulated by osmolality in fish gill (6, 7). Although mRNA stabilization is a potential mechanism that could increase transcript levels during hyperosmotic stress, it is more likely that changes in mRNA abundance result from transcriptional regulation. However, this conclusion is based primarily on mechanisms of osmotic regulation in mammalian cells (8). Because increased abundances of ion and water transporters are generally apparent after at least 12-18 h of exposure to hyperosmotic stress, it is possible that they are mediated by immediate early gene (IEG) transcription factors.
The only known animal transcription factor that is specifically involved in osmoregulation is the tonicity-response element binding protein TonEBP (9). This transcription factor has been characterized only in mammals and is not an IEG because it is induced relatively slowly, reaching maximal abundance at 18 h (10). How transcriptional regulation is orchestrated during salinity adaptation of euryhaline fishes is unknown. We have shown (11) that the teleost homolog of c-jun may be involved in osmosensory signal transduction via posttranslational modification. However, c-jun abundance does not change during osmotic stress in euryhaline Gillichthys mirabilis, and c-jun is part of the AP-1 general stress-response transcription factor and unlikely to mediate osmotic stress specific regulation. In this study, we have screened mRNA isolated from gill epithelium of Oreochromis mossambicus to identify IEGs that are induced in vivo >3-fold at 4 h after transfer of fish from FW to SW. We report the cloning, rapid and osmotic stress-specific coinduction, and structural analysis of two transcription factors, osmotic stress transcription factor 1 (OSTF1) and the tilapia homolog of general transcription factor II B (TFIIB).
Materials and Methods
Animals and Acclimation Procedures. Tilapia (O. mossambicus) were maintained in large tanks (diameter, 4 feet) supplied with flow-through heated (25-28°C) well water (FW) at the Center for Aquatic Biology and Aquaculture (University of California, Davis). At 2 days before acclimation, fish were transferred to 20-gallon recirculation aquaria containing FW at 25-28°C to minimize handling stress during experiments. Experiments were started by transferring tilapia to 20-gallon recirculation aquaria containing either full-strength SW (1,000 mosmol/kg), diluted SW (300, 600, and 900 mosmol/kg), FW (handling stress control), 1 mM H2O2 at 28°C (oxidative stress), or FW at 36°C (heat shock). Fish were sampled at the indicated times, gills were perfused, and gill epithelia were collected by scraping from individual gill arches as described (12). Gill epithelium was used for subsequent RNA or protein extraction.
RNA Isolation, Suppression Subtractive Hybridization (SSH), RACE, and DNA Sequencing. Total RNA was extracted by lysing cells in 1,000 μl of tissue per 100 mg of denaturing solution (4 M guanidine isothiocyanate/0.02 M sodium citrate/0.5% sarcosyl/0.7% β-mercaptoethanol) and grinding in a glass homogenizer (Wheaton Scientific), followed by phenol-chloroform extraction (13). Purity of RNA was confirmed by reading absorbance at 260, 280, and 320 nm. The ratio of background-free absorbance at 260/280 nm was always 2.0-2.2. SSH was performed with the PCR-Select cDNA subtraction kit (Clontech). SMART cDNA from control (FW) fish was used as the “driver” sample and the corresponding cDNA from fish exposed for 4 h to SW as the “tester” sample. The final PCR product was cloned into pGEM-T vector (Promega) to generate a subtracted cDNA library. Full-length sequences of SSH clones were obtained by using degenerate primers and SMART RACE cDNA amplification (Clontech). PCR products were extracted from agarose gels and plasmids isolated from bacterial clones by using a kit (Qiagen, Valencia, CA). They were then double-pass sequenced on an ABI 3730 automated DNA sequencer. Primer sequences used for RACE-PCR are given in Table 1, which is published as supporting information on the PNAS web site.
RT-PCR Analysis. The methods are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Briefly, total RNA (1 μg) was reverse-transcribed in the presence of oligo(dT) primers into cDNA by using Super-Script III first-strand synthesis kit for RT-PCR (Invitrogen). Semiquantitative PCR was performed with a MasterCycler (Eppendorf) by using the following parameters: 94°C for 1 min; and 30 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 1 min, and 72°C for 5 min. Quantitative PCR (qPCR) was performed in a PRISM 7700 real-time thermal cycler (Applied Biosystems) by using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) with the following parameters: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. All data were normalized to β-actin content and are expressed as fold change over controls as described (14). Gene-specific primer sequences were designed with Primer Express (Applied Biosystems) and are given in Table 1. Primer design for tilapia HSP70 was based on a published sequence (GenBank accession no. AJ001312).
