Abstract
Protein transport systems are fundamentally important for maintaining mitochondrial function. Nevertheless, mitochondrial protein translocases such as the kinetoplastid ATOM complex have recently been shown to vary in eukaryotic lineages. Various evolutionary hypotheses have been formulated to explain this diversity. To resolve any contradiction, estimating the primitive state and clarifying changes from that state are necessary. Here, we present more likely primitive models of mitochondrial translocases, specifically the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) complexes, using scrutinized phylogenetic profiles. We then analyzed the translocases’ evolution in eukaryotic lineages. Based on those results, we propose a novel evolutionary scenario for diversification of the mitochondrial transport system. Our results indicate that presequence transport machinery was mostly established in the last eukaryotic common ancestor, and that primitive translocases already had a pathway for transporting presequence-containing proteins. Moreover, secondary changes including convergent and migrational gains of a presequence receptor in TOM and TIM complexes, respectively, likely resulted from constrained evolution. The nature of a targeting signal can constrain alteration to the protein transport complex.
Keywords: mitochondria, TOM complex, TIM complex, protein transport, eukaryotes
Introduction
Mitochondria are crucially important to sustain cellular activity and viability because they are the main source of cellular ATP and are the location for other metabolic pathways such as amino acid metabolism, lipid metabolism, and formation of iron–sulfur clusters. It is now widely accepted that mitochondria were established by the endosymbiosis of an ancestral α-proteobacterium in a methane-producing archaea (Yang et al. 1985; Andersson et al. 1998; Martin and Müller 1998). It has been hypothesized that restructuring of energy generation from the endosymbiont sustains the complexity of eukaryotes (Lane and Martin 2010). Conversion into an organelle progressed with massive endosymbiotic gene transfer into the nucleus and deletion of genes that are no longer necessary for the host (Adams and Palmer 2003; Timmis et al. 2004). Establishment of protein transport systems was necessary in mitochondrial evolution because appropriate transport of nuclear encoded proteins is necessary to maintain the present mitochondrial function. The origin of the transport system is regarded as being as old as the origin of eukaryotes. The system evolved along with mitochondrial evolution. Revealing the evolutionary history of the transport systems in eukaryotic lineages is therefore useful for elucidating the evolution of diverse mitochondria.
Mitochondrial proteins are translocated across the mitochondrial double membrane by protein translocases: Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) complexes (Chacinska et al. 2009; Endo and Yamano 2009; Stojanovski et al. 2012; Schulz et al. 2015). The most common targeting signal is an N-terminal cleavable presequence. Actually, about half of mitochondrial proteins are synthesized with a presequence. The TOM complex comprises the channel-forming β-barrel protein Tom40 and six other subunits, each containing a single α-helical transmembrane (TM) segment: Tom20, Tom22, and Tom70, and the regulatory small Tom proteins (Tom5, Tom6 and Tom7). One segment that is loosely associated with the TOM complex, Tom20, acts as a receptor for the presequence. Together with Tom20, Tom22 also functions as a presequence receptor (Yamano et al. 2008). The inner membrane has translocase of two types: TIM23, a presequence translocase-associated motor (PAM) complex and the TIM22 complex. The TIM23–PAM complex delivers the presequence-containing precursor proteins to the inner membrane or matrix in a membrane potential-dependent manner. Two forms of the TIM23 complex have been identified. One form, which comprises a channel formed by Tim23 and Tim17 with accessory components, Tim50, Mgr2 and Tim21, is involved in the early stage of precursor protein transfer from the TOM complex. Tim50 functions as a presequence receptor subunit of the TIM23 complex. Tim21 binds to the TOM complex and contributes to the transient connection between the outer and inner membrane translocases. Another form is the TIM23–PAM complex, which functions in the translocation of precursor proteins into the matrix. PAM drives translocation into the matrix in an ATP-powered manner. The PAM complex comprises the molecular chaperone, mitochondrial Hsp70 (mtHsp70), Tim44, and membrane-associated co-chaperones (Pam16 and Pam18). Tim44 contributes to the connection between TIM23 and PAM. Its N-terminal and C-terminal domains respectively contribute to interaction with the PAM and TIM23 complexes, (Banerjee et al. 2015). The TIM22 complex imports hydrophobic membrane proteins, particularly mitochondrial carrier proteins with cryptic internal targeting signals (Chacinska et al. 2009; Endo and Yamano 2009; Stojanovski et al. 2012). Tim22 forms the complex pore and shares a common ancestor with Tim17 and Tim23. The hexameric chaperone complex consisting of Tim9–Tim10 delivers hydrophobic precursor proteins to the TIM22 complex in the aqueous environment of the intermembrane space (IMS).
The features of translocases presented above are mainly detected in mammals and fungi. In eukaryotic lineages, translocases can have independent differences while maintaining presequence transport. For example, a functionally analogous translocase complex named ATOM was discovered in the kinetoplastid mitochondrial outer membrane (Mani et al. 2016, 2015). Presequence receptors in the outer membrane complex have been acquired independently in Opisthokonta, Viridiplantae and Kinetoplastida (Perry et al. 2006; Eckers et al. 2012; Mani et al. 2015). However, evolutionary changes to inner membrane translocases have not been clarified. To explain differences in mitochondrial translocases among eukaryotic organisms, Dolezal et al. (2006) proposed a pioneering model of primitive forms of the translocases. Thereafter, various models for their evolution have been proposed (Lithgow and Schneider 2010; Eckers et al. 2012; Mani et al. 2016). However, these models mutually conflict. Elucidating the evolution of translocases persists as a conundrum. To investigate the evolutionary history of translocases, one must fix a primitive translocase form to define a “start point” and then estimate evolutionary changes of various aspects in eukaryotic lineages (e.g. incorporating loss of protein translocase components with loss of the transported proteins and their targeting signals). Here, we fixed primitive translocase forms based on scrutinized phylogenetic profiles of the mitochondrial translocase genes. Then we investigated their alterations in eukaryotic lineages and developed a novel evolutionary model.
Results
Evolution of the TOM Complex
Discovery of Excavata Tom7 Supports an Old Hypothesis for the Primitive TOM Complex
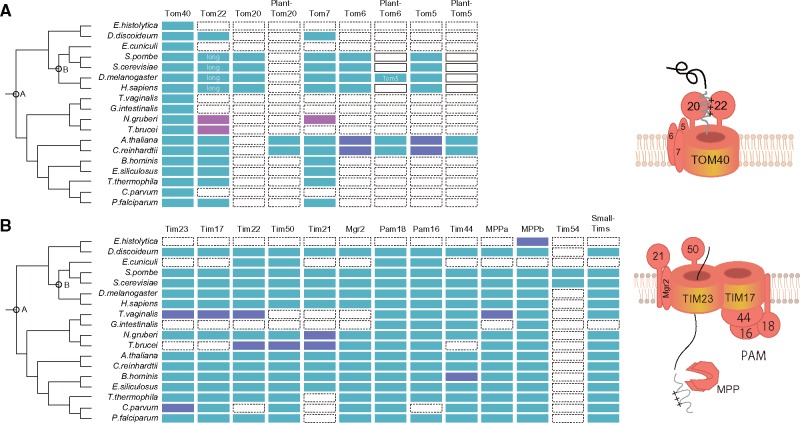
The primitive form of the mitochondrial transport system is key information for determining rational mitochondrial evolution because the transport system enabled massive gene transfer during endosymbiosis. To estimate the primitive form of mitochondrial translocase, it is necessary to determine gain, conservation, or loss of the genes coding components of the translocases. This determination depends largely on the quality of phylogenetic profiles. Therefore, we generated reliable phylogenetic profiles against 54 eukaryotic species covering five eukaryotic supergroups (supplementary table S1, Supplementary Material online, for details) using a reference tree reconstructed from 143 evolutionarily conserved genes (Hampl et al. 2009) (supplementary fig. S1, Supplementary Material online, for details). To produce reliable phylogenetic profiles, we carefully checked orthology (see Materials and Methods for details). Experimental evidence, if available, was also considered. We then estimated timing of gene gain and loss using CLIME (Li et al. 2014). Figure 1 presents phylogenetic profiles of the components of the outer and inner membrane translocases for representative organisms.
