Abstract
The size of proteins that can be studied by solution NMR spectroscopy has increased significantly because of recent developments in methodology. Important experiments include those that make use of approaches that increase the lifetimes of NMR signals or that define the orientation of internuclear bond vectors with respect to a common molecular frame. The advances in NMR techniques are strongly coupled to isotope labeling methods that increase sensitivity and reduce the complexity of NMR spectra. We show that these developments can be exploited in structural studies of high-molecular-weight, single-polypeptide proteins, and we present the solution global fold of the monomeric 723-residue (82-kDa) enzyme malate synthase G from Escherichia coli, which has been extensively characterized by NMR in the past several years.
Keywords: isotope labeling, protein NMR, transverse relaxation-optimized spectroscopy
NMR spectroscopy is one of the most powerful techniques for the study of protein structure and dynamics (1). In addition, NMR spectroscopy has emerged as an important tool for the investigation of protein–ligand interactions (2), and their quantification in terms of structure, dynamics, kinetics, and thermodynamics. However, a drawback of the methodology is the size limitation of the molecules that can be studied. The short lifetimes of NMR signals and the complexity of spectra generated in applications involving high-molecular-weight systems still limit many NMR applications to studies of relatively small biomolecules.
In this regard, important advances have been made in the past several years that have significantly extended the range of molecules that are now amenable to investigation. Large gains in both the sensitivity and the resolution of NMR spectra of large molecules can be achieved by using the so-called transverse relaxation-optimized spectroscopy (TROSY) approach (3), in which only the slowly decaying components of nuclear magnetization contribute to the final signal. Since the original pioneering developments involving studies of amide (3) and aromatic moieties (4), more recent applications with methyl (5) and methylene groups (6) have appeared. A second major advance has involved the “reintroduction” of magnetic interactions that would normally average to zero in isotropic solution by means of the use of media leading to a weak alignment of the macromolecule of interest (7). The resulting orientational restraints, such as dipolar couplings and changes in chemical shifts that are generated by such alignment, are extremely valuable in structural studies, in particular for large proteins in which the number of restraints per residue is significantly less than what is normally obtained in studies of small (≤30-kDa) proteins. A third important contribution has been the development of isotopic-labeling approaches, such as those involving uniform 15N, 13C labeling along with high levels of deuteration, which maximize the lifetimes of NMR signals and optimize the H—N TROSY effect. NMR experiments that have emerged to exploit these labeling schemes are equally important.
Building on the developments mentioned above, studies of proteins and protein complexes in the 100-kDa range have been reported, focusing primarily on backbone chemical-shift assignments (8, 9). In a number of cases, solution NMR structures of β-barrel membrane-spanning proteins have been described in which the overall aggregate molecular mass of the protein–lipid complex is on the order of 50–60 kDa and the protein component is <200 residues (10–12). Solution structures of proteins have been limited to molecules with ≈400 residues or less (13–15).
Here, we show that it is possible to obtain well defined global folds of proteins that are considerably larger than 400 residues by solution NMR spectroscopy. By using recently introduced labeling, experimental, and data-processing approaches, the global backbone fold of the monomeric 723-residue enzyme malate synthase G (MSG, 82 kDa) has been obtained from experimental NMR restraints exclusively. MSG catalyzes the Claisen condensation of glyoxylate with an acetyl group of acetyl–CoA, producing malate, which is an intermediate in the citric acid cycle. This glyoxylate pathway enzyme is exclusive to a number of pathogenic organisms (16), and therefore, structural studies of MSG are an important first step in the design of inhibitory compounds as potential antimicrobial agents. To this end, crystal structures of MSG complexed with magnesium and glyoxylate (17) (at 2.0 Å) and a ternary abortive MSG–pyruvate–acetyl–CoA complex (18) (at 1.95 Å) have been solved. The apo form of MSG has been extensively characterized by our laboratory using NMR, including assignments of backbone (9) and methyl resonances (19), studies of domain orientation and ligand binding (20), and most recently, side-chain dynamics (21), setting the groundwork for the structural study reported here.
Materials and Methods
MSG Samples and NMR Spectroscopy. A number of samples of MSG have been prepared as described (9, 21, 22) (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for more details, including the acquisition parameters of all nuclear Overhauser effect (NOE) data sets collected in this work). Measurements of 1H—15N dipolar couplings and 13CO chemical-shift changes upon alignment in Pf1 phage have been described (20). NMR spectra were processed by using nmrpipe/nmrdraw software (23) and analyzed by using the program nmrview (24) and home-written tcl/tk scripts for visualization of spectra. Processing of the sparsed 4D methyl—methyl NOE data set acquired with nonlinear exponentially biased sampling was performed by using multidimensional decomposition techniques (25), as described in detail elsewhere (26).