Protein Extraction and Western Immunodetection. These methods are described in Supporting Materials and Methods. Briefly, epithelial tissue was lysed and homogenized. Protein was measured by bicinchoninic acid (BCA) protein assay (Pierce). Proteins were separated by SDS/PAGE as described (15). We loaded 20 μg of protein in each lane, and after separation, proteins were blotted onto Immobilon P membrane (Millipore). Membranes were incubated for 2 h in blocking buffer containing either TFIIB (1:500 dilution; c-18; Santa Cruz Biotechnology) or OSTF1 polyclonal antibody (1:75 dilution), which was raised by immunizing rabbits with a synthetic peptide corresponding to 19 aa in the C-terminal portion of OSTF1. After washing, blots were incubated for 1 h in blocking buffer containing secondary antibody coupled to horseradish peroxidase (1:1,000 dilution), developed with SuperSignal Femto (Pierce), and imaged with a ChemiImager (Alpha Innotech, San Leandro, CA). Densitometry was performed with imagequant software (Bio-Rad).
Bioinformatics and Statistical Analysis. Multiple-sequence alignments and phylogentic trees were done with alignx software (Informax, Bethesda). Sequences were evaluated for putative phosphorylation sites and protein domains by using netphos 2.0 (www.cbs.dtu.dk/services/netphos), motifscan (http://hits.isb-sib.ch/cgi-bin/pfscan), scansite (http://scansite.mit.edu/motifscan_seq.phtml), elm (http://elm.eu.org), scan-prosite (www.expasy.org/cgi-bin/prosite), interproscan (www.ebi.ac.uk/interproscan), pfa m (www.sanger.ac.uk/software/pfam), and smart (http://smart.embl-heidelberg.de). Sequence features were mapped and visualized with vectornti 9.0 (Informax). Data analysis was carried out with sigmaplot 9.0 (Systat, Evanston, IL). Differences between pairs of data were analyzed by unpaired t test. Significance threshold was set at P < 0.05, and data are presented as mean ± SEM.
Results
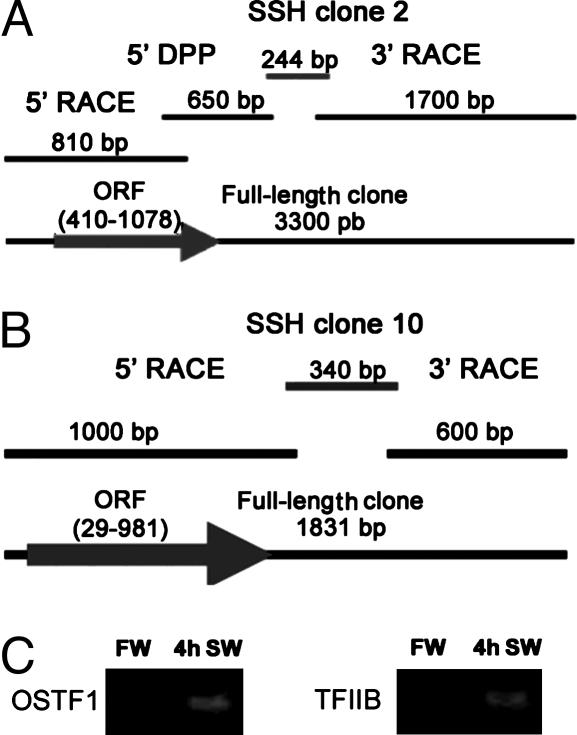
Cloning of Two Transcription Factors That Are Regulated by Osmolality. We used SSH for identification of IEGs that are induced during SW acclimation in gill cells of euryhaline tilapia. Among significantly induced transcripts are two previously uncharacterized transcription factors, osmotic stress transcription factor 1 (OSTF1) and the tilapia homolog of general TFIIB. Full-length coding sequences were obtained by RACE-PCR and degenerate primer PCR, starting with original SSH clones 2 and 10 (Fig. 1 A and B). Semiquantitative PCR using gill epithelial mRNA samples from FW and 4-h SW-acclimated fish confirms induction of OSTF1 and TFIIB transcripts during hypertonic stress (Fig. 1C). Both sequences were deposited in GenBank (accession nos. AAT84345 and AAT84346, for OSTF1 and TFIIB, respectively).
Fig. 1.
Isolation of OSTF1 and TFIIB as transcriptionally induced clones identified by suppression subtractive hybridization (SSH). SSH clones 2 (A) and 10 (B) were fully extended by using PCR-based methods. (C) Semiquantitative RT-PCR from samples of FW fish (control) and fish acclimated to SW for 4 h.