Fig. 1.
Phylogenetic profiles for (A) TOM and (B) TIM complexes. Light blue cells show the presence of components. Magenta cells show orthologs detected from a profile–profile search. Purple cells show presence with notifications: Tim23 in C. parvum is fragmental and has weak similarity; Tim17/Tim22/Tim23 family sequences of T. vaginalis cannot be classified reliably; and MPP β in E. histolytica is not localized in mitosomes (Makiuchi et al. 2013). A and B respectively, denote the left tree point LECA and the last common ancestor of opisthokonts.
For the TOM complex, our profile estimates that Tom40, Tom22, and Tom7 were gained in the last eukaryotic common ancestor (LECA) (node A in fig. 1), suggesting that the primitive TOM complex consists of at least these three proteins. This estimation is also consistent with pioneering work on this matter (Dolezal et al. 2006). However, the recent discovery of a kinetoplastid mitochondrial outer membrane translocase, the ATOM complex, casts doubt on which components are included in the primitive forms of the TOM complex. The ATOM complex comprises six subunits: ATOM40, ATOM14 (divergent Tom22 family), ATOM46, ATOM69, ATOM11, and ATOM12 (Mani et al. 2015, 2016). A channel protein of the ATOM complex, ATOM40, was first identified as a homolog of bacterial Omp85-like protein (Perry et al. 2006). However, another view holds that ATOM40 and Tom40 are members of the eukaryotic porin family (Zarsky et al. 2012). Moreover, the ATOM complex lacked a Tom7 counterpart: A homolog sequence of Tom7 has not yet been found in Excavata, including kinetoplastids and Naegleria gruberi (Mani et al. 2015, 2016). Key points in this argument are therefore whether ATOM40 belongs to the eukaryotic porin family and whether the primitive TOM complex includes Tom7.
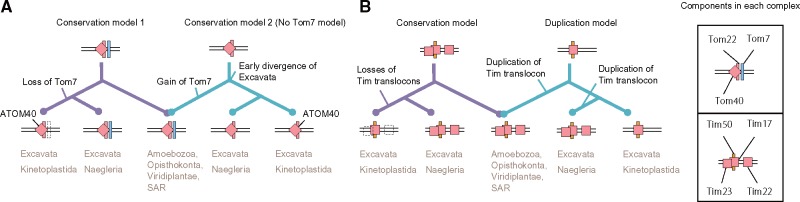
Our profile-sequence search detected ATOM40 as a member of the eukaryotic porin family with a significant score. This result strongly supports a previous view, that ATOM40 is likely to be a member of the eukaryotic porin family (Zarsky et al. 2012). Therefore, we conclude that ATOM40 is a member of the eukaryotic porin family. The origin of Tom7 is rather a settled issue. Two “conserved models” should be considered with the primitive TOM complex, including ATOM40 belonging solely to the eukaryotic porin family (fig. 2A). In the conservation models, a crucial issue is whether the primitive TOM complex includes Tom7, or not, because no Tom7 homolog has been found in Excavata (Mani et al. 2015, 2016). Moreover, the difference in the models relies on arguments related to segregation timing of Excavata in eukaryotic evolution, which are related to the root of eukaryotic phylogeny (Hampl et al. 2009; Fritz-Laylin et al. 2010; Derelle et al. 2015). “Conservation model 1” says that the primitive TOM complex consists of Tom40, Tom22 and Tom7; then Tom7 was lost in divergence to kinetoplastids. However, in “conservation model 2” the primitive TOM complex consists of Tom40 and Tom22; then Tom7 is integrated into the TOM complex after divergence of Excavata. However, the Excavata (or sub category Discoba) root is not supported by a recently reported reliable phylogenetic tree (Derelle et al. 2015) suggesting that conservation model 2 is unsuitable. It can therefore be confirmed that Excavata Tom7 is supportive of conservation model 1. However, sequenced genomes are still limited for Excavata, which engenders poor sensitivity of sequence profile searches, even with profile–profile methods. In the profile generating process, we therefore added the proteome from the recently sequenced Naegleriafowleri genome (Zysset-Burri et al. 2014). To our surprise, the sequence profile search revealed previously unidentified sequences having fused features of Tom7 and Tom22 in both N. gruberi and N. fowleri (NAEGRDRAFT_69998 and NF00098930, respectively). The first and second predicted TM segment of the genes respectively showed sequence similarity to Tom7 and Tom22 (supplementary fig. S2, Supplementary Material online, for details). Consequently, conservation model 1 predicts the most likely form of the primitive TOM complex: The primitive TOM complex consisted of at least Tom40, Tom22, and Tom7. It is particularly interesting that the three components form a structural core in the recently revealed TOM complex architecture. In fact, they are tightly and symmetrically packed as a complex (Shiota et al. 2015). Additionally, two important points can be discussed: 1) existence of the fused gene supports the root of the current phylogenetic tree and 2) divergence of Tom22 or Tom7 seems to be earlier than divergence of Tom40 in the ATOM complex evolution.
Fig. 2.
Proposed models for evolution of (A) TOM and (B) TIM complexes.
Primitive Tom40 and Its Paths for Precursor Protein Transport
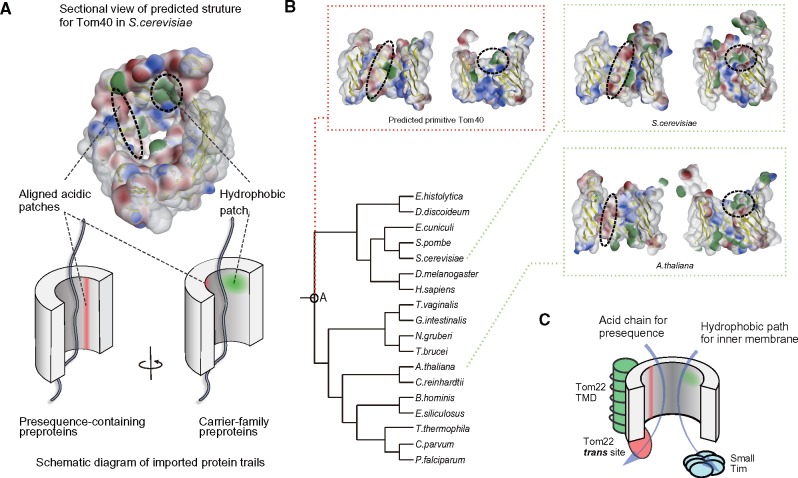
Presequence receptors have been acquired convergently: Tom20, plant-type Tom20, and ATOM46 were gained independently in eukaryotic lineages (Perry et al. 2006; Mani et al. 2015). The last common ancestral form of TOM complex consisted of at least Tom40, Tom22, and Tom7 and therefore probably imported precursor proteins without the aid of a specific receptor. How does the primitive translocase conduct this fundamental task? Our recent interaction mapping of yeast Tom40 using a photo cross-linking approach revealed different protein transport paths for positively charged presequence precursors and hydrophobic precursors by membrane carrier proteins on the inner wall of the Tom40 pore; aligned acidic patches are located near the residues that cross-link with presequences, whereas hydrophobic patches are located near the residues that cross-link with the carrier precursor (Shiota et al. 2015) (fig. 3A). To investigate the conservation of the physicochemical properties on the inner wall of Tom40, we generated 3D structure models of Tom40s using unikont and bikont (unikont and bikont models from Saccharomyces cerevisiae and Arabidopsisthaliana, respectively) (fig. 3B). The aligned acidic patches were observed at similar positions on the inside of pores of ScTom40 and AtTom40, suggesting that the primitive TOM complex may import protein directly using the acidic patches. Hydrophobic patches also showed clearer conservation. The two transport paths might have been maintained from primitive mitochondria. To explore this hypothesis, we conducted ancestral sequence reconstruction from multiple sequence alignments of Tom40 orthologs; then we modeled the 3D structure of the ancestral Tom40. As expected, both aligned acidic and hydrophobic patches were observed in the ancestral Tom40 model (fig. 3B). The existence of the aligned acidic patch in primitive TOM40 is consistent with conservation of the negatively charged IMS domain of TOM22 (supplementary fig. S3, Supplementary Material online, for details), which is important for presequence importation in yeast (Moczko et al. 1997). Conservation of the hydrophobic patch is also consistent with conservation of IMS chaperone proteins, small Tims, which contribute to the import of carrier proteins (Chacinska et al. 2009; Endo and Yamano 2009; Stojanovski et al. 2012) (fig. 1). Considering conservation of the aligned acidic patch, acidic IMS domain of Tom22, hydrophobic patch and small Tims, we modeled transport of the primitive TOM complex (fig. 3C). The model suggests that a fundamental import system was already established before divergence of supergroups. A previously proposed acid chain model for presequence transport (Schatz 1997), in which precursors containing positively charged presequence are relayed from the TOM to the TIM23 complex following acidic feature of the complexes, is likely to have been developed mostly in a primitive TOM complex.
