Abstract
Hepatitis B virus (HBV) causes acute and chronic hepatitis and hepatocellular carcinoma. Although a preventive vaccine is available, the therapeutic options for chronically infected patients are limited. It has been shown that RNA interference can prevent HBV gene expression and replication in vivo when HBV expression vectors are delivered simultaneously with small interfering RNA (siRNA) or siRNA expression constructs. However, the therapeutic potential of siRNAs to interrupt ongoing HBV replication in vivo has not been established. Here, we show that expression of HBV-specific siRNAs in the liver of HBV transgenic mice by recombinant adenoviruses can suppress preexisting HBV gene expression and replication to almost undetectable levels for at least 26 days. These results demonstrate that efficiently delivered siRNAs should be able to silence HBV in chronically infected patients.
Keywords: adenovirus vector, RNA interference
Chronic hepatitis B virus (HBV) infection causes >1 million deaths each year due to cirrhosis of the liver and hepatocellular carcinoma (www.cdc.gov/hepatitis and www.who.int/mediacentre/factsheets/fs204/en). Hence, there is a need for alternative ways to treat this persistent infection. RNA interference (RNAi) has rapidly emerged as a technology for regulating mammalian gene expression because it provides a means of sequence-directed degradation of specific RNAs (1–8). In the case of HBV, published in vivo hydrodynamic transfection studies have shown that simultaneous delivery of HBV expression plasmids and HBV-specific small interfering RNAs (siRNAs) (or siRNA-expressing constructs) to the mouse liver can prevent the induction of HBV gene expression and replication (9–11). To expand on those studies, we have examined the therapeutic potential of RNAi for the treatment of chronic HBV infection where ongoing viral gene expression and replication are established in the liver before siRNA delivery.
Several variables that have not been previously addressed could affect the utility of RNAi for the treatment of an ongoing HBV infection. First, in the context of an established infection, viral RNAs may be protected within complexes or nucleocapsid structures as has been reported for hepatitis D virus (1), Rous sarcoma virus (12), and respiratory syncytial virus (RSV) (13). This is particularly relevant to HBV, which replicates in capsids after encapsidation of the viral pregenomic RNA. A second variable is how efficiently preexisting viral RNA can be reduced and to what extent RNA suppression can block viral DNA replication. Finally, established viral gene expression may allow for the induction of viral RNAi-defense mechanisms as reported for plant viruses (14) and flockhouse nodavirus (15).
It has been shown that RNAi is capable of reducing mammalian gene expression in vivo. Genes such as fas (16), caspase-8 (17), agouti-related peptide (18), tyrosine hydroxylase (19), and ras (20, 21) have been targeted for degradation in mice, and corresponding changes in phenotype were observed. Yet, the actual degree of gene suppression achieved in each case varied, sometimes with as little as a 36% reduction in mRNA. Although this magnitude of suppression was sufficient to reduce the effects of the endogenous genes being studied, the degree to which viral RNA must be suppressed to disrupt viral replication in vivo is not known. In a recent study, it was demonstrated that influenza virus titers could be reduced 2–3 logarithms in the lungs of mice if 120 μg of virus-specific siRNA was delivered 5 h after infection. Interestingly, however, <1 logarithmic decrease was observed when the same siRNA was delivered 24 h after infection (22).
In this study, we identified HBV RNAi target sequences capable of mediating the degradation of HBV transcripts and used recombinant adenovirus delivery to test the ability of these target sequences to inhibit constitutive HBV replication in vivo in the liver of HBV transgenic mice. The data show that ongoing HBV replication in vivo can be cleared by RNAi-targeted suppression of viral RNA for at least 26 days. Importantly, the potent, long-lasting inhibition of HBV gene expression and replication achieved in this model suggests that an RNAi-based treatment strategy could be successfully used clinically in patients with chronic HBV infection.
Methods
Cell Lines. Huh7 GFP–HBV cells were created by transfecting Huh7 hepatocytes (from the American Type Culture Collection) with a selectable SV40-driven expression vector (Invitrogen) containing a GFP–HBV fusion construct. Zeocin-resistant cells constitutively express GFP from a fusion transcript containing the GFP ORF followed by HBV sequences 2630–0-1986 (Fig. 1a). Cells were maintained in DMEM supplemented with 10% FCS and penicillin–streptomycin–glutamine (Invitrogen).
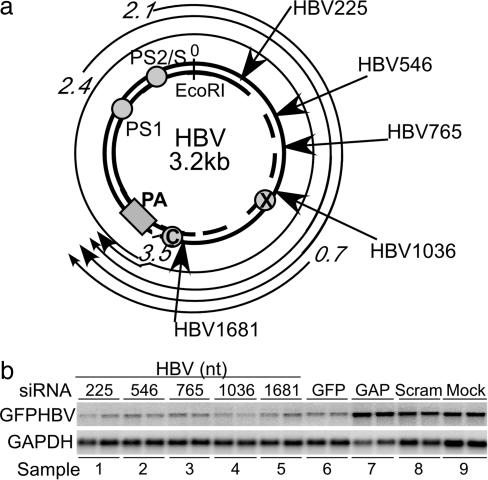
Fig. 1.