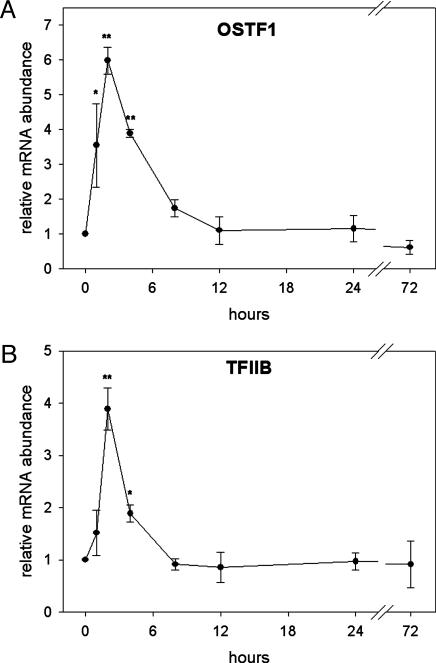
OSTF1 and TFIIB are IEGs During Hyperosmotic Stress. To determine whether hyperosmotic induction of OSTF1 and TFIIB is transient or permanent and identify the time at which they are maximally induced we performed another experiment in which tilapia were transferred from FW to SW for various times. qPCR was used to study the kinetics of expression of OSTF1 and TFIIB transcripts during SW acclimation. Both transcription factors show a similar time course of mRNA induction, reaching maxima between 1 and 4 h, and gradually declining to baseline levels within 12 h (see Fig. 3). Maximal mRNA induction at 2 h is 6-fold for OSTF1 (Fig. 2A) and 4-fold for TFIIB (Fig. 2B). These data show that OSTF1 and TFIIB are coinduced during hyperosmotic stress with induction kinetics characteristic of IEGs.
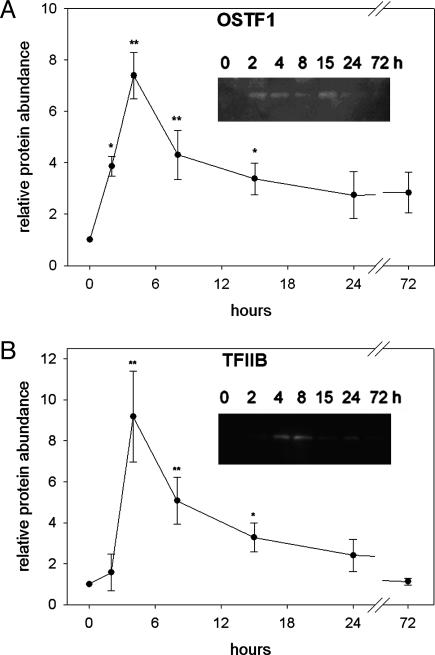
Fig. 3.
Time-course analysis of hyperosmotic induction of OSTF1 and TFIIB protein abundance. OSTF1 (A) and TFIIB (B) protein levels were determined by Western blot analysis and quantified by densitometry. (Inset) A typical Western blot image. Experiments were performed with n = 4. Data are given as means ± SEM. *, P < 0.05; **, P < 0.01, compared with control sample (0 h, FW).
Fig. 2.
Time-course analysis of hyperosmotic induction of OSTF1 and TFIIB mRNA. OSTF1 (A) and TFIIB (B) transcript abundance was analyzed by using quantitative RT-PCR. Experiments were performed with n = 4. Data are given as means ± SEM. *, P < 0.05; **, P < 0.01, compared with control sample (0 h, FW).
OSTF1 and TFIIB Are Regulated by Osmolality at the Protein Level. Next, we sought to determine whether OSTF1 and TFIIB are also regulated by osmolality at the protein level. Another acclimation experiment was performed in which tilapia were transferred from FW to SW for various times, followed by protein extraction from gills. Kinetics of OSTF1 and TFIIB protein abundance after SW transfer was assessed by SDS/PAGE and Western blot immunodetection with specific antibodies. Both transcription factors increase rapidly in abundance during hyperosmotic stress with similar kinetics to the mRNAs. Highest protein levels are reached after 4-8 h of SW acclimation, being 7.5-fold for OSTF1 (Fig. 3A) and 9-fold for TFIIB (Fig. 3B). Therefore, hyperosmotic induction of OSTF1 and TFIIB is likely to be functionally significant because it is strong at the protein level.
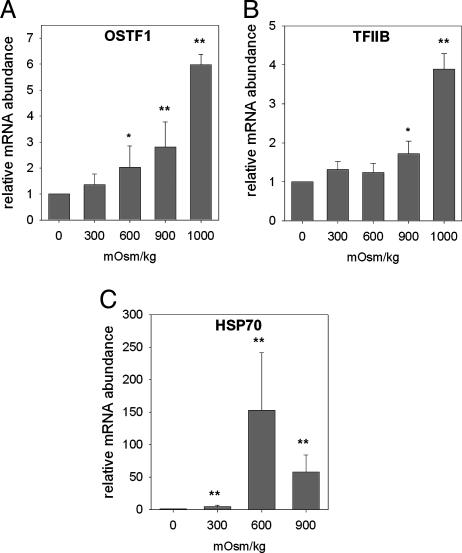
OSTF1 and TFIIB Induction Depends on the Severity of Hyperosmolality. To investigate the osmotic concentration dependence of hyperosmotic OSTF1 and TFIIB induction, another experiment was performed in which tilapia were transferred from FW to a range of intermediate salinities. OSTF1 and TFIIB mRNAs in gill were quantified by qPCR. The hyperosmotic induction of both transcription factors clearly depends on the degree of increase in salinity (Fig. 4 A and B). However, it is statistically insignificant at 300 mosmol/kg for OSTF1 (Fig. 4A) and at 300 and 600 mosmol/kg for TFIIB (Fig. 4B). The mRNA level of tilapia Hsp70 was monitored as a control and increases dramatically at all salinities (Fig. 4C). In contrast to OSTF1 and TFIIB, Hsp70 induction is highest at 600 mosmol/kg and declines at 900 mosmol/kg (Fig. 4C).