Conformational Restraints. The following three types of distance restraints were used in the generation of structures: (i)HN—HN restraints acquired on the U-[15N, 2H]-labeled MSG sample; (ii) methyl—methyl restraints from NOE spectra recorded on U-[15N, 2H], Ileδ1-[13CH3], Leu,Val-[13CH3, 12CD3]-labeled MSG dissolved in D2O; and (iii) methyl—HN restraints obtained from spectra recorded on the same sample as in ii dissolved in H2O (see Table 2, which is published as supporting information on the PNAS web site). A total of 746 HN—HN (99 long-range), 428 methyl—methyl (386 long-range), and 357 methyl—HN (142 long-range) restraints were obtained from analyses of NOE data. A qualitative approach was used in the derivation of distance bounds based on earlier structural studies of MBP (42 kDa, ref. 14). In particular, HN—HN distance bounds of 1.8–3.5 and 1.8–5.0 Å were used for strong and weak crosspeaks, respectively, except for contacts between HNi and HNi±1,(HNi±2) in helices and turns in which the distances were constrained to a maximum of 3.0 (4.0) Å. Methyl—methyl contacts were constrained between 1.8–8.0 Å, except for a few very intense crosspeaks where upper distance bounds of 4.0 Å were used. Distances of 1.8–6.0 Å were used for methyl—HN NOEs, except for intraresidue HN—methyl contacts in which upper bounds of 4.0 Å were used.
A total of 415 1H—15N dipolar couplings (1DHN) and 300 13CO chemical-shift changes upon alignment of MSG in Pf1 phage were used in the structure calculations. Alignment parameters Da and R were determined from the least-squares fit of the extreme (Dzz, Dyy) and the most populated (Dxx) values of the couplings in the experimental 1DHN histogram, as described by Clore et al. (27). Values of Da = -18.5 Hz and R = 0.45 obtained from the fit were used in all calculations. Note that these values differ slightly from those reported in our previous study of domain orientation in MSG [Da = -17 Hz and R = 0.45 (20)], in which the x-ray coordinates of glyoxylate-bound MSG (17) were used to determine the alignment tensor parameters (order parameters and orientation) of individual domains. Values of σxx = -74.7, σyy = -11.8, and σzz = 86.5 ppm were used for the 13CO chemical-shielding tensor in all calculations.
Dihedral-backbone-angle (ϕ,Ψ) predictions were made by using the backbone 15N, 13Cα, and 13CO chemical shifts of MSG with the program talos (28) after the chemical shifts of MSG were removed from the database and the chemical shifts were corrected for 2H isotope effects. Restraints consisting of the average ϕ,Ψ values ± 2 SDs (or at least ±20° from the average predicted value) were used for 533 residues of MSG. Also, χ1 angles of 35 Val residues have been constrained to one of the preferred rotameric states (180, 60, -60) ±15°, as established in our earlier study from measurement of 3JCγCO and 3JCγN scalar couplings for the side chains that do not experience rotamer averaging (21).
Structure Calculations. Structure calculations were performed with National Institutes of Health x-plor (version 2.9.3) software (29) by using a combination of torsion-angle and Cartesian dynamics and employing a protocol very similar to that described (14) in our studies of maltose-binding protein. See Supporting Materials and Methods for details.
Only a small subset of unambiguous long-range NOE restraints [263 long-range (|i-j| > 3) restraints] originating from correlations that could be assigned with certainty were used in initial structure calculations. Regions of the molecule that were predicted to adopt helical secondary structure based on the chemical shift index (CSI) of Wishart et al. (30) were “fixed” to an α-helical conformation by using (artificial) hydrogen-bond restraints between HN of residue i and the carbonyl of residue i-4, excluding the first three residues of the (predicted) helix. In conjunction with the complete set of 1H—15N dipolar and carbonyl chemical-shift anisotropy restraints, dihedral restraints from the talos database, and the RG potential (see Supporting Materials and Methods), an approximate backbone fold was obtained [pairwise rms deviation (rmsd) of backbone atom coordinates of 5.2–5.6 Å for the 10 lowest-energy structures]. These initial structures were used as a reference for the assignment of additional NOE crosspeaks. The remaining NOE restraints were incorporated gradually into the structure calculations. At the final step of the calculations, all hydrogen bond restraints were removed and dihedral angle (ϕ,Ψ) restraints from the CSI secondary structure predictions were introduced for those residues in which the angles found by talos did not satisfy the acceptance criteria. Values of ϕ = -75 (±35)°, Ψ = -25 (±45)° and ϕ = -110 (±50)°, Ψ = +135 (±35)° have been used for residues in α-helices and β-sheets, respectively, when the CSI predictions were used (17 residues in total). The final 10 structures with the lowest overall energy are shown in Figs. 2 and 3, and their statistical parameters are given in Table 1. A small subset of the 30 structures (of a total of 60 structures produced in the final calculation) that are well converged in the calculations are shown for ease of visualization; all 30 structures have similar global folds. It is worth emphasizing that any of the published x-ray structures of MSG (all are ligated) were not used at any point in any of the calculations.
