Abstract
Purpose
Philadelphia chromosome (Ph) –like acute lymphoblastic leukemia (ALL) is a high-risk subtype of childhood ALL characterized by kinase-activating alterations that are amenable to treatment with tyrosine kinase inhibitors. We sought to define the prevalence and genomic landscape of Ph-like ALL in adults and assess response to conventional chemotherapy.
Patients and Methods
The frequency of Ph-like ALL was assessed by gene expression profiling of 798 patients with B-cell ALL age 21 to 86 years. Event-free survival and overall survival were determined for Ph-like ALL versus non–Ph-like ALL patients. Detailed genomic analysis was performed on 180 of 194 patients with Ph-like ALL.
Results
Patients with Ph-like ALL accounted for more than 20% of adults with ALL, including 27.9% of young adults (age 21 to 39 years), 20.4% of adults (age 40 to 59 years), and 24.0% of older adults (age 60 to 86 years). Overall, patients with Ph-like ALL had an inferior 5-year event-free survival compared with patients with non–Ph-like ALL (22.5% [95% CI, 14.9% to 29.3%; n = 155] v 49.3% [95% CI, 42.8% to 56.2%; n = 247], respectively; P < .001). We identified kinase-activating alterations in 88% of patients with Ph-like ALL, including CRLF2 rearrangements (51%), ABL class fusions (9.8%), JAK2 or EPOR rearrangements (12.4%), other JAK-STAT sequence mutations (7.2%), other kinase alterations (4.1%), and Ras pathway mutations (3.6%). Eleven new kinase rearrangements were identified, including four involving new kinase or cytokine receptor genes and seven involving new partners for previously identified genes.
Conclusion
Ph-like ALL is a highly prevalent subtype of ALL in adults and is associated with poor outcome. The diverse range of kinase-activating alterations in Ph-like ALL has important therapeutic implications. Trials comparing the addition of tyrosine kinase inhibitors to conventional therapy are required to evaluate the clinical utility of these agents in the treatment of Ph-like ALL.
INTRODUCTION
Despite remarkable improvements in the treatment outcome for children with acute lymphoblastic leukemia (ALL),1 the prognosis of adults with ALL is poor, with 5-year survival rates of 40% overall2 and approximately 7% for patients who experience relapse.3 Current chemotherapy regimens are limited in adults as a result of toxicity. Compared with children, adult patients with ALL have an increased frequency of high-risk genetic alterations, such as BCR-ABL1, IGH, or MLL rearrangements, and complex karyotypes.4,5 However, the genetic basis of adult ALL is incompletely understood.
Philadelphia chromosome (Ph) –like or BCR-ABL1–like ALL6,7 is a high-risk subtype of ALL characterized by rearrangements, sequence mutations, and copy number alterations involving kinase or cytokine receptor genes.8,9 The prevalence of Ph-like ALL increases from 10% in standard-risk childhood ALL to more than 25% in young adults.8 Preclinical studies and case reports indicate that the addition of tyrosine kinase inhibitors (TKIs) to current chemotherapeutic regimens may improve the poor outcome of patients with Ph-like ALL.10-13
The prevalence of Ph-like ALL in adults is unclear. A recent report from the Dutch-Belgium Hemato-Oncology Cooperative Group identified Ph-like ALL in 17% of patients age 16 to 71 years.14 Another report from the German Multicenter ALL group observed a peak prevalence of Ph-like ALL of approximately 20% in adolescents and young adults, which decreased to less than 10% in adults age 40 to 84 years.15 Both studies examined small numbers of patients, used different methods for identifying the Ph-like ALL gene signature, and lacked genomic characterization. A comprehensive understanding of the prevalence, outcome, and genetic basis of Ph-like ALL in adults is needed to identify patients who may benefit from TKI therapy and to determine the optimal TKIs to use. Here, we demonstrate that the incidence of Ph-like ALL exceeds 20% in patients age 21 to 86 years at diagnosis, is associated with a poor outcome, and is composed of genetic alterations that activate tyrosine kinase or cytokine receptor signaling. These results suggest that the identification of patients with Ph-like ALL should be incorporated into the design of future clinical trials.
PATIENTS AND METHODS
Patients and Treatment
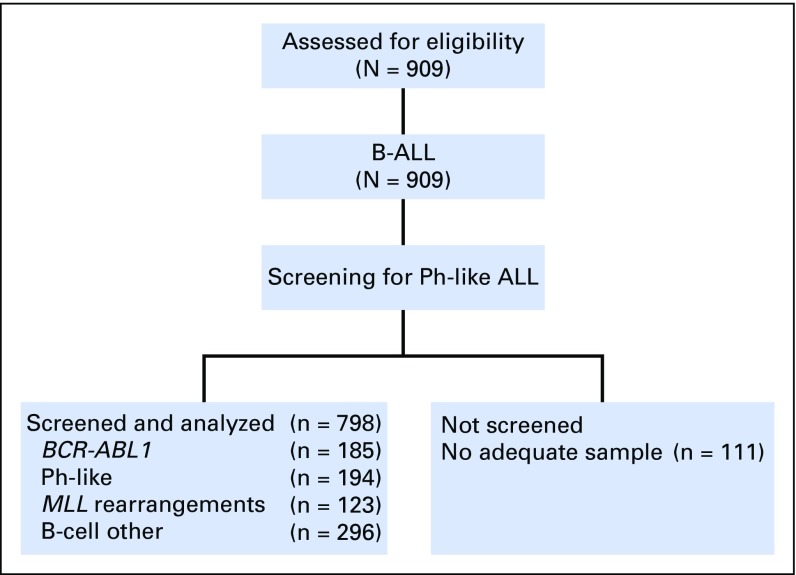
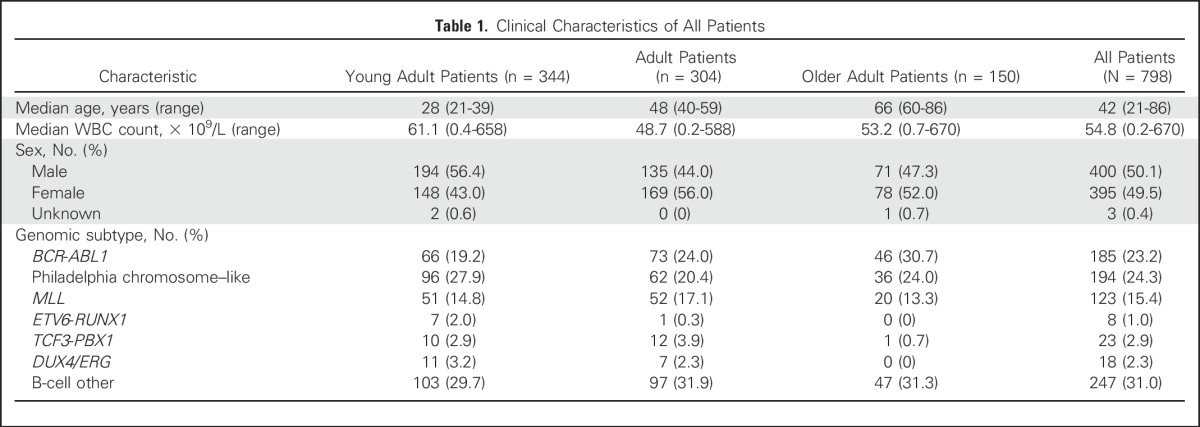
We studied leukemia samples from banked material obtained at diagnosis from 909 patients with precursor B-cell ALL (B-ALL), 798 of whom had suitable material for genomic analysis (Appendix Fig A1, online only). Patients were enrolled onto clinical trial protocols of the following groups or centers: Alliance for Clinical Trials in Oncology Group (Cancer and Leukemia Group B) protocols C19802 (ClinicalTrials.gov identifier: NCT00003700),16 C10102 (NCT00061945), and C10403 (NCT00558519); the Eastern Cooperative Oncology Group–American College of Radiology Imaging Network protocols E29932 (NCT00002514) and E1910 (NCT02003222); The University of Texas MD Anderson Cancer Center (MDACC)17-20; Northern Italy Leukemia Group protocols 09-200021 and 10/07 (NCT00795756); Princess Margaret Cancer Centre22; Southwest Oncology Group protocols S0333 (NCT00109837) and S9400 (NCT00002665)23; and City of Hope. The cohort was divided into the following three age groups: young adults (n = 344; age 21 to 39 years), adults (n = 304; age 40 to 59 years), and older adults (n = 150; age 60 to 86 years; Table 1; Data Supplement). Details for patient samples are provided in the Data Supplement. Patients gave written informed consent for sample collection and research. The study was approved by the St Jude Institutional Review Board.
