Abstract
Insects with restricted diets rely on symbiotic bacteria to provide essential metabolites missing in their diet. The blood-sucking lice are obligate, host-specific parasites of mammals and are themselves host to symbiotic bacteria. In human lice, these bacterial symbionts supply the lice with B-vitamins. Here, we sequenced the genomes of symbiotic and heritable bacterial of human, chimpanzee, gorilla, and monkey lice and used phylogenomics to investigate their evolutionary relationships. We find that these symbionts have a phylogenetic history reflecting the louse phylogeny, a finding contrary to previous reports of symbiont replacement. Examination of the highly reduced symbiont genomes (0.53–0.57 Mb) reveals much of the genomes are dedicated to vitamin synthesis. This is unchanged in the smallest symbiont genome and one that appears to have been reorganized. Specifically, symbionts from human lice, chimpanzee lice, and gorilla lice carry a small plasmid that encodes synthesis of vitamin B5, a vitamin critical to the bacteria-louse symbiosis. This plasmid is absent in an old world monkey louse symbiont, where this pathway is on its primary chromosome. This suggests the unique genomic configuration brought about by the plasmid is not essential for symbiosis, but once obtained, it has persisted for up to 25 My. We also find evidence that human, chimpanzee, and gorilla louse endosymbionts have lost a pathway for synthesis of vitamin B1, whereas the monkey louse symbiont has retained this pathway. It is unclear whether these changes are adaptive, but they may point to evolutionary responses of louse symbionts to shifts in primate biology.
Keywords: Ca. Riesia, endosymbiont replacement, Anoplura, pantothenate, thiamin, plasmid
Introduction
Insect species with nutritionally incomplete diets (e.g., phloem or blood) often harbor mutualistic bacteria that synthesize missing nutrients (Baumann 2005; Lopez-Sanchez et al. 2009; Manzano-Marin et al. 2015). Many of these symbiotic bacteria (herein endosymbionts) are intracellular, occupy specialized host cells (bacteriocytes; reviewed by Buchner 1965), and are transmitted vertically from mother to offspring (reviewed by Bright and Bulgheresi 2010). Generally, endosymbionts of insects are maintained over evolutionary time-scales and many cospeciate with their insect hosts (Clark et al. 2000; Sauer et al. 2000; Thao et al. 2000).
Symbiosis between blood-feeding lice and heritable endosymbionts has been well-studied (Ries 1931, 1932, 1933, 1935; Aschner and Ries 1933; Ries and van Weel 1934; Buchner 1965; Eberle and Mclean 1982, 1983; Sasaki-Fukatsu et al. 2006; Allen et al. 2007, 2009, 2016; Perotti et al. 2007; Boyd and Reed 2012; Boyd et al. 2014, 2016). The most familiar is the human head louse (Pediculus humanus), which has a large visible mass of endosymbiont cells in the abdomen (Hooke 1665; Buchner 1965; Perotti et al. 2007). The role of human louse endosymbionts has been studied experimentally (Puchta 1955) and these endosymbionts supply the lice with essential B-vitamins (Puchta 1955; Perotti et al. 2009). Central to the symbiosis is the synthesis of vitamin B5 (pantothenate; Puchta 1955; Perotti et al. 2009), which is a precursor of coenzyme A and can be transported between the endosymbiont and host cells (Vallari and Rock 1985). Synthesis of vitamin B5 requires both the louse and the endosymbiont, a process known as metabolic complementation (Wilson and Duncan 2015). Here, the louse synthesizes precursors needed for the synthesis of vitamin B5 and the pathway is completed by the endosymbiont. Vitamin B5 is then available for both the endosymbiont and host.
Human lice belong to the insect suborder Anoplura, more commonly known as the sucking lice (Durden and Musser 1994). This suborder consists of 532 species of lice and each species parasitizes one or a few closely related species of mammals (Durden and Musser 1994). Many of these louse species have been shown to harbor endosymbionts (Ries 1931, 1932, 1933, 1935; Aschner and Ries 1933; Ries and van Weel 1934; Buchner 1965). Because all sucking lice feed on mammal blood, it seems likely that other louse endosymbionts have similar roles of providing B-vitamins. However, it appears that symbioses between lice and bacteria have arisen multiple times in different louse species (Buchner 1965; Hypsa and Krizek 2007; Allen et al. 2009, 2016; Boyd and Reed 2012; Boyd et al. 2016), rather than being derived from a common ancestor.