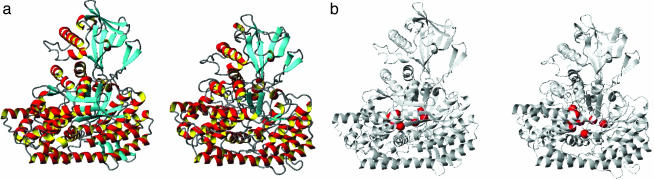
Fig. 2.
Comparison of x-ray and NMR-derived structures of MSG. (a) Ribbon diagrams of the x-ray structure of MSG (Left, PDB ID code 1D8C; ref. 17) and the lowest energy NMR structure (Right) calculated on the basis of 1,531 NOE, 1,101 dihedral angle, 415 residual dipolar couplings, and 300 carbonyl-shift restraints. (b) Ribbon representations of MSG (Left shows x-ray structure, and Right shows the lowest-energy NMR structure) are shown with the Cα carbons of residues that either contact or are proximal to glyoxylate (D270, E272, R338, E427, F453, L454, D455, and D631) in the active site of the protein, indicated with red spheres. The image was prepared by using molmol (39).
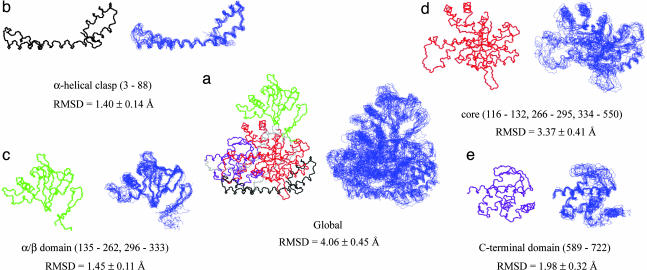
Fig. 3.
Comparison of x-ray and NMR structures on a per-domain basis. (a) The x-ray structure (PDB ID code 1D8C; ref. 17) and the 10 lowest-energy NMR structures of MSG calculated on the basis of experimental restraints. Backbone traces of the x-ray structure (Left) and NMR structures (Right) are displayed and superimposed by aligning residues in elements of regular secondary structure. The α-clasp, α/β, core, and C-terminal domains are shown in black, green, red, and purple, respectively, in the x-ray structure, with the linkers shown in gray. Individual domains [α-clasp (b), α/β (c), core (d), and C-terminal (e)] are shown and superimposed by fitting over residues in regular secondary structure. The rmsd of the NMR ensemble (10 structures) and the x-ray are indicated for heavy backbone atoms of regular secondary structure elements for the entire molecule and individual domains.
Table 1. Structural statistics for the 10 final structures of MSG.