Table 1.
Clinical Characteristics of All Patients
Genomic Profiling
Gene expression profiling was performed at the University of New Mexico Cancer Center on 798 RNA samples using either U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA)8 or a 15-gene Taqman (Thermo Fisher, Waltham, MA) quantitative reverse transcriptase polymerase chain reaction (PCR) low-density array (LDA) that identifies the Ph-like ALL gene signature, P2RY8-CRLF2, BCR-ABL1, ETV6-ABL1, TCF3-PBX1, and DUX4/ERG-deregulated ALL (Data Supplement).24 Analysis of the 15 genes results in an integrated value between 0 and 1, termed the Ph-like coefficient. Patients with a coefficient of 0.5 to 1 were designated as having Ph-like ALL. All other genomic assays were performed at St Jude Children’s Research Hospital (Memphis, TN). High expression of CRLF2 was determined by the LDA card, and CRLF2 rearrangements (IGH-CRLF2 or P2RY8-CFRLF2) were confirmed using fluorescence in situ hybridization (FISH) as described in the Data Supplement. PCR and Sanger sequencing of tumor cDNA was performed to detect sequence mutations in JAK1 and JAK2 (Data Supplement). A flowchart outlining the genomic assays performed is provided in the Data Supplement. Single nucleotide polymorphism (SNP) analysis was performed on 165 of 194 Ph-like samples with available DNA using either SNP 6.0 microarrays (Affymetrix; n = 49) or the Infinium Omni2.5Exome-8 BeadChip Kit (Illumina, San Diego, CA; n = 116). Details for the LDA algorithm and SNP processing are provided in the Data Supplement.
Transcriptome Sequencing
Transcriptome sequencing (RNA-seq) was performed using the TruSeq library preparation on the Illumina HISeq 2000 platform. All sequencing was paired end and performed using either total RNA and stranded RNA sequencing (n = 63) or polyadenylated-selected nonstranded sequencing (n = 23).8 Sequencing metrics and processing details are provided in the Data Supplement. Samples were analyzed by CICERO8 and FusionCatcher25 to detect fusions. Sequence mutations were analyzed from RNA-seq according to the Genome Analysis Toolkit pipeline.26
Statistical Analysis
Associations between categorical values were examined using Fisher’s exact test. Event-free survival and overall survival from diagnosis were estimated using the Kaplan-Meier method. All groups were compared using the log-rank test.27 An event was defined as a failure to achieve remission, a relapse after remission, or the development of a second malignancy. A multivariable analysis of event-free and overall survival was performed using the Cox proportional hazards regression model.28 Analyses were performed using Prism software version 6.0 (GraphPad Software, La Jolla, CA), R software (www.r-project.org), and SAS software version 9.1.2 (SAS Institute, Cary, NC).
RESULTS
Clinical Characteristics
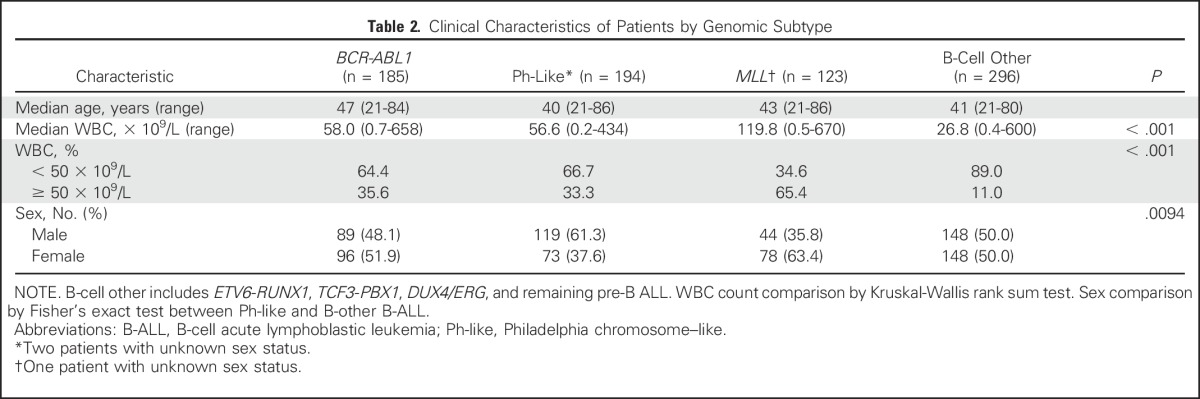
Overall, 194 (24.3%) of 798 patients with B-ALL were classified as having Ph-like ALL. The prevalence of ETV6-RUNX1, TCF3-PBX1, and DUX4/ERG ALL was low (1.0%, 2.9%, and 2.3%, respectively), whereas the prevalence of patients with BCR-ABL1 and MLL-rearranged ALL was 23.2% and 15.4%, respectively. The prevalence of Ph-like ALL exceeded 20% in all age groups (27.9% in young adults, 20.4% in adults, and 24.0% in older adults; Table 1; Data Supplement). Patients with Ph-like ALL had higher WBC counts at diagnosis compared with patients with non–Ph-like ALL (excluding BCR-ABL1 and MLL), both overall (mean, 56.6 v 26.8 × 109/L, respectively; P < .001) and in the different age cohorts (Table 2; Data Supplement). A higher proportion of patients with Ph-like ALL were male compared with patients with non–Ph-like ALL (61.3% v 50%, respectively; P = .0094; Table 2).
