Abstract
The NMR structure of the recombinant elk prion protein (ePrP), which represents the cellular isoform (ePrPC) in the healthy organism, is described here. As anticipated from the highly conserved amino acid sequence, ePrPC has the same global fold as other mammalian prion proteins (PrPs), with a flexibly disordered “tail” of residues 23–124 and a globular domain 125–226 with three α-helices and a short antiparallel β-sheet. However, ePrPC shows a striking local structure variation when compared with most other mammalian PrPs, in particular human, bovine, and mouse PrPC. A loop of residues 166–175, which links the β-sheet with the α2-helix and is part of a hypothetical “protein X” epitope, is outstandingly well defined, whereas this loop is disordered in the other species. Based on NMR structure determinations of two mouse PrP variants, mPrP[N174T] and mPrP[S170N,N174T], this study shows that the structured loop in ePrPC relates to these two local amino acid exchanges, so that mPrP[S170N,N174T] exactly mimics ePrPC. These results are evaluated in the context of recent reports on chronic wasting disease (CWD) in captive and free-ranging deer and elk in the U.S. and Canada, and an animal model is proposed for support of future research on CWD.
Keywords: transmissible spongiform encephalopathy, chronic wasting disease
Chronic wasting disease (CWD) is a neurological disorder in cervids that has been shown to be a transmissible spongiform encephalopathy (TSE) or “prion disease”; other prion diseases include scrapie in sheep, bovine spongiform encephalopathy (BSE), and Creutzfeldt-Jakob disease in humans (1–3). CWD was diagnosed in 1978 as a TSE in a captive mule deer (Odocoileus hemionus) (4, 5) and was diagnosed in 1981 in a free-ranging elk (Cervus elaphus nelsoni) (6). Today, CWD is known to affect captive and free-ranging elk, mule deer, and white-tailed deer (O. virginianus), and the disease seems to be endemic in areas of the western United States and Canada.
CWD is unique among TSEs by the fact that is has been studied in free-ranging species (7). The natural route of exposure appears to be oral, possibly through direct interaction between animals or through environmental contamination (8, 9). Although there is evidence for transmission to different mammalian species by intracerebral inoculation (7, 10, 11), domestic animals such as cattle, sheep, and goats are not known to be naturally susceptible to CWD (12). Compared with bovine spongiform encephalopathy (BSE), CWD transmission to cattle, goats, and laboratory animals has been reported to be inefficient, suggesting that there is a rather stringent species barrier (13–15). Cell-free conversion experiments (15) led to the prediction that TSE transmission efficiency from cervids to humans might be similar to that from cattle to humans, which is clearly of serious concern. Overall, the potential threat to livestock and the human population from the recent spread of CWD in free-ranging cervids in the U.S. and Canada is still difficult to assess, and further research on CWD is a high priority (16).
Here, we present the NMR structure of the recombinant elk prion protein (ePrP), which corresponds to the cellular form (ePrPC) found in the healthy organism (17). The global architecture of ePrPC is similar to that of other mammalian prion proteins (PrPs) solved to date (18–22). However, a loop of residues 166–175 linking a β-sheet and a helix (α2) is outstandingly precisely defined, which contrasts with pronounced structural disorder of this loop in PrPC of other species, such as mice, humans, and bovines (18–20). This molecular region has independently been identified as a probable epitope for disease-related interaction with a hypothetical, species-specific “protein X” (23).
To identify the origin of this outstanding local structural feature, which could be a consequence of either local species-specific variations of the amino acid sequence in the loop or longer-range effects from mutations in other parts of the polypeptide chain, we prepared mouse/elk hybrid PrPs. Starting with the stable fragment 121–231 of the mouse PrP (mPrP) (18), we introduced individual residues from the ePrP sequence (Fig. 1A). We then solved the NMR structures of mPrP[N174T] [the variant of recombinant mPrP(121–231) with Asn-174 replaced by Thr] and mPrP[S170N,N174T] [the variant of recombinant mPrP(121–231) with Ser-170 replaced by Asn and Asn-174 replaced by Thr], and, as a reference, we redetermined the NMR structure of mPrP(121–231) (18).