Fig. 4.
Analysis of OSTF1, TFIIB, and Hsp70 mRNA expression during hyperosmotic stress. OSTF1 (A), TFIIB (B), and Hsp70 (C) transcript abundances of fish exposed to different salinities for 2 h were quantified by using quantitative RT-PCR. Experiments were performed with n = 6. Data are given as means ± SEM. *, P < 0.05; **, P < 0.01, compared with control sample (FW).
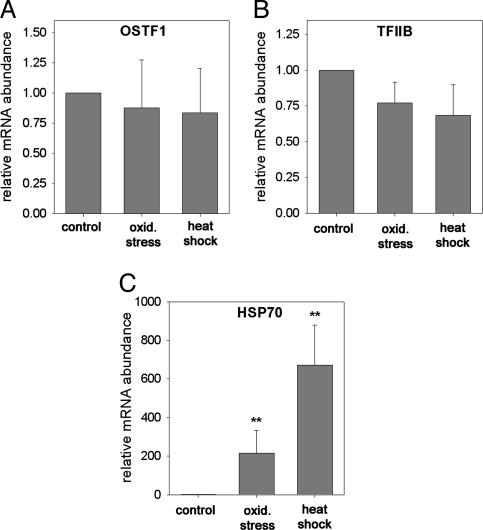
OSTF1 and TFIIB Induction Is Osmotic Stress-Specific. To determine the stressor specificity of induction of OSTF1 and TFIIB, tilapia were exposed to other types of environmental stress for 2 h. Exposure of fish to oxidative stress (1 mM H2O2) or heat shock (36°C) does not increase mRNA abundance of OSTF1 (Fig. 5A) or TFIIB (Fig. 5B). To confirm that oxidative stress and heat shock represented stressful conditions to the fish, we assayed induction of Hsp70 in the same samples. Hsp70 is highly induced during oxidative stress and heat shock (Fig. 5C). Hsp70 induction during oxidative stress and heat shock is even higher than during hyperosmotic stress (Figs. 4C and 5C). These data indicate that OSTF1 and TFIIB induction is specific for osmotic stress and does not occur during other common types of stress.
Fig. 5.
Analysis of OSTF1, TFIIB, and Hsp70 mRNA expression during oxidative stress (1 mM H2O2) and heat shock (+10°C). OSTF1 (A), TFIIB (B), and Hsp70 (C) transcript levels of fish exposed to oxidative stress or heat shock for 2 h were measured by using quantitative RT-PCR. Experiments were performed with n = 6. Data are shown as means ± SEM. *, P < 0.05; **, P < 0.01, compared with control sample (FW, 26°C).
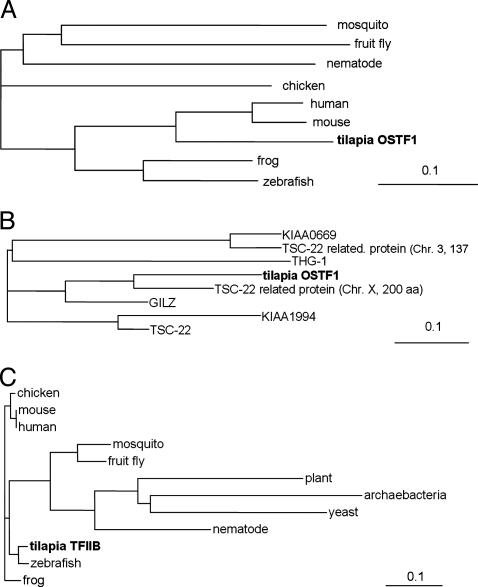
OSTF1 Has Similarity to TGF-β-Stimulated Clone 22 (TSC-22)-Related Transcription Factors, and TFIIB Is Highly Conserved. Multiple-sequence alignment with the most similar proteins identified by mpsrch and blastp programs was carried out for OSTF1 (Fig. 6 A and B) and TFIIB (Fig. 6C). OSTF1 is a transcription factor with a theoretical mass of 29,969 Da, an isoelectric point of 5.06, and relatively low degree of homology to other known transcription factors. It is similar to TSC-22-related mammalian proteins, but it has a longer and unique C terminus. Within this protein family, OSTF1 is most similar to an unnamed 200-aa alternative splice variant of glucocorticoid induced leucine zipper (GILZ, 53.0% sequence identity; Fig. 6B). Sequence identity is lower when compared with GILZ (29.1%), TSC-22 (25.0%), and a 137-aa TSC-22-related protein (22.4%).
Fig. 6.