Table 2.
Clinical Characteristics of Patients by Genomic Subtype
Treatment Outcomes
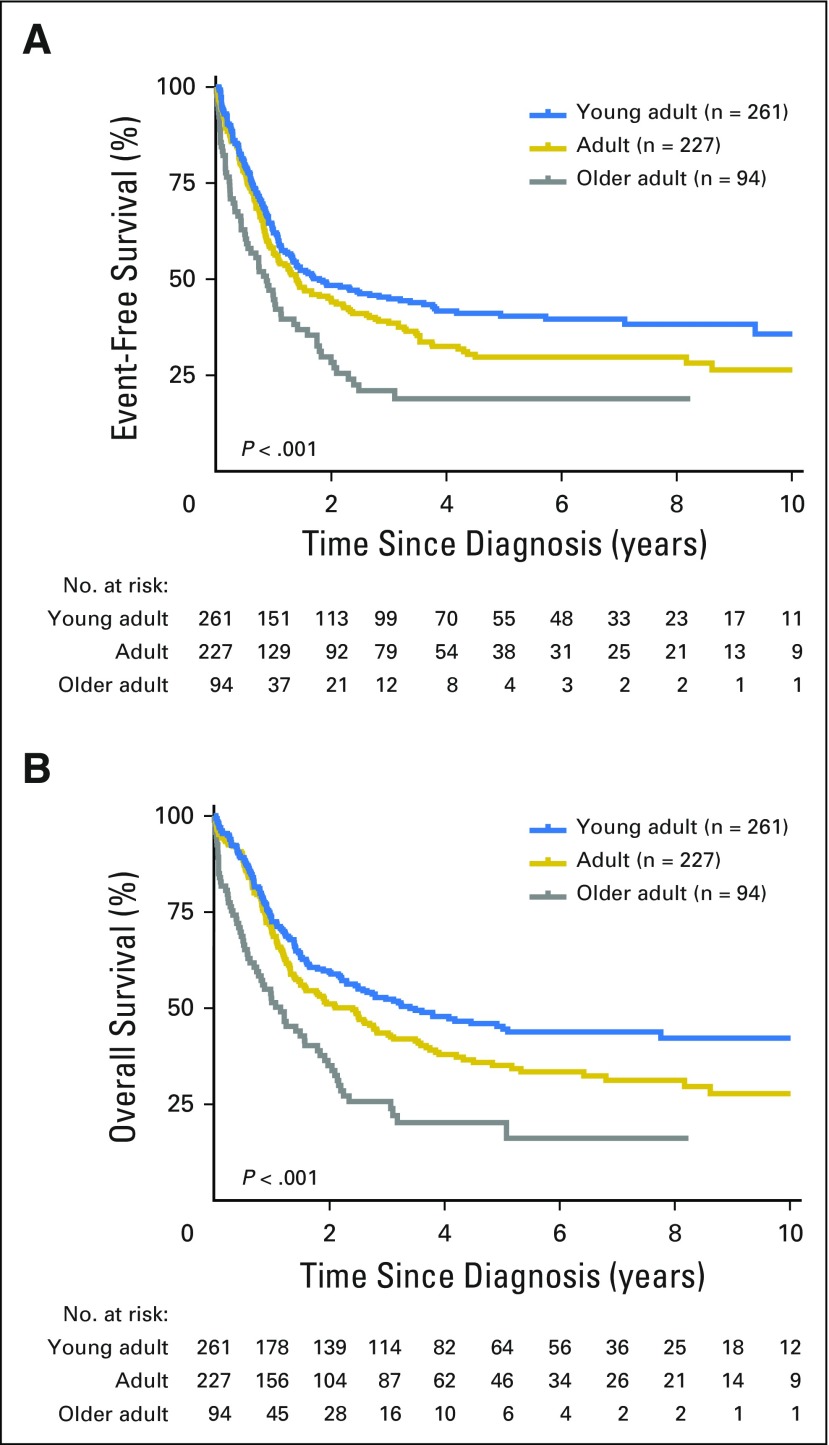
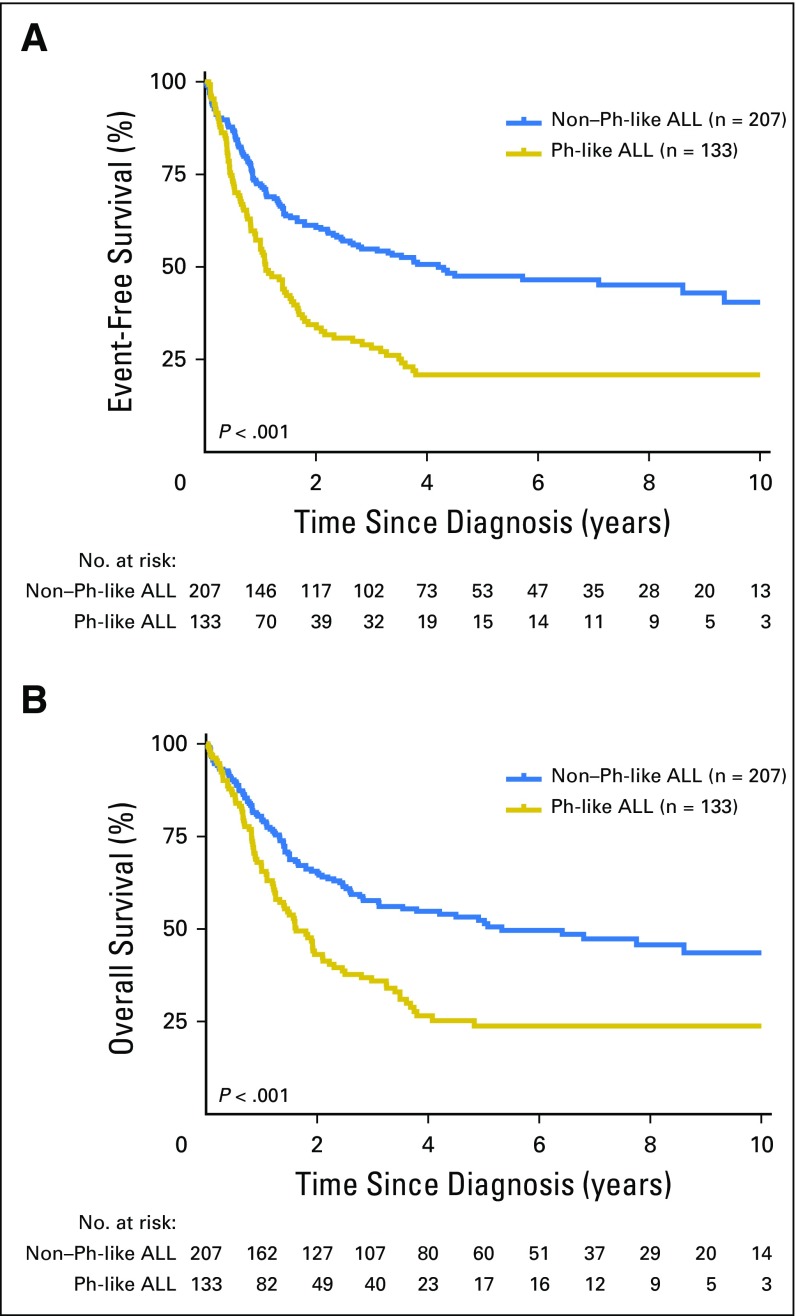
Among all patients, increasing age was associated with inferior outcome. The 5-year event-free survival rates for young adults, adults, and older adults were 40.4% (95% CI, 34.1% to 46.8%), 29.8% (95% CI, 23.3% to 36.0%), and 18.9% (95% CI, 8.9% to 27.1%; P < .001), respectively, with 5-year overall survival rates of 45.2% (95% CI, 38.8% to 51.9%), 35.1% (95% CI, 28.4% to 41.8%), and 16.2% (95% CI, 4.7% to 24.8%; P < .001; Fig 1), respectively. Overall, the outcome of patients with Ph-like ALL was inferior compared with the outcome of patients with non–Ph-like ALL (excluding BCR-ABL1 and MLL), with 5-year event-free survival rates of 22.5% (95% CI, 14.9% to 29.3%) compared with 49.3% (95% CI, 42.8% to 56.2%; P < .001) and overall survival rates of 23.8% (95% CI, 16.0% to 32.4%) compared with 52.4% (95% CI, 45.4% to 59.9%; P < .001; Fig 2 and Data Supplement), respectively. In multivariable analyses, age (hazard ratio [HR], 2.55; P < .001), sex (HR, 1.97; P < .001), and the presence of Ph-like ALL (HR, 1.48; P = .014) were independently associated with poor survival (Data Supplement). Minimal residual disease at the end of induction was assessed by six-color flow cytometry for patients treated on The University of Texas MD Anderson Cancer Center protocols. Of those tested, patients with Ph-like ALL were less likely to achieve minimal residual disease–negative remission (< 0.01% minimal residual disease) compared with patients with non–Ph-like ALL (47% [n = 19] v 94% [n = 17], respectively; P = .002). Outcome after hematopoietic stem-cell transplantation was available for patients enrolled onto E2993. There was no significant difference in survival between patients who did (n = 7) and did not receive transplantation (n = 14; P = .2; Data Supplement).
Fig 1.
Overall outcome of adult acute lymphoblastic leukemia. Kaplan-Meier estimates of (A) event-free survival and (B) overall survival of young adults, adults, and older adults combined. Outcome data were available for 582 of 798 patients studied.
Fig 2.