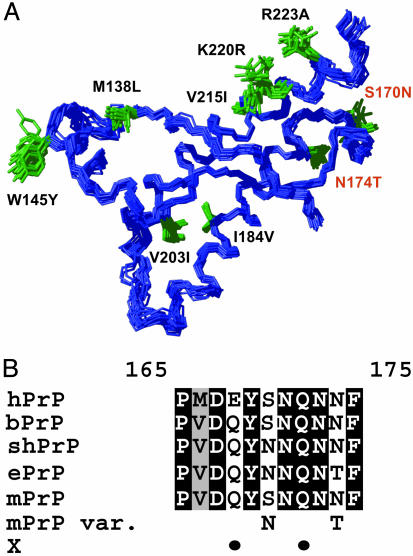
Fig. 1.
Prion protein amino acid sequence and 3D structure of the elk and other mammalian species. (A) NMR structure of ePrP(121–231) presented as a bundle of 20 energy-minimized conformers. The backbone is blue, and the amino acid side chains that are different from mPrP are shown in green and identified with black or red lettering, starting with the amino acid one-letter code for the corresponding amino acid in mPrP and followed by the sequence position and the amino acid in ePrP. The 20 conformers were superimposed for best fit of the backbone heavy atoms of residues 125–225. (B) Amino acid sequence alignment of the loop region 165–175, which connects the regular secondary structures β2 and α2, for the following species: hPrP, bPrP, shPrP, ePrP, mPrP, and “mPrP var.” (mPrP with designed amino acid replacements in positions 170 and 174). “X” indicates with black dots the positions 168 and 172 that have been proposed to be part of an epitope for interactions with a species-specific protein X (23). Identical amino acids in the five proteins are shown on a black background, conservative substitutions are shown on gray, and nonconservative replacements are on a white background.
Materials and Methods
Protein Preparation. The ePrP DNA was provided to us by Dr. A. Bossers (Central Institute for Animal Disease Control, Lelystad, The Netherlands) and the mPrP(121–231) gene was obtained from Dr. R. Zahn (Eidgenössische Technische Hochschule Zürich, Zürich). The variant proteins mPrP[S170N,N174T] and mPrP[N174T] were prepared by using the QuikChange site-directed mutagenesis protocol (Stratagene). All four genes were cloned into the vector pRSETA, and the proteins were expressed in Escherichia coli. For the purification of the recombinant proteins, we followed procedures described in ref. 24.
NMR Measurements and Structure Calculation. NMR measurements were performed at 20°C on DRX500, DRX600, DRX750, and DRX900 spectrometers (Bruker, Billerica, MA). The protein samples used were uniformly 15N-labeled ePrP(23–231), 15N-labeled and 13C, 15N-labeled ePrP(121–231), mPrP(121–231), mPrP[N174T](121–231) and mPrP[S170N, N174T](121–231). The NMR samples were either in 95% H2O/5% D2O or in 99.9% D2O and contained 5 mM sodium acetate buffer at pH 4.5, and the protein concentration was 0.6–1.0 mM. The program xeasy (25) was used for the spectral analysis.
