Abstract
Purpose
The major causes of mortality after allogeneic hematopoietic-cell transplantation (allo-HCT) are relapse, graft-versus-host disease (GVHD), and infection. We have reported previously that alterations in the intestinal flora are associated with GVHD, bacteremia, and reduced overall survival after allo-HCT. Because intestinal bacteria are potent modulators of systemic immune responses, including antitumor effects, we hypothesized that components of the intestinal flora could be associated with relapse after allo-HCT.
Methods
The intestinal microbiota of 541 patients admitted for allo-HCT was profiled by means of 16S ribosomal sequencing of prospectively collected stool samples. We examined the relationship between abundance of microbiota species or groups of related species and relapse/progression of disease during 2 years of follow-up time after allo-HCT by using cause-specific proportional hazards in a retrospective discovery-validation cohort study.
Results
Higher abundance of a bacterial group composed mostly of Eubacterium limosum in the validation set was associated with a decreased risk of relapse/progression of disease (hazard ratio [HR], 0.82 per 10-fold increase in abundance; 95% CI, 0.71 to 0.95; P = .009). When the patients were categorized according to presence or absence of this bacterial group, presence also was associated with less relapse/progression of disease (HR, 0.52; 95% CI, 0.31 to 0.87; P = .01). The 2-year cumulative incidences of relapse/progression among patients with and without this group of bacteria were 19.8% and 33.8%, respectively. These associations remained significant in multivariable models and were strongest among recipients of T-cell–replete allografts.
Conclusion
We found associations between the abundance of a group of bacteria in the intestinal flora and relapse/progression of disease after allo-HCT. These might serve as potential biomarkers or therapeutic targets to prevent relapse and improve survival after allo-HCT.
INTRODUCTION
Greater than 18,000 patients undergo allogeneic hematopoietic-cell transplantation (allo-HCT) yearly with curative intent for hematologic neoplasms.1 In recent years, major strides have been made in the reduction of its treatment-related toxicity.2 Relapse of primary malignancy, however, remains the leading cause of death after allo-HCT.3
The efficacy of allo-HCT in the treatment of hematologic malignancies has been attributed to both the cytotoxic effects of conditioning chemotherapy and the control or elimination of residual malignant cells through graft-versus-tumor (GVT) activity mediated by the donor immune system. Several risk factors of relapse after allo-HCT have been identified, including somatic genetic alterations, depth of response to pretransplantation treatment,4-6 conditioning intensity,7 graft source,8,9 mismatch at the human leukocyte antigen (HLA) –DPB1 locus,10,11 donor KIR haplotype,12 graft-versus-host disease (GVHD),13 and GVHD prophylaxis modalities.14 These factors, however, provide only a partial explanation of relapse risk.
The intestinal microbiota interact with and regulate host immunity.15 Several important outcomes after allo-HCT—in particular, bloodstream infections,16 GVHD,17-22 organ toxicity,23 and overall survival24—have been associated with intestinal bacterial composition. Recent studies have demonstrated that the intestinal microbiota modulate antitumor immunity.25-31 Whether immune receptors that recognize bacteria can modulate the risk of relapse after allo-HCT is an unresolved question.32
Because allo-immunity plays an important role in the prevention of posttransplantation relapse, we hypothesized that the composition of the intestinal microbiota might be associated with GVT activity. In this study, we retrospectively examined a single-center population for features of the intestinal microbiota composition that might be associated with post–allo-HCT relapse or progression of disease (POD).
METHODS
Patients and Samples
Stool samples were collected, and genomic 16S ribosomal sequencing was performed as described.16,18 The patients were admitted to our center for allo-HCT from 2009 to 2015 after they were prospectively enrolled in a fecal-collection protocol approved by the institutional review board. Patients with at least one sequenced sample collected between days 0 and 21 of a first allo-HCT were included.
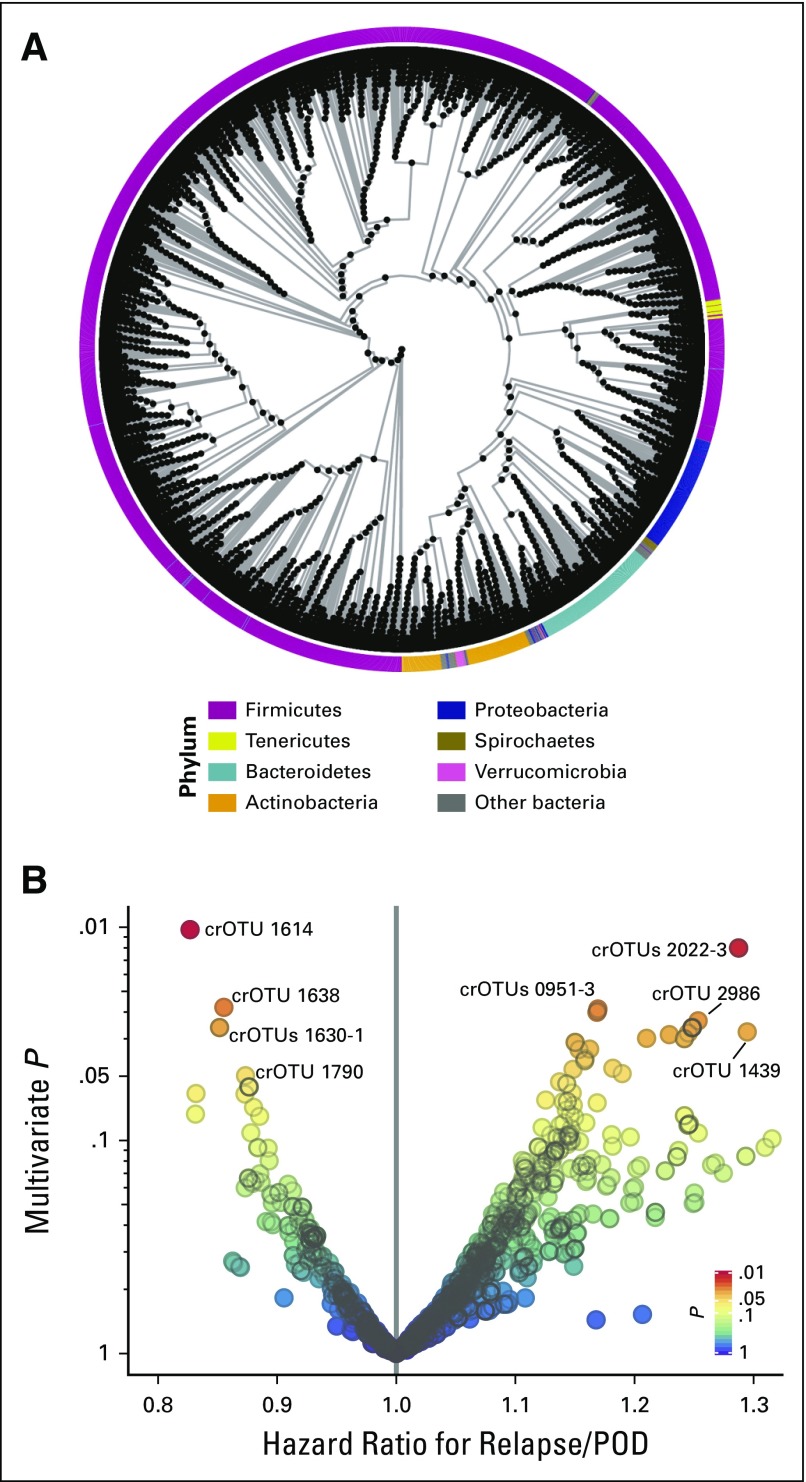