| Average backbone rmsd to 1D8C(17), Å | |
| α-Clasp (3–88) | 1.40 ± 0.14* (1.58 ± 0.17) |
| α/β (135–262, 296–333)† | 1.45 ± 0.11 (2.06 ± 0.31) |
| C-terminal (589–722) | 1.98 ± 0.32 (3.57 ± 0.60) |
| Core (116–132, 266–295, 334–550) | 3.37 ± 0.41 (3.73 ± 0.35) |
| Global (3–722)† | 4.06 ± 0.45 (4.64 ± 0.45) |
| Average pairwise backbone rmsd, Å | |
| α-Clasp (3–88) | 1.48 ± 0.25 (1.67 ± 0.29) |
| α/β (135–262, 296–333)† | 1.40 ± 0.24 (2.34 ± 0.40) |
| C-terminal (589–722) | 1.83 ± 0.24 (2.96 ± 0.46) |
| core (116–132, 266–295, 334–550) | 2.98 ± 0.52 (3.27 ± 0.45) |
| Global (3–722)† | 2.92 ± 0.31 (3.40 ± 0.30) |
| ϕ/ψ Space‡ | |
| Most favored region, % | 77.0 ± 1.3 |
| Additionally allowed region, % | 18.0 ± 1.3 |
| Generously allowed region, % | 3.8 ± 0.6 |
| Disallowed region, % | 1.2 ± 0.4 |
| Deviations from idealized geometry§ | |
| Bond, Å | 0.0022 ± 0.00003 |
| Angles, ° | 0.322 ± 0.005 |
| Impropers, ° | 0.324 ± 0.009 |
| Deviations from experimental restraints | |
| NOEs, Å | 0.108 ± 0.005 |
| Dihedral angles, ° | 0.14 ± 0.03 |
| Dipolar couplings, Hz¶ | 3.2 ± 0.2 |
| CSA, parts per billion¶ | 10.6 ± 0.7 |
Averages are over heavy backbone nuclei and are calculated from residues in regions of secondary structure only. Numbers in parentheses refer to calculations including all residues
Residues of a surface loop (300–310) were excluded from the rmsd calculation due to missing coordinates in the x-ray structure
Calculated with procheck-nmr (40)
Evaluated by xplor-nih (29)
A Q factor (41) of 0.18 ± 0.01 was obtained for both dipolar coupling and chemical-shift anisotropy (CSA) restraints (all included in structure calculations)
Results
An Isotope-Labeling Strategy. A highly deuterated MSG sample has been used with selective reincorporation of protons into methyl positions of Ile(δ1), Leu, and Val residues. Methyls are abundant in protein molecules, and they are frequently located in the hydrophobic cores of macromolecular structures. Therefore, methyl—methyl NOEs are a rich source of long-range distance information. This labeling strategy represents a compromise between the need for high levels of deuteration to enhance the sensitivity of the resulting NMR spectra with practical concerns reflecting the measurement of a sufficient number of distance restraints (in the form of 1H—1H NOEs) to obtain structures.
Because methyl (13CH3) groups are the major probes of structure (see below), attempts have been made to design experiments that exploit their unique NMR properties. In certain classes of experiments involving methyl groups (heteronuclear multiple quantum correlation-based) half of the signal traverses a pathway in which magnetic fields from the intramethyl spins cancel so that the signal is long-lived (5). Therefore, many of the experiments that have been used for studies of small proteins have been modified to include this so-called methyl—TROSY effect, thereby maximizing sensitivity and resolution. We have recently proposed an isotope-labeling scheme that optimizes methyl TROSY and involves 13C1H3 labeling of only a single methyl group in Val and Leu, whereas the other methyl is 12CD3 (22). This labeling pattern may be used in combination with uniform 13C-labeling, which is necessary for side-chain assignments and 3JCγCO-coupling measurements (19, 21). Alternatively, a strategy in which selective incorporation of 13C1H3 into Ile δ1 and one of the methyl groups of Val and Leu, with the remaining positions 12CD, can be used, as for the NOE experiments in this work. Below, we briefly describe experimental procedures that have used these labeling strategies to derive as many dihedral angle and distance restraints in MSG as possible.
Stereospecific Assignments of Prochiral Methyls Using Selectively Labeled Samples. Near complete stereospecific assignments of the prochiral methyl groups of Val and Leu in MSG have been achieved (21) based on a fractional (10%) 13C-labeling strategy developed by Wüthrich and coworkers (31) and, in the case of Val residues, a series of new methyl—TROSY quantitative J experiments for measuring 3JCγN and 3JCγCO scalar couplings. These couplings are related by Karplus-type equations to the side-chain χ1 torsion angle in Val (32), providing both the stereospecific assignment as well as the χ1 rotamer of ordered Val side chains (32) that can be used as dihedral restraints in subsequent structure calculations.
NOE Spectroscopy of MSG. A series of 3D and 4D TROSY-based data sets have been used to measure CH3—CH3, HN—HN, and HN—CH3 distance restraints in MSG. Fig. 1 a, c, and d show regions of 2D planes from the 4D data sets that have been recorded to quantify distances. Extensive use has been made of methyl—TROSY (5) and H—N—TROSY (3) in concert with appropriate isotope labeling schemes (22), as described in Materials and Methods.
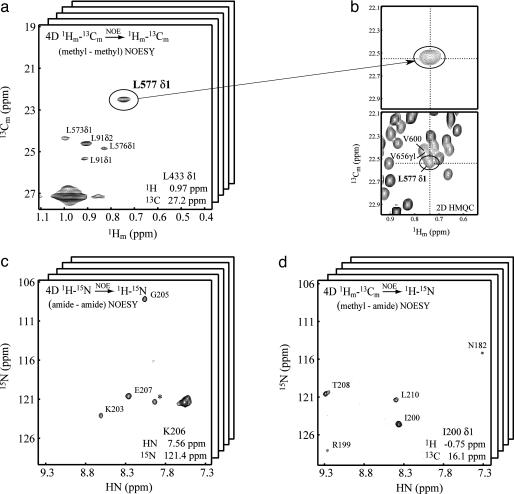
Fig. 1.