Inhibition of GFP–HBV RNA after transient siRNA transfection in vitro. (a) Map of the HBV genome, with location of siRNA target sequences indicated. (HBV-specific siRNA numbers refer to beginning nucleotide of siRNA relative to the unique viral EcoRI site.) (b) Huh7 GFP–HBV hepatocytes were transfected in duplicate at a concentration of 285 nM of the indicated siRNAs. Two days after transfection, total RNA was isolated for Northern blot analysis. Blots were probed for GFP and GAPDH.
HBVMet hepatocytes were derived from HBV transgenic mice as described in ref. 23. Cells were plated on collagen I-coated dishes (Becton Dickinson) in RPMI medium 1640 supplemented with 10% FCS (Invitrogen), 55 ng/ml EGF (Becton Dickinson), 16 ng/ml insulin-like growth factor II (Calbiochem), 10 μg/ml insulin (Sigma), and penicillin–streptomycin–glutamine (Invitrogen), as described in ref. 23.
Mice. HBV transgenic mouse lineages 1.3.32 and 1.3.46 are described in ref. 24. Both lineages express and replicate HBV in the liver from an integrated greater-than-genome-length transgene at levels comparable to that seen during HBV infection. Inbred 1.3.32 C57BL/6 mice were mated with B10D2 mice to produce the F1 hybrids that were used. To produce IFN-α/β and IFN-γ signaling-deficient HBV transgenic mice, 1.3.46 B10D2 mice were backcrossed onto an IFN-γ-deficient genetic background (25), and those mice were then crossed onto an IFN-α/β receptor-deficient genetic background (26). IFN-α/β receptor-deficient mice and IFN-γ-/- mice were provided by Timothy Stewart (Genentech) and Michel Aguet (University of Zurich, Zurich), respectively.
Mice were matched by serum hepatitis B e antigen (HBeAg) levels before experimental manipulation with the EBK 125I RIA Kit (DiaSorin, Stillwater, MN). Mice were intravenously injected by means of the tail vein with adenovirus diluted in saline for a final volume of 200 μl and were bled retroorbitally at indicated time points for serum analysis.
Animal protocols were performed in accordance with the National Institutes of Health and The Scripps Research Institute Guidelines for Animal Care. Animals were housed in pathogen-free rooms under strict barrier conditions.
siRNAs. HBV target sequences were chosen in regions overlapping the viral 3.5-kb, 2.4-kb, and 2.1-kb RNAs, according to the parameters indicated on the siRNA Target Finder web site (www.ambion.com). RNAs were synthesized by using the Silencer siRNA Construction Kit (Ambion, Austin, TX). Scrambled negative-control and GFP positive-control siRNAs were chemically synthesized (Qiagen, Valencia, CA). RNAs were transfected at 285 nM by using oligofectamine (Invitrogen).
Mouse Polymerase III Cloning. Primers were designed to clone the mouse U6 promoter by one-step RT-PCR (Qiagen), and the PCR product was ligated into the pGEMTEasy vector (Promega). Clones were verified by sequence analysis (The Scripps Research Institute, MEM DNA core facility). Individual short hairpin RNAs (shRNAs) were cloned at the +1 position behind the mouse PolIII promoter analogous to the pSUPER human PolIII vector (27).
Recombinant Adenoviruses. PolIII-siRNA cassettes were cloned into the AdEasy shuttle vector pShuttle (Quantum Biotechnologies, Montreal), in which the insert is flanked by portions of the adenovirus genome (28). PolIII-siRNA shuttle constructs were cotransfected into 293 cells with a plasmid containing the rest of the adenoviral genome (pacAd5 9.2–100), including regions of homology to the pShuttle vector to allow for homologous recombination (B. Davidson, Gene Transfer Vector Core, University of Iowa, Iowa City). Because neither plasmid is able to produce adenovirus independently, only DNAs that have recombined generate virus (29). Virus stocks were amplified in 293 cells, purified over two CsCl gradients, and dialyzed with PBS/3% glucose according to the shuttle vector manufacturer's protocols.
Hepatitis B Surface Antigen (HBsAg) and HBeAg ELISA. Secretion of HBsAg into the blood was measured by quantitative sandwich ELISA (Abbott). HBeAg in the blood was measured by sandwich ELISA (International Immuno-Diagnostics, Foster City, CA) and quantified relative to a standard curve of serial dilutions of recombinant HBeAg (Fitzgerald Industries International, Concord, MA).