Focusing on human and primate louse endosymbionts, the classification of these endosymbionts reflects that they may have independent origins. The endosymbionts of lice that parasitize hominids (humans, chimpanzees, and gorillas) are classified into the genus Candidatus Riesia (hereafter Riesia; Sasaki-Fukatsu et al. 2006; Allen et al. 2007, 2009). Riesia appears to have been derived from an Arsenophonous-like bacteria (Allen et al. 2007, 2016; Novakova et al. 2009) 12.95–25 Ma (Allen et al. 2009). This is relatively young compared with similar endosymbionts in different insects that have persisted from 25 to 270 My (reviewed by Gosalbes et al. 2010). However, once established, Riesia appears to have cospeciated with their louse hosts (Allen et al. 2007, 2009) and louse speciation was largely driven by cospeciation with hominids (Reed et al. 2007; Light and Reed 2009). The lice that parasitize old world monkeys, the sister group of hominid lice (Light and Reed 2009; Light et al. 2010), appear to have an unrelated endosymbiont, Candidatus Puchtella (hereafter Puchtella; studied in red colobus monkeys and macaques thus far and named in macaques; Allen et al. 2009, 2016; Fukatsu et al. 2009). This suggests that there was a replacement of an endosymbiont in the common ancestor of hominid lice.
Riesia species have small genomes (Kirkness et al. 2010; Boyd et al. 2014). The two genomes sequenced thus far from endosymbionts of human and chimpanzee lice consist of a linear chromosome of ∼0.57 Mb and a circular plasmid ∼8 kb (Kirkness et al. 2010; Boyd et al. 2014). This small genome is typical of many endosymbionts (reviewed by McCutcheon and Moran 2012; Moran and Bennett 2014). However, a unique feature of the Riesia genome is that the genes underlying vitamin B5 synthesis, a pathway key to the symbiosis, are encoded on the plasmid (Kirkness et al. 2010; Boyd et al. 2014). In closely related bacteria, these same genes are encoded the primary chromosome, with genes in two different regions of the chromosome (e.g., Escherichia coli). Encoding essential metabolic functions on a plasmid is unusual (Villasenor et al. 2011), however a similar occurrence has been observed in aphids and their endosymbionts (Buchnera; Baumann et al. 1999; Wernegreen and Moran 2001). The movement of essential functions on to a plasmid may have evolutionary consequences for the endosymbiont. For example, extra steps during cell division may be required to ensure all daughter cells receive the plasmid (e.g., see Summer 1996 for review of segregation instability). However, there may be benefits of this genome organization as well. For example, uniting these genes on a small plasmid brings them into close physical proximity in the genome. This configuration could be beneficial by increasing gene copy number, facilitating increased pantothenate synthesis.
Because Riesia has speciated with hominid primates, this group presents a unique opportunity study the timing and persistence of genomic changes. In this study we sequenced and assembled genomes of endosymbionts from human, chimpanzee, gorilla, and red colobus monkey lice. This includes endosymbionts of three louse ecotypes on humans that have different feeding strategies and ability to transmit disease (Raoult and Roux 1999; Anderson and Anderson 2000; Raoult et al. 2006). We examine the characteristics of each endosymbiont genome and focus on their capacity for B-vitamin synthesis; including in which species pantothenate synthesis is located on a plasmid. We then re-evaluate the phylogenetic placement of louse endosymbionts, Riesia and Puchtella using genomic data and compare with past phylogenetic studies of these endosymbionts that were based on only one or two genes (Sasaki-Fukatsu et al. 2006; Allen et al. 2007, 2016; Fukatsu et al. 2009). Finally, we evaluate potential symbions replacement.
Results
Species Relationships
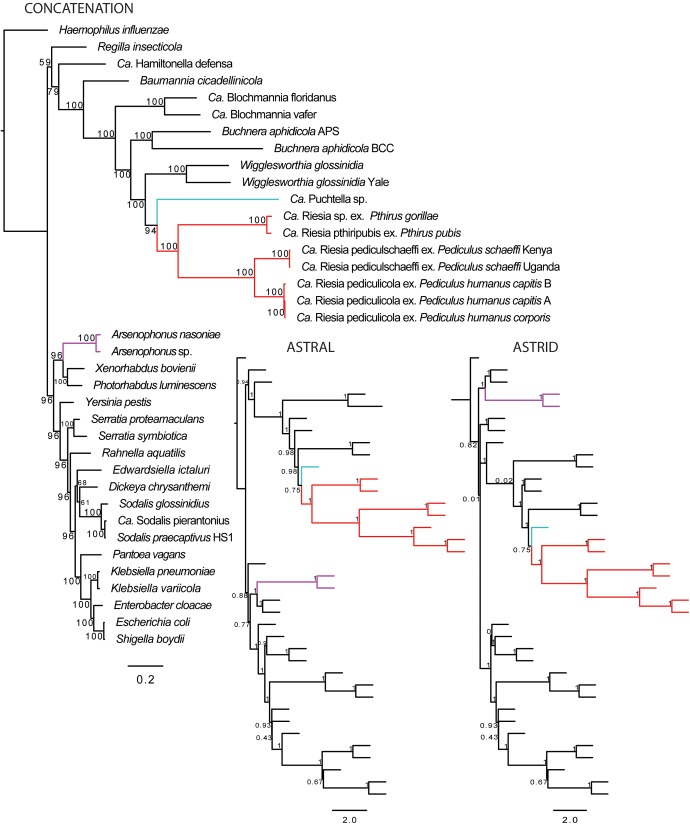