Representative planes from 4D NOE data sets. (a)F1(1H)—F2(13C) plane from the 4D CH3—CH3 NOESY spectrum showing correlations to L433δ1. (b) The correlation involving L577δ1 can be assigned despite the fact that this residue is in a very crowded region of the 2D 1H—13C correlation map. (c)F3(15N)—F4(1HN) plane from the HN—HN 4D data set showing correlations to Lys 206 HN. (d)F3(15N)—F4(1HN) plane from the methyl—HN 4D matrix, showing NOEs between I200δ1 and proximal amide protons.
As mentioned above, the major advantage of TROSY derives from the increase in the lifetime of the NMR signal. However, gains can be realized only in spectra in which acquisition times are sufficiently long to exploit the decreased signal decay. The use of sufficient acquisition times is very often possible in 2D and 3D spectra recorded by using conventional schemes, but for 4D data sets, the need to limit acquisition to within reasonable measuring times (<1 week) places severe restrictions on the maximum evolution times in each of the three indirectly detected dimensions. As a result, the potential resolution gains associated with increased signal lifetimes are not realized in conventional 4D spectra. Therefore, the 4D CH3—CH3 NOE data set was measured by using a nonlinear sampling procedure, recording only a fraction (≈30%) of the data that would normally be obtained in a conventional experiment (26). Subsequently, the 4D spectrum was reconstructed as a series of shapes whose products reproduce the data set by using a nonlinear least-squares fitting procedure discussed in ref. 25. The resolution of the 4D data set is shown in Fig. 1b, where the NOE connecting L577δ1 with L433δ1 could be readily assigned, despite the fact that the 1H—13C correlation for L577δ1 is in a very crowded region of the 2D 1H—13C heteronuclear multiple quantum correlation spectrum. It would not be possible to make this assignment from a 4D data set recorded and processed by using conventional approaches. In total, 1,531 approximate distance restraints (627 long-range; i.e., NOEs between residues more than three apart in sequence) were assigned from 3D and 4D data sets and incorporated step-wise into structure calculations (see Materials and Methods). Notably, 386, 99, and 142 long-range CH3—CH3, HN—HN, and HN—CH3 restraints were obtained, emphasizing the important role of methyl groups in this procedure.
NMR-Derived Global Fold of MSG. Fig. 2a compares the x-ray-derived structure of the glyoxylate loaded form of MSG (17) (Left) with the global fold of the apo form of the enzyme established on the basis of the restraints described above (Right). In addition, orientational restraints in the form of 1H—15N residual dipolar couplings (415), changes in 13CO chemical shifts upon alignment of the protein (300), and ϕ,Ψ dihedral angle restraints from chemical shifts (1,066) have also been included in structure calculations.
MSG is composed of four domains, including (i) a centrally located β8/α8 core, (ii) an N-terminal α-helical domain (α-helical clasp) linked to the first strand of the barrel by a long extended loop, (iii) an α/β domain appended to the molecular core, and (iv) the C-terminal end of the enzyme consisting of a five-helix “plug” connected to the barrel by an extended loop. The core folds to form a triose phosphate isomerase (TIM) barrel arranged such that the eight strands form a parallel β-sheet that wraps in a cylinder surrounded by the eight α-helices. The center of the barrel is highly hydrophobic and composed of side chains from alternate residues of the strands. It is clear that the main topological features of the enzyme are reproduced in the solution global fold, including the direction of the polypeptide chain, the domain organization, the position and the orientation of the helical elements, and the locations of most of the β-strands. The pairwise rmsd of the backbone heavy atom coordinates from regions of secondary structure between x-ray and NMR derived models is 4.1 Å, averaged over the lowest 10 energy NMR structures (4.15 Å for the top 30 structures). Note that the helices are well defined (in structure and orientation) in the NMR ensemble, whereas regions of β-sheet (in particular, the eight-strand parallel β-sheet in the center of the core) are not reproduced entirely by the NMR data, with most of the strands being significantly shorter than their counterparts in the x-ray structure (Fig. 2a). The α-helices are readily defined by sequential and HN(i)—HN(i ± 3) NOEs that can be identified easily in 15N edited NOE spectra. In contrast, the labeling scheme that we have chosen (high levels of deuteration) eliminates NOEs between proximal Hα protons across strands that form β-sheets. The closest HN(i)—HN(j) distances across planar parallel β-sheets (4.0 Å) are generally longer than in their planar antiparallel counterparts (3.3 Å), and they can be even longer than 4.0 Å in cases of strong deviations from planarity. Therefore, some of the β-strands in the core of the molecule have been defined primarily because of dihedral restraints from secondary structure predictions on the basis of chemical shifts (28, 30), rather than distance restraints.