The genomic 16S ribosomal RNA V4/V5 variable region was amplified and sequenced on the Illumina MiSeq (San Diego, CA) platform, as previously described.18,24 Quality-filtered sequences with > 97% identity were grouped into operational taxonomic units (OTUs).33 Time-weighted average abundances (Data Supplement) were calculated for each OTU. Similar metrics of area-under-the-curve of concentration over time have been used in other biomarker time-series studies.34,35 A phylogenetic tree was constructed from a sequence alignment of all of the OTUs. The branch points (nodes) of the phylogenetic tree were considered to be clusters of related OTUs (crOTUs; Fig 1A; Data Supplement). The abundance of each crOTU was calculated as the sum of the abundances of its member OTUs. Additional details are in the Data Supplement.
Fig 1.
(A) Phylogenetic tree of operational taxonomic units (OTUs) and clusters of related OTUs (crOTUs). Each black point is a crOTU. Phylum is color coded along the circumference. A schematic of how the tree was constructed and a detailed tree are presented in the Data Supplement. (B) Volcano plot of multivariable P values of crOTUs against the multivariable hazard ratios for relapse/progression of disease (POD) in the discovery set. crOTUs are color coded by P value. Multivariable adjustment was performed for refined disease risk index, graft source, and conditioning intensity. The most abundant species in each of the labeled crOTUs are as follows: Eubacterium limosum in crOTU 1614; Streptococcus sinensis in crOTU 2022-3; E. limosum in crOTU 1638; E. limosum in crOTU 1630-1; Parvimonas micra in crOTU 1790; Leptotrichia hongkongensis in crOTU 0951-3; Flavonifractor plautii in crOTU 2986; and Actinomyces odontolyticus in crOTU 1439. OTU-level analysis and univariable analysis results are presented in the Data Supplement. crOTU composition at the species level and numeric values are tabulated for the top candidates and for all candidates in the Data Supplement.
Statistical Analysis
The primary outcome was time to relapse or time to POD by disease-specific criteria. Detection of minimal residual disease was scored as a relapse/POD event when flow cytometry, radiographic, or molecular results were acted upon clinically by initiation of therapy, infusion of donor lymphocytes, or withdrawal of immunosuppression. The 541-patient cohort was partitioned temporally into discovery (n = 271) and validation (n = 270) sets. Cause-specific Cox proportional hazards multivariable regression models were used to assess associations between microbiota and outcomes. The cumulative incidences of relapse/POD, transplant-related mortality, and GVHD were determined with the competing-risks method. Additional details are in the Data Supplement.
RESULTS
Patient Characteristics
Among patients who underwent transplantation at our center between 2009 and 2015, a cohort of 541 patients met the inclusion criteria and had at least one sample collected in the 3 weeks after allo-HCT (Data Supplement). This was a heterogeneous population with a variety of underlying primary hematologic malignancies (Table 1). The patients had a range of pretransplantation relapse/POD risks, as assessed by the refined disease risk index (DRI), a validated disease-focused prognostic instrument.4 The conditioning regimens and types of allografts administered varied: 50.6% of the patients received T-cell–depleted grafts, 31.8% received unmodified grafts of peripheral-blood stem cells (PBSCs) or bone marrow (BM) stem cells, and 17.6% received double umbilical cord blood transplantations. Relapse/POD events occurred in 138 of the patients (incidence, 25.5%) during the 2-year period of analysis. Characteristics of these 138 patients are tabulated in the Data Supplement.
Table 1.
Baseline Characteristics of the Patients

Microbiota Features
Genomic 16S ribosomal RNA sequences were obtained from 2,303 stool samples from 541 patients (mean, 4.3 per patient). Of these, 1,186 had been collected in the 3 weeks after stem-cell infusion (Data Supplement). According to a standard approach in microbiota studies, 16S sequences > 97% identical were grouped into bins known as OTUs.33 The relative abundance of each OTU was determined for each sample. To accommodate variegated collection time points and numbers of samples for each patient, the cumulative exposure of each patient to each OTU was calculated as a time-weighted average abundance during the 3-week period after stem-cell infusion (Data Supplement).