Outcome of adults with Philadelphia chromosome (Ph) –like acute lymphoblastic leukemia (ALL). Kaplan-Meier estimates of (A) event-free survival and (B) overall survival of all patients with Ph-like ALL compared with patients with non–Ph-like ALL (including ETV6-RUNX1, TCF3-PBX1, DUX4/ERG, and all other ALLs). Outcome data were available for 340 of 490 patients with Ph-like and non–Ph-like ALL.
Inferior outcome was observed for patients with Ph-like ALL compared with patients with non–Ph-like ALL among young adults and adults, with 5-year event-free survival rates of 24.1% (95% CI, 12.6% to 34.1%) compared with 60.5% (95% CI, 51.2% to 71.4%; P < .001) and 21.4% (95% CI, 7.6% to 32.5%) compared with 38.7% (95% CI, 27.6% to 50.0%; P = .0021; Data Supplement), respectively. The difference in outcomes was not significant for older adults, with 3-year event-free survival rates of 8.0% (95% CI, 0.5% to 21.6%) and 33.3% (95% CI, 15.8% to 52.2%; P = .47) for patients with Ph-like ALL and non–Ph-like ALL, respectively. Likewise, the 5-year overall survival rates for patients with Ph-like ALL were inferior to those of patients with non–Ph-like ALL in young adults (25.4% [95% CI, 14.8% to 37.5%] v 64.2% [95% CI, 53.1% to 73.3%], respectively; P < .001) and adults (27.0% [95% CI, 13.3% to 42.7%] v 44.9% [95% CI, 33.5% to 55.7%], respectively; P = .023), but the difference in 3-year survival rates in older adults was not significant between the two groups (13.5% [95% CI, 2.3% to 34.3%] v 37.9% [95% CI, 19.2% to 56.6%], respectively; P = .64; Data Supplement). The lack of significance in older adults may be a result of the small number of patients available for outcome analysis (n = 44); therefore, screening larger cohorts of patients older than age 60 years will be important to assess the outcome of Ph-like ALL in the elderly.
Genomic Landscape of Ph-Like ALL in Adults
Genomic alterations activating kinase signaling were identified in 171 (88.1%) of 194 patients with Ph-like ALL and 171 (95.0%) of 180 patients with materials enabling genomic analysis. Overall, 99 (51%) of 194 patients with Ph-like ALL had high CRLF2 expression. The frequency was 51.1% in young adults, 46.8% in adults, and 58.4% in older adults. We identified IGH-CRLF2 (n = 57; 57.6%) and P2RY8-CRLF2 (n = 21; 21.2%) rearrangements; for 21 patients, there was insufficient material for analysis. Concomitant Janus kinase (JAK1/JAK2) mutations were identified in 25 (25%) of 99 patients with high CRLF2 expression (Data Supplement).