Steady-state 15N{1H}-nuclear Overhauser effects (NOEs) of 15N-labeled ePrP(23–231) were measured following the methods of Dayie and Wagner (26). Sequence-specific resonance assignments for the proteins were obtained by using standard triple-resonance experiments (27). Distance constraints for the structure determination were obtained from 3D 13C-resolved [1H, 1H]-NOESY, 3D 15N-resolved [1H, 1H]-NOESY, and 2D [1H, 1H]-NOESY spectra recorded at a proton frequency of 750 MHz or 900 MHz with mixing times of 40 or 50 ms. For ePrP (121–231), the automatic NOE assignment module candid (28) implemented in the program dyana (29) was used for the structure determination. For the structure determinations of mPrP (121–231), mPrP[N174T], and mPrP[S70N,N174T], the programs atnos (30) and candid (28) implemented in the program dyana (29) were used. dyana (29) also was used to convert NOE intensities into upper distance constraints according to an inverse sixth power peak volume-to-distance relationship, to remove meaningless constraints, and to derive constraints for the backbone torsion angles ϕ and Ψ from the Cα chemical shift values (31, 32). The final round of dyana structure calculation was started with 100 randomized conformers. The 20 conformers with the lowest residual target function values were energy-minimized in a water shell with the program opalp (33, 34) by using the AMBER force field (35). The program molmol (36) was used to analyze the results of the structure calculations (Table 1) and to prepare the drawings of the structures.
Table 1. Input for the structure calculation and characterization of the energy-minimized NMR structures of ePrP(121–231), mPrP(121–231), and two variants of mPrP(121–231).
| Quantity* | ePrP | mPrP[S170N,N174T] | mPrP[N174T] | mPrP |
|---|---|---|---|---|
| NOE upper distance limits | 2,254 | 2,183 | 2,027 | 2,264 |
| Dihedral angle constraints | 110 | 110 | 110 | 110 |
| Residual target function value, Å2 | 1.77 ± 0.41 | 1.67 ± 0.22 | 1.68 ± 0.32 | 1.93 ± 0.20 |
| Residual distance constraint violations | ||||
| No. >0.1 Å | 29 ± 4 | 35 ± 4 | 26 ± 4 | 26 ± 4 |
| Maximum, Å | 0.13 ± 0.01 | 0.15 ± 0.01 | 0.15 ± 0.02 | 0.15 ± 0.01 |
| Residual dihedral angle constraint violations | ||||
| No. >2.5° | 0 | 0 | 0 | 1 ± 0 |
| Maximum, ° | 1.3 ± 0.4 | 1.9 ± 0.7 | 1.7 ± 0.6 | 2.6 ± 0.4 |
| amber energies, kcal/mol | ||||
| Total | –4,821 ± 57 | –4,978 ± 80 | –4,879 ± 97 | –4,911 ± 71 |
| Van der Waals | –320 ± 14 | –339 ± 13 | –351 ± 15 | –306 ± 13 |
| Electrostatic | –5,044 ± 56 | –5,598 ± 84 | –5,470 ± 89 | –5,523 ± 75 |
| rms deviation to the mean coordinates, ņ | ||||
| N, Cα, and C′ (125–228) | 0.57 ± 0.10 | 0.54 ± 0.07 | 0.67 ± 0.08 | 0.60 ± 0.09 |
| All heavy atoms (125–228) | 1.01 ± 0.09 | 0.95 ± 0.07 | 1.03 ± 0.09 | 1.00 ± 0.08 |
| N, Cα, and C′ (127–166 and 172–225) | 0.53 ± 0.09 | 0.51 ± 0.06 | 0.54 ± 0.07 | 0.46 ± 0.06 |
| All heavy atoms (127–166 and 172–225) | 0.96 ± 0.10 | 0.93 ± 0.08 | 0.92 ± 0.07 | 0.88 ± 0.06 |
Except for the top two entries, the average for the 20 conformers with the lowest residual dyana target function values and the standard deviation among them are given
The numbers in parentheses identify the polypeptide segments for which the rms deviation was calculated
Results
The full-length mature protein ePrP(23–231) is clearly divided into two domains. A flexibly disordered N-terminal tail consisting of residues 23–124 is characterized by negative heteronuclear 15N{1H}-NOEs (data not shown), and there is a well structured globular domain consisting of residues 125–226. The C-terminal pentapeptide is again flexibly disordered. This global architecture coincides with all other known mammalian PrPC structures (18–22, 37–39). In the following, we focus on the C-terminal globular domain studied in the polypeptide fragment of residues 121–231.