Fig. 3.
Analyses of physicochemical properties of the inner wall of predicted Tom40 structures. (A) Schematic diagram of acidic and hydrophobic patches on the interior of the Tom40 model (B) analysis of electrostatic and hydrophobic patches of the interior surface of the LECA, S. cerevisiae, and A. thaliana Tom40 models (C) Transport model for the primitive form of the TOM complex.
Determination of Tom20 Gain Supports Extension of Tom22 as a Secondary Event
The TOM complex evolved independently into various complex forms in eukaryotic lineages. The most important alteration is the gain of a presequence receptor. It was reported recently that an Amoebozoa, Acanthamoeba castellanii TOM complex, has Tom20 and Tom22 (Wojtkowska et al. 2015; Buczek et al. 2016; Mani et al. 2016). The orthology of the Amoebozoa Tom20 is important to ascertain the gain timing of Tom20. Based on the amoeba Tom20 gene, two alternative scenarios now exist for Tom20 gain (supplementary fig. S4, Supplementary Material online, for details). Known presequence receptors have helical repeat domains such as tetratricopeptide repeats (TPRs) or armadillo (ARM) repeats for presequence recognition; the Amoebozoa Tom20 has a TPR. However, searching for helical repeats requires careful investigation because of local and strong similarities over different protein families. Therefore, careful ortholog search is necessary to ascertain whether the TPR emerge from same origin of known Tom20 TPRs. Although we conducted careful ortholog searches (see Materials and Methods), we were unable to show that the Amoebozoa Tom20 shares a common origin with the known presequence receptors. In addition, the Amoebozoa Tom20 does not have a predictable TM domain (TMD) although a TMD is predicted in both opisthokont and plant Tom20s. We conclude that Tom20 was conserved only in opisthokonts, plant-type Tom20 was conserved among green plants, and ATOM46 was conserved only in kinetoplastids (fig. 1; supplementary table S1, Supplementary Material online, for details). We infer that Amoebozoa Tom20 was gained independently and that the TPR motif is a result of convergent evolution in Amoebozoa.
In yeast, Tom22 also contributes to recognition of the presequence: Tom20 interacts with the hydrophobic face of the presequence, whereas Tom22 interacts with the positively charged face (Shiota et al. 2011). Consequently, collaborative recognition was developed during evolution from the LECA to yeast. Coevolutionary change in Tom22 is therefore important to explain Tom20 evolution. Intriguingly, Tom22s in fungi and metazoa have an acidic N-terminal extension that contributes to recognition of positively charged presequences (fig. 1 and supplementary fig. S3, Supplementary Material online, for details). Two possibilities exist for the evolution of the domain: The domain was acquired as a result of elongation of the Tom22 ancestor (Maćašev et al. 2004) or it was truncated (Carrie et al. 2010). To settle the argument, determination of the Tom22 state in Amoebozoa is crucial because of its phylogenetic position. Genes annotated as Tom22 in the Amoebozoa TOM complex (Wojtkowska et al. 2015; Buczek et al. 2016) share no sequence similarity to known Tom22, as argued by Mani et al. (2016). In contrast to the obscure Tom22, we found more reliable Tom22 orthologs in two sequenced Amoebozoa genomes (Dictyostelium discoideum and Polysphondyliumpallidum): DDB0219756 and PPL_03846. These genes conserve invariant tryptophan and proline in a predicted TM region (supplementary fig. S3, Supplementary Material online, for details). Moreover, mitochondrial localization of the Tom22 ortholog, ACA1_095640, was confirmed previously in amoeba A. castellanii (Gawryluk et al. 2014). The gene has a short N-terminal domain and no acidic cluster, suggesting that primitive Tom22 had a short N-terminal domain. The estimated elongation timing of Tom22 is consistent with gain timing of the opisthokont Tom20. Therefore, it is likely that the composite recognition of presequences by Tom20 and Tom22 is established after speciation from Amoebozoa on the phylogenetic tree (node B in fig. 1). This is also consistent with the fact that the cytosolic domain of plant short Tom22 (Tom9) lacks the ability to bind presequence (Rimmer et al. 2011).
Evolution of the TIM Complex
Estimation of the Primitive TIM Complex
The TIM23–PAM and TIM22 complexes in the inner membrane receive precursor proteins from the TOM complex and further transport them to the matrix or inner membrane. Many components of inner membrane translocase complexes are widely conserved, implying the early gain and functional importance of the complexes (fig. 1). Three channel proteins (Tim17, Tim22, and Tim23) are present in the TIM23–PAM and the TIM22 complexes. These channel proteins, which belong to the same protein family, are considered to be a result of early duplications occurred before the appearance of the LECA. Which is a true ancestral gene remains obscure. Reliable determination of the ancestor in this family requires an outgroup such as a bacterial counterpart. However, we did not find a significant outgroup sequence of this family from sequenced genomes. The establishment of the TIM22 complex seems to be a newer event than that of TIM23–PAM complex. Subunits of yeast TIM22 complex, Tim54 and Tim18, acquired in fungi, and a novel subunit Tim29 was recently discovered in human TIM22 complex (Kang et al. 2016). Tim29 is conserved only in metazoan. Therefore, TIM22 complexes in yeast and human independently evolved in each lineage.