Serum Alanine Aminotransaminase (sALT). sALT activity was assayed with ThermoTrace Infinity ALT Reagent (ThermoDMA, Louisville, CO) by using a SpectraMax Plus spectrophotometer (Molecular Devices). Values are given as units/liter; units/liter × 16.67 × 103 = μkat/liter.
RNA Analysis. Total RNA was isolated by the guanidine thiocyanate method (30). For Northern blot analysis, RNA was resolved in formaldehyde agarose gels and transferred to nytran nylon membranes (Schleicher & Schuell). Transcripts were detected by hybridization with 32P-labeled DNA probes and then analyzed by using a storage phosphor system (Cyclone, Packard). For RNase protection assay (RPA), RNA was hybridized to 32P-labeled RNA probes in solution with the mirVana miRNA Detection Kit (Ambion). RNA probes were generated by using the mirVana microRNA Probe Construction Kit (Ambion).
DNA Analysis. Tissue was digested overnight in lysis buffer (50 mM Tris·HCL, pH 8/20 mM EDTA/1% SDS/1 mg/ml proteinase K), and DNA was phenol-chloroform-extracted. Twenty micrograms of DNA was analyzed by Southern blot hybridization after HindIII digestion.
Immunohistochemical Hepatitis B Core Antigen (HBcAg) Staining. HBV core protein was detected by immunohistochemical staining of 10% zinc-buffered formalin-fixed tissue as described in ref. 24.
Results
Identification of Effective HBV siRNA Targets. A series of HBV-specific siRNAs were tested by transient transfection in a human hepatocyte cell line constitutively expressing a stable GFP–HBV fusion transcript (Fig. 1a). Duplicate cultures were transfected with the indicated siRNA, and total RNA was extracted for Northern blot analysis 2 days posttransfection (Fig. 1b).
When normalized for loading differences by using GAPDH mRNA levels, HBV transcript levels were reduced an average of 10-fold by all of the HBV-specific siRNAs (Fig. 1b, lanes 1–5) relative to the scrambled and mock siRNA transfection controls (Fig. 1b, lanes 8–9). Similarly, a commercially purchased GFP-specific siRNA control induced a 7.5-fold reduction in the level of the GFP–HBV transcript (Fig. 1b, lane 6), whereas transfection of a GAPDH-specific siRNA did not affect the level of the GFP–HBV mRNA (Fig. 1b, lane 7). Because all HBV-specific siRNAs tested reduced GFP–HBV transcripts levels to the same degree or more than the GFP-specific siRNA control, we pursued all these target options for use as expressed shRNA structures in future experiments.
Clearance of HBV RNA by Adenovirus-Mediated Delivery of shRNAs in Vitro. As a means of producing shRNAs intracellularly, we cloned the mouse U6 RNA Polymerase III (mPolIII) promoter to express short hairpin versions of our HBV target sequences, as well as the control scrambled sequence (27). To efficiently deliver these mPolIII-driven shRNAs to hepatocytes, we transferred our expression cassettes into an adenovirus shuttle vector and produced recombinant adenoviruses.
To test the effectiveness of the resulting adenovirus-expressed shRNAs, we infected duplicate cultures of a highly differentiated HBV-positive hepatocyte cell line derived from the 1.3.46 HBV transgenic mice (23). Total RNA was extracted from cells 72 h postinfection to determine the level of HBV RNA. The results for the two best-performing HBV-specific shRNA adenoviruses are shown (Fig. 2).
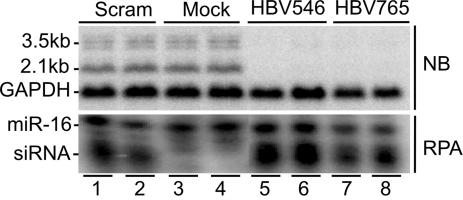
Fig. 2.
Clearance of HBV RNA after adenovirus delivery of shRNAs in vitro. Duplicate cultures of HBVMet hepatocytes were infected at a multiplicity of infection of 100 with adenoviruses expressing the indicated shRNA. Total cellular RNA was isolated 72 h postinfection for analysis. (Upper) Northern blot of HBV and GAPDH RNA. (Lower) RPA detection of cellular miR-16 RNA and the indicated adenovirus-expressed siRNA.
By Northern blot, virtually all detectable HBV RNA was eliminated from cells infected with adenoviruses expressing the HBV-specific shRNAs, shHBV546 or shHBV765 (Fig. 2 Upper, lanes 5 and 6 and 7 and 8, respectively), and this effect lasted for 9 days, at which time HBV RNA levels began to rise (data not shown). Cells infected with the adenovirus expressing the scrambled shRNA, shSCRAM, had HBV RNA levels comparable to those of mock infected cells (Fig. 2 Upper, lanes 3 and 4 vs. 1 and 2). Similarly, an adenovirus expressing a GFP reporter did not reduce HBV RNA levels in these cells (data not shown).