Resonance Assignment and NMR Structure Determination of ePrP(121–231). Complete polypeptide backbone assignments and nearly complete side-chain assignments were obtained for all residues of ePrP(121–231). The chemical shift list has been deposited with the BioMagResBank (www.bmrb.wisc.edu) with the entry code 6383. The subsequent NMR structure determination is described in the first column in Table 1.
ePrP(121–231) has the shape of a flattened sphere containing three α-helices of residues 144–155, 172–190, and 200–225 and a short, two-stranded antiparallel β-sheet of residues 128–131 and 160–163. The second strand of the β-sheet precedes a 310-helical turn made up of residues 164–170 (Figs. 1 A and 2 A).
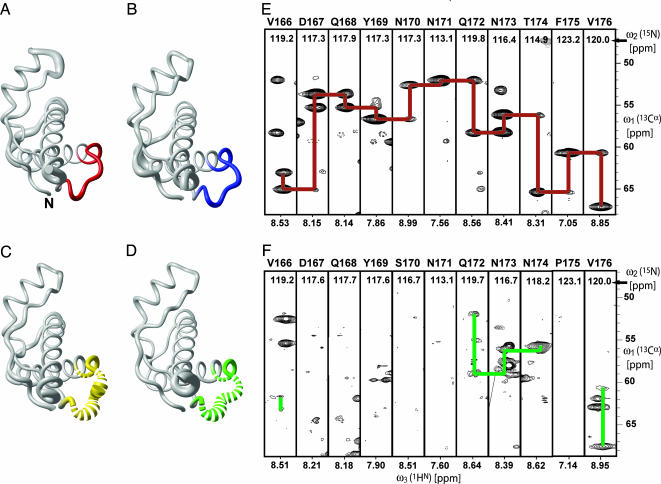
Fig. 2.
Conformation of the loop of residues 166–176 in PrPC from different species and corresponding NMR spectra. (A) Polypeptide backbone of ePrP (121–231) represented by a spline function through the Cα positions. The radius of the gray cylindrical rod is proportional to the mean global backbone displacement per residue, as evaluated after superposition for best fit of the atoms N, Cα, and C′ of the residues 125–228 in the bundle of 20 energy-minimized conformers used to represent the NMR structure (Fig. 1 A). The region comprising residues 166–175 is highlighted in red. (B) Same as A for mPrP[S170N,N174T], with the residues 166–175 in blue. (C) Same as A for mPrP[N174T]. For residues 166–175, the cylindrical rod is yellow; where no backbone resonances could be observed, the cylindrical rod is drawn as a broken line. (D) Same as C for wild-type mPrP, with residues 166–175 in green. (E) 3D HNCA spectrum of ePrP(121–231). Strips are displayed for the residues 166–176, with sequential and intraresidual Cα–Cα connectivities indicated with red lines. The strip containing Phe-175 has been drawn with lower contour levels to show the weak sequential signal. (F) Same presentation as in E for mPrP(121–231), with the observed sequential and intraresidual Cα–Cα connectivities indicated with green lines.
Unique Structural Feature in ePrP. In ePrP(121–231), the loop of residues 166–175 connecting the β-strand 2 and the helix α2 is well structured (Fig. 2A) and shows sharp resonance lines in the NMR spectra (Fig. 2E). This feature contrasts with other mammalian PrPs, such as mPrP(121–231), where this loop is disordered (Fig. 2D) and the NMR lines are missing because of conformational exchange (Fig. 2F).
NMR Structure Determination of mPrP(121–231), mPrP[N174T], and mPrP[S170N,N174T]. These structure determinations were based on complete or nearly complete (mPrP and mPrP[N174T]) resonance assignments for the polypeptide constructs with residues 121–231. They are described in the second, third, and fourth columns in Table 1.