The active site of MSG is formed from residues at the C-termini of a number of the β-strands of the core barrel, not unlike many of the enzymes sharing the triose phosphate isomerase (TIM) barrel fold. Residues that coordinate Mg2+ and glyoxylate and the neighboring residues in the active site are predominately acidic (three Asp and two Glu) or basic (one Arg), and include a pair of hydrophobic (one Leu and one Phe) amino acids as well (17). There are no side-chain restraints for any of these residues, and NOEs are measured only for three of them, yet interestingly, they localize to a region in the NMR structure that is similar to the one found by x-ray. The placement of these residues is shown in Fig. 2b, where red spheres indicate the Cα atoms of those residues that have been identified in the x-ray structure to make contact with or that are proximal to the bound glyoxylate.
Fig. 3a compares the x-ray structure (Left, color-coded according to domain) with the ensemble of the 10 lowest NMR structures (Right), whereas the individual domains are shown in Fig. 3 b–e. The listed rmsd values compare the ensemble of NMR-derived structures with the x-ray (backbone heavy atoms), considering only regions of secondary structure. It is clear that the individual domains are reasonably well defined, despite the large size of the enzyme.
Discussion
Over the past 15 years, protocols have been developed for the structure determination of proteins of less than ≈40 kDa. The basic approach involves obtaining complete chemical-shift assignments (all protons in the molecule) and, subsequently, using the assignments in concert with NOE measurements to compile a list of pairwise distances between sites in the protein. These distances are then used as input to molecular mechanics programs that attempt to satisfy them during the course of folding the protein to its correct 3D structure. Distance restraints are often supplemented by orientational restraints in the form of dipolar couplings and by torsion angles that are obtained from the assigned chemical shifts and by measurement of scalar couplings.
For studies of high-molecular-weight proteins, such as MSG, a different approach is needed because spectral overlap and sensitivity considerations preclude a strategy in which every site in the protein is assigned. Therefore, we have used a labeling scheme in which high levels of deuteration are used, with selective protonation at the level of methyl groups of Ile(δ1), Leu, and Val (33). In this manner, assignments of chemical shifts are restricted to the backbone spins and to methyl groups. The importance of including protonation at methyl sites is made clear by noting that 84% of the long-range (|i–j| > 3) NOE restraints in MSG are derived from HN—methyl and methyl—methyl contacts. The small number of long-range amide—amide NOEs is due to the large fraction of helical structure in the enzyme (≈75% of the regular secondary structure is helix) and the fact that amide distances across helices are generally too large to be observed. In fact, only a total of six HN—HN long-range NOEs were quantified from the N-terminal α-helical clasp and the largely helical C-terminal domain together, and notably, none of these NOEs involved residues that were in helices. In cases in which only a small number of NOEs are available (on average, ≈1 long-range NOE per residue for MSG), the use of residual dipolar couplings in the structure determination becomes critical. Structures calculated on the basis of NOEs and dihedral-angle restraints exclusively were not as well defined, with increases in average rmsd from the x-ray structure of ≈1.5 Å, relative to folds obtained with orientational restraints. By contrast, inclusion of even a small number of orientational restraints can have a substantial effect. Here, a total of 415 1H—15N dipolar couplings were used, along with 300 13CO chemical-shift changes upon alignment. However, because the conformational space available to a given secondary structure element is reduced significantly in the context of the intact protein, even a few couplings per element can be sufficient to align it correctly, with a small number of NOEs providing translational restraints.
Although the global fold of MSG determined by solution NMR is clearly not of high resolution, it is evident that, even at this level of resolution, very useful information has been obtained. First, a number of the domains are well defined and potentially could be useful for the interpretation of weak electron-density maps during the course of crystallographic studies. In this regard, it is of interest that MSG is notoriously difficult to crystallize (D. M. Anstrom and S. J. Remington, personal communication) and is susceptible to inactivation and aggregation under x-ray irradiation (17, 34). Second, an analysis of the relative orientation of domains in the apo-solution form and the glyoxylate-loaded x-ray state shows that the there is no significant rearrangement of domains upon ligand binding, in contrast to what has been observed for other related enzymes (35). Third, despite the fact that very few NOE restraints are available for residues contacting or neighboring glyoxylate, the location of these active-site residues was, nonetheless, reasonably well reproduced in the solution fold. Further improvements can be obtained through the use of various molecular-modeling techniques, such as fold recognition and comparative modeling (36); however, the goal of this work is to demonstrate the feasibility of de novo protein folding of high-molecular-weight proteins solely from experimental NMR data.