Although adenovirus infection itself did not affect HBV RNA levels (Fig. 2, lanes 1 and 2), it was theoretically possible that activation of the RNAi–RNA-induced silencing complex pathway, which would only have occurred in cells expressing the HBV-specific shRNAs, may have resulted in IFN induction that could have led to cytokine-mediated inhibition of HBV in those cultures. Hence, to determine whether the reduction of HBV RNA could have been due to induction of IFN in shHBV-adenovirus-infected cells, we performed an RPA for the presence of a panel of cytokines, including TNF-α, IFN-γ, and 2′5′-oligoadenylate synthetase as a marker of IFN-α/β expression. None of the cytokines previously shown to inhibit HBV gene expression was induced in any of the adenovirus-infected cells, supporting the conclusion that the reduction in HBV RNA observed was specifically mediated by RNAi (data not shown). Additionally, RPA analysis verified expression of the different siRNAs in the adenovirus-infected cells (Fig. 2 Lower). Detection of the cellular miR-16 RNA served as a loading control for the siRNA RPA samples.
Inhibition of Preexisting HBV Gene Expression in the Liver of Mice. To determine whether these shRNAs could inhibit ongoing HBV gene expression in vivo, we infected HBV 1.3.32 transgenic mice with 2 × 109 plaque-forming units (pfus) of recombinant adenovirus expressing shHBV546, shHBV765, or shSCRAM and analyzed HBV gene expression (Fig. 3).
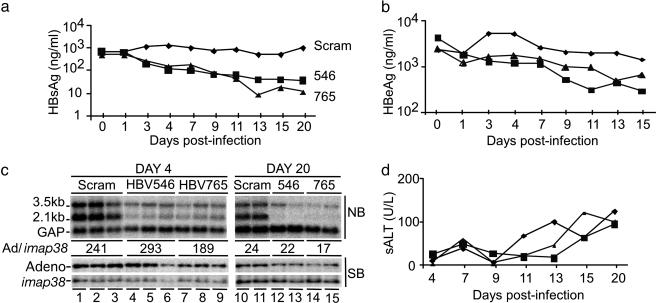
Fig. 3.
Inhibition of HBV gene expression in vivo after adenovirus delivery of shRNAs. (a, b, and d) HBV transgenic mice were infected with 2 × 109 pfu of recombinant adenovirus expressing the HBV546 (▪), HBV765 (▴), or scrambled (♦) sRNAs. (a) Serum HBsAg was measured by quantitative ELISA, and the average ng/ml serum is plotted for each group at the indicated times postadenovirus delivery. (b) Serum HBeAg was measured by quantitative ELISA, and the average ng/ml serum is plotted for each group at the indicated times postadenovirus delivery. (c) (Upper) Northern blot analysis of HBV and GAPDH liver RNA at days 4 and 20 postinfection. (Lower) Southern blot analysis of adenovirus DNA relative to the single-copy genomic imap38 gene in the liver at days 4 and 20 postinfection. (d) Average sALT plotted for each group at the indicated times postadenovirus delivery.
Levels of HBsAg and HBeAg were measured in the serum of mice as a downstream indicator of HBV gene expression. By day 4 postinfection, mice receiving either the shHBV546- or the shHBV765-expressing adenovirus had 5- to 6-fold less HBsAg in their serum relative to mice infected with the shSCRAM control virus. HBsAg levels continued to decline in those mice until ≈day 13 and subsequently remained suppressed at up to 100-fold reduced levels (Fig. 3a). At day 4, the average level of HBeAg in the mice receiving either the shHBV546- or the shHBV765-expressing adenovirus had decreased 3- to 4-fold relative to mice infected with the shSCRAM virus, with the greatest reduction in HBeAg being observed at day 11 (6.5-fold) and day 13 (4-fold), respectively (Fig. 3b).
At days 4 and 20 postinfection, mice from each group were killed to examine HBV RNA and adenovirus DNA levels in the liver (Fig. 3c). Corresponding with the serum antigen results, Northern blot analysis showed an average 5-fold reduction of HBV RNA 4 days postdelivery of shHBV546 (lanes 4–6) or shHBV765 (lanes 7–9) compared with mice receiving shSCRAM (lanes 1–3). By day 20, the inhibition of total HBV RNA had increased to 9-fold and 7-fold in shHBV546 and shHBV765 adenovirus infections, respectively (Fig. 3c, lanes 12–15 vs.10 and 11). Hence, in the context of ongoing viral replication, HBV viral RNAs can be targeted for inhibition by shRNAs. However, consistent with the different magnitude of suppression observed for serum HBsAg and HBeAg, both HBV-specific shRNAs reduced the 2.1-kb envelope mRNA to a greater extent than the 3.5-kb transcript, despite the fact that the target sequences are present in both RNAs. Specifically, by day 20 postinfection, the 2.1-kb envelope mRNA was reduced ≈50-fold, whereas the 3.50-kb viral RNA was reduced only 4- to 5-fold.