The globular domains of the two mouse/elk hybrid PrPs have the same overall fold as ePrP(121–231) (Fig. 1A) and mPrP(121–231) (18). For mPrP[N174T], no resonances of the backbone amide groups of Asp-167, Gln-168, Tyr-169, Ser-170, Asn-171, and Phe-175 were detected in any of the NMR spectra recorded. As a result, the loop comprising residues 166–175 is poorly defined (Fig. 2C). This finding coincides with the corresponding data for mPrP (18), which were, in this paper, recorded under identical conditions as the mouse/elk hybrids to have a proper reference for the present project (Fig. 2 D and F). The absence of the NMR lines in both proteins is due to slow conformational exchange.
The protein mPrP[S170N,N174T] exhibits the same precise definition of the loop 166–175 as was seen in ePrP(121–231) (Fig. 2 A and B), and the numbers of observable NOE distance constraints involving the loop segment were comparable for both proteins. There is also a long-range effect of the amino acid replacements in mPrP[S170N,N174T] in that the helix α3 is well defined over its entire length up to residue 226.
Hydrogen Bonding Polymorphism in the Loop 166–175 of mPrP[S170N,N174T]. The nonregular secondary structure formed by residues 166–175 includes two separate, local hydrogen-bonding networks. The first part of the loop in mPrP[S170N,N174T] forms a 310-helical turn comprising residues 165–169. It is defined by two backbone hydrogen bonds, which are recognized in 19 of the 20 energy-minimized dyana conformers (Fig. 3, red broken lines).
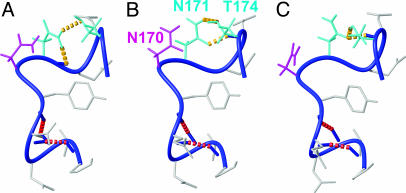
Fig. 3.
Local hydrogen-bonding polymorphisms in the loop comprising residues 165–175 in mPrP[S170N,N174T]. Three of the 20 dyana conformers used to represent the NMR structure (Table 1) are displayed. The backbone is a dark blue spline function through the Cα atoms, and side chains are gray, except for the following: The side chain (including hydrogen atoms) of N170 is shown in magenta, and the side chains of the hydrogen-bonded residues N171 and T174 are in cyan. The following hydrogen bonds in the three conformers are indicated with dashed yellow lines: Asn-171 Hδ1–Asn-171 O′ and Asn-171 Hδ2–Thr-174 Oγ (A), Asn-171 Hδ2–Thr-174 Oγ and Thr-174 HN–Asn-171 Oδ (B), and Thr-174 Hγ–Asn-171 Oδ and Thr-174 HN–Asn-171 Oδ (C). Two additional hydrogen bonds from Pro-165 O′ to Gln-168 HN and from Val-166 O′ to Tyr-169 HN (red broken lines), which define the 310-helical turn, are present in all 3 conformers [and in 19 of the 20 conformers used to represent the NMR structure (Table 1)].
A second network of hydrogen bonds is formed by the side chains of the residues Asn-171 and Thr-174. One or more hydrogen bonds between these residues prevail in 19 of the 20 dyana conformers used to represent the NMR structure (Table 1). Because of the bivalent function of the amide group of the side chain of Asn-171 as a hydrogen donor or acceptor, different combinations of hydrogen bonds are observed. Three of these local networks are displayed in Fig. 3. Within the structure defined by the NOE distance constraints, multiple different local hydrogen bonds can thus be formed, all of which are compatible with the experimental input for the structure calculation.
Discussion
In this study, we discovered a so far unique local structural feature in ePrPC, where a polypeptide loop of residues 166–175 is precisely defined. This polypeptide loop is disordered in other mammalian species.
There are two amino acid exchanges in this loop of ePrP, compared with bovine PrP (bPrP), human PrP (hPrP), and mPrP (Fig. 1B). Through studies of the variant protein mPrP[S170N,N174T], where the loop amino acid sequence of ePrP is introduced into mPrP, we then showed that the structured loop of ePrP can be related entirely to this localized exchange of two amino acids. These observations indicate intriguing follow-ups with regard to CWD and protein structural biology.