It is likely that the approach of using highly deuterated proteins with Ile(δ1), Leu, Val methyl protonation will be useful for global fold determination of many other proteins as well. In the case of MSG, the fraction of Ile, Leu, and Val residues is 22%, whereas in a survey of >1,000 unrelated proteins of known sequence, the fraction was found to be very similar (21%) (37). Simulations that we have done previously with a set of four proteins with different secondary structures suggest that the folds obtained for MSG are consistent with expectations for a multidomain protein with tight contacts between the domains (14). Similar conclusions about the usefulness of the Ile(δ1), Leu, Val labeling approach were obtained by Montelione and coworkers (38) based on both experiment and computation.
Together with isocitrate lyase, malate synthase is unique to the glyoxylate shunt bypass that converts the two carbon unit acetate into malate (four carbons) for energy production and biosynthesis. This pathway is used by bacteria, yeast, and fungi when the organisms are exposed to low-oxygen conditions. Enzymes of the glyoxylate shunt have been implicated as virulance factors in a number of pathogens (16), and because the cycle has not been found in humans, MSG is a promising drug target. NMR spectroscopy is ideally suited to play an important role in these studies. The recent development of methodologies allows the quantification of the kinetics and thermodynamics of binding, changes in chemical shifts can be used to establish the site(s) of interaction, and structural studies as a function of ligand can be undertaken as necessary. This work describes the methodology that we have found to be necessary for structural studies of high-molecular-weight proteins, and it demonstrates that useful information can be obtained from global fold determination of such molecules.
Supplementary Material
Acknowledgments
We thank Dr. Philipp Neudecker (University of Toronto) for helpful discussions. This work was supported by a grant from the Canadian Institutes of Health Research (to L.E.K.). V.T. was supported by the Human Frontiers Science Program. L.E.K. holds a Canada Research Chair in Biochemistry.
Author contributions: L.E.K. designed research; V.T., W.-Y.C., V.Y.O., and L.E.K. performed research; V.Y.O processed the 4D methyl NOESY data set; V.T. and W.-Y.C. analyzed data; and V.T. and L.E.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MSG, malate synthase G; NOE, nuclear Overhauser effect; TROSY, transverse relaxation-optimized spectroscopy; rmsd, rms deviation.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1Y8B).
References
- 1.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 2.Lian, L. Y., Barsukov, I. L., Sutcliffe, M. J., Sze, K. H. & Roberts, G. C. (1994) Methods Enzymol. 239, 657-700. [DOI] [PubMed] [Google Scholar]
- 3.Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. (1997) Proc. Natl. Acad. Sci. USA 94, 12366-12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pervushin, K., Riek, R., Wider, G. & Wüthrich, K. (1998) J. Am. Chem. Soc. 120, 6394-6400. [Google Scholar]
- 5.Tugarinov, V., Hwang, P. M., Ollerenshaw, J. E. & Kay, L. E. (2003) J. Am. Chem. Soc. 125, 10420-10428. [DOI] [PubMed] [Google Scholar]
- 6.Miclet, E., Williams, D. C., Jr., Clore, G. M., Bryce, D. L., Boisbouvier, J. & Bax, A. (2004) J. Am. Chem. Soc. 126, 10560-10570. [DOI] [PubMed] [Google Scholar]
- 7.Tjandra, N. & Bax, A. (1997) Science 278, 1111-1114. [DOI] [PubMed] [Google Scholar]
- 8.Salzmann, M., Pervushin, K., Wider, G., Senn, H. & Wüthrich, K. (2000) J. Am. Chem. Soc. 122, 7543-7548. [Google Scholar]