Intestinal microbial diversity, as assessed by the inverse Simpson index, was not associated with time to relapse/POD (P = .16; Data Supplement). This is in keeping with our prior observation of no association between diversity and relapse-related mortality.24 Next, the association of abundance of particular bacterial subsets with time to relapse/POD was considered. We hypothesized that evolutionarily related species exhibit functional similarities. Therefore, OTUs were grouped by evolutionary distance by the construction of a phylogenetic tree empirically from a sequence alignment of all OTUs identified in the whole cohort. Members of the same phyla were grouped together (Fig 1A; Data Supplement), which indicated that the tree was broadly concordant with standard taxonomy. The branch points (nodes) of the tree were defined as clusters of related OTUs (crOTUs; Data Supplement). The crOTUs were then evaluated for associations with clinical outcomes.
Association of Microbiota With Time to Relapse/POD in the Discovery Set
The analysis was limited to taxa that met an abundance threshold of > 0.01% in > 10% of patients (Data Supplement). Associations between abundance and time to relapse/POD in the discovery set were evaluated for 208 OTUs and 1,343 crOTUs (Fig 1B; Data Supplement). Abundance was considered as a log10-transformed continuous variable (Data Supplement). Multivariable models were adjusted for DRI, conditioning intensity, and graft source. The candidate most closely associated with relapse/POD risk in the discovery set was crOTU 1614 (multivariable hazard ratio [HR], 0.84; 95% CI, 0.73 to 0.96; P = .01). This HR can be interpreted as a 0.84-fold change (reduction) in risk of relapse/POD for every 10-fold increase in relative abundance of the crOTU. crOTU 1614 was composed mostly of Eubacterium limosum as well as other related species (Fig 1B; Data Supplement).
Association of Microbiota With Time To Relapse/POD in the Validation Set
The top candidate crOTU identified in the discovery set was evaluated for the reproducibility of its association with the outcome in the validation set. The abundance of crOTU 1614 was significantly associated with reduced risk of relapse/POD in the validation set (HR, 0.82; 95% CI, 0.71 to 0.95; P = .009; Table 2). This HR can be interpreted as a 0.82-fold change (reduction) in risk of relapse/POD for every 10-fold increase in relative abundance of the crOTU. This association remained significant after multivariable adjustment for conditioning intensity, graft source, and DRI (HR, 0.82; 95% CI, 0.70 to 0.96; P = .01; Table 2). Of note, the second-best candidate from the discovery set (crOTU 2022; Fig 1B; Data Supplement) was associated with an increased risk of relapse/POD in the discovery set but did not have any association with the outcome in the validation set (HR, 0.96; 95% CI, 0.8 to 1.16; P = .66).
Table 2.
Univariable and Multivariable Associations of crOTU 1614 With the Risk of Relapse/POD After Allo-HCT
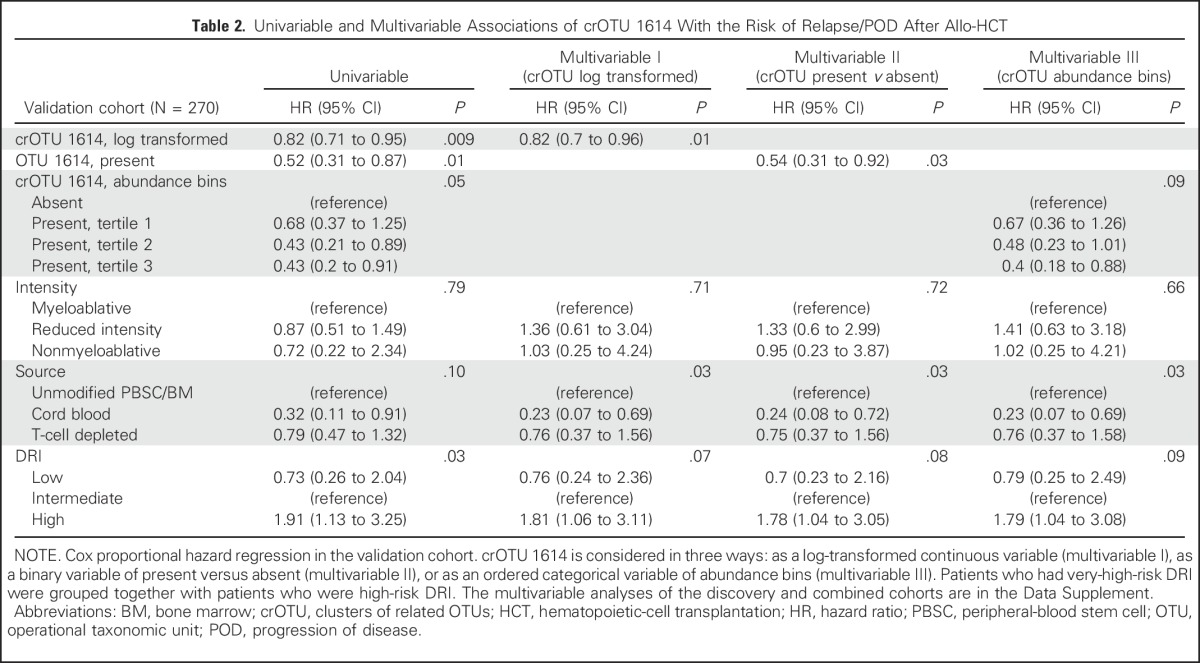
We next stratified the patients in the discovery and validation sets according to presence or absence of crOTU 1614. Presence was defined as any detectable amount, as detailed in the Data Supplement. Intestinal presence of crOTU 1614 was associated with reduced risk of relapse/POD in both the discovery (HR, 0.49; 95% CI, 0.30 to 0.82; P = .006) and the validation (HR, 0.52; 95% CI, 0.31 to 0.87; P = .01) sets (Fig 2A). In the validation set, the 2-year cumulative incidence of relapse/POD in patients with intestinal presence of crOTU 1614 was 19.8%; in patients without crOTU 1614, it was 33.8%. This association remained significant after adjustment for DRI, graft source, and conditioning intensity in both the discovery set (HR, 0.46; 95% CI, 0.27 to 0.78; P = .004; Data Supplement) and the validation set (HR, 0.54; 95% CI, 0.31 to 0.92; P = .03; Table 2).