Phylogenetic trees for OSTF1 and TFIIB. (A) Multiple-sequence alignment of tilapia OSTF1 and homologs from mouse (Mus musculus, AAG41222.1), human (Homo sapiens, BAC03934.1), zebrafish (Danio rerio, AAH56586.1), frog (Xenopus laevis, AAH43841.1), chicken (Gallus gallus, BAA11565.1), mosquito (Anopheles gambiae, EAA14371.1), fruit fly (Drosophila melanogaster,XP_395024.1), and nematode (Caenorhabditis elegans, NP_510086). (B) Multiple-sequence alignment of tilpia OSTF1 and human homologs GILZ (AAG12456), THG-1 (CAB43491), KIAA0669 protein (AAH44643), chr3 TSC-22 related protein (AAG41224), TSC-22 (AAG53077), KIAA1994 protein (AB082525.1), and chr X TSC-22 related protein (BAC03934.1). (C) Multiple-sequence alignment of tilapia TFIIB and TFIIB homologs from zebrafish (NP_955991), human (NP_001505), mouse (AAH16637.1), frog (CAA44668), mosquito (XP_310128), fruit fly (NP_476888), nematode (AAG24202), yeast (Schizosaccharomyces pombe, CAB11044), plant (Arabidopsis thaliana (AAF02810), and archaebacteria (Halobacterium sp. NRC-1, AAG18850). GenBank accession numbers are given in parentheses above.
Tilapia TFIIB has a theoretical mass of 34,957 Da, an isoelectric point of 8.79, and it is highly similar to other vertebrate TFIIB proteins (89.6% sequence identity; Fig. 6C). These results demonstrate that OSTF1 may have functions that are similar to those of the mammalian family of TSC-22-related transcription factors and that TFIIB is the true tilapia homolog of general TFIIB.
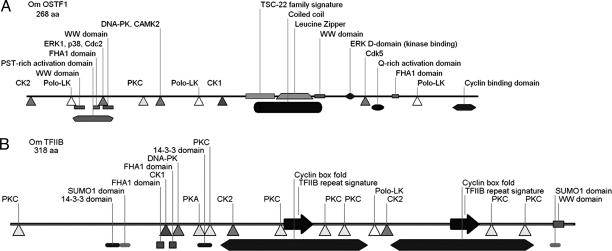
OSTF1 and TFIIB Have Many Consensus Sites for Additional Posttranslational Regulation. OSTF1 contains the TSC-22 family signature sequence and a leucine-zipper domain, indicating that it is a member of this protein family (Fig. 7A). The TSC-22 family signature (amino acids 131-149) and the leucine-zipper domain (amino acids 149-183) are the most highly conserved regions of OSTF1. OSTF1 contains putative phosphorylation sites for casein kinases 1 and 2 (CK1/2), PKC, Polo-like kinases (Polo-LK), MAP kinases [p38 and extracellular signal-regulated kinase (ERK)], cyclin-dependent kinases (cdk; Cdc2 and Cdk5), DNA-PK and calcium-calmodulin-dependent kinase 2 (CAMK2) (Fig. 7A). These kinases are critical elements of stress-signaling pathways. In addition to putative phosphorylation sites, OSTF1 also contains binding domains for ERK and cyclin-cdk complexes in its unique C terminus. OSTF1 contains two transcriptional activation domains: a proline-serine-threonine-rich domain in the N terminus and a glutamine-rich domain in the unique C terminus. (Fig. 7A). Its predicted intracellular localization is nuclear (psort ii, http://psort.nibb.ac.jp/form2.html).
Fig. 7.
Putative phosphorylation sites, motifs, and interaction domains in OSTF1 (A) and TFIIB (B). The N terminus is shown on the left, and the C terminus is shown on the right. For methods of analysis, please refer to the text.
OSTF1 contains two prominent types of protein-interaction modules, FHA1 and WW domains, which may facilitate posttranslational regulation. FHA1 domains are forkhead transcription factor-associated protein-interaction domains that bind to phosophothreonine and mediate DNA repair and cell-cycle-checkpoint signaling during stress (16). WW domains are small protein-interaction modules that recognize proline-containing ligands and are regulated by tyrosine phosphorylation (17).
TFIIB contains putative phosphorylation sites for a similar set of protein kinases as OSTF1 (Fig. 7B). In addition, TFIIB includes two small ubiquitin-related modifier 1 (SUMO1) domains, a WW domain, two FHA1 domains, and two 14-3-3 binding domains (Fig. 7B). SUMO domains bind SUMO, leading to modulation of protein interactions, subcellular localization, and protein stability (18). These results suggest that OSTF1 and TFIIB are substrates of a similar set of protein kinases and subject to posttranslational regulation by means of protein-protein interaction.