- 9.Tugarinov, V., Muhandiram, R., Ayed, A. & Kay, L. E. (2002) J. Am. Chem. Soc. 124, 10025-10035. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez, C., Adeishvili, K. & Wüthrich, K. (2001) Proc. Natl. Acad. Sci. USA 98, 2358-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora, A., Abildgaard, F., Bushweller, J. H. & Tamm, L. K. (2001) Nat. Struct. Biol. 8, 334-338. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, P. M., Choy, W.-Y., Lo, E. I., Chen, L., Forman-Kay, J. D., Raetz, C. R. H., Prive, G. G., Bishop, R. E. & Kay, L. E. (2002) Proc. Natl. Acad. Sci. USA 99, 13560-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu, L., Petros, A. M., Schnuchel, A., Zhong, P., Severin, J. M., Walter, K., Holzman, T. F. & Fesik, S. W. (1997) Nat. Struct. Biol. 4, 483-489. [DOI] [PubMed] [Google Scholar]
- 14.Mueller, G. A., Choy, W. Y., Yang, D., Forman-Kay, J. D., Venters, R. A. & Kay, L. E. (2000) J. Mol. Biol. 300, 197-212. [DOI] [PubMed] [Google Scholar]
- 15.Williams, D. C., Jr., Cai, M. & Clore, G. M. (2004) J. Biol. Chem. 279, 1449-1457. [DOI] [PubMed] [Google Scholar]
- 16.Lorentz, M. C. & Fink, G. R. (2001) Nature 412, 83-86. [DOI] [PubMed] [Google Scholar]
- 17.Howard, B. R., Endrizzi, J. A. & Remington, S. J. (2000) Biochemistry 39, 3156-3168. [DOI] [PubMed] [Google Scholar]
- 18.Anstrom, D. M., Kallio, K. & Remington, S. J. (2003) Protein Sci. 12, 1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tugarinov, V. & Kay, L. E. (2003) J. Am. Chem. Soc. 125, 13868-13878. [DOI] [PubMed] [Google Scholar]
- 20.Tugarinov, V. & Kay, L. E. (2003) J. Mol. Biol. 327, 1121-1133. [DOI] [PubMed] [Google Scholar]
- 21.Tugarinov, V. & Kay, L. E. (2004) J. Am. Chem. Soc. 126, 9827-9836. [DOI] [PubMed] [Google Scholar]
- 22.Tugarinov, V. & Kay, L. E. (2004) J. Biomol. NMR 28, 165-172. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, B. A. & Blevins, R. A. (1994) J. Biomol. NMR 4, 603-614. [DOI] [PubMed] [Google Scholar]
- 25.Orekhov, V. Y., Ibraghimov, I. V. & Billeter, M. (2001) J. Biomol. NMR 20, 49-60. [DOI] [PubMed] [Google Scholar]
- 26.Tugarinov, V., Kay, L. E., Ibraghimov, I. & Orekhov, V. Y. (2005) J. Am. Chem. Soc., in press. [DOI] [PubMed]
- 27.Clore, G. M., Gronenborn, A. M. & Bax, A. (1998) J. Magn. Reson. 113, 216-221. [DOI] [PubMed] [Google Scholar]
- 28.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289-302. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. (2003) J. Magn. Reson. 160, 65-73. [DOI] [PubMed] [Google Scholar]
- 30.Wishart, D. S. & Sykes, B. D. (1994) J. Biomol. NMR 4, 171-180. [DOI] [PubMed] [Google Scholar]
- 31.Neri, D., Szyperski, T., Otting, G., Senn, H. & Wüthrich, K. (1989) Biochemistry 28, 7510-7516. [DOI] [PubMed] [Google Scholar]
- 32.Bax, A., Vuister, G. W., Grzesiek, S., Delaglio, F., Wang, A. C., Tschudin, R. & Zhu, G. (1994) Methods Enzymol. 239, 79-105. [DOI] [PubMed] [Google Scholar]
- 33.Goto, N. K., Gardner, K. H., Mueller, G. A., Willis, R. C. & Kay, L. E. (1999) J. Biomol. NMR 13, 369-374. [DOI] [PubMed] [Google Scholar]
- 34.Dürchschlag, H. & Zipper, P. (1985) Radiat. Environ. Biophys. 24, 99-111. [DOI] [PubMed] [Google Scholar]
- 35.Remington, S. J., Wiegand, G. & Huber, R. (1982) J. Mol. Biol. 158, 111-152. [DOI] [PubMed] [Google Scholar]
- 36.Rohl, C. A., Strauss, C. E., Misura, K. M. & Baker, D. (2004) Methods Enzymol. 383, 66-93. [DOI] [PubMed] [Google Scholar]
- 37.McCaldon, P. & Argos, P. (1988) Proteins 4, 99-122. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, D., Huang, Y. J., Moseley, H. N., Xiao, R., Aramini, J., Swapna, G. V. & Montelione, G. T. (2003) Protein Sci. 12, 1232-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-55. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski, R. A., Rullman, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1998) J. Biomol. NMR 8, 477-486. [DOI] [PubMed] [Google Scholar]
- 41.Cornilescu, G., Marquardt, J., Ottiger, M. & Bax, A. (1998) J. Am. Chem. Soc. 120, 6836-6837. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.