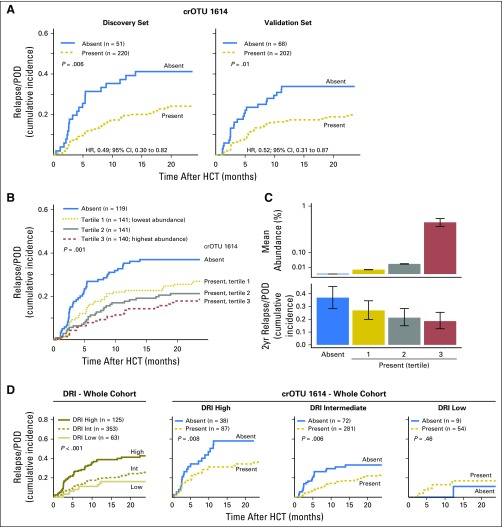
Fig 2.
Cluster of related operational taxonomic units (crOTU) 1614, which includes members of the family Eubacteriaceae, is associated with decreased relapse/progression of disease (POD) after allogenic hematopoietic-cell transplantation (allo-HCT). (A) Cumulative incidence of relapse/POD in the discovery (n = 271) and validation (n = 270) sets stratified by presence or absence of crOTU 1614. (B) Cumulative incidence of relapse/POD in the whole cohort (N = 541) stratified by crOTU 1614 abundance. (C) Upper panel, mean abundance of crOTU 1614 in the four strata (error bars are standard error of the mean) and, lower panel, cumulative incidence of relapse/POD at 2 years in the four strata (error bars are 95% CIs). (D) Far left: refined disease risk index (DRI) alone stratifies the relapse/POD risk in this cohort. Three right panels: crOTU 1614 presence further stratifies relapse among patients with high-risk and intermediate-risk DRIs. Patients with very-high-risk DRIs were grouped together with those who had high-risk DRIs. HR, hazard ratio.
Discriminatory Ability of the Biomarker Relative to Clinical Predictors: Combined Cohorts
After an association of crOTU 1614 with a lower risk of relapse/POD was reproducibly observed, the discovery and validation sets were combined for additional exploratory analyses. crOTU 1614 was present in 422 (78%) of the 541 patients in the combined cohort; the mean abundance was 0.16%, and the maximum abundance was 8%. A progressively lower risk of relapse/POD was observed across the combined cohort when it was stratified into four abundance bins (P = .001; Figs 2B and 2C), and this association remained significant after multivariable adjustment (P = .004; Data Supplement).
Examination of the clinical features of patients according to presence or absence of crOTU 1614 (Data Supplement) demonstrated no significant differences in disease type, conditioning intensity, or graft source. There was a moderate preponderance of higher-risk DRI scores in patients who had an absence of intestinal crOTU 1614 (P = .02; Data Supplement), but the association remained significant when the analysis was adjusted for DRI in a multivariable model (HR, 0.54; 95% CI, 0.38 to 0.78; P < .001; Data Supplement). Patients who had a presence of crOTU 1614 were slightly older (mean age, 54.4 years v 51.7 years; P = .04). We also investigated the association between crOTU 1614 and relapse/POD risk within each of the three DRI categories. Intestinal presence of crOTU 1614 was associated with less relapse/POD risk among patients with high DRIs (HR, 0.45; 95% CI, 0.25 to 0.81; P = .008) and intermediate DRIs (HR, 0.52; 95% CI, 0.33 to 0.83; P = .006; Fig 2D). There was no association, however, between crOTU 1614 presence and relapse/POD risk in patients with low DRIs (HR, 2.16; 95% CI, 0.28 to 16.95; P = .46).
To evaluate the discriminatory ability of crOTU 1614 in relation to known clinical risk factors of relapse/POD, we used a concordance index (c-index), in which a value closer to 1 indicates greater accuracy (Data Supplement).36 The discriminatory ability of crOTU 1614 (c-index, 0.572) was comparable to that of the DRI (c-index, 0.569). The combination of crOTU 1614, DRI, graft source, and conditioning intensity produced a moderately stronger discriminatory power (c-index, 0.650) than the three clinical factors alone (c-index, 0.619). This degree of predictive power is comparable to established models for other outcomes after allo-HCT37 and indicates that an intestinal microbiota biomarker can add to currently known clinical risk assessments of relapse/POD.