Discussion
Role of IEG Transcription Factors for Salinity Adaptation. Many molecular adaptations of euryhaline fishes to salinity change have been determined, including induction and transcriptional activation of ATPases, secondary active transporters, and ion channels. However, not much is known about osmotic regulation of gene expression in fish. In mammalian cells, hyperosmotic induction of osmoprotective genes depends on the TonEBP transcription factor (9). TonEBP is induced during hyperosmotic stress, but its induction kinetics is slower than for a typical IEG (maximum at 18 h) (10). Zebrafish and pufferfish genomes contain a distant homolog of TonEBP but currently nothing is known about its role in teleost osmoregulation (GenBank accession nos. CAG03377 and NP_956189). Tilapia OSTF1 and TFIIB are induced more rapidly and transiently during hyperosmotic stress, which is typical for IEGs (19). Because of their rapid induction OSTF1 and TFIIB are suitable as early transducers of osmosensory signals and potential key regulators of transcriptional regulation during salinity adaptation of euryhaline fish. Virtually identical induction kinetics of OSTF1 and TFIIB suggests that these two transcription factors are coregulated and fulfill a common function for osmosensory signal transduction.
Significance of Osmotic Regulation of General Transcription Factor TFIIB. Because TFIIB is a general transcription factor, it was unexpected that hyperosmotic stress induces this protein. The reason for TFIIB induction is currently unknown, although it may be required to compensate for chromatin compaction and decreased efficiency of transcription during hyperosmotic stress (20). TFIIB is a general subunit of the active polymerase II preinitiation complex. It binds preferentially (but not exclusively) to a specific DNA sequence called the B-factor-response element (BRE) and promotes assembly of Pol II complexes on promoters. Stress-mediated induction of TFIIB transcripts has been observed in archaea and yeast (21, 22). Because of its coinduction with TFIIB in tilapia gills, OSTF1 may recruit TFIIB preferentially to osmoprotective genes. Consistent with this hypothesis, TFIIB interacts with other transcription factors during stress (including NF-κB, c-jun, and p53), suggesting that it is targeted to stress response genes to facilitate their transcription (23).
Of interest, based on cDNA microarray experiments the yeast (Saccharomyces cerevisiae) genome can be divided into two classes of genes (24). One class is preferentially targeted by the Spt-Ada-GCN5-acetyltransferase (SAGA) transcriptional complex and contains many stress-inducible genes (≈10% of the genome), whereas the other class is preferentially targeted by the TFIID transcriptional complex and associated with house-keeping functions (≈90% of the genome). TFIIB cooperates with the yeast SAGA complex (25), and it is possible that similar interactions direct TFIIB function during osmotic stress in vertebrates.
Possible Functional Consequences of OSTF1 Induction During Hyperosmolality. The TSC-22/GILZ family consists of multiple leucine zipper-containing transcription factors that form homodimers and heterodimers with other family members (at least seven in mammals and 5-10 in teleosts). These proteins are known by multiple names, including TSC-22, GILZ, TSC-22 homologous gene (THG), and δ-sleep-inducing immunoreactive peptide (DSIPI). TSC-22 was isolated based on rapid and transient transcriptional induction by TGF-β (26). TSC-22 transcription is increased by many stimuli, including anticancer drugs, progesterone, and growth inhibitors, and it may play a suppressive role in tumorigenesis (27). Because we have shown (28) that hypertonicity causes DNA damage, and OSTF1 contains putative phosphorylation and binding sites for DNA-PK, cdk, and MAP kinases, it may be involved in regulating DNA-damage check-points and apoptosis during osmotic stress.
The closest human homolog of OSTF1 is a splice variant of GILZ. Originally, GILZ was identified as a 134-aa protein whose expression was induced after treatment of thymocytes with dexamethasone (29). GILZ mRNA increases rapidly after aldosterone treatment in principal cells of the renal collecting duct (30). GILZ interacts with NF-κB and Raf and inhibits AP-1, FoxO3, and NF-κB-mediated apoptotic pathways (31-33). Because OSTF1 contains putative ERK phosphorylation and binding sites, it may be involved in feedback regulation of MAP kinase pathways during hyperosmotic stress.
Glucocorticoid receptors are up-regulated in gills of tilapia exposed to hyperosmolality (34), and tilapia cortisol receptor was partially cloned (35). Cortisol is a central hormone in fish osmoregulation that promotes SW adaptation by regulating gill cell differentiation, Na+/K+-ATPase, cotransporters, and ion channels, resulting in overall salt secretion to maintain body-water and ion homeostasis. Plasma cortisol levels increase very rapidly (within minutes) after SW transfer of tilapia (36). These data and structural similarity of OSTF1 to mammalian GILZ suggest that hypertonic induction of OSTF1 may be (at least partly) due to increased cortisol and that OSTF1 mediates cortisol effects on ion transport and cell differentiation in gills.
It is uncertain how much functional conservation exists between OSTF1 and other vertebrate members of the TSC-22/GILZ family because many functions of these proteins are conferred by their weakly conserved N- and C-terminal domains. However, based on what is known about functions of TSC-22/GILZ family members, OSTF1 likely plays a dominant role in the immediate response to hyperosmotic stress.