Most eukaryotic organisms have the three channel proteins, but only one of the three genes is found in kinetoplastid genomes where ATOM components are coded. Similar to estimation of the primitive TOM complex, the existence of a one-channel protein and ambiguous phylogenetic position of kinetoplastids suggest two models for the common ancestral form of TIM complexes (Schneider et al. 2008; Eckers et al. 2012) (fig. 2B). The conservation model is based on the hypothesis that the three channel proteins existed at the LECA and that two of them have been lost during divergence to kinetoplastids. The duplication model emerged from the hypothesis that a channel protein existed at the LECA and that the Excavata lineage branched first in eukaryotic evolution and that the remaining two proteins were gained by gene duplications in and out of the Excavata clade. As described above, a recent phylogenetic tree (Derelle et al. 2015) does not support the early branching of Excavata. Therefore, the duplication model is less likely. In addition, the Excavata genome of N. gruberi has three channel proteins, supporting the last common ancestor having the three channels. Based on our phylogenetic profile and the conservation model, the last common ancestral form of the TIM23 complex is estimated as containing all known components of the complex: Tim23, Tim17, Tim50, Tim21, and Mgr2. It is particularly interesting that the primitive TIM form has almost identical components to those observed in yeast (Dolezal et al. 2006). Similarly, the primitive TIM22 complex was estimated as having contained the main components for protein translocation: Tim22 and small Tims. The TOM complex hands over the precursor protein with a positively charged presequence to the TIM23 complex and also hydrophobic precursors to the TIM22 complex. Considering that the estimated primitive Tom40 has acidic and hydrophobic patches in its pore, cooperative protein sorting by TOM and TIM complexes using acidic chains and hydrophobic paths was likely established at the LECA.
Analogous Functional Inner Membrane Translocase in Kinetoplastids
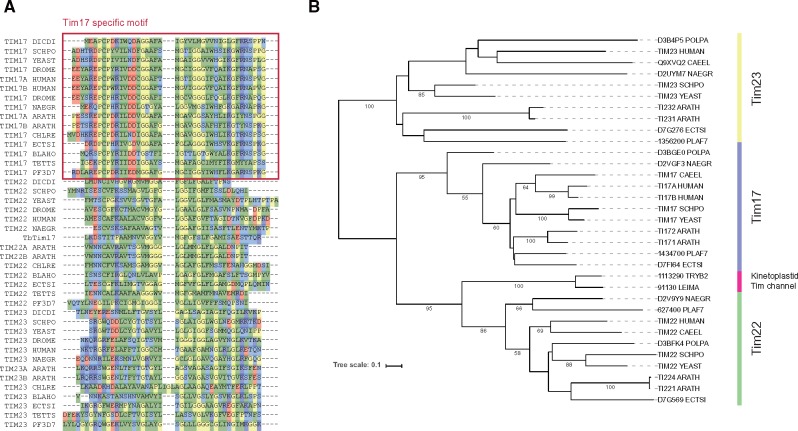
In contrast to TOM complexes, evolution of TIM complexes has not been clarified. The loss pattern of the Tim17/Tim22/Tim23 family appears to be stochastic in eukaryotic lineages: There is no well-conserved subfamily as there is for Tom40 (fig. 1). Like the TIM23 complex members, components in the PAM complex/module are also widely conserved. The conservation pattern of Tim44, which tethers the TIM23 and the PAM complex, is particularly noteworthy. Tim44 has a bacterial origin (Clements et al. 2009) and consists mainly of three parts: Presequence, N-terminal, and C-terminal domains (supplementary fig. S5, Supplementary Material online, for details). Importance of the N-terminal region of mature Tim44 for interaction with PAM was confirmed genetically (Schilke et al. 2012). Recently, interaction of the C-terminal domains of Tim44 and Tim17 was reported (Banerjee et al. 2015). The conservation of Tim17 and Tim44 are closely correlated (fig. 1). Actually, E. hystolytica and microsporidia have no Tim44 orthologs and have also lost Tim17, although the former has no a detectable TIM23 complex and the latter has only conserved Tim22. Curiously, kinetoplastids have no homologous sequences for Tim44 (Martincová et al. 2015), although Tim17 is reportedly conserved (Singha et al. 2008). The TIM complex of Trypanosomabrucei, named Tim17, consists of a 16.2 kD channel protein (TbTim17) and several subunits, including lineage specific genes (Singha et al. 2012). TbTim17 does not have well-conserved Tim17 specific N-terminal motif (fig. 4A), although its function and mass are similar to that of Tim17. Therefore, we reexamined the orthology of the Tim17/Tim22/Tim23 family carefully using two distinct methods (phylogenetic analysis and clustering based on reciprocal best-hits) with a literature search because of mutual similarity among the family. It is particularly interesting that results of our phylogenetic analysis suggest that TbTim17 is potentially diverged from Tim22 (fig. 4B), which is responsible for importing hydrophobic membrane proteins. Orthology to Tim22 is also supported by clustering (e.g. the kinetoplastid Tim17 genes are clustered with Tim22 orthologs in OrthoMCL, OG5_128586). Considering clear conservation of the Tim17-Tim44 pair in other organisms, the kinetoplastid TIM complex channel protein is likely to be a diverged Tim22. The kinetoplastid TIM complex is unlikely to be a canonical TIM23 complex, although it retains presequence transport ability (Häusler et al. 1997).
Fig. 4.
Analyses of the Tim17/Tim22/Tim23 family. (A) Extracted N-terminal Tim17 motif region following multiple sequence alignment of the Tim17/Tim22/Tim23 family. (B) Reconciled gene tree for the Tim17/Tim22/Tim23 family with selected taxons by PhyML in the tree best package. Numbers on branches are bootstrap values in 100 replications (<50 are hidden).
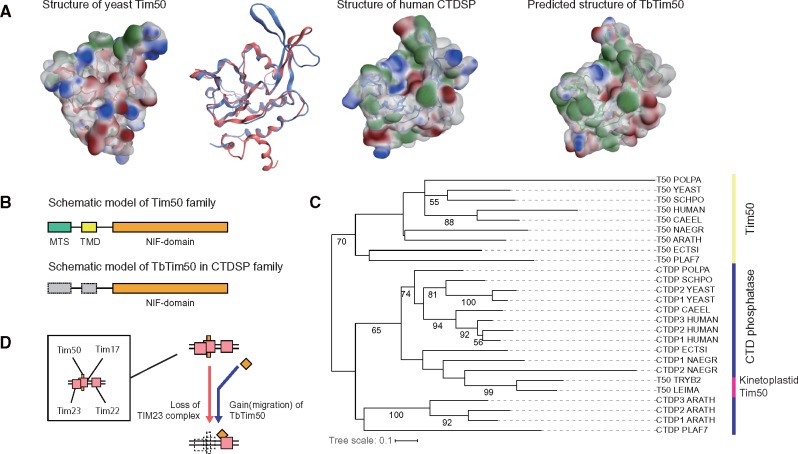
Reportedly, the T. brucei TIM complex retains a homolog of Tim50, presequence receptor in the TIM23 complex (Duncan et al. 2013). Tim50 has a presequence, a TMD, and an NIF domain, which is known for the phosphatase activity (Pfam: PF03031). The existence of Tim50 is inconsistent with loss of the TIM23 complex. However, we found that T. brucei Tim50 (TbTim50) is also distinct from canonical Tim50; Clustering based on reciprocal best-hits suggests that TbTim50 is a member of a different gene cluster (OG5_127075 in OrthoMCL), which includes C-terminal domain RNA polymerase II polypeptide A small phosphatase 1 (CTDSP1) and no canonical Tim50 genes. Structural features of TbTim50 support a different origin: 1) the surface properties of the NIF domain model of TbTim50 appears closer to human CTDSP1 than to yeast Tim50 (fig. 5A) and 2) TbTim50 probably lacks TMD because a predicted TMD (Duncan et al. 2013) is located in the NIF domain (fig. 5B and supplementary fig. S6, Supplementary Material online, for details). The reconciled gene tree also supports TbTim50 as an ortholog of the CTDSP family (fig. 5C). Collectively, the original Tim50 was lost and evolutionary migration of the CTDSP family into the Tim17 complex happened on the evolutionary path to the kinetoplastids. Considering the Tim22 orthology of the channel protein, the Tim44-Tim17 pair and the replacement of Tim50, the T. brucei TIM complex is not a diverged TIM23 complex. We conclude that the inner membrane complex of T. brucei, and possibly other kinetoplastids, is dramatically altered. We summarize the evolutionary alteration in figure 5D. The kinetoplastid TIM complex could evolve to retain presequence transport by the replacement and gain of novel subunits.