Other Clinical Outcomes and Subset Analyses: Combined Cohorts
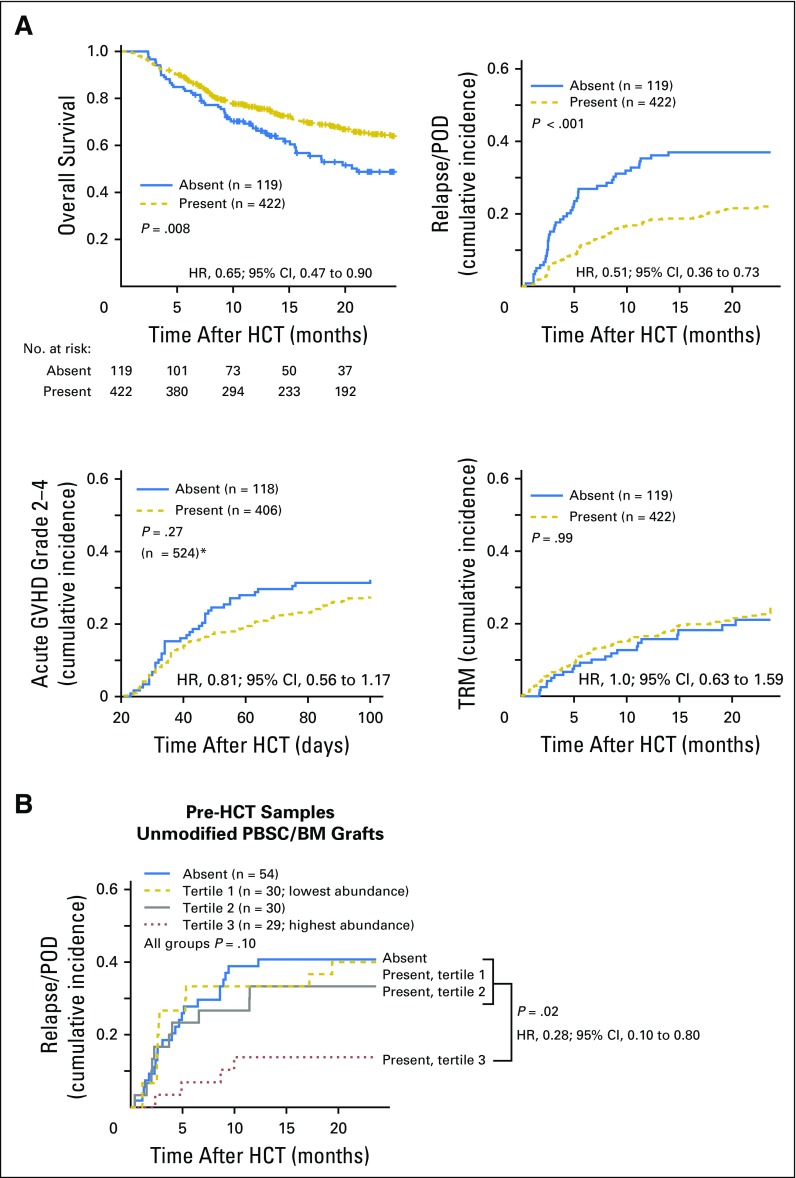
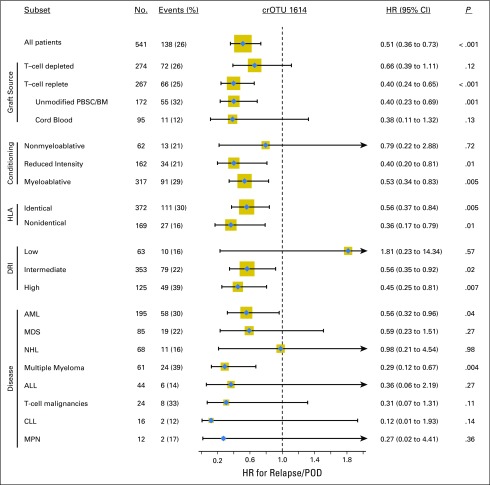
Intestinal presence of crOTU 1614 was associated with an increase in overall survival (HR, 0.65; 95% CI, 0.47 to 0.90; P = .008; Fig 3A). The crOTU was not significantly associated with acute GVHD (grades 2 to 4; HR, 0.81; 95% CI, 0.56 to 1.17; P = .27) or with transplantation-related mortality (HR, 1.0; 95% CI, 0.63 to 1.59; P = .99; Fig 3A). In light of the heterogeneity of the population under study, we explored the association of crOTU 1614 with relapse/POD in patient subsets according to graft source, conditioning intensity, extent of HLA match, DRI, and disease type (Fig 4). With respect to disease type, the association of crOTU 1614 with a reduced risk of relapse/POD was significant among patients with acute myeloid leukemia (HR, 0.56; 95% CI, 0.32 to 0.96; P = .04) and multiple myeloma (HR, 0.29; 95% CI, 0.12 to 0.67; P = .004) and not statistically significant for other disease types. For graft source (Fig 4; Data Supplement), the association of crOTU 1614 with reduced risk of relapse/POD was significant in recipients of T-cell–replete grafts (HR, 0.40; 95% CI, 0.24 to 0.65; P < .001), particularly among recipients of unmodified PBSC/BM grafts (HR, 0.40; 95% CI, 0.23 to 0.69; P = .001). No significant association was observed in recipients of T-cell–depleted grafts (HR, 0.66; 95% CI, 0.39 to 1.11; P = .12) or in recipients of cord-blood grafts (HR, 0.38; 95% CI, 0.11 to 1.32; P = .13; Data Supplement).
Fig 3.
(A) Presence of cluster of related operational taxonomic units (crOTUs) 1614 in the 3 weeks after allogenic hematopoietic-cell transplantation (allo-HCT) is associated with increased overall survival and decreased cumulative incidence of relapse/progression of disease (POD), but it is not associated with cumulative incidence of acute grades 2 to 4 graft-versus-host disease (GVHD) or with transplantation-related mortality (TRM). (*)Seventeen patients developed grades 2 to 4 GVHD before landmark day 21 and were excluded from this panel. (B) Among 143 recipients of unmodified peripheral-blood stem-cell (PBSC)/bone marrow (BM) stem-cell grafts with available pretransplantation microbiota data, those with the highest abundance of crOTU 1614 in a single sample collected before allo-HCT had a reduced risk of relapse/POD (P = .02; for all four groups P = .10). The pretransplantation analysis for recipients of all graft types is plotted in the Data Supplement. HR, hazard ratio.
Fig 4.
Association of the presence of cluster of related operational taxonomic units (crOTU) 1614 with relapse/progression of disease (POD) in patient subsets according to graft source, conditioning intensity, degree of HLA match, refined disease index (DRI), and disease type. The size of the gold box is proportional to number of patients in the subgroup. Cumulative incidence curves by graft source are plotted in the Data Supplement. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BM, bone marrow stem cells; CLL, chronic lymphocytic leukemia; HR, hazard ratio; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; NHL, non-Hodgkin lymphoma; PBSC, peripheral-blood stem cells.