Stressor Specificity and Potential Posttranslational Regulation of OSTF1 and TFIIB. TFIIB is a general transcription factor and mammalian TSC-22 family members are involved in apoptotic signaling and transcriptional regulation in response to diverse stimuli. However, tilapia OSTF1 and TFIIB are induced only during hyperosmotic stress but not during oxidative stress or heat shock. This finding indicates that their induction in tilapia gill cells depends on sensors that are specifically activated by osmotic stress. Recently, hyperosmolality was shown to trigger oxidative DNA and protein damage (37), and because H2O2 is a common intermediate of oxidative stress, the lack of OSTF1 and TFIIB induction by H2O2 suggests that the sensors and pathways responsible for hyperosmotic regulation of these transcription factors are distinct from oxidative stress pathways triggered during hyperosmolality. Thus, hyperosmotic induction of OSTF1 and TFIIB is not part of the cellular stress response that is caused by macromolecular damage (38) but part of a cellular homeostasis response prompted by events that are unique to hyperosmolality, possibly by salt-induced chromatin compaction in combination with activation of specific osmosensors and rapid increase in plasma cortisol.
Transcription factors are almost always activated by multiple mechanisms. For example, hyperosmotic activation of mammalian TonEBP is due to phosphorylation, dimerization, nuclear translocation, and induction (39). OSTF1 and TFIIB have numerous putative phosphorylation sites and protein-interaction domains that may modulate their transcriptional activity. Of particular interest are putative phosphorylation sites for MAP kinases (ERK1 and p38) and cdk (Cdc2 and Cdk5) in OSTF1 because they are accompanied by binding domains for ERK and cyclin-cdk complexes, suggesting that these kinases are important for OSTF1 regulation. Based on the nature of putative phosphorylation sites and protein-interaction domains, multiple signals of varying stressor specificity may converge at the level of posttranslational OSTF1 modulation and potentiate functional consequences of induction.
TFIIB may not only be induced but also stabilized during hyperosmotic stress because protein levels rise more than mRNA levels. This phenomenon is true also for OSTF1 but to a lesser degree. TFIIB stabilization could be mediated by means of protein-interaction domains (e.g., by decreased sumoylation). In addition, 14-3-3 is regulated by osmolality in euryhaline fishes (6) and may act in concert with SUMO to modulate protein-protein interactions, nuclear-cytoplasmic compartmentation, and the stability of TFIIB.
In summary, we have cloned and characterized two tilapia transcription factors that are rapidly induced during hyperosmotic stress. OSTF1 may represent the molecular link between the SW adaptation hormone cortisol and osmotic regulation of gene expression. OSTF1 and TFIIB are likely to be key elements for transcriptional regulation of transport proteins and gill cell differentiation during osmotic adaptation.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB 0244569.
Author contributions: D.K. designed research; D.F.F. performed research; D.F.F. and D.K. analyzed data; and D.F.F. and D.K. wrote the paper.
Abbreviations: FW, fresh water; SW, seawater; OSTF1, osmotic stress transcription factor 1; IEG, immediate early gene; TFIIB, transcription factor II B; TSC-22, TGF-β-stimulated clone 22; cdk, cyclin-dependent kinase(s); ERK, extracellular signal-regulated kinase; SUMO, small ubiquitin-related modifier; qPCR, quantitative PCR.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AAT84345 and AAT84346).
References
- 1.Laurent, P. & Dunel, S. (1980) Am. J. Physiol. 238, R147-R159. [DOI] [PubMed] [Google Scholar]
- 2.Evans, D. H. (2002) J. Exp. Zool. 293, 336-347. [DOI] [PubMed] [Google Scholar]
- 3.Tipsmark, C. K., Madsen, S. S., Seidelin, M., Christensen, A. S., Cutler, C. P. & Cramb, G. (2002) J. Exp. Zool. 293, 106-118. [DOI] [PubMed] [Google Scholar]
- 4.Mistry, A. C., Honda, S., Hirata, T., Kato, A. & Hirose, S. (2001) Am. J. Physiol. 281, R1594-R1604. [DOI] [PubMed] [Google Scholar]
- 5.Cutler, C. P. & Cramb, G. (2002) J. Exp. Biol. 205, 2643-2651. [DOI] [PubMed] [Google Scholar]
- 6.Kültz, D., Chakravarty, D. & Adilakshmi, T. (2001) J. Exp. Biol. 204, 2975-2985. [DOI] [PubMed] [Google Scholar]
- 7.Mistry, A. C., Honda, S. & Hirose, S. (2001) Biochem. J. 360, 107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burg, M. B., Kwon, E. D. & Kültz, D. (1996) FASEB J. 10, 1598-1606. [DOI] [PubMed] [Google Scholar]
- 9.Woo, S. K., Lee, S. D. & Kwon, H. M. (2002) Pflügers Arch. 444, 579-585. [DOI] [PubMed] [Google Scholar]
- 10.Miyakawa, H., Woo, S. K., Chen, C. P., Dahl, S. C., Handler, J. S. & Kwon, H. M. (1998) Am. J. Physiol. 43, F753-F761. [DOI] [PubMed] [Google Scholar]
- 11.Kültz, D. (1996) Am. J. Physiol. 271, C1181-C1193. [DOI] [PubMed] [Google Scholar]
- 12.Kültz, D. & Somero, G. N. (1995) J. Exp. Biol. 198, 1883-1894. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 14.Pfaffl, M. W. (2001) Nucleic Acids Res. 29, 2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kültz, D., Madhany, S. & Burg, M. B. (1998) J. Biol. Chem. 273, 13645-13651. [DOI] [PubMed] [Google Scholar]
- 16.Durocher, D., Taylor, I. A., Sarbassova, D., Haire, L. F., Westcott, S. L., Jackson, S. P., Smerdon, S. J. & Yaffe, M. B. (2000) Mol. Cell 6, 1169-1182. [DOI] [PubMed] [Google Scholar]
- 17.Ilsley, J. L., Sudol, M. & Winder, S. J. (2002) Cell Signal. 14, 183-189. [DOI] [PubMed] [Google Scholar]
- 18.Verger, A., Perdomo, J. & Crossley, M. (2003) EMBO Rep. 4, 137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams, M., Lyu, M. S., Yang, Y. L., Lin, E. P., Dunbrack, R., Birren, B., Cunningham, J. & Hunter, K. (1999) Genomics 55, 327-334. [DOI] [PubMed] [Google Scholar]
- 20.Kültz, D. (2000) in Environmental Stressors and Gene Responses, eds. Storey, K. B. & Storey, J. (Elsevier, Amsterdam), pp. 157-179.