We investigated the species relationships of primate and human louse endosymbionts using phylogenomic methods. First, we identified 177 single-copy orthologous protein-coding genes across 38 eubacterial taxa (37 Enterobacteriaceae and one outgroup taxon), including 8 louse endosymbionts. Third codon positions, which had higher frequency of AT bases in endosymbionts, were excluded from the analysis. Using two different software packages, two separate species trees were then estimated using coalescent-gene tree summary methods. In both cases, all of the louse endosymbionts included in this study were found to be monophyletic (fig. 1; supplementary figs. S2 and S3, Supplementary Material online). Endosymbionts of tsetse flies (Wigglesworthia) were identified as the closest relative of louse endosymbionts in our data set. Support for the monophyly of louse endosymbionts and bipartitions within the louse endosymbiont clade were high (0.75–1.0). Next, we again estimated the species relationships based on simultaneous analysis of all genes as a single data matrix. The concatenation of all gene alignments provided 128,006 sites for this analysis, and a Maximum Likelihood (ML) tree was estimated from the concatenated alignment. The resulting ML species tree agreed with coalescent analysis finding that louse endosymbionts were monophyletic and sister to Wigglesworthia (fig. 1). Support for this relationship was again high, 94% bootstrap for monophyly of louse endosymbionts, and 100% bootstrap supporting bipartitions within the louse endosymbiont clade. While it was previously suggested that Riesia was closely related to Arsenophonus, we failed to recover that relationship in any phylogenetic analysis.
Fig. 1.
Comparison of phylogenetic trees of gamma-proteobacteria including louse endosymbionts obtained from different phylogenomic methods. Left is ML tree based on concatenation of 177 orthologous protein-coding genes. Support is presented as percent of times each bipartition was recovered from 100 bootstrap replicates. Middle and right are coalescent trees (ASTRAL and ASTRID) based on analyses of 177 individual ML gene trees. Red = Riesia; light blue = Puchtella, and purple = Arsenophonus.
Within the louse endosymbiont tree, we find phylogenetic patterns of the endosymbionts are similar to the lice they inhabit. The endosymbiont Puchtella, from the lice of red colobus monkeys, is sister to the hominid louse endosymbionts, which is consistent with the louse phylogeny described by Light and Reed (2009) and Allen et al. (2017). Also, inclusion of the gorilla louse endosymbiont shows that it is closely related to the human pubic louse endosymbionts. This was expected as gorilla lice and human pubic lice are closely related (a pattern resulting from lice switching from gorillas to humans; Reed et al. 2007; Light and Reed 2009). Also as expected, human and chimpanzee louse endosymbionts are sister to each other. Within human louse endosymbionts, again the tree reflects phylogenetic patterns seen in the lice (Reed et al. 2004). Within human head and body lice there are three distinct clades of lice identified by their mitochondrial haplotypes, potentially resulting from exchange of lice between modern and archaic humans (Reed et al. 2004). We examined endosymbionts from human lice belonging to two of these clades; “Clade A” which is found worldwide and includes both human head and body lice and “Clade B” which is found primarily in western Europe and the New World and only includes head lice (Reed et al. 2004). Again the phylogeny of the endosymbionts reflect their host’s mitochondrial history, with Clade A louse endosymbionts being most closely related to each other and a Clade B louse endosymbiont sister to Clade A strains. Unlike human louse endosymbionts, we see no divergence in chimpanzee louse endosymbionts. This is despite known population structure and limited gene flow in chimpanzees (Becquet et al. 2007). Perhaps additional sampling will reveal structure within this species.