The recently registered wide propagation of CWD in the free-range elk and deer herds in the U.S. indicates ease of transmission of the disease among cervids, and possible horizontal transmission has been discussed (9, 40). In contrast, infectious transmission of CWD to other species seems to be inefficient (10, 15, 41, 42). Overall, however, when compared with bovine spongiform encephalopathy (BSE) and Creutzfeldt-Jakob disease, information of the routes of CWD propagation is sparse (9, 43). Considering further the hypothesis that the loop 166–175 forms part of a disease-related epitope for protein X, which has been proposed to include the residues 168, 172, 215, and 218 (23), it seems of considerable interest to investigate possible correlations between the well defined structure of the loop 166–175 in ePrPC and propagation of CWD. Experiments with transgenic mice expressing mPrP[S170N,N174T] would now appear to be a promising animal model for such investigations.
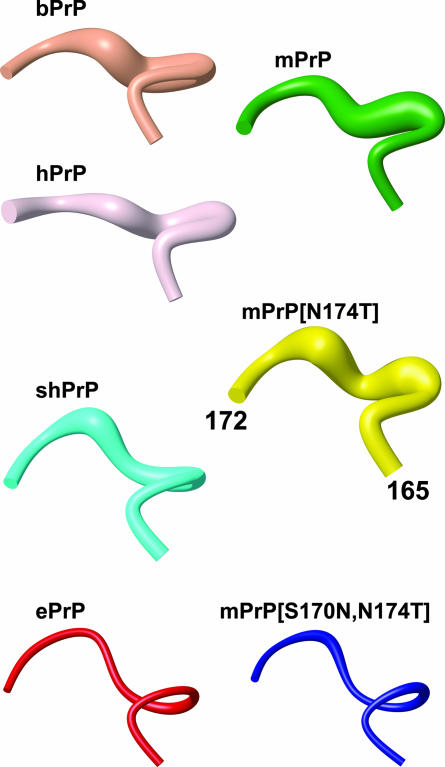
It is quite remarkable that the change from a disordered mPrP-type structure of the loop 166–175 to the precise spatial arrangement in ePrP depends on exchange of both residues 170 and 174, although these two side chains do not appear to interact in PrPC (Fig. 3). Asn-170 alone, as it occurs in Syrian hamster PrP (shPrP) (Fig. 1B), partially stabilizes the loop structure, and all backbone amide resonances of the loop have been observed in shPrP (21, 37, 38) as well as in a designed variant of human PrP, hPrP[S170N] (44). Fig. 4 shows that there is also a closer structural resemblance of the loop 166–175 in shPrP and ePrP than in mPrP and ePrP. In contrast, introduction of Thr in position 174 of mPrP has no noticeable effects on either structure or NMR spectra of mPrP, and only mPrP[S170N,N174T] shows properties identical to ePrP in the loop region (Fig. 4). Finally, there is also a so-far unexplained long-range effect from this two-amino acid substitution, in that the helix α3 is significantly better defined in mPrP[S170N,N174T] than in wild-type mPrP (18, 39).
Fig. 4.
Polypeptide segment 165–172 in the energy-minimized NMR structures of bPrP, hPrP, mPrP, shPrP, and ePrP and in mPrP[N174T] and mPrP[S170N,N174T]. The backbone is represented by a spline function through the Cα positions. The radius of the cylindrical rods is proportional to the mean backbone displacement per residue, as evaluated after superposition for best fit of the atoms N, Cα, and C′ of the residues 166–172 in the 20 energy-minimized conformers used to represent the NMR structures. For the amino acid sequences, see Fig. 1B.
Acknowledgments
We thank Mrs. C. von Schroetter for help with the protein preparation. This work was supported by the Schweizerischer Nationalfonds and the Eidgenössische Technische Hochschule Zürich through the National Center of Competence in Research, Structural Biology. S.B. is the recipient of a Fellowship from Hoffmann–La Roche AG.