To confirm that the titers of the different adenovirus infections were comparable, we performed Southern blot analysis to determine the level of adenovirus DNA in the liver. Detection of the single-copy genomic mouse imap38 gene served as a DNA loading control (31). When quantified relative to imap38, the level of adenovirus DNA in the livers of mice infected with the different viruses was found to be roughly equivalent at day 4 postinfection (Fig. 3c). Interestingly, despite 90% clearance of the adenovirus template by day 20 (Fig. 3c), sALTs remained relatively low throughout infection with all three viruses (Fig. 3d).
Inhibition of Ongoing HBV Replication in the Liver of Mice. We were interested to determine whether RNAi-mediated degradation of viral RNA is sufficient to disrupt the viral life cycle and inhibit HBV DNA replication. However, unlike HBV RNA, which is unaffected by adenovirus infection in HBV transgenic mice (Fig. 3) (32, 33), in vivo adenovirus infection does induce interferons that clear HBV DNA from the liver (32, 33). Therefore, we repeated our shRNA-adenovirus delivery experiment in HBV transgenic mice that are genetically deficient for the expression of IFN-γ and the IFN-α/β receptor. In the absence of signaling by these cytokines, adenovirus infection did not induce cytokine-mediated inhibition of HBV DNA replication (Fig. 4a, lanes 5–9, 13–15, and 19 and 20), thus allowing us to determine the extent to which HBV replication is inhibited by the shRNAs delivered. Groups of IFN-α/β/γ signaling-deficient HBV transgenic mice were infected with 5 × 109 pfus of recombinant adenovirus expressing shHBV765 or shSCRAM, and mice were analyzed at days 4, 17, and 26 postinfection (Fig. 4).
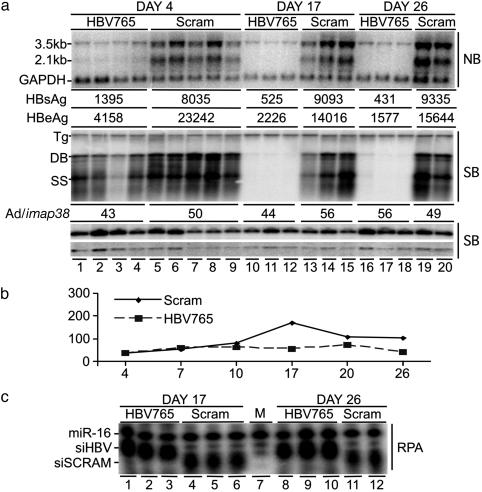
Fig. 4.
Inhibition of HBV DNA replication in vivo after adenovirus delivery of shRNAs. HBV transgenic mice deficient in IFN-α,-β, and -γ signaling were infected with 5 × 109 pfu of recombinant adenovirus expressing the indicated shRNA. Mice from both groups were analyzed at days 4, 17, and 26 postinfection. (a)(Top) Total liver RNA was analyzed by Northern blot analysis for HBV and GAPDH. Serum HBsAg and HBeAg were measured by quantitative ELISA and are displayed as the average ng/ml serum for each group. (Middle and Bottom) Total liver DNA was isolated for HBV Southern blot analysis. The blot was subsequently hybridized for adenovirus and imap38. Relative adenovirus DNA levels were calculated as a ratio of adenovirus/imap38. Group averages are shown. (b) Average sALTs plotted for each group at the indicated times postadenovirus delivery. (c) RPA detection of siRNA expression at days 17 and 26 postadenovirus infection. (Relative to the HBV765 siRNA probe, the cellular miR-16 probe was designed to be 2 nt longer, and the siSCRAM probe was designed to be 1 nt shorter.)
Similar to the previous experiment, by day 4 postinfection, both serum HBsAg and HBeAg levels were 5.5-fold lower in mice infected with the shHBV765 adenovirus compared with mice that received the control shSCRAM adenovirus (Fig. 4a). Likewise, HBV RNA levels were 8-fold lower in shHBV765 adenovirus-infected mice (Fig. 4a Top, lanes 1–4 vs. 5–9). As seen in the wild-type HBV transgenic mice, RNAi-mediated suppression of viral gene expression increased over time such that by day 17 postinfection, the majority of detectable HBV RNA had been eliminated from the liver, except for a residual 10% of the 3.5-kb species (Fig. 4a, lanes 10–12), and this pattern of HBV RNA inhibition was maintained through day 26 post-delivery (Fig. 4a, lanes 16–18).