Endosymbiont Genomes
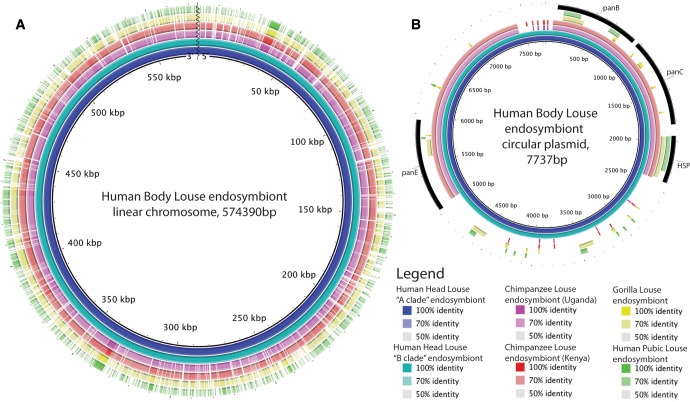
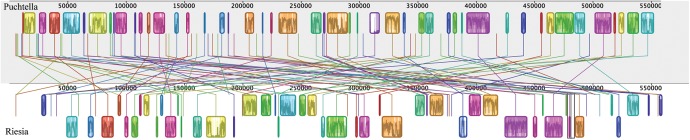
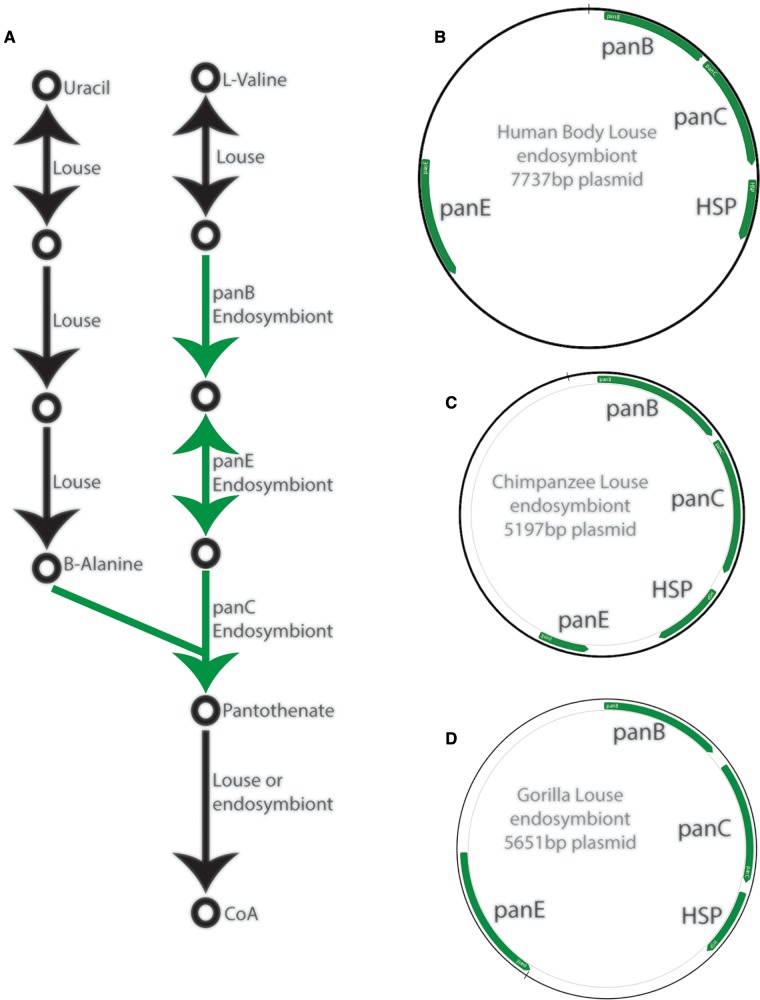
We completely or partially sequenced the genomes of six new strains of louse endosymbionts (Puchtella and Reisia species; table 1). Similar to the two published Riesia genomes from human body and chimpanzee lice, the newly sequenced Riesia genomes (from human head lice, human pubic lice, chimpanzee lice, and gorilla lice) consist of a primary chromosome ranging from 0.529–0.574 Mb in length and a small circular plasmid 5.2–7.8 kb (table 2 and fig. 2). The previously sequenced endosymbiont genome from a human louse was the USDA strain of Riesia pediculicola, which is derived from an inbred strain of lice adapted to feed on rabbits in the laboratory (Kirkness et al. 2010). Here, we find the genome of the endosymbiont derived from these rabbit-adapted lice is similar to the genomes of endosymbiont in wild populations of human head lice (fig. 2). The genomes of A clade human head lice and body lice were nearly identical (99.9% of bases were the same in both genomes) with no large insertions or deletions (indels). There was slightly more variation between endosymbiont genomes of head lice, than between the endosymbionts genomes of the body louse and the most closely related head louse endosymbiont. Compared with the body louse endosymbiont, the B clade endosymbiont genome is more divergent (97.8% of bases were identical) and has three larger insertions (22–100 bp) and three deletions (51–26 bp). Five of the six indels occurred in noncoding parts of the genome, with one deletion in the pabA gene that encodes a protein involved in folate synthesis. However, this deletion is near the 3’ end of the gene and may not have disrupted gene function. The genome of the endosymbiont (Riesia pediculschaeffi) from chimpanzee lice was slightly smaller than the human body louse genome, however we identified a similar number and order of genes. The gorilla louse endosymbiont (Riesia sp.) had the smallest genome of any investigated and showed some loss of genes compared with the endosymbiont of the human body louse. Throughout Riesia genomes, the general order of genes is conserved between genomes, with minor exceptions occurring near the 3’ end of the primary chromosome, where endosymbionts of human lice have a duplicated region, and small duplications in the plasmid (found between the heat shock protein encoding and panE genes). We were able to recover only part of the genome from the endosymbiont of the human pubic louse; however it is likely most similar to the gorilla louse endosymbiont genome, as these species are closely related. In Puchtella the genome was assembled into two scaffolds totaling 0.559 Mb (table 2). While the genome of Puchtella is similar in size to Riesia genomes, the organization and order of genes is different than that of Riesia (fig. 3). There was no evidence of the plasmid in Puchtella.