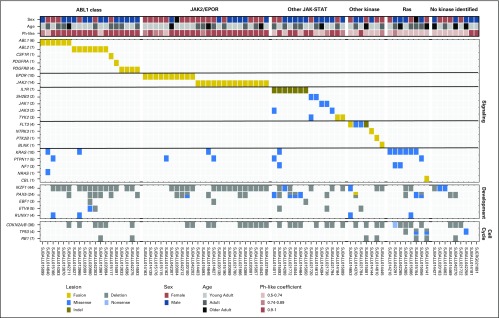
Transcriptome sequencing was performed in 86 of the remaining 95 patients with Ph-like ALL. Genetic alterations were classified into subgroups of kinase and cytokine receptor signaling pathways (Fig 3). These included rearrangements of CRLF2 (51% of patients), ABL class fusions (ABL1, ABL2, CSF1R, PDGFRA, and PDGFRB; 9.8%), JAK2 (7.2%), or EPOR (5.2%); sequence mutations of IL7R, SH2B3, JAK1, or JAK3 and rearrangement of TYK2 (termed other JAK-STAT; 7.2%); Ras pathway mutations (KRAS, NF1, PTPN11, and CBL1; 3.6%); and uncommon fusions (FLT3, NTRK3, BLNK, and PTK2B; 4.1%). A subset of patients did not have a kinase-activating alteration identified by transcriptome analysis (4.6%) or lacked material for genomic analysis (7.2%).
Fig 3.
Kinase alterations identified in patients with Philadelphia chromosome (Ph) –like acute lymphoblastic leukemia (ALL). Data are shown for 86 patients with Ph-like ALL who underwent transcriptome sequencing. The cohort is divided into patients with ABL class fusions (ABL1, ABL2, CSF1R, PDGFRA, and PDGFRB); EPOR or JAK2 rearrangements; other JAK-STAT sequence mutations (IL7R, SH2B3, JAK1, JAK3, and TYK2); other kinase alterations (FLT3, NTRK3, PTK2B, and BLNK); RAS pathway alterations (KRAS, PTPN11, NF1, NRAS, and CBL); or no kinase identified. The Ph-like coefficient was obtained from the low-density array card using an algorithm including 15 genes highly expressed in patients with Ph-like ALL.
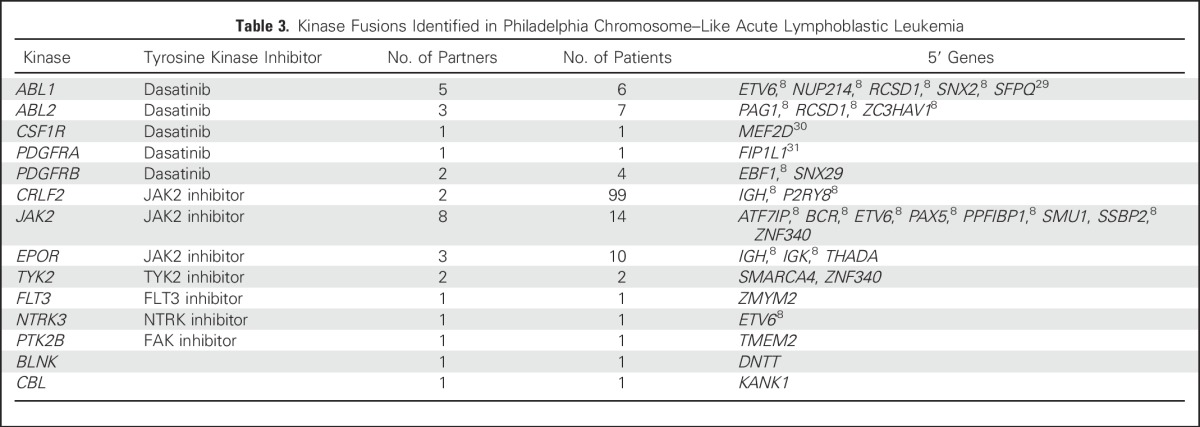
Rearrangements activating kinase signaling were identified in 126 (65%) of 194 patients, including 32 different rearrangements (10 of which were recurrent) in the following 14 kinase or cytokine receptor genes (Table 3; Data Supplement): JAK2 (eight fusion partners), ABL1 (n = 5), ABL2 (n = 3), EPOR (n = 3), CRLF2 (n = 2), PDGFRB (n = 2), TYK2 (n = 2), BLNK (n = 1), CBL (n = 1), CSF1R (n = 1), FLT3 (n = 1), NTRK3 (n = 1), PDGFRA (n = 1), and PTK2B (n = 1). The following 11 rearrangements had not previously been identified in B-ALL: FIP1L1-PDGFRA, SNX29-PDGFRB, SMU1-JAK2, ZNF340-JAK2, THADA-EPOR, SMARCA4-TYK2, ZNF340-TYK2, ZYMM2-FLT3, DNTT-BLNK, TMEM2-PTK2B, and KANK1-CBL1. All kinase fusions retained an intact tyrosine kinase domain (Data Supplement) and showed increased expression of the kinase gene downstream of the exon breakpoint (Data Supplement). With the exception of EPOR rearrangements, we did not observe significant differences in the frequency of kinase subtypes across the age groups (Data Supplement).
Table 3.
Kinase Fusions Identified in Philadelphia Chromosome–Like Acute Lymphoblastic Leukemia
Sequence mutations in genes activating JAK-STAT signaling were identified in 14 patients with low CRLF2 expression, including IL7R (seven patients), JAK1 (two patients), JAK3 (one patient), and SH2B3 (two patients) and two patients with rearrangements involving TYK2. Seven patients harbored alterations within the Ras pathway only, including sequence mutations in KRAS (five patients) and PTPN11 (one patient) and one patient with a novel rearrangement involving CBL. In addition, two patients with KRAS mutations also harbored sequence mutations within NF1 (Data Supplement).
Patients with Ph-like ALL were classified on the basis of an LDA coefficient between 0.5 and 1.24 Within this classification, we observed differences between the distribution of kinase groups on the basis of the coefficient. For patients with Ph-like ALL with a rearrangement involving ABL class genes, CRLF2, or JAK2/EPOR, we observed 90%, 68%, and 79% of patients, respectively, with a coefficient between 0.9 and 1. In contrast, 41% of patients with other kinase or JAK-STAT alterations exhibited a coefficient between 0.9 and 1. Patients harboring Ras pathway alterations had a weaker coefficient, with 86% of patients classified in the 0.5 to 0.74 category (Data Supplement). Altogether, patients harboring a kinase rearrangement were more likely to have a coefficient greater than 0.9 than patients with a sequence mutation (73% v 31%, respectively; P < .001). Patients harboring rearrangements of CRLF2 or JAK2/EPOR had a trend toward inferior 5-year event-free and overall survival compared with patients harboring ABL class or other JAK-STAT alterations (P = .057 and P = .072, respectively; Data Supplement).