- 21.Thompson, D. K., Palmer, J. R. & Daniels, C. J. (1999) Mol. Microbiol. 33, 1081-1092. [DOI] [PubMed] [Google Scholar]
- 22.Hoopes, B. C., Bowers, G. D. & DiVisconte, M. J. (2000) Nucleic Acids Res. 28, 4435-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espinosa, J. M., Verdun, R. E. & Emerson, B. M. (2003) Mol. Cell 12, 1015-1027. [DOI] [PubMed] [Google Scholar]
- 24.Huisinga, K. L. & Pugh, B. F. (2004) Mol. Cell 13, 573-585. [DOI] [PubMed] [Google Scholar]
- 25.Dietz, M., Heyken, W. T., Hoppen, J., Geburtig, S. & Schuller, H. J. (2003) Mol. Microbiol. 48, 1119-1130. [DOI] [PubMed] [Google Scholar]
- 26.Shibanuma, M., Kuroki, T. & Nose, K. (1992) J. Biol. Chem. 267, 10219-10224. [PubMed] [Google Scholar]
- 27.Kester, H. A., Blanchetot, C., den Hertog, J., van der Saag, P. T. & van der, B. B. (1999) J. Biol. Chem. 274, 27439-27447. [DOI] [PubMed] [Google Scholar]
- 28.Kültz, D. & Chakravarty, D. (2001) Proc. Natl. Acad. Sci. USA 98, 1999-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Adamio, F., Zollo, O., Moraca, R., Ayroldi, E., Bruscoli, S., Bartoli, A., Cannarile, L., Migliorati, G. & Riccardi, C. (1997) Immunity 7, 803-812. [DOI] [PubMed] [Google Scholar]
- 30.Robert-Nicoud, M., Flahaut, M., Elalouf, J. M., Nicod, M., Salinas, M., Bens, M., Doucet, A., Wincker, P., Artiguenave, F., Horisberger, J. D., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 2712-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mittelstadt, P. R. & Ashwell, J. D. (2001) J. Biol. Chem. 276, 29603-29610. [DOI] [PubMed] [Google Scholar]
- 32.Ayroldi, E., Zollo, O., Macchiarulo, A., Di Marco, B., Marchetti, C. & Riccardi, C. (2002) Mol. Cell. Biol. 22, 7929-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asselin-Labat, M. L., David, M., Biola-Vidamment, A., Lecoeuche, D., Zennaro, M. C., Bertoglio, J. & Pallardy, M. (2004) Blood 104, 215-223. [DOI] [PubMed] [Google Scholar]
- 34.Dean, D. B., Whitlow, Z. W. & Borski, R. J. (2003) Gen. Comp. Endocrinol. 132, 112-118. [DOI] [PubMed] [Google Scholar]
- 35.Tagawa, M., Hagiwara, H., Takemura, A., Hirose, S. & Hirano, T. (1997) Gen. Comp. Endocrinol. 108, 132-140. [DOI] [PubMed] [Google Scholar]
- 36.Abo, H. S. & Hanke, W. (1984) Gen. Comp. Endocrinol. 54, 409-417. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Z., Dmitrieva, N. I., Park, J. H., Levine, R. L. & Burg, M. B. (2004) Proc. Natl. Acad. Sci. USA 101, 9491-9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kültz, D. (2005) Annu. Rev. Physiol. 67, in press. [DOI] [PubMed]
- 39.Lee, S. D., Colla, E., Sheen, M. R., Na, K. Y. & Kwon, H. M. (2003) J. Biol. Chem. 278, 47571-47577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.