Fig. 5.
Analyses of TbTim50. (A) Comparisons among predicted structures of TbTim50, ScTim50 and HsCTDSP (B) Schematic models for primary structures of the canonical Tim50 gene and TbTim50 (C) Reconciled gene tree for Tim50 and the CTDP family. The procedure used is the same as that used for the Tim17/Tim22/Tim23 family. Numbers on branches are bootstrap values of 100 replications (<50 are hidden) (D) reported model for evolution of Tim50 in kinetoplastids.
The Targeting Signal is Likely to Function as a Constraint to Translocase Evolution
Although the mitochondrial translocases are remarkably different among organisms, their presequences are rather conserved in terms of properties and functions (Eckers et al. 2012; Mani et al. 2015). The existence of our estimated primitive TOM complex, the TIM23–PAM complex, and the acidic patches in the primitive Tom40 pore all indicate that numerous presequence-containing genes are likely to exist in the LECA. Consequently, the evolution of translocases is likely to retain presequence recognition. Indeed, presequence receptors in the TOM complex were convergently gained in Opisthokonta, Viridiplantae, and Kinetoplastida. Moreover, Tom22 was elongated independently to recognize presequences in opisthokonts, whereas kinetoplastid ATOM and TIM complexes independently gained analogous receptors for presequences through convergent evolution or migrational gains. If translocases evolved from the LECA to retain recognition of presequence properties, then the following two points must be confirmed: 1) whether presequences are conserved from the LECA to extant organisms, or not; and 2) whether newly gained genes acquire a presequence, or not, even after alteration of the translocases. Kinetoplastids are appropriate to discuss this hypothesis because kinetoplastid presequences function in other organisms (Eckers et al. 2012) and because their translocases are altered dynamically.
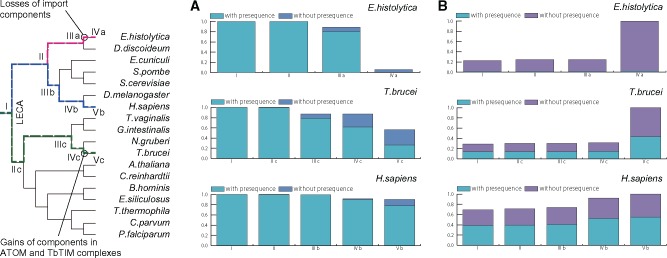
To examine the conservation of presequences, we first estimated the primitive presequence-containing genes. To extract primitive presequence-containing orthologs, we developed PhyloFates using phylogenetic profiles with explicit error rate consideration. PhyloFates estimated 245 primitive presequence clusters (supplementary table S2, Supplementary Material online, for details, and see Materials and Methods; supplementary Materials and Methods, Results and Discussion, Supplementary Material online). We investigated conservation rates of the estimated gene clusters in each evolutionary stage of T. brucei and compared the results with those of Homo sapiens and Entamoeba histolytica (fig. 6A). E. histolytica has reductively evolved mitochondria named mitosomes, which are altered drastically in terms of their inability to produce ATP using oxidative phosphorylation, and in modified metabolite transport processes and biochemical pathways (Makiuchi and Nozaki 2014). It was also discussed that the loss of the mitochondrial membrane potential led to the loss of presequence in their genomes (Garg and Gould 2016). Mitochondria in H.sapiens rarely lost primitive genes or their presequences during evolution, although mitosomes in E. histolytica lost almost all the genes and presequences between stage IIIa and IVa (fig. 6A). Our primitive presequence cluster analysis showed shrinkage of mitochondrial proteins and loss of presequence in E. histolytica. It is particularly interesting that T. brucei mitochondria show intermediate evolution. Kinetoplastid mitochondria lost numerous primitive genes and their presequences. However, a considerably large number of genes with presequences are still conserved after alterations of translocases occurred between stage IVc and Vc (fig. 6A).
Fig. 6.
Gains and losses of orthogroups and their presequences in representative organisms. The Y-axis shows the fraction of ortholog states (absence, existence without presequence, and existence with presequence). The X-axis shows evolutionary stages, which are represented in the schematic tree. (A) Losses of estimated 245 primitive presequence-containing orthogroups. Bars show fractions of estimated ancestral states of 245 orthogroups at each stage. (B) Gains in mitochondrial or mitosomal proteomes. Fractions of genes with and without predicted presequences are shown in bars. Presequence prediction was performed by MitoFates (Fukasawa et al. 2015). Genes gained during stage I are primitive genes; genes gained after stage IV or V are more lineage-specific.
Secondly, we estimated gain timings of mitochondrial genes and then analyzed whether newly gained genes can acquire presequences, or not, after translocase alteration in three representative organelle proteomes (Mi-Ichi et al. 2009; Panigrahi et al. 2009; Calvo et al. 2015). Most T. brucei mitochondrial genes are estimated as gained after divergence to Kinetoplastida. A large fraction of those lineage specific genes obtained presequences, even after translocase alteration (Vc in fig. 6B). Most human mitochondrial genes are estimated as gained in the LECA. New presequence gains were also observed. A large fraction of mitosomal genes were estimated to have gained lineage specifically in E. histolytica. Such gained genes contain only one predicted presequence, which is within the range of the expected error rate of our presequence predictor, MitoFates (Fukasawa et al. 2015). Therefore, the loss of presequence in E. histolytica was also observed in this estimation using known mitosomal genes (fig. 6B).
Given the conservation of primitive presequences and the continuous gain of presequences after translocase alteration, the properties of presequences are likely to constrain the evolution of mitochondrial translocases. Apparently for this reason, the exchangeability of presequences between distant organisms (Eckers et al. 2012). If this constraint is lost, then more drastic changes (losses of most components) can occur in the transport system, as exemplified by the case of the E. histolytica mitosome.