Southern blot analysis of HBV DNA showed that at day 4, before HBV RNA was completely cleared from the liver of shHBV765-adenovirus-infected mice, viral DNA replicative intermediates were reduced only 50% compared with shSCRAM-adenoviruses-infected mice (Fig. 4a Middle, lanes 1–4 vs. 5–9). However, by day 17, HBV replicative intermediates were virtually undetectable in the livers of mice that received the shHBV765 RNA (Fig. 4a, lanes 10–12), and this inhibition was maintained through day 26 postinfection (Fig. 4a, lanes 16–18).
Again, Southern blot detection of adenovirus DNA quantified relative to the level of the single-copy genomic imap38 gene verified that delivery of the shHBV765 and shSCRAM adenoviruses was equivalent (Fig. 4a). Consistent with the low sALTs detected, adenovirus DNA levels did not decrease during the 26-day infection (Fig. 4b). Additionally, RPA analysis confirmed the presence of both shHBV765 and shSCRAM RNA at day 17 (Fig. 4c, lanes 1–3 and 4–6) and day 26 (Fig. 4c, lanes 7–9 and 10 and 11).
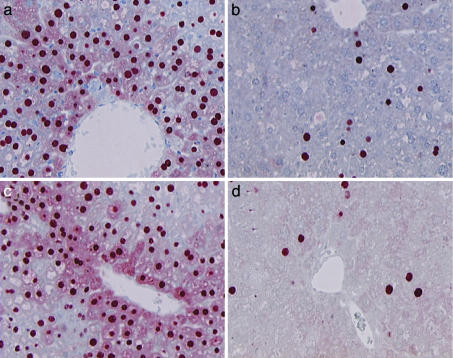
HBsAg is secreted too rapidly for intracellular detection, but immunohistochemical staining for 3.5-kb-derived gene product HBcAg in liver tissue from shHBV765- and shSCRAM-adenovirus-infected mice allowed us to directly observe HBV clearance from hepatocytes (Fig. 5). Like serum antigen levels, staining for HBcAg was greatly reduced in the hepatocytes of shHBV765-infected mice on day 17 (Fig. 5b) and day 26 (Fig. 5d) compared with shSCRAM-infected mice (Fig. 5a and 5c). Consistent with the presence of cytoplasmic HBcAg being an indicator of active viral replication, cytoplasmic HBcAg staining in the hepatocytes of shHBV765-infected mice on day 17 or 26 (Fig. 5 b and d) could not be detected.
Fig. 5.
Immunohistochemical analysis of HBcAg in vivo. Pieces of liver from the animals described in Fig. 4 were fixed overnight in 10% buffered zinc formalin and were sliced for staining of HBcAg. Representative fields are shown. (a and b) siSCRAM and siHBV765 adenovirus infection, respectively, on day 17. (c and d) siSCRAM and siHBV765 adenovirus infection, respectively, on day 26.
Discussion
In this study, we show that the majority of preexisting HBV transcripts in the liver during ongoing viral replication are susceptible to RNAi inhibition and that clearance of viral transcripts is sufficient to abolish HBV DNA replication. Within 4 days after delivery of HBV-specific shRNAs to the liver, steady-state levels of HBV gene expression were reduced. By day 17 after a 5 × 109-pfu dose, only evidence of a residual 3.5-kb viral RNA species was detected. As a result of HBV RNA depletion, viral replication had stopped by day 17 and had not begun to return by the end of our 26-day experiment. The magnitude and duration of suppression attained emphasizes the potential this approach may have for treating chronic HBV and other persistent liver infections, such as hepatitis C virus (34).
Notably, there was a relatively stable pool of viral 3.5-kb RNA that persisted in the liver despite the presence of HBV-specific shRNAs. Because the sequences targeted by the shRNAs in this study are present in the 3.5-kb RNA, these data suggest that there is a population of HBV RNA protected from direct RNAi-mediated degradation. Two distinct 3.5-kb RNAs are transcribed from the HBV genome, the pregenomic RNA (pgRNA) and the 31-nt-longer HBeAg mRNA (35). Although our observations initially may appear to suggest that some HBV pgRNA is protected inside viral capsids, this is unlikely because encapsidated pgRNA present in the cell would be expected to be rapidly degraded as it is reverse transcribed into DNA. On the other hand, the observation that shHBV-mediated suppression of serum HBeAg was not as dramatic as the suppression of serum HBsAg suggests that at least some of the residual 3.5-kb RNA is the 31-nt-longer HBeAg mRNA species. Because this pattern of inhibition was observed with multiple shRNAs, the RNAi resistance of this subset does not appear to be due to secondary structure masking of a particular target sequences; rather, it appears to be due to a more global protection of a subset of the HBeAg mRNA within RNA–protein complexes, perhaps polyribosome complexes on the rough endoplasmic reticulum. Further investigation will be needed to confirm whether the HBeAg 3.5-kb transcript is less sensitive to RNAi-mediated clearance and why.