Analysis of DNA copy number alterations confirmed a high frequency of IKZF1 alterations (deletion and mutation) in patients with Ph-like ALL (120 of 165 patients; 73%). IKZF1 alterations were also more common in patients with a kinase or cytokine receptor rearrangement (109 of 137 patients; 80%) than a sequence mutation (eight of 17 patients; 47%; P < .001). Other common targets of genetic alteration included CDKN2A/B (51%), PAX5 (38%), BTG1 (33%), and EBF1 (22%; Data Supplement). We also observed enrichment of copy number alterations of IKZF1, PAX5, EBF1, and CDKN2A/B in patients harboring a Ph-like coefficient greater than 0.9 (Data Supplement). A summary of all copy number alterations identified in Ph-like ALL is provided in the Data Supplement. Overall, 142 (86%) of 165 patients with Ph-like ALL assessed by SNP profiling harbored alterations affecting lymphoid development (IKZF1, PAX5, EBF1, ETV6, and RUNX1), and 89 (54%) of 165 patients had alterations of the cell cycle (CDKN2A/B, TP53, and RB1).
DISCUSSION
In this comprehensive retrospective study of 798 patients with B-ALL age 21 to 86 years, we have demonstrated that the frequency of Ph-like ALL exceeds 20% across the adult age spectrum. With the combined analysis of patients treated on different protocols, Ph-like ALL is independently associated with a poor prognosis. We show that Ph-like ALL in adults is characterized by a variety of genetic alterations that activate kinase or cytokine receptor signaling and that the majority of lesions are targetable with US Food and Drug Administration–approved TKIs.
A similar spectrum of kinase-activating alterations was identified in adults with Ph-like ALL as that found in children. The overall frequency of CRLF2 rearrangement (51%) is similar to that identified in previous reports, with older adults showing the highest frequency at 58%. However, we observed a 25% frequency of JAK mutations in CRLF2-rearranged adult patients, which is lower than the frequency reported for childhood ALL (50%).8 Additional targets of mutation in CRLF2-rearranged patients include IL7R, FLT3, and SH2B3.8 Identification of the other co-operating lesions in adult patients will provide important insights into the mechanisms of oncogenic signaling by CRLF2 in ALL.
We also show a high frequency of JAK2 and EPOR rearrangements in the adult ALL population. Overall, approximately 70% of adult patients with Ph-like ALL harbor genetic alterations that may be targetable with JAK inhibitors, such as ruxolitinib. These include CRLF2 rearrangements with or without JAK1/JAK2 mutation, rearrangements involving JAK2 and EPOR, and sequence mutations of JAK1, JAK3, IL7R, and SH2B3. The only exception is TYK2, which is not sensitive to ruxolitinib and will require development of selective TYK2 inhibitors.32 Multiple studies have demonstrated potent activity of ruxolitinib in preclinical models of Ph-like ALL or early T-cell precursor ALL. Although variations in response were observed in CRLF2-rearranged cells, this was independent of JAK mutation12,33 and is likely a result of the presence of other genetic alterations activating JAK-STAT signaling (eg, IL7R mutations).8 These preclinical studies have shown that the efficacy of ruxolitinib is determined by the degree of JAK-STAT activation rather than JAK mutational status. To determine the efficacy of JAK inhibition in patients harboring lesions activating the JAK-STAT pathway, ruxolitinib in combination with conventional chemotherapy will be assessed in trials from the Children’s Oncology Group and The University of Texas MD Anderson Cancer Center.
Another major Ph-like ALL genetic subgroup involves kinases that are targeted by ABL inhibitors (imatinib and dasatinib), including ABL1, ABL2, CSF1R, PDGFRA, and PDFGRB. Rearrangements resulting in FIP1L1-PDGFRA have been identified in eosinophilic leukemia and are responsive to ABL inhibitors.31 We report this fusion for the first time, to our knowledge, in Ph-like ALL. An additional new target of rearrangement is B-cell linker (BLNK), an adaptor protein that transduces signals from the B-cell receptor and activates the Ras pathway via ERK.34 Another putative mechanism of Ras activation was identified through the KANK1-CBL fusion. CBL is a negative regulator of signal transduction pathways, including Ras. Fusion with KANK1 likely inactivates CBL by disrupting the open reading frame, leading to hyperactivation of Ras signaling. Overall, we identified 11 new rearrangements that had not previously been identified in Ph-like ALL. These data further highlight the genetic heterogeneity of Ph-like ALL and the importance of incorporating comprehensive sequencing to identify all genetic alterations.
To enable prompt delivery of TKIs, accurate and fast diagnosis of patients with Ph-like ALL is a critical factor when designing clinical trials for TKI therapy. In the research setting, we identified patients with the Ph-like ALL signature using a TaqMan LDA card, which is rapidly performed in a Clinical Laboratory Improvement Amendments–compliant laboratory. This is only a screening tool, and additional approaches are required for the identification of specific kinase alterations. FISH was performed on patients with high CRLF2 expression to identify CRLF2 rearrangements, and RNA-seq was performed on the remainder of patients to identify additional kinase alterations. Although effective for research studies, this tiered approach may delay identification of the kinase-activating lesions in the diagnostic setting. A number of approaches are being evaluated for the clinical identification of Ph-like ALL and underlying kinase-activating alterations. As a result of the genetic complexity of Ph-like ALL, assays to identify rearrangements that are agnostic of the 5′ fusion partner are ideal. A simple and cost-effective approach is to perform FISH for break apart of the kinase gene, which will identify most fusions involving key kinase genes.9 Reverse transcriptase PCR may be used as a screening or confirmatory approach but will only confirm known fusions. Another option is the digital molecular barcoding platform NanoString (NanoString, Seattle, WA), which can multiplex more than 200 different genetic alterations. This assay is simple and requires little material but also only identifies known fusions. Capture-based RNA sequencing (eg, Archer FusionPlex Oncology Research Kit, Archer DX, Boulder, CO; Foundation One Heme, Foundation Medicine, Cambridge, MA35) effectively identifies kinase fusions and is agnostic of the 5′ fusion partner but requires Clinical Laboratory Improvement Amendments–approved sequencing capability. Thus, although FISH or targeted approaches are satisfactory for a subset of alterations, it is likely that next-generation sequencing technologies in diagnostic laboratories are required for the comprehensive and timely identification of patients with Ph-like ALL. Although these assays may not be feasible for all clinical laboratories at present, they will be increasingly available in the future.
In summary, Ph-like ALL is common across the age spectrum of adult ALL, including more than 20% of patients age 21 to 86 years, with a notably high prevalence of JAK-STAT activating alterations. These findings warrant the development of clinical trials in adults that assess the efficacy of TKIs, similar to those that are being established for pediatric ALL.
ACKNOWLEDGMENT
Genomic data have been deposited at the European Genome Phenome archive (Accession No. EGAS00001000654). We thank Joshua Stokes of Biomedical Communications and the staff of the Biorepository, Hartwell Centre for Bioinformatics and Biotechnology, Flow Cytometry and Cell Sorting Core Facility, and Cytogenetics Core Facility of St Jude Children’s Research Hospital.
Appendix
Fig A1.