Biomarker Performance in Pretransplantation Samples
A biomarker of relapse risk might have utility if it could be assessed prior to transplantation while treatment decisions are being made. Among the 172 recipients of unmodified PBSC/BM (T-cell–replete) grafts in whom the association of crOTU 1614 was most clear, 143 patients (83%) also had a sample collected in the 3 weeks before stem-cell infusion. These patients were stratified into four bins on the basis of the abundance of crOTU 1614 in a single stool sample collected during the 3 weeks before allo-HCT. The patients in the highest abundance bin had a lower risk of relapse/POD than the combined three lower-abundance groups (HR, 0.28; 95% CI, 0.10 to 0.80; P = .02; Fig 3B). Thus, intestinal presence of crOTU 1614 either before or after allo-HCT can be used as a biomarker of posttransplantation relapse/POD risk.
Bacterial Composition of the Biomarker
The composition of crOTU 1614 (Data Supplement) includes 30 OTUs, of which 5 OTUs, 7 OTUs, and a single OTU were identified as, respectively, E. limosum, Anaerofustis stercorihominis, and Pseudoramibacter alactolyticus. These are all members of the family Eubacteriaceae. An additional 15 OTUs were identified as Peptococcus niger, a member of the related family Peptococcaceae. In the whole cohort, the majority (67%) of the abundance of crOTU 1614 was attributable to E. limosum, with lesser contributions from A. stercorihominis and P. niger (15% each; Data Supplement). The OTU sequences that define the composition of this cluster are presented in FASTA format (Data Supplement). Thus, in this cohort of allo-HCT recipients, a cluster of species predominantly comprising E. limosum and other related bacteria was associated with a decreased risk of relapse/POD.
DISCUSSION
In this retrospective, observational, single-center study, the intestinal microbiota composition of patients who underwent allo-HCT was analyzed. To our knowledge, this is the largest cohort of allo-HCT patients assembled for this type of analysis to date. A discovery-validation approach was used to identify an association (Fig 2) between the abundance of a particular subset of intestinal bacteria and a decreased risk of relapse/POD after allo-HCT. In contrast, the overall diversity of the intestinal microbiota was not associated with relapse/POD. (Data Supplement). This is consistent with our previous observation in a much smaller cohort that diversity of the intestinal flora measured at the time of posttransplantation neutrophil engraftment was not associated with relapse-related mortality.24
The cohorts in which this biomarker was identified and validated were heterogeneous with respect to graft source and disease type. In an exploratory subset analysis, the association of the microbial biomarker with a lower risk of relapse/POD was strongest among recipients of grafts containing T cells and other mature lymphocytes (Fig 4). In these patients, there is a greater role for donor-cell–mediated GVT activity compared with recipients of T-cell–depleted grafts, which suggests that the composition of the intestinal microbiota could modulate GVT activity. This interpretation is interesting in light of an emerging appreciation of interactions between the intestinal microbiota and systemic immunity,38 which include antitumor immune reactions.25-31
E. limosum is the most abundant species in this group of bacteria identified as a biomarker of GVT. This bacterium is an anaerobic, non–spore-forming Gram-positive rod that is a common member of the human intestinal microbiota.39 It can ameliorate an experimental model of colitis, perhaps through production of immunomodulatory short-chain fatty acids,40 and it has been reported to characterize a microbiota signature prevalent in centenarians.41
Two aspects of the methodologic approach were particularly helpful in this analysis. First, consideration of cumulative microbial exposures as time-weighted averages of abundance allowed inclusion of many more samples per patient and potentially reduced biases that could have occurred by sampling only single time points. This is particularly relevant in the setting of the rapid shifts in microbiota composition that can occur in the weeks after allo-HCT.16,18
Another useful technique was the empirical derivation of crOTUs from the branch points (nodes) of a phylogenetic tree (Fig 1A; Data Supplement). One limitation of a traditional OTU-level analysis is that the association strength of a single species can be distributed among multiple OTUs. Conversely, in an analysis of higher taxonomic levels, such as genus or family, potential associations may be lost when dozens or hundreds of OTUs are grouped together. The approach used in this study overcame these issues by combining the abundances of evolutionarily related OTUs, as determined by 16S sequence similarity.
Most of the bacteria in the crOTU biomarker identified in this study are members of the family Eubacteriacae (Data Supplement). There are, however, an additional 15 species in the data set that belong to the family Eubacteriaceae that are not part of crOTU 1614. In fact, when standard taxonomic classifications were evaluated, neither the family Eubacteriaceae as a whole, nor the genus Eubacterium, had a significant association with relapse/POD (data not shown). Of note, the main OTU that represented E. limosum was itself associated with reduced risk of relapse (Data Supplement). This illustrates the utility of this approach in the study of associations between microbiota and clinical outcomes. Moreover, it is in line with an increasing appreciation of 16S rRNA sequence similarity as a better measure of evolutionary proximity than phenotypic traits.42
Limitations of this study include its retrospective nature and the collection of samples from patients at a single center. Prospective and multicenter approaches in strictly defined patient populations would be important next steps to confirm this association between microbiota and relapse after allo-HCT. In this study, a feature of the microbiota was identified as a potential biomarker; whether components of the microbiota have the potential to become modifiable risk factors for relapse after allo-HCT remains to be seen. With a detailed understanding of how the microbiota and the immune system interact, microbiota-based strategies43 might be designed to improve patient outcomes after allo-HCT.
ACKNOWLEDGMENT
We thank the patients who provided the samples and the nurses and clinical practitioners who collected them. We also thank Joao Xavier for helpful discussions.