Importantly, delivering shRNA effectors to a high percentage of hepatocytes in the liver may not be necessary to silence chronic HBV infection. The hypothesis that viral clearance may only initially require viral gene suppression in a limited number of cells is based on the fact that patients chronically infected with HBV maintain an HBV-specific cytotoxic T lymphocyte (CTL) response throughout the infection. Although the immune response of chronic carriers is not strong enough to clear the virus, it does continue to destroy cells expressing HBV antigens, and this results in hepatocyte turnover (36). In this milieu, any cells in which HBV has been silenced would have a significant survival advantage and be selected over time to repopulate the liver. This hypothesis can be tested by adoptive transfer of HBV-specific CTLs into HBV transgenic mice after infection with siRNA-expressing lentiviral vectors that stably transduced only a limited number of hepatocytes.
In terms of the potential for viral clearance, RNAi-based therapies would be predicted to have significant advantages over the currently licensed HBV treatments. Besides the ability to target multiple conserved viral elements, minimizing the chances of viral escape, RNAi-mediated viral suppression is fundamentally distinct in that it inhibits viral gene expression. The weak host immune response associated with chronic HBV infection is rarely sufficient to achieve sustained viral clearance, even after long-term suppression of HBV DNA replication with lamivudine (nucleoside analogue) or adefovir (nucleotide analogue). This is probably because these treatments do not reduce the high viral antigenemia that is thought to blunt the T cell response in chronically infected patients and, hence, help HBV avoid immune clearance (36). In contrast, the reduction in viral antigen levels attained after RNAi-mediated degradation of viral RNA may relieve the negative impact of chronic antigen stimulation on the T cell response and thus allow for recovery of that response. Alternatively, the use of RNAi to temporarily clear overwhelming levels of HBV antigen may provide a treatment window during which the host could mount a more vigorous response than it otherwise would to a therapeutic vaccine. In either case, the strengthened immune response would be more likely to clear HBV after treatment. To further investigate this possibility, we are currently using HBV transgenic mice to test whether therapeutic immunization can break tolerance if administered during a period when endogenous HBV is silenced by RNAi.
In summary, we have demonstrated that adenovirus-mediated delivery of shRNAs to the liver of mice is an extremely effective and long-lasting approach to suppress ongoing viral gene expression and replication in vivo. Although this approach should prove to be very useful as a research tool for transient gene suppression in vivo, alternative delivery options for clinical use still need to be examined. Nonetheless, the data presented here provide the justification needed to pursue the challenge of developing RNAi as an effective treatment for chronic HBV and hepatitis C infection.
Acknowledgments
We thank Sadie Medrano for maintenance and screening of rodent colonies and Dr. Chiaho Shih for critical review of the manuscript. This work was supported by National Institutes of Health Grant CA40489 (to F.V.C.). This is manuscript 16684-MEM from The Scripps Research Institute.
Author contributions: S.L.U. and F.V.C. designed research; S.L.U., B.B., and A.A. performed research; S.L.U. and F.V.C. analyzed data; S.L.U. wrote the paper; and F.V.C. obtained funding to support the research.
Abbreviations: HBV, hepatitis B virus; siRNA, small interfering RNA; RNAi, RNA interference; HBcAg, HBV core antigen; HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e antigen; sALT, serum alanine aminotransaminase; shRNA, short hairpin RNA; RPA, RNase protection assay; pfu, plaque-forming unit.