Author contributions: K.W. designed research; A.D.G., S.B., D.A.L., and F.F. performed research; and A.D.G., S.B., D.A.L., and K.W. wrote the paper.
Abbreviations: PrP, prion protein; PrPC, cellular form of PrP; ePrP, elk PrP; hPrP, human PrP; bPrP, bovine PrP; mPrP, mouse PrP; shPrP, Syrian hamster PrP; CWD, chronic wasting disease; TSE, transmissible spongiform encephalopathy; NOE, nuclear Overhauser effect.
Data deposition: The coordinates of ensembles of the 20 conformers have been deposited in the Protein Data Bank, www.pdb.org {PDB ID codes 1XYW [ePrP(121–231)], 1XYX [mPrP(121–231)], 1Y15 (mPrP[N174T]), and 1Y16 (mPrP[S170N,N174T])}.
References
- 1.Weissmann, C. (1996) FEBS Lett. 389, 3-11. [DOI] [PubMed] [Google Scholar]
- 2.Griffith, J. S. (1967) Nature 215, 1043-1044. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams, E. S. & Young, S. (1980) J. Wildl. Dis. 16, 89-98. [DOI] [PubMed] [Google Scholar]
- 5.Williams, E. S. & Young, S. (1982) J. Wildl. Dis. 18, 465-471. [DOI] [PubMed] [Google Scholar]
- 6.Spraker, T. R., Miller, M. W., Williams, E. S., Getzy, D. M., Adrian, W. J., Schoonveld, G. G., Spowart, R. A., O'Rourke, K. I., Miller, J. M. & Merz, P. A. (1997) J. Wildl. Dis. 33, 1-6. [DOI] [PubMed] [Google Scholar]
- 7.Williams, E. S. & Miller, M. W. (2002) Rev. Sci. Tech. 21, 305-316. [DOI] [PubMed] [Google Scholar]
- 8.Williams, E. S. & Miller, M. W. (2003) Rev. Sci. Tech. 22, 145-156. [DOI] [PubMed] [Google Scholar]
- 9.Miller, M. W. & Williams, E. S. (2003) Nature 425, 35-36. [DOI] [PubMed] [Google Scholar]
- 10.Williams, E. S. & Young, S. (1992) Rev. Sci. Tech. 11, 551-567. [DOI] [PubMed] [Google Scholar]
- 11.Williams, E. S., Miller, M. W., Kreeger, T. J., Kahn, R. H. & Thorne, E. T. (2002) J. Wildl. Manage. 66, 551-563. [Google Scholar]
- 12.Miller, M. W. & Williams, E. S. (2004) Curr. Top. Microbiol. Immunol. 284, 193-214. [DOI] [PubMed] [Google Scholar]
- 13.Hamir, A. N., Cutlip, R. C., Miller, J. M., Williams, E. S., Stack, M. J., Miller, M. W., O'Rourke, K. I. & Chaplin, M. J. (2001) J. Vet. Diagn. Invest. 13, 91-96. [DOI] [PubMed] [Google Scholar]
- 14.Bruce, M., Chree, A., Williams, E. S. & Fraser, H. (2000) Brain Pathol. 10, 662-663. [Google Scholar]
- 15.Raymond, G. J., Bossers, A., Raymond, L. D., O'Rourke, K. I., McHolland, L. E., Bryant, P. K., Miller, M. W., Williams, E. S., Smits, M. & Caughey, B. (2000) EMBO J. 19, 4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enserink, M. (2001) Science 294, 978-979. [DOI] [PubMed] [Google Scholar]
- 17.Hornemann, S., Schorn, C. & Wüthirch, K. (2004) EMBO Rep., 5, 1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R. & Wüthrich, K. (1996) Nature 382, 180-182. [DOI] [PubMed] [Google Scholar]