Footnotes
Supported by National Institutes of Health (NIH) Awards No. R01-HL069929, R01-AI100288, R01-AI080455, and R01-AI101406 (M.R.M.v.d.B.), 5KL2-TR001115-03 (A.D.S), P01-CA023766 (R.J.O.), R01-HL124112 (R.R.J.); P30CA008748 (Memorial Sloan Kettering Cancer Center [MSKCC] support/core grant), Project 4 of Award No. P01-CA023766 (M.R.M. v.d.B.), and NIH Training Grants No. 2T32CA009207-36A1 and UL1TR00457 (J.U.P.). Also supported by the Lymphoma Foundation, the Susan and Peter Solomon Divisional Genomics Program, MSKCC Cycle for Survival, and the Tow Foundation/Lucille Castori Center for Microbes, Inflammation, and Cancer (E.G.P.). J.U.P. is a recipient of a Merck/Society for Immunotherapy of Cancer Clinical Fellowship Award, a Young Investigator Award of the Conquer Cancer Foundation, and an MSKCC Clinical Scholars Biomedical Research Training Program Award of the Charles A. Dana Foundation.
Presented previously at the Annual Meeting of the American Society of Hematology, Orlando, FL, December 7, 2015; the Annual Meeting of the American Society of Blood and Marrow Transplantation, Honolulu, HI, February 20, 2016; the Annual Meeting of the International Human Microbiota Consortium, Houston, TX, November 20, 2016; and the Annual Meeting of the Society for Immunotherapy of Cancer, National Harbor, MD, November 12, 2016.
The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institutes of Health.
See accompanying Editorial on page 1636
AUTHOR CONTRIBUTIONS
Conception and design: Jonathan U. Peled, Sean M. Devlin, Anna Staffas, Melissa Lumish, Raya Khanin, Anthony D. Sung, Daniela Weber, Amin M. Alousi, Boglarka Gyurkocza, Doris M. Ponce, Miguel-Angel Perales, Eric G. Pamer, Robert R. Jenq, Marcel R.M. van den Brink
Collection and assembly of data: Jonathan U. Peled, Sean M. Devlin, Melissa Lumish, Eric R. Littmann, Lilan Ling, Satyajit Kosuri, Molly Maloy, John B. Slingerland, Katya F. Ahr, Kori A. Porosnicu Rodriguez, Doris M. Ponce, Juliet N. Barker, Miguel-Angel Perales, Sergio A. Giralt, Ying Taur, Eric G. Pamer, Robert R. Jenq, Marcel R.M. van den Brink
Data analysis and interpretation: Jonathan U. Peled, Sean M. Devlin, Anna Staffas, Raya Khanin, Yusuke Shono, Ann E. Slingerland, Melissa D. Docampo, Ying Taur, Robert R. Jenq, Marcel R.M. van den Brink
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Jonathan U. Peled
Stock or Other Ownership: Sequenom (I)
Research Funding: Merck, Seres Therapeutics
Patents, Royalties, Other Intellectual Property: Patent on Methods and Compositions for Detecting Risk of Cancer Relapse, provisional application filed.
Other Relationship: Seres Therapeutics (licensing fee)
Sean M. Devlin
No relationship to disclose
Anna Staffas
No relationship to disclose
Melissa Lumish
No relationship to disclose
Raya Khanin
No relationship to disclose
Eric R. Littmann
No relationship to disclose
Lilan Ling
No relationship to disclose
Satyajit Kosuri
No relationship to disclose
Molly Maloy
No relationship to disclose
John B. Slingerland
No relationship to disclose
Katya F. Ahr
No relationship to disclose
Kori A. Porosnicu Rodriguez
No relationship to disclose
Yusuke Shono
Patents, Royalties, Other Intellectual Property: Seres Therapeutics
Ann E. Slingerland
No relationship to disclose
Melissa D. Docampo
No relationship to disclose
Anthony D. Sung
Research Funding: Novartis, Cellective
Daniela Weber
No relationship to disclose
Amin M. Alousi
No relationship to disclose
Boglarka Gyurkocza
No relationship to disclose
Doris M. Ponce
Consulting or Advisory Role: Guidepoint Global
Juliet N. Barker
No relationship to disclose
Miguel-Angel Perales
Honoraria: Incyte, Merck
Consulting or Advisory Role: Seattle Genetics
Sergio A. Giralt
Honoraria: Takeda, Sanofi, Jazz Pharmaceuticals, Genzyme, Amgen, Celgene
Research Funding: Takeda, Sanofi, Celgene, Jazz Pharmaceuticals
Ying Taur
No relationship to disclose
Eric G. Pamer
Patents, Royalties, Other Intellectual Property: Methods and compositions for reducing Clostridium difficile infection (Application No. WPO2015179437A1)
Travel, Accommodations, Expenses: Seres Therapeutics, Bristol-Myers Squibb, Celgene
Robert R. Jenq
Consulting or Advisory Role: Ziopharm Oncology, Seres Therapeutics, Evelo Therapeutics
Patents, Royalties, Other Intellectual Property: Intestinal blautia and reduced GVHD-related mortality (United States Provisional Application Serial No. 62/084,219, filed on November 25, 2014); Seres (licensing fees)
Marcel R.M. van den Brink
Consulting or Advisory Role: Novartis, Seres Therapeutics, Evelo Therapeutics
Patents, Royalties, Other Intellectual Property: Non–patient-specific use of hematopoietic precursor cells committed to the T cell lineage for off-the-shelf immunotherapy in immunosuppressed individuals (pending: application date, February 19, 2016); Use of IL22 as a thymic growth factor (pending: application date, June 15, 2015); T-cell receptor repertoire analysis (pending, January 5, 2015); Method of sex steroid ablation to promote immune function after radiation injury (published November 3, 2014); IL-22 as a stem cell growth factor (pending: application date, December 19, 2015); Methods of treating post-myeloproliferative neoplasms (MPNs) and post-MPN acute myeloid leukemia (pending October 31, 2014); Intestinal abundance of Blautia/Clostridiales as prognostic factors and therapeutic targets for GVHD (pending November 25, 2015); Method of use for BMP4 promoting thymus function (pending October 29, 2015); A novel small molecule inducing oxidative stress in lymphoma cells while modulating the oxidative stress defense (pending October 30, 2015); The abundance of certain bacteria in the intestinal flora is associated with relapse after allogeneic hematopoietic stem-cell transplantation (pending February 22, 2016)
Travel, Accommodations, Expenses: Seres
REFERENCES
- 1.Gratwohl A, Baldomero H, Aljurf M, et al. Hematopoietic stem cell transplantation: A global perspective. JAMA. 2010;303:1617–1624. doi: 10.1001/jama.2010.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gooley TA, Chien JW, Pergam SA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363:2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anasetti C, Logan BR, Lee SJ, et al. Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med. 2012;367:1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armand P, Kim HT, Logan BR, et al. Validation and refinement of the disease risk index for allogeneic stem cell transplantation. Blood. 2014;123:3664–3671. doi: 10.1182/blood-2014-01-552984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet S, Labopin M, Esteve J, et al. Impact of FLT3 internal tandem duplication on the outcome of related and unrelated hematopoietic transplantation for adult acute myeloid leukemia in first remission: A retrospective analysis. J Clin Oncol. 2012;30:735–741. doi: 10.1200/JCO.2011.36.9868. [DOI] [PubMed] [Google Scholar]
- 6.Walter RB, Gooley TA, Wood BL, et al. Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J Clin Oncol. 2011;29:1190–1197. doi: 10.1200/JCO.2010.31.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoudjhane M, Labopin M, Gorin NC, et al. Comparative outcome of reduced intensity and myeloablative conditioning regimen in HLA identical sibling allogeneic haematopoietic stem cell transplantation for patients older than 50 years of age with acute myeloblastic leukaemia: A retrospective survey from the Acute Leukemia Working Party (ALWP) of the European group for Blood and Marrow Transplantation (EBMT) Leukemia. 2005;19:2304–2312. doi: 10.1038/sj.leu.2403967. [DOI] [PubMed] [Google Scholar]
- 8.Brunstein CG, Gutman JA, Weisdorf DJ, et al. Allogeneic hematopoietic cell transplantation for hematologic malignancy: Relative risks and benefits of double umbilical cord blood. Blood. 2010;116:4693–4699. doi: 10.1182/blood-2010-05-285304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milano F, Gooley T, Wood B, et al. Cord-blood transplantation in patients with minimal residual disease. N Engl J Med. 2016;375:944–953. doi: 10.1056/NEJMoa1602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw BE, Gooley TA, Malkki M, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110:4560–4566. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 11.Fleischhauer K, Shaw BE, Gooley T, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: A retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venstrom JM, Pittari G, Gooley TA, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. 2012;367:805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- 14.Pavletic SZ, Kumar S, Mohty M, et al. NCI first international workshop on the biology, prevention, and treatment of relapse after allogeneic hematopoietic stem cell transplantation: Report from the committee on the epidemiology and natural history of relapse following allogeneic cell transplantation. Biol Blood Marrow Transplant. 2010;16:871–890. doi: 10.1016/j.bbmt.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenq RR, Taur Y, Devlin SM, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21:1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holler E, Butzhammer P, Schmid K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: Loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20:640–645. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shono Y, Docampo MD, Peled JU, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8:339ra71. doi: 10.1126/scitranslmed.aaf2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathewson ND, Jenq R, Mathew AV, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17:505–513. doi: 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriguchi Y, Takashima S, Oka H, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of α-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 23.Harris B, Morjaria SM, Littmann ER, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med. 2016;194:450–463. doi: 10.1164/rccm.201507-1491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taur Y, Jenq RR, Perales M-A, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulos CM, Wrzesinski C, Kaiser A, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zitvogel L, Galluzzi L, Viaud S, et al. Cancer and the gut microbiota: An unexpected link. Sci Transl Med. 2015;7:271ps1. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daillère R, Vétizou M, Waldschmitt N, et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Holler E, Hahn J, Andreesen R, et al. NOD2/CARD15 polymorphisms in allogeneic stem-cell transplantation from unrelated donors: T depletion matters. J Clin Oncol. 2008;26:338–339, author reply 339. doi: 10.1200/JCO.2007.14.1325. [DOI] [PubMed] [Google Scholar]
- 33.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 34.Mano A, Falcão A, Godinho I, et al. CA-125 AUC as a new prognostic factor for patients with ovarian cancer. Gynecol Oncol. 2005;97:529–534. doi: 10.1016/j.ygyno.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 35.Oudard S, Banu E, Thiounn N, et al: Corrected area under serum PSA curve as a predictor of survival after chemotherapy for hormone-refractory prostate cancer (HRPC) patients. J Clin Oncol 22:9579, 2004 (suppl; abstr 9579) [Google Scholar]
- 36.Wolbers M, Blanche P, Koller MT, et al. Concordance for prognostic models with competing risks. Biostatistics. 2014;15:526–539. doi: 10.1093/biostatistics/kxt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorror ML, Storb RF, Sandmaier BM, et al. Comorbidity-age index: A clinical measure of biologic age before allogeneic hematopoietic cell transplantation. J Clin Oncol. 2014;32:3249–3256. doi: 10.1200/JCO.2013.53.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajilić-Stojanović M, de Vos WM. The first 1,000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanauchi O, Fukuda M, Matsumoto Y, et al. Eubacterium limosum ameliorates experimental colitis and metabolite of microbe attenuates colonic inflammatory action with increase of mucosal integrity. World J Gastroenterol. 2006;12:1071–1077. doi: 10.3748/wjg.v12.i7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peled JU, Jenq RR, Holler E, et al. Role of gut flora after bone marrow transplantation Nat Microbiol 116036, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]