References
- 1.Chang, J. & Taylor, J. (2003) J. Virol. 77, 9728-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanda, T., Kusov, Y., Yokosuka, O. & Gauss-Muller, V. (2004) Biochem. Biophys. Res. Commun. 318, 341-345. [DOI] [PubMed] [Google Scholar]
- 3.Wiebusch, L., Truss, M. & Hagemeier, C. (2004) J. Gen. Virol. 85, 179-184. [DOI] [PubMed] [Google Scholar]
- 4.Ying, C., De Clercq, E. & Neyts, J. (2003) Biochem. Biophys. Res. Commun. 309, 482-484. [DOI] [PubMed] [Google Scholar]
- 5.Hamasaki, K., Nakao, K., Matsumoto, K., Ichikawa, T., Ishikawa, H. & Eguchi, K. (2003) FEBS Lett. 543, 51-54. [DOI] [PubMed] [Google Scholar]
- 6.Shlomai, A. & Shaul, Y. (2003) Hepatology 37, 764-770. [DOI] [PubMed] [Google Scholar]
- 7.Arias, C., Dector, M., Segovia, L., Lopez, T., Camacho, M., Isa, P., Espinosa, R. & Lopez, S. (2004) Virus Res. 102, 43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joost Haasnoot, P., Cupac, D. & Berkhout, B. (2003) J. Biomed. Sci. 10, 607-616. [DOI] [PubMed] [Google Scholar]
- 9.Giladi, H., Ketzinel-Gilad, M., Rivkin, L., Felig, Y., Nussbaum, O. & Galun, E. (2003) Mol. Ther. 8, 769-767. [DOI] [PubMed] [Google Scholar]
- 10.McCaffrey, A., Nakai, H., Pandey, K., Huang, Z., Salazar, F., Xu, H., Wieland, S., Marion, P. & Kay, M. (2003) Nat. Biotechnol. 21, 639-644. [DOI] [PubMed] [Google Scholar]
- 11.Klein, C., Bock, C., Wedemeyer, H., Wustefeld, T., Locarnini, S., Dienes, H., Kubicka, S., Manns, M. & Trautwein, C. (2003) Gastroenterology 125, 9-18. [DOI] [PubMed] [Google Scholar]
- 12.Hu, W., Myers, C., Kilzer, J., Pfaff, S. & Bushman, F. (2002) Curr. Biol. 12, 1301-1311. [DOI] [PubMed] [Google Scholar]
- 13.Bitko, V. & Barik, S. (2001) BMC Microbiol. 1, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, W. & Ding, S. (2001) Curr. Opin. Biotechnol. 12, 150-154. [DOI] [PubMed] [Google Scholar]
- 15.Li, H., Li, W. X. & Ding, S. W. (2002) Science 296, 1319-1321. [DOI] [PubMed] [Google Scholar]
- 16.Song, E., Lee, S. K., Wang, J., Ince, N., Ouyang, N., Min, J., Chen, J., Shankar, P. & Lieberman, J. (2003) Nat. Med. 9, 347-351. [DOI] [PubMed] [Google Scholar]
- 17.Zender, L., Hutker, S., Liedtke, C., Tillmann, H., Zender, S., Mundt, B., Waltemathe, M., Gosling, T., Flemming, P., Malek, N., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 7797-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makimura, H., Mizuno, T., Mastaitis, J., Agami, R. & Mobbs, C. (2002) BMC Neurosci. 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hommel, J., Sears, R., Georgescu, D., Simmons, D. & DiLeone, R. (2003) Nat. Med. 9, 1539-1544. [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp, T., Bernards, R. & Agami, R. (2002) Cancer Cell 2, 243-247. [DOI] [PubMed] [Google Scholar]
- 21.Yang, G., Thompson, J., Fang, B. & Liu, J. (2003) Oncogene 22, 5694-5701. [DOI] [PubMed] [Google Scholar]
- 22.Ge, Q., Filip, L., Bai, A., Nguyen, T., Eisen, H. & Chen, J. (2004) Proc. Natl. Acad. Sci. USA 101, 8676-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasquetto, V., Wieland, S. F., Uprichard, S. L., Tripodi, M. & Chisari, F. V. (2002) J. Virol. 76, 5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guidotti, L. G., Matzke, B., Schaller, H. & Chisari, F. V. (1995) J. Virol. 69, 6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalton, D. K., Pitts-Meek, S., Keshav, S., Figari, I. S., Bradley, A. & Stewart, T. A. (1993) Science 259, 1739-1742. [DOI] [PubMed] [Google Scholar]
- 26.van den Broek, M., Muller, U., Huang, S., Zinkernagel, R. & Aguet, M. (1995) Immunol. Rev. 148, 5-18. [DOI] [PubMed] [Google Scholar]
- 27.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 28.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson, R., Haskell, R., Xia, H., Roessler, B. & Davidson, B. (2000) Gene Ther. 7, 1034-1038. [DOI] [PubMed] [Google Scholar]
- 30.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 31.Krucken, J., Stamm, O., Schmitt-Wrede, H., Mincheva, A., Lichter, P. & Wunderlich, F. (1999) J. Biol. Chem. 274, 24383-24391. [DOI] [PubMed] [Google Scholar]
- 32.McClary, H., Koch, R., Chisari, F. & Guidotti, L. (2000) J. Virol. 74, 2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavanaugh, V. J., Guidotti, L. G. & Chisari, F. V. (1997) J. Virol. 71, 3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radhakrishnan, S., Layden, T. & Gartel, A. (2004) Virology 323, 173-181. [DOI] [PubMed] [Google Scholar]
- 35.Kosovsky, M., Qadri, I. & Siddisqui, A. (1995) in Hepatitis B Virus: Molecular Mechanisms in Disease and Novel Strategies for Therapy, eds. Koshy, R. & Caselmann, W. (Imperial College Press, London), pp. 21-49.
- 36.Rehermann, B. & Chisari, F. V. (1995) in Hepatitis B Virus: Molecular Mechanisms in Disease and Novel Strategies for Therapy, eds. Koshy, R. & Caselmann, W. (Imperial College Press, London), pp. 85-117.