- 19.López García, F., Zahn, R., Riek, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8334-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahn, R., Liu, A., Lührs, T., Riek, R., von Schroetter, C., López García, F., Billeter, M., Calzolai, L., Wider, G. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James, T. L., Liu, H., Ulyanov, N. B., Farr-Jones, S., Zhang, H., Donne, D. G., Kaneko, K., Groth, D., Mehlhorn, I., Prusiner, S. B., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 10086-10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lysek, D. A., Schorn, C., Nivon, L. G., Esteve-Moya, V., Christen, B., Calzolai, L., von Schroetter, C., Fiorito, F., Herrmann, T., Güntert, P. & Wüthrich, K. (2005) Proc. Natl. Acad. Sci. USA 102, 640-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko, K., Zulianello, L., Scott, M., Cooper, C. M., Wallace, A. C., James, T. L., Cohen, F. E. & Prusiner, S. B. (1997) Proc. Natl. Acad. Sci. USA 94, 10069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahn, R., von Schroetter, C. & Wüthrich, K. (1997) FEBS Lett. 417, 400-404. [DOI] [PubMed] [Google Scholar]
- 25.Bartels, C., Xia, T. H., Billeter, M., Güntert, P. & Wüthrich, K. (1995) J. Biomol. NMR 6, 1-10. [DOI] [PubMed] [Google Scholar]
- 26.Dayie, K. T. & Wagner, G. (1994) J. Magn. Reson. 111, 121-126. [Google Scholar]
- 27.Bax, A. & Grzesiek, S. (1993) Acc. Chem. Res. 26, 131-138. [Google Scholar]
- 28.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Mol. Biol. 319, 209-227. [DOI] [PubMed] [Google Scholar]
- 29.Güntert, P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273, 283-298. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Biomol. NMR 24, 171-189. [DOI] [PubMed] [Google Scholar]
- 31.Spera, S. & Bax, A. (1991) J. Am. Chem. Soc. 113, 5490-5492. [Google Scholar]
- 32.Luginbühl, P., Szyperski, T. & Wüthrich, K. (1995) J. Magn. Reson. 109, 229-233. [Google Scholar]
- 33.Luginbühl, P., Güntert, P., Billeter, M. & Wüthrich, K. (1996) J. Biomol. NMR 8, 136-146. [DOI] [PubMed] [Google Scholar]
- 34.Koradi, R., Billeter, M. & Güntert, P. (2000) Comput. Phys. Commun. 124, 139-147. [Google Scholar]
- 35.Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179-5197. [Google Scholar]
- 36.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-57. [DOI] [PubMed] [Google Scholar]
- 37.Donne, D. G., Viles, J. H., Groth, D., Mehlhorn, I., James, T. L., Cohen, F. E., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13452-13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, H., Farr-Jones, S., Ulyanov, N. B., Llinas, M., Marqusee, S., Groth, D., Cohen, F. E., Prusiner, S. B. & James, T. L. (1999) Biochemistry 38, 5362-5377. [DOI] [PubMed] [Google Scholar]
- 39.Riek, R., Hornemann, S., Wider, G., Glockshuber, R. & Wüthrich, K. (1997) FEBS Lett. 413, 282-288. [DOI] [PubMed] [Google Scholar]
- 40.Miller, M. W., Wild, M. A. & Williams, E. S. (1998) J. Wildl. Dis. 34, 532-538. [DOI] [PubMed] [Google Scholar]
- 41.Bartz, J. C., Marsh, R. F., McKenzie, D. I. & Aiken, J. M. (1998) Virology 251, 297-301. [DOI] [PubMed] [Google Scholar]
- 42.Sigurdson, C. J. & Miller, M. W. (2003) Br. Med. Bull. 66, 199-212. [DOI] [PubMed] [Google Scholar]
- 43.Miller, M. W., Williams, E. S., Hobbs, N. T. & Wolfe, L. L. (2004) Emerg. Infect. Dis. 10, 1003-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calzolai, L., Lysek, D. A., Güntert, P., von Schroetter, C., Riek, R., Zahn, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8340-8345. [DOI] [PMC free article] [PubMed] [Google Scholar]