CONSORT diagram. Among the 909 eligible patients with B-cell acute lymphoblastic leukemia (B-ALL), 798 had adequate samples to screen for the Philadelphia chromosome (Ph) –like ALL signature.
Footnotes
Supported in part by the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital; a Stand Up to Cancer Innovative Research Grant and St Baldrick’s Foundation Scholar Award (C.G.M.); an American Society of Hematology Scholar Award (K.G.R.); a Lady Tata Memorial Trust Award (I.I.); the Leukemia Research Foundation (C.D.B.); National Cancer Institute Grants No. CA21765 (St Jude Cancer Center Support grant), HHSN261200800001E (C.G.M.), U01 CA157937 (C.L.W.), U10 CA180861 (G.M. and C.D.B.), U24 CA196171 (The Alliance National Clinical Trials Network Biorepository and Biospecimen Resource), and CA145707 (C.L.W. and C.G.M.); and the following National Institutes of Health grants to the Eastern Cooperative Oncology Group–American College of Radiology Imaging Network Cancer Research Group: Grants No. CA180820, CA180827, CA21115, CA180791, CA14958, CA189859, CA15488, CA17145, CA13650, and CA180790.
Clinical trial information: NCT00003700, NCT00061945, NCT00558519, NCT00002514, NCT02003222, NCT00795756, NCT00109837, NCT00002665.
AUTHOR CONTRIBUTIONS
Conception and design: Kathryn G. Roberts, Charles G. Mullighan
Collection and assembly of data: Kathryn G. Roberts, Debbie Payne-Turner, Kelly McCastlain, Richard C. Harvey, I-Ming Chen, Ilaria Iacobucci, Marcus Valentine, Alessandro Rambaldi, Manuela Tosi, Orietta Spinelli, Jerald P. Radich, Mark D. Minden, Jacob M. Rowe, Selina Luger, Mark R. Litzow, Martin S. Tallman, Peter H. Wiernik, Ravi Bhatia, Ibrahim Aldoss, Jessica Kohlschmidt, Krzysztof Mrózek, Guido Marcucci, Clara D. Bloomfield, Wendy Stock, Stephen Kornblau, Hagop M. Kantarjian, Marina Konopleva, Elisabeth Paietta, Cheryl L. Willman, Charles G. Mullighan
Data analysis and interpretation: Kathryn G. Roberts, Zhaohui Gu, Richard C. Harvey, I-Ming Chen, Deqing Pei, Stanley B. Pounds, Lei Shi, Yongjin Li, Jinghui Zhang, Cheng Cheng, Charles G. Mullighan
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
High Frequency and Poor Outcome of Philadelphia Chromosome–Like Acute Lymphoblastic Leukemia in Adults
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Kathryn G. Roberts
Patents, Royalties, Other Intellectual Property: Inventor on a pending patent application related to gene-expression signatures for detection of underlying Philadelphia chromosome–like events and therapeutic targeting in leukemia (PCT/US2012/069228)
Zhaohui Gu
No relationship to disclose
Debbie Payne-Turner
No relationship to disclose
Kelly McCastlain
No relationship to disclose
Richard C. Harvey
Patents, Royalties, Other Intellectual Property: Inventor on a pending patent application related to gene-expression signatures for detection of underlying Philadelphia chromosome–like events and therapeutic targeting in leukemia (PCT/US2012/069228)
I-Ming Chen
No relationship to disclose
Deqing Pei
No relationship to disclose
Ilaria Iacobucci
No relationship to disclose
Marcus Valentine
No relationship to disclose
Stanley B. Pounds
No relationship to disclose
Lei Shi
No relationship to disclose
Yongjin Li
No relationship to disclose
Jinghui Zhang
No relationship to disclose
Cheng Cheng
No relationship to disclose
Alessandro Rambaldi
No relationship to disclose
Manuela Tosi
No relationship to disclose
Orietta Spinelli
No relationship to disclose
Jerald P. Radich
Consulting or Advisory Role: Bristol-Myers Squibb, Novartis, ARIAD, Pfizer, Incyte
Research Funding: Novartis
Mark D. Minden
Honoraria: Astellas Pharma
Consulting or Advisory Role: Astellas Pharma
Travel, Accommodations, Expenses: Celgene
Jacob M. Rowe
No relationship to disclose
Selina Luger
Honoraria: Novartis, Sigma-Tau, Karyopharm Therapeutics, Tolero Pharmaceuticals, Astellas Pharma
Consulting or Advisory Role: Pfizer
Research Funding: Amgen (Inst), Cyclacel (Inst), ARIAD (Inst), Onconova (Inst), Seattle Genetics (Inst)
Mark R. Litzow
No relationship to disclose
Martin S. Tallman
No relationship to disclose
Peter H. Wiernik
Honoraria: Novartis
Travel, Accommodations, Expenses: Novartis
Ravi Bhatia
No relationship to disclose
Ibrahim Aldoss
Consulting or Advisory Role: Helocyte
Jessica Kohlschmidt
No relationship to disclose
Krzysztof Mrózek
No relationship to disclose
Guido Marcucci
No relationship to disclose
Clara D. Bloomfield
No relationship to disclose
Wendy Stock
Honoraria: Sigma-Tau, ADC Therapeutics, Gilead Sciences, Amgen, Pfizer
Research Funding: Sigma-Tau (Inst)
Patents, Royalties, Other Intellectual Property: Royalties for a chapter in UpToDate
Stephen Kornblau
No relationship to disclose
Hagop M. Kantarjian
No relationship to disclose
Marina Konopleva
Stock or Other Ownership: Reata Pharmaceuticals
Honoraria: Calithera Biosciences, AbbVie, Genentech
Consulting or Advisory Role: AbbVie
Research Funding: Stemline Therapeutics (Inst), Novartis (Inst), AbbVie (Inst), Calithera Biosciences (Inst), Eli Lilly (Inst), Genentech (Inst), Bristol-Myers Squibb (Inst), Cellectis (Inst)
Elisabeth Paietta
No relationship to disclose
Cheryl L. Willman
Patents, Royalties, Other Intellectual Property: Inventor on a pending patent application related to gene-expression signatures for detection of underlying Philadelphia chromosome–like events and therapeutic targeting in leukemia (PCT/US2012/069228)
Charles G. Mullighan
Honoraria: Amgen
Speakers' Bureau: Amgen
Research Funding: Loxo
Patents, Royalties, Other Intellectual Property: Inventor on a pending patent application related to gene-expression signatures for detection of underlying Philadelphia chromosome–like events and therapeutic targeting in leukemia (PCT/US2012/069228)
REFERENCES
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 2.Goldstone AH, Richards SM, Lazarus HM, et al. In adults with standard-risk acute lymphoblastic leukemia, the greatest benefit is achieved from a matched sibling allogeneic transplantation in first complete remission, and an autologous transplantation is less effective than conventional consolidation/maintenance chemotherapy in all patients: Final results of the International ALL Trial (MRC UKALL XII/ECOG E2993) Blood. 2008;111:1827–1833. doi: 10.1182/blood-2007-10-116582. [DOI] [PubMed] [Google Scholar]