Table 1.
Collection Data for Louse Endosymbionts.
| Endosymbiont | Louse Host | Description | Mammal Host | Collection | Source |
|---|---|---|---|---|---|
| Riesia pediculicola str. USDA | Pediculus humanus humans “Mt clade A” | Rabbit adapted clothing louse | “Homo sapiens” | Lab strain | Kirkness et al. (2010) |
| Riesia pediculicola str. HHAC | Pediculus humanus capitus “Mt clade A” | Human head louse | Homo sapiens | Cambodia | This study |
| Riesia pediculicola str. HHAN | Pediculus humanus capitus “Mt clade A” | Human head louse | Homo sapiens | Netherlands | This study |
| Riesia pediculicola str. HHBH | Pediculus humanus capitus “Mt clade B” | Human head louse | Homo sapiens | Honduras | This study |
| Riesia pediculschaeffi str. PTSU | Pediculus schaeffi | Chimpanzee louse | Pan troglodytes schweinfurthii | Uganda | Boyd et al. (2014) |
| Riesia pediculschaeffi str. PTSK | Pediculus schaeffi | Chimpanzee louse | Pan troglodytes cf. schweinfurthii | Kenya | This study |
| Riesia sp. str. GBBU | Pthirus gorillae | Gorilla louse | Gorilla beringei beringei | Uganda | This study |
| Puchtella sp. str. PRUG | Pedicinus badii | Red colobus monkey louse | Procolobus rufomitratus | Uganda | This study |
Note.—Mt, Mitochondira.
Table 2.
Genome Assembly Summary of Louse Endosymbionts.
| Taxonomy | Seq. Technology | Seq. Depth | SD Depth | Genome Length | Genome %GC | Plasmid Length | Plasmid %GC | Genes |
|---|---|---|---|---|---|---|---|---|
| Riesia USDA | Sanger | NA | NA | 574,390 | 28.5 | 7737 | 35.2 | 557 |
| Riesia HHAC | Illumina HiSeq 2000 | 22.9.2 | 51.5 | 574,389 | 28.5 | 7737 | 35.3 | 566 |
| Riesia HHAN | Illumina HiSeq 2500 | 2.3 | 1.5 | NA | NA | NA | NA | NA |
| Riesia HHBH | Illumina HiSeq 2000 | 424.6 | 107 | 574,386 | 28.5 | 7737 | 35.9 | 573 |
| Riesia PTSU | Illumina HiSeq 2000 | 147.2 | 36.4 | 566,667 | 31.6 | 5197 | 37 | NA |
| Riesia PTSK | Illumina HiSeq 2500 | 164.1 | 64.4 | 566,667 | 31.6 | 5197 | 37 | 594 |
| Riesia HPNS | Illumina HiSeq 2500 | 9 | 99.7 | NA | NA | NA | NA | NA |
| Riesia GBBU | Illumina HiSeq 2500 | 83 | 39.1 | 528,693 | 25 | 5651 | 29 | 476 |
| Puchtella | Illumina HiSeq 2000 | 396.4 | 71.3 | 558,106 | 24.2 | NA | NA | 564 |
Note.—SD, standard deviation; Seq, sequencing.
Fig. 2.
Comparison of Riesia species genomes, endosymbiont of human, chimpanzee, and gorilla lice. Image generated using Blast Ring Image Generator (Alikhan et al. 2011). Color intensity based on percent identify of reference genome (Riesia pediculicola str. USDA) and other Riesia species. Squiggly line identifies the ends of the linear primary chromosome.
Fig. 3.
Comparison of endosymbiont of the human body louse (Riesia pediculicola str. USDA) genome and the genome of the endosymbiont of the red colobous monkey louse (Puchtella sp.) using MAUVE (Darling et al. 2004). Blocks indicate continuous genome alignments or candidate orthologous genome regions.
Between 476 and 594 genes were predicted in endosymbiont genomes (supplementary table S1, Supplementary Material online), with the endosymbiont of the gorilla louse having the fewest genes. Bi-directional best blast hits were used to estimate how many genes were shared between the endosymbiont of the human head louse and other endosymbionts. Nearly all of the 557 predicted genes in the endosymbiont genome of the human body louse were detected in the most similar endosymbiont genome of the Human head louse (553 genes) using blast. We were able to detect progressively fewer candidate orthologs in more distantly related endosymbiont to this reference strain (526, 450, 416, and 387, respectively) using blast. This gradual drop-off in the number of identified orthologs with phylogenetic distance may reflect an artifact of sequence divergence and the ability of blast to detect a significant hit. Therefore, we compared functional prediction in each endosymbiont. Functionally, the endosymbiont genomes from all lice were similar. Much of the coding capacity of the endosymbiont genomes was devoted to vitamin and protein metabolism (supplementary table S1, Supplementary Material online). There was some reduction of fatty acid and amino acid metabolism in the smallest genome (from the endosymbiont of the gorilla louse) and in Puchtella, which has seen extensive reorganization of its genome when compared with Riesia species.
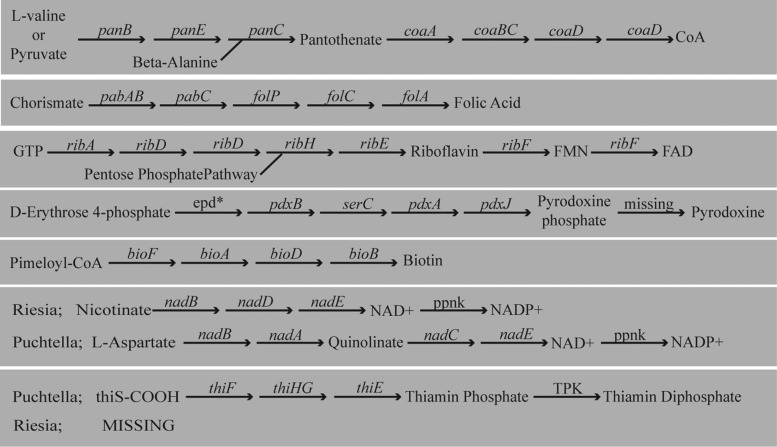
Synthesis of B-vitamins is likely the primary role of endosymbionts in blood sucking lice. We compared B-vitamin biosynthesis in Puchtella with Riesia (focusing the human body louse endosymbiont as gene content was similar across strains; fig. 4). Both species possess the de novo pathway for synthesis of patothenate from exogenous substrates. They can also synthesize Coenzyme A (CoA) from pantothenate. Synthesis of folic acid, riboflavin, pyrodoxine phosphate, and biotin also appear intact. The only difference between the two genera is that Riesia appears to be missing the gene encoding d-erythrose 4-phosphate dehydrogenase (epd) required for pyridoxine biosynthesis, while Puchtella retains this function. However, because the remainder of the pyridoxine synthesis pathway is intact in Riesia, the missing epd encoding gene could be due to an assembly or annotation error. Metabolism of nicotinamide was different in Riesia and Puchtella with each using different starting compound (fig. 4). Finally, we found that Puchtella can synthesize thiamin, while Riesia cannot (fig. 4).
Fig. 4.
Comparison of B vitamin synthesis pathways present in Riesia pediculicola and Puchtella. Pathways are the same for Riesia and Puchtella unless otherwise noted. *Gene missing in Riesia likely due to assembly or annotation error, but present in Puchtella.
In Riesia, proteins involved in pantothenate synthesis (a pathway critical to symbiosis with lice) were encoded on the plasmid by genes panB, panC, and panE (fig. 5). The genes required for pantothenate synthesis in the Puchtella genome were located on the primary chromosome scaffolds (genomic positions: panE = scaffold-95: 17,590–18,486, panC = scaffold-96: 278,331–279,170, panB = scaffold-96: 279,173–279,970). The panD gene (a gene involved in pantothenate biosynthesis in nonhost associated bacteria and redundant with host metabolism) was not identified in any of the louse endosymbiont genomes. In all Riesia species the plasmid encoded four genes. Three genes previously mentioned encode proteins involved in pantothenate biosynthesis, and the fourth encodes a chaperone protein (HSP20 family, Locus WP_041855293 in RefSeq; fig. 2). This chaperone protein is highly conserved across all of the Riesia species examined in this study (fig. 2); however, we were unable to locate a homolog in the Puchtella genome. This chaperone may have some important, but as-yet undetermined role. There is a small repeated region between the chaperonin protein and the panE gene in plasmids recovered from human louse endosymbionts. This repeat region is missing in other Riesia genomes.
Fig. 5.
(A) Schematic of pantothenate-CoA synthesis pathway with steps that can only be done by the endosymbiont highlighted in green and (B–D) representative plasmid sequences from Hominid louse endosymbionts with genes annotated in green. HSP, heat shock protein; CoA, coenzyme A.
Gene Tree–Species Tree Conflict
A phylogenetic analysis of the genes on the plasmid and their orthologs in other taxa was conducted. An ML tree based on concatenation of plasmid-based genes produced a topology different from that of the species trees described above (genes in the Riesia plasmid appear mostly closely related to orthologs in Arsenophonus; supplementary fig. S4, Supplementary Material online). However, bootstrap support for this plasmid tree was low and therefore difficult to interpret. The plasmid tree was based on only four genes and it is possible that gene trees/species tree conflict or a lack of informative bases within each gene is driving this alternate typology. To better understand how gene trees contribute to phylogenetic signal in isolation, we compared individual gene trees with the ML species tree that was based 177 genes. When gene trees and the species tree were collapsed leaving only bipartitions with greater than 50% bootstrap support, a mean of 20% of bipartitions in the gene trees conflicted with the species tree (mean = 0.20, Q1 = 0.13, Q3 = 0.27; range = 0.0–0.71; supplementary fig. S5, Supplementary Material online). However, when collapsing bipartitions at higher values, the conflict decreased considerably (collapsing at 75% bootstrap mean = 0.06, Q1 = 0.0, Q3 = 0.14; range = 0.0–0.5; and at 95% bootstrap mean = 0.03, Q1 = 0.0, Q3 = 0.0; range = 0.0–0.5; supplementary fig. S5, Supplementary Material online). These results show that conflict between individual gene trees and the species tree is pervasive, but these deviations are not well supported. Those genes on the plasmid showed a similar pattern of conflict that is not supported by bootstrap replicates (50%, mean = 0.31, Q1 = 0.26, Q3 = 0.39; range = 0.16–0.4; 75%, mean = 0.14, Q1 = 0.12, Q3 = 0.18; range = 0.07–0.18; 95%, mean = 0.02, Q1 = 0.0, Q3 = 0.02; range = 0.0–0.08; supplementary fig. S5, Supplementary Material online). These comparisons suggest that lack of support limits phylogenetic tree estimation and that conflict between the species tree and individual gene trees is not unique to the plasmid genes.
Previous phylogenetic studies examining primate louse endosymbionts have provided evidence for frequent endosymbiont replacement (Sasaki-Fukatsu et al. 2006; Allen et al. 2007, 2016; Hypsa and Krizek 2007; Fukatsu et al. 2009). It had been suggested that the plasmid could move during such replacements (Kirkness et al. 2010) through horizontal exchange (HGT). To test for evidence of HGT, we determined if plasmid-based genes had a phylogenetic topology that significantly differed from the presumed species tree when compared with genes found on the primary chromosome (where HGT is not expected). A Welch two-sampled t-test failed to reject the null hypothesis that the plasmid genes have a conflict that is significantly higher than genes on the primary chromosome as would be expected under HGT. Results of t-test are as follows when collapsing tree bipartitions at 50% bootstrap t(3) =−1.9095, P > 0.05; collapsing at 75% bootstrap t(3) = −1.8504, P > 0.05; and collapsing at 90% bootstrap t(3) = −0.3851, P > 0.05). These results indicate that phylogenetic trees estimated from plasmid-based genes are statistically compatible to phylogenetic trees estimated from other genes. Therefore we find no evidence of HGT.
Discussion
Here we examine the phylogenetic relationships and genome structure of diverse primate louse endosymbionts. Our results agree with previous phylogenetic studies that suggested endosymbionts have cospeciated with human and chimpanzee which diverged 5.4 Ma (Allen et al. 2009) and gorilla lice which diverged 7.4–17.4 Ma (Allen et al. 2007, 2009). We also describe new evidence that endosymbionts co-diverged along with their gorilla and human pubic louse hosts, which diverged following a host switch ∼3–4 Ma (Reed et al. 2007). We also find that endosymbionts in human head and body lice have similar phylogenetic patterns to human louse mitochondria. The endosymbionts of human head and body lice, both belonging to the louse mitochondrial clade A have nearly identical genomes. This is not surprising as human body lice diverged from head lice ∼80–100 ka, after humans began wearing clothes (Toups et al. 2011). Therefore, the lab-strain body lice have endosymbionts that are representative of endosymbionts from A clade head lice, a common strain of lice found world-wide (Reed et al. 2004). However, we did find there was more variation between human head louse endosymbiont genomes that corresponded to different louse mitochondrial clades (clades A and B). Reed et al. (2004) found evidence that these louse strains diverged up to 1.18 Ma. They proposed that these lice diverged on extinct humans and were acquired by modern humans via a host switch in Europe. Our finding that the genomes of endosymbionts of head lice are more divergent than A clade head and body lice fits with what is known about louse evolution.
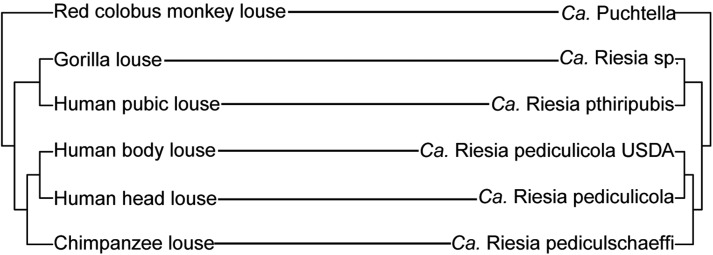
However, our findings diverge from previous studies in two ways. First, we find no evidence that the human and other hominid louse endosymbionts, Riesia, are closely related to the genus Arsenophonus. Instead, we find they are closely related to the endosymbionts of a primate louse, Puchtella, which is found in the lice of red colobus monkeys (fig. 1; supplementary figs. S2 and S3, Supplementary Material online). Second, the close relationships of Riesia and Puchtella suggests there was no endosymbiont replacement in the common ancestor of hominid lice; instead the phylogenetic pattern of the louse endosymbionts reflects the species history of the lice themselves (fig. 6; see Light and Reed 2009; Light et al. 2010; and Allen et al. 2017 for the louse phylogeny). Our results support a scenario of repeated cospeciation of endosymbionts and lice. These findings also push back the date of the association of these endosymbionts with lice. Based on host divergence information, it appears hominid lice and old world monkey lice diverged between 20 and 25 Ma (Light and Reed 2009; Light et al. 2010), suggesting the louse-endosymbiont association is at least that old. Essentially, this constrains the association between lice and this clade of endosymbionts to the upper end of a previously reported data range of 12.9–25 Ma (Allen et al. 2009). Future work should critically evaluate the relationship of the louse endosymbionts studied here and endosymbionts from other old world monkey lice not included in this study. Possibly the association between Riesia species and lice has persisted for more than 25 My.
Fig. 6.
Comparison of louse and endosymbiont phylogenies. Left is the phylogenetic tree of lice parasitizing primates and on the right is their endosymbionts. Lines connect endosymbionts to their louse hosts.
Correctly placing insect-endosymbionts, including endosymbionts of lice, into the phylogenentic tree of Enteroabacteriaceae has been the focus of many studies (e.g., Herbeck et al. 2005; Williams et al. 2010; Sach et al. 2011, 2014; Allen et al. 2016). The reason that previous phylogenetic studies based on only one or two genes arrived at a different result for the placement of Riesia and Puchtella (e.g., Hypsa and Krizek 2007; Allen et al. 2007, 2009, 2016; Fukatsu et al. 2009; Novakova et al. 2009 using 16SrRNA or groEL), may be due to the use of single gene trees that show a history incongruent with the species history, or that provide an insufficient amount of phylogenetically informative data. Our results show there is often conflicting signals between gene trees, but support for this conflict is low. Therefore, phylogenies built on one or two genes may not be sufficient to resolve the louse endosymbiont evolutionary tree. Phylogenetic studies seeking to more broadly evaluate polyphyly of endosymbionts in sucking lice should be based on many genes.
Additional issues arise beyond gene tree–species tree conflict. The endosymbiont of the human louse, Riesia, has a DNA substitution rate higher than most other endosymbionts (Allen et al. 2009). Like in other endosymbionts, mutation favors AT bases (Moran 2002; Van Leuven and McCutcheon 2011). This high rate of substitution and AT bias may cause endosymbionts to artificially cluster together in a phylogenetic tree, despite them being derived from independent lineages (long-branch attraction; Felsenstein 1978). One approach that provides opportunity to overcome some phylogenetic issues is implementing a nonhomogeneous model of sequence evolution (Herbeck et al. 2005). Unfortunately, this approach cannot yet be implemented with large phylogenomic data sets and relies on limited methods for assessing alternative trees (nearest neighbor interchange). To attempt to limit the effect of AT base bias, we eliminated third codon positions. This position showed the greatest bias in endosymbionts compared with other bacteria. In an attempt to detect methodological shortcomings of tree building with Enterobacteriaceae, we used two different methods of summarizing the species tree. First, using the concatenation method, all the sequence data were analyzed as a single matrix to estimate a species tree. This approach was appealing as bacterial genes are shorter than eukaryotic genes and individual genes may provide limited phylogenetically informative variants. The second approach summarized species trees from gene trees. This approach is appealing as different classes of genes might experience different rates of substitution or mutation biases. Therefore, each gene tree can contribute to the resulting species tree equally. These methods both agree that Wigglesworthia and louse symbionts are the closest relatives in our analysis. It seems unlikely that Wigglesworthia was directly exchanged between insects. One possibility is that Wigglesworthia and louse symbionts were both derived from a closely related “progenitor strain” of bacteria. This scenario would be like Sodalis species, which have been detected in distantly related insects, including lice (Fukatsu et al. 2007; Smith et al. 2013; Boyd et al. 2016).
The genomes of all louse symbionts sampled are small and much of the genome is devoted to vitamin synthesis. In two instances, we noted the loss of metabolic function potentially facilitated by genome reduction and reorganization. These losses largely impacted fatty acid and amino acid metabolism, while leaving vitamin metabolism intact. The synthesis of B-vitamins is important to the symbiosis between endosymbionts and lice. A comparison of Riesia and Puchtella found that both have similar B-vitamin synthesis capabilities. One major difference is that Puchtella retains the pathway for synthesis of thiamin while Riesia does not. The loss of thiamin biosynthesis was also noted in the Sodalis-like endosymbiont of seal lice (Boyd et al. 