Discussion
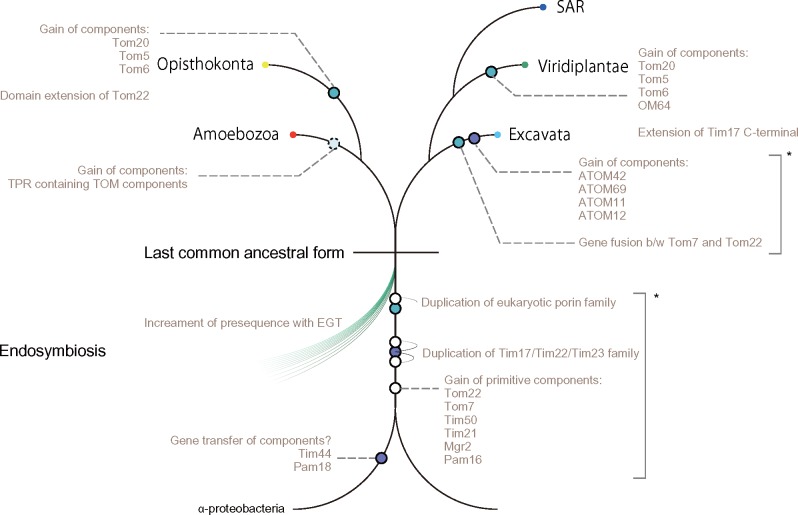
Estimation of the primitive translocase form is a challenging and fundamental problem, but it is one that must be resolved to elucidate its evolution. We suggest primitive forms of TOM and TIM complexes, which are estimated as mostly established in the LECA, based on carefully made phylogenetic profiles of their components and a structure model of primitive Tom40. This suggestion is supported by the 245 estimated primitive presequence-containing gene clusters. The primitive model helps to explain which part of the translocase is conserved and altered. Our modeled evolutionary alterations to TOM and TIM complexes are shown in figure 7. In our model, the primitive TOM complex consists of Tom40, Tom22, and Tom7, whereas the primitive TIM23–PAM complex consists of Tim17, Tim23, Tim50, Tim21, Tim44, Mgr2, Pam18, and Pam16 in the LECA. Our model is consistent with that presented in pioneering work on this matter (Dolezal et al. 2006). Moreover, the primitive model for the TOM complex is the same. Our primitive TIM23 complex model additionally contained Tim21, Tim50, and Mgr2. Even though the protein transport system is a core factor for mitochondrial evolution, this system is evolving independently to transport precursors effectively. Gain timing of Tom5 and Tom6 remains unclear because it is difficult to detect orthologs of these families and ambiguous orthology exists between the two protein families. For example, yeast Tom5 appears to be similar to plant Tom6; yeast Tom6 is similar to plant Tom5 (Mani et al. 2016). We were unable to detect significant homology between fungi Tom5 and plant Tom6 or between fungi Tom6 and plant Tom5, even though we used highly sensitive sequence searches (fig. 1). At present, it remains difficult to ascertain definitively whether the primitive TOM complex contains Tom5 and Tom6, or not. In Excavata, the highly diverged ATOM complex has garnered interest, but the TOM complex in Naegleria also seems to be atypical. The TOM complex in this lineage remains an open question (see supplementary Results and Discussion, Supplementary Material online for details). This point demands further experimental confirmation.
Fig. 7.
Summarized evolutionary events of mitochondrial translocases. Asterisks denote that the chronological order of events is elusive.
The primitive model also sheds light on functions of the primitive component Tom22: Assuming the receptor function of the cytosolic domain of Tom22 is limited in opisthokonts, what is the conserved role of Tom22 in the TOM complex? One conserved feature is an acidic IMS domain (supplementary fig. S3, Supplementary Material online, for details), which contributes to presequence import (Moczko et al. 1997). Additionally, we recently found that Tom22 tethers Tom40s to form a trimeric TOM complex (Shiota et al. 2015). A hypothesized model for the interaction of Tom22 with Tom40 is that the TM segment of Tom22, bent at the proline, tethers two Tom40s through the interactions of its N-terminal basic residue cluster and C-terminal acidic residue cluster. Indeed, the TM region of Tom22 has been conserved through evolution, with an invariant proline, flanked by a basic residue cluster and an acidic residue cluster (supplementary fig. S3, Supplementary Material online, for details). The primitive TOM complex might adopt a trimer form.
Decades ago, the acid chain hypothesis was suggested: Presequence-containing precursor proteins are relayed from the TOM to the TIM23 complex following acidic features of the complexes (Schatz 1997). A recent study supports the hypothesis (Shiota et al. 2015). Our primitive translocase model supports the relay system being developed mostly by the LECA. The protein complex would be functionally expanded by adding new subunit(s). Mutational proximity in the protein interaction space can explain such expansion (Levy 2010). In fact, a Pam18-like gene in α-proteobacteria is functional in the TIM23 complex with manual mutation (Clements et al. 2009). In bacteria, even very specific interaction can appear by few mutations (Aakre et al. 2015). As described above, the kinetoplastid TIM complex acquired novel subunits by the replacement of similar folded proteins under the constraint of presequence transport. This proximity is apparently fundamentally important for generation and expansion of the protein complex.
Although the transport system is fundamentally important to maintain mitochondrial function, the protein translocases are altered independently in eukaryotic lineages. This flexible alteration is apparently a strategy to sustain a system that transports proteins of various types in different environments. In contrast to the flexibility, the properties of presequences are conserved among organisms. The existence of presequences can be expected to constrain the evolution of mitochondrial translocases, as in the case of the kinetoplastid analogous translocase. The loss of the constraint can be expected to result in great losses of import machinery components (e.g. mitosomal translocases). What is a major driving force of loss of the presequence constraint? It was recently discussed that the loss of membrane potential leads to the loss of presequences (Garg and Gould 2016). Considering that the transport pathway depends on the membrane potential and positively charged feature of presequence, it seems reasonable. In addition to E. histolytica, other species containing MRO losing membrane potential, such as Encephalitozooncuniculi, Trichomonasvaginalis, and Cryptosporidiumparvum, indeed lost all subunits including presequence receptors in the canonical presequence transport, except Tom40 (fig. 1), as previously discussed for microsporidia (Waller et al. 2009).
Moreover, recent studies investigating protein transport in another MRO, hydrogenosome in T. vaginalis have revealed the existence of N-terminal independent matrix targeting (Garg et al. 2015; Rada et al. 2015). It is noteworthy that the N-terminal independent targeting signal is also functional in yeast, supporting its early acquisition: The recognition system for this targeting signal is established at least in LECA. Combining the N-terminal independent targeting with the primitive presequence clusters and two characteristics (acidic and hydrophobic) patches of primitive Tom40 pore, the LECA mitochondria are likely to have already imported proteins using multiple targeting systems. It seems that those targeting systems had been established between first eukaryotic common ancestor (FECA) and LECA. However, it is difficult to estimate the transition of the import system between FECA and LECA from current methodologies using available sequenced genomes. Garg and Gould recently discussed that the early mitochondrial protein import was likely positive charge-independent if the early mitochondrion imported proteins in the presence of ATP-synthesis at the host’s plasma membrane (Garg and Gould 2016). Details of the N-terminal independent targeting signal remain unknown, but the feature of this signal and import pathway are interesting topics in the sense that primitive mitochondrial protein targeting is expected to performed under low membrane potential (Garg et al. 2015; Garg and Gould 2016). Recently, a single-pass membrane protein, subunit e of –ATP synthase (Su e) is reported to be transported by membrane potential-independent import in yeast (Turakhiya et al. 2016). Its N-terminal hydrophobic region including TMD was also reported to be crucial in mitochondrial targeting (Everard-Gigot et al. 2005). It is an interesting question whether the hydrophobic patch of primitive Tom40 pore is involved with the N-terminal independent targeting signal, which is putatively important for ancient membrane proteins.
Materials and Methods
Proteome and Genome Data Sets
Predicted amino acid sequences for 54 species, and in addition N. fowleri, were retrieved from either UniProt (UniProt Consortium 2015) or EupathDB (Aurrecoechea et al. 2010). Genome sequences for the same species were retrieved from NCBI. These data sources are presented in supplementary table S3, Supplementary Material online, for details. For computational efficiency, 100% identical sequences of the same length were removed using CD-HIT (Li and Godzik 2006). Suspicious entries were filtered out by cross-referencing the NCBI gene DB if reliable mapping was available for proteomes (Maglott et al. 2005).
Phylogenetics
Phylogenetic analysis was conducted by maximum likelihood using RAxML ver. 8.1.17 (Stamatakis 2014) and the LG model (Le and Gascuel 2008). Rate heterogeneity among sites was considered using a discrete gamma distribution with four categories. For a reference tree of selected taxons in this research, 143 conserved protein sequences were extracted from all eukaryotic species in the data set (Hampl et al. 2009). Homologous sequences for species not listed in the data set were selected with an automatic pipeline (Grant and Katz 2014) and manual curation. The original data set consisted of sequences from multiple species of a few taxons. Such sequences were replaced with orthologous sequences from the 54 species. Amino acid sequences were aligned using MAFFT (Katoh and Standley 2013). Trimming was conducted automatically using trimAl (Capella-Gutiérrez et al. 2009). Alignment for the TIM transport channel requires high accuracy. For that reason, the merge option was first applied to each subfamily. Sequences with unclear subfamily assignment, were not integrated at this step but were added to the merged alignment using the add option.
Ortholog Identification
Homologous sequences were searched using the Blast suite (Camacho et al. 2009), HMMER-2.2.0 for glocal alignment (Eddy 1998), HMMER-3.1b1 for iterative local alignment (Johnson et al. 2010) and the HHsuite for profile–profile alignment (Soding 2005; Remmert et al. 2012). Fundamentally, a modified jackhmmer, run on a local computer cluster using hmmpgmd in the HMMER-3.1b1, was applied to query sequences with five iterations. For Tom20, where glocal profile–sequence alignment is required, HMM was generated from a multiple sequence alignment calculated using MAFFT with manually curated orthologous sequences. The candidate list of Tom20 generated by glocal search was further filtered with TM prediction (Hessa et al. 2007). For profile–profile alignment cases, HHblits was applied. Each protein in target proteomes was converted to HMM with HHblits iteration two times. After searching protein sequences in a translated proteome set, qualified orthologs were used to search genome sequences with tblastn to reduce gene prediction errors. We used (Hessa et al. 2007) to predict TM helices for collected homologous sequences. The window parameter of was tried from 15 to 31. Manual curation against phylogenetic reconstruction using the ML method implemented in RAxML was performed if necessary. The TreeBeST algorithm was applied to homologous sequences in Tim21/Coa1 and Tim17/Tim22/Tim23 family for classification. Because of mutational saturation, tree reconciliation was performed using PhyML (TreeBeST version) with only amino acid sequences constrained on the species tree. For large-scale orthologous sequence analysis, sequence search-based ortholog identification was conducted automatically using OrthoMCL ver. 2.0.9 (Li et al. 2003) with our proteome data sets.
Homology Modeling
To build structural models of Tom40s and a NIF domain of TbTim50, we used the crystal structures of mouse VDAC1 (PDB ID: 3EMN:X) (Ujwal et al. 2008) and human SCP1 (2GHT:A) (Zhang et al. 2006), respectively, as templates. For each protein, 100 structural models were built using the Homology Model application of the Molecular Operating Environment software package (MOE 2014.0901) (Chemical Computing Group Inc. 2016), based on the alignments calculated using FORTE (Tomii and Akiyama 2004) and HHpred (Soding 2005; Remmert et al. 2012), which are profile–profile comparison methods for protein structure prediction. Then we chose the best models based on GB/VI scores (Labute 2008). Calculation of hydrophobic, positive and negative patches on the protein surfaces was performed using the Patch Analyzer application of MOE.
Ancestral Sequence Reconstruction of Tom40
The alignment for reconstruction of the ancestral Tom40 sequence was computed using MAFFT with structural alignment constraint, which was used to predict three-dimensional structures. Ancestral sequence reconstruction was conducted by FastML (Ashkenazy et al. 2012) using a phylogenetic tree for selected taxons (highly diverged sequences observed in MROs or kinetoplastids were filtered out), an LG substitution matrix. The ancestral Tom40 sequence of the LECA was estimated from marginal probability. Indel determination was conducted using the maximum likelihood model in FastML with threshold 0.7.
Determination of Primitive Presequence-Containing Orthologs
Phylogenetic profiles for each ortholog cluster were generated by OrthoMCL. Presequence prediction was performed using MitoFates (Fukasawa et al. 2015). PhyloFates computes parameters of signal evolution from these phylogenetic profiles and the species tree (see supplementary Materials and Methods, Supplementary Material online for details). The source code is available at https://github.com/foxyiris/PhyloFates (last accessed March 3, 2017). A parameter in PhyloFates is signal gain node, . This parameter was used for screening of primitive presequence-containing ortholog clusters. Because the candidate gene clusters for primitive presequences-containing genes are numerous, a rough filter was defined to reduce the search. The number of candidate clusters (at least one predicted ortholog) was 8,960. It is unlikely to be primitive if the number of predicted presequences is small and sparse. Therefore, the minimum number of predicted presequences was set as three in each cluster. After the initial filtering, was used as a second filter. Filtered out clusters that had not in the root branch, caused 327 clusters. For these clusters, the expected value for distribution was computed. Then we kept a gene cluster as a primitive presequence-containing cluster if the value was higher than 0.5.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (Matryoshka-type evolution no. 3308) to Y.F., T.O., K.T., and K.I. (grant no. 26117733) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Grants-in-Aid for Scientific Research to Y.F. (grant no. 12J06550) and K.I. (grant no. 16K21680) from Japan Society for the Promotion of Science (JSPS), and the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED) to Y.F., T.O., K.I., and K.T. We thank Dr Martin Frith for helpful discussion and comments.
References
- Aakre CD, Herrou J, Phung TN, Perchuk BS, Crosson S, Laub MT.. 2015. Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell 163:594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Palmer JD.. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenetics Evol 29:380–395. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UCM, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG.. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133–140. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Penn O, Doron-Faigenboim A, Cohen O, Cannarozzi G, Zomer O, Pupko T.. 2012. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res 40:W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. 2010. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res 38:D415–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Gladkova C, Mapa K, Witte G, Mokranjac D.. 2015. Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein. eLife 4:e11897.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek D, Wojtkowska M, Suzuki Y, Sonobe S, Nishigami Y, Antoniewicz M, Kmita H, Makałowski W.. 2016. Protein import complexes in the mitochondrial outer membrane of Amoebozoa representatives. BMC Genomics 17:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK.. 2015. Mitocarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44:D1251–D1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 101: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Whelan J.. 2010. An in silico analysis of the mitochondrial protein import apparatus of plants. BMC Plant Biol 10:249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N.. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemical Computing Group Inc. 2016. Molecular Operating Environment (MOE), 2013.08.
- Clements A, Bursac D, Gatsos X, Perry AJ, Civciristov S, Celik N, Likic VA, Poggio S, Jacobs-Wagner C, Strugnell RA, et al. 2009. The reducible complexity of a mitochondrial molecular machine. Proc Natl Acad Sci U S A 106:15791–15795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Torruella G, Klimeš V, Brinkmann H, Kim E, Vlček Č, Lang BF, Eliáš M.. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A 112:E693–E699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T.. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318. [DOI] [PubMed] [Google Scholar]
- Duncan MR, Fullerton M, Chaudhuri M.. 2013. Tim50 in Trypanosoma brucei possesses a dual specificity phosphatase activity and is critical for mitochondrial protein import. J Biol Chem 288:3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckers E, Cyrklaff M, Simpson L, Deponte M.. 2012. Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol Chem 393:513.. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K.. 2009. Multiple pathways for mitochondrial protein traffic. Biol Chem 390:723–730. [DOI] [PubMed] [Google Scholar]
- Everard-Gigot V, Dunn CD, Dolan BM, Brunner S, Jensen RE, Stuart RA.. 2005. Functional analysis of subunit e of the F1F0-ATP synthase of the yeast Saccharomyces cerevisiae: importance of the N-terminal membrane anchor region. Eukaryot Cell 4:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642. [DOI] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K.. 2015. Mitofates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14:1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Stölting J, Zimorski V, Rada P, Tachezy J, Martin WF, Gould SB.. 2015. Conservation of transit peptide-independent protein import into the mitochondrial and hydrogenosomal matrix. Genome Biol Evol 7:2716–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SG, Gould SB.. 2016. The role of charge in protein targeting evolution. Trends Cell Biol 26:894–905. [DOI] [PubMed] [Google Scholar]
- Gawryluk RM, Chisholm KA, Pinto DM, Gray MW.. 2014. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J Proteomics 109:400–416. [DOI] [PubMed] [Google Scholar]
- Grant JR, Katz LA.. 2014. Building a phylogenomic pipeline for the eukaryotic tree of life-addressing deep phylogenies with genome-scale data. PLoS Curr Tree Life Edition 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ.. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic supergroups. Proc Natl Acad Sci U S A 106:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler T, Stierhof Y, Blattner J, Clayton C.. 1997. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes crithidia, trypanosoma and trichomonas. Eur J Cell Biol 73:240–251. [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G.. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030. [DOI] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, Portugaly E.. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 111: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Baker MJ, Liem M, Louber J, McKenzie M, Atukorala I, Ang CS, Keerthikumar S, Mathivanan S, Stojanovski D.. 2016. Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. eLife 5:e17463.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labute P. 2008. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem 29:1693–1698. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W.. 2010. The energetics of genome complexity. Nature 467:929–934. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Levy ED. 2010. A simple definition of structural regions in proteins and its use in analyzing interface evolution. J Mol Biol 403:660–670. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Li Y, Calvo SE, Gutman R, Liu JS, Mootha VK.. 2014. Expansion of biological pathways based on evolutionary inference. Cell 158:213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T, Schneider A.. 2010. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B: Biol Sci 365:799–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maćašev D, Whelan J, Newbigin E, Silva-Filho MC, Mulhern TD, Lithgow T.. 2004. Tom22, an 8-kda trans-site receptor in plants and protozoans, is a conserved feature of the TOM complex that appeared early in the evolution of eukaryotes. Mol Biol Evol 21:1557–1564. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T.. 2005. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 33:D54–D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiuchi T, Nozaki T.. 2014. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie 100:3–17. [DOI] [PubMed] [Google Scholar]
- Makiuchi T, Mi-Ichi F, Nakada-Tsukui K, Nozaki T.. 2013. Novel TPR-containing subunit of TOM complex functions as cytosolic receptor for entamoeba mitosomal transport. Sci Rep 3:1129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J, Desy S, Niemann M, Chanfon A, Oeljeklaus S, Pusnik M, Schmidt O, Gerbeth C, Meisinger C, Warscheid B, et al. 2015. Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat Commun 6:6646.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J, Meisinger C, Schneider A.. 2016. Peeping at TOM-Diverse entry gates to mitochondria provide insights into the evolution of eukaryotes. Mol Biol Evol 33:337–351. [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M.. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392:37–41. [DOI] [PubMed] [Google Scholar]
- Martincová E, Voleman L, Pyrih J, Žárskỳ V, Vondráčková P, Kolísko M, Tachezy J, Doležal P.. 2015. Probing the biology of Giardia intestinalis mitosomes using in vivo enzymatic tagging. Mol Cell Biol 35:2864–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Ichi F, Yousuf MA, Nakada-Tsukui K, Nozaki T.. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A 106:21731–21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N.. 1997. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol 17:6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ogata Y, Zíková A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD.. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9:434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AJ, Hulett JM, Likić VA, Lithgow T, Gooley PR.. 2006. Convergent evolution of receptors for protein import into mitochondria. Curr Biol 16:221–229. [DOI] [PubMed] [Google Scholar]
- Rada P, Makki AR, Zimorski V, Garg S, Hampl V, Hrdỳ I, Gould SB, Tachezy J.. 2015. N-terminal presequence-independent import of phosphofructokinase into hydrogenosomes of Trichomonas vaginalis. Eukaryot Cell 14:1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Soding J.. 2012. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. [DOI] [PubMed] [Google Scholar]
- Rimmer KA, Foo JH, Ng A, Petrie EJ, Shilling PJ, Perry AJ, Mertens HD, Lithgow T, Mulhern TD, Gooley PR.. 2011. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J Mol Biol 405:804–818. [DOI] [PubMed] [Google Scholar]
- Schatz G. 1997. Just follow the acid chain. Nature 388:121–122. [DOI] [PubMed] [Google Scholar]
- Schilke BA, Hayashi M, Craig EA.. 2012. Genetic analysis of complex interactions among components of the mitochondrial import motor and translocon in saccharomyces cerevisiae. Genetics 190:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Bursać D, Lithgow T.. 2008. The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol 18:12–18. [DOI] [PubMed] [Google Scholar]
- Schulz C, Schendzielorz A, Rehling P.. 2015. Unlocking the presequence import pathway. Trends Cell Biol 25:265–275. [DOI] [PubMed] [Google Scholar]
- Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T.. 2011. In vivo protein-interaction mapping of a mitochondrial translocator protein tom22 at work. Proc Natl Acad Sci U S A 108:15179–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M, et al. 2015. Molecular architecture of the active mitochondrial protein gate. Science 349:1544–1548. [DOI] [PubMed] [Google Scholar]
- Singha UK, Peprah E, Williams S, Walker R, Saha L, Chaudhuri M.. 2008. Characterization of the mitochondrial inner membrane protein translocator Tim17 from trypanosoma brucei. Mol Biochem Parasitol 159:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Hamilton V, Duncan MR, Weems E, Tripathi MK, Chaudhuri M.. 2012. Protein translocase of mitochondrial inner membrane in Trypanosoma brucei. J Biol Chem 287:14480–14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Bohnert M, Pfanner N, van der Laan M.. 2012. Mechanisms of protein sorting in mitochondria. Cold Spring Harb Perspect Biol 4:a011320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W.. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5:123–135. [DOI] [PubMed] [Google Scholar]
- Tomii K, Akiyama Y.. 2004. FORTE: a profile-profile comparison tool for protein fold recognition. Bioinformatics 20:594–595. [DOI] [PubMed] [Google Scholar]
- Turakhiya U, von der Malsburg K, Gold VA, Guiard B, Chacinska A, van der Laan M, Ieva R.. 2016. Protein import by the mitochondrial presequence translocase in the absence of a membrane potential. J Mol Biol 428:1041–1052. [DOI] [PubMed] [Google Scholar]
- Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J.. 2008. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci U S A 105:17742–17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Jabbour C, Chan NC, Celik N, Likić VA, Mulhern TD, Lithgow T.. 2009. Evidence of a reduced and modified mitochondrial protein import apparatus in microsporidian mitosomes. Eukaryot Cell 8:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowska M, Buczek D, Stobienia O, Karachitos A, Antoniewicz M, Slocinska M, Makałowski W, Kmita H.. 2015. The TOM complex of Amoebozoans: the cases of the amoeba Acanthamoeba castellanii and the slime mold Dictyostelium discoideum. Protist 166:349–362. [DOI] [PubMed] [Google Scholar]
- Yamano K, ichi Yatsukawa Y, Esaki M, Hobbs AEA, Jensen RE, Endo T.. 2008. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem 283:3799–3807. [DOI] [PubMed] [Google Scholar]
- Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR.. 1985. Mitochondrial origins. Proc Natl Acad Sci U S A 82:4443–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarsky V, Tachezy J, Dolezal P.. 2012. Tom40 is likely common to all mitochondria. Curr Biol 22:R479–R481. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP.. 2006. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell 24:759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zysset-Burri DC, Müller N, Beuret C, Heller M, Schürch N, Gottstein B, Wittwer M.. 2014. Genome-wide identification of pathogenicity factors of the free-living amoeba Naegleria fowleri. BMC Genomics 15:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.