- 3.Fielding AK, Richards SM, Chopra R, et al. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL): An MRC UKALL12/ECOG 2993 study. Blood. 2007;109:944–950. doi: 10.1182/blood-2006-05-018192. [DOI] [PubMed] [Google Scholar]
- 4.Moorman AV, Ensor HM, Richards SM, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 5.Moorman AV, Harrison CJ, Buck GA, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): Analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109:3189–3197. doi: 10.1182/blood-2006-10-051912. [DOI] [PubMed] [Google Scholar]
- 6.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lengline E, Beldjord K, Dombret H, et al. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica. 2013;98:e146–e148. doi: 10.3324/haematol.2013.095372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weston BW, Hayden MA, Roberts KG, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31:e413–e416. doi: 10.1200/JCO.2012.47.6770. [DOI] [PubMed] [Google Scholar]
- 12.Maude SL, Tasian SK, Vincent T, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi K, Miyagawa N, Mitsui K, et al. TKI dasatinib monotherapy for a patient with Ph-like ALL bearing ATF7IP/PDGFRB translocation. Pediatr Blood Cancer. 2015;62:1058–1060. doi: 10.1002/pbc.25327. [DOI] [PubMed] [Google Scholar]
- 14.Boer JM, Koenders JE, van der Holt B, et al. Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica. 2015;100:e261–e264. doi: 10.3324/haematol.2014.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold T, Baldus CD, Gökbuget N. Ph-like acute lymphoblastic leukemia in older adults. N Engl J Med. 2014;371:2235. doi: 10.1056/NEJMc1412123#SA1. [DOI] [PubMed] [Google Scholar]
- 16.Stock W, Johnson JL, Stone RM, et al. Dose intensification of daunorubicin and cytarabine during treatment of adult acute lymphoblastic leukemia: Results of Cancer and Leukemia Group B Study 19802. Cancer. 2013;119:90–98. doi: 10.1002/cncr.27617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantarjian H, Thomas D, O’Brien S, et al. Long-term follow-up results of hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone (Hyper-CVAD), a dose-intensive regimen, in adult acute lymphocytic leukemia. Cancer. 2004;101:2788–2801. doi: 10.1002/cncr.20668. [DOI] [PubMed] [Google Scholar]
- 18.Ravandi F, O’Brien S, Thomas D, et al. First report of phase 2 study of dasatinib with hyper-CVAD for the frontline treatment of patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia. Blood. 2010;116:2070–2077. doi: 10.1182/blood-2009-12-261586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas DA, Faderl S, Cortes J, et al. Treatment of Philadelphia chromosome-positive acute lymphocytic leukemia with hyper-CVAD and imatinib mesylate. Blood. 2004;103:4396–4407. doi: 10.1182/blood-2003-08-2958. [DOI] [PubMed] [Google Scholar]
- 20.Thomas DA, O’Brien S, Faderl S, et al. Chemoimmunotherapy with a modified hyper-CVAD and rituximab regimen improves outcome in de novo Philadelphia chromosome-negative precursor B-lineage acute lymphoblastic leukemia. J Clin Oncol. 2010;28:3880–3889. doi: 10.1200/JCO.2009.26.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassan R, Spinelli O, Oldani E, et al. Improved risk classification for risk-specific therapy based on the molecular study of minimal residual disease (MRD) in adult acute lymphoblastic leukemia (ALL) Blood. 2009;113:4153–4162. doi: 10.1182/blood-2008-11-185132. [DOI] [PubMed] [Google Scholar]
- 22.Storring JM, Minden MD, Kao S, et al. Treatment of adults with BCR-ABL negative acute lymphoblastic leukaemia with a modified paediatric regimen. Br J Haematol. 2009;146:76–85. doi: 10.1111/j.1365-2141.2009.07712.x. [DOI] [PubMed] [Google Scholar]
- 23.Pullarkat V, Slovak ML, Kopecky KJ, et al. Impact of cytogenetics on the outcome of adult acute lymphoblastic leukemia: Results of Southwest Oncology Group 9400 study. Blood. 2008;111:2563–2572. doi: 10.1182/blood-2007-10-116186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey RC, Kang H, Roberts KG: Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukemia (ALL) patients with a Philadelphia chromosome-like (“Ph-like” or “BCR-ABL1-like”) signature for therapeutic targeting and clinical intervention. Blood 122:826, 2013 (abstr) [Google Scholar]
- 25.Nicorici D, Satalan M, Edgren H, et al: FusionCatcher: A tool for finding somatic fusion genes in paired-end RNA-sequencing data. http://biorxiv.org/content/early/2014/11/19/011650 [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 28.Cox DR. Regression models and life-tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 29.Duhoux FP, Auger N, De Wilde S, et al. The t(1;9)(p34;q34) fusing ABL1 with SFPQ, a pre-mRNA processing gene, is recurrent in acute lymphoblastic leukemias. Leuk Res. 2011;35:e114–e117. doi: 10.1016/j.leukres.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Lilljebjörn H, Ågerstam H, Orsmark-Pietras C, et al. RNA-seq identifies clinically relevant fusion genes in leukemia including a novel MEF2D/CSF1R fusion responsive to imatinib. Leukemia. 2014;28:977–979. doi: 10.1038/leu.2013.324. [DOI] [PubMed] [Google Scholar]
- 31.Gotlib J, Cools J, Malone JM, III, et al. The FIP1L1-PDGFRalpha fusion tyrosine kinase in hypereosinophilic syndrome and chronic eosinophilic leukemia: Implications for diagnosis, classification, and management. Blood. 2004;103:2879–2891. doi: 10.1182/blood-2003-06-1824. [DOI] [PubMed] [Google Scholar]
- 32.Liang J, van Abbema A, Balazs M, et al. Lead optimization of a 4-aminopyridine benzamide scaffold to identify potent, selective, and orally bioavailable TYK2 inhibitors. J Med Chem. 2013;56:4521–4536. doi: 10.1021/jm400266t. [DOI] [PubMed] [Google Scholar]
- 33.Maude SL, Dolai S, Delgado-Martin C, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015;125:1759–1767. doi: 10.1182/blood-2014-06-580480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamura Y, Oda A, Katahira T, et al. BLNK binds active H-Ras to promote B cell receptor-mediated capping and ERK activation. J Biol Chem. 2009;284:9804–9813. doi: 10.1074/jbc.M809051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J, Abdel-Wahab O, Nahas MK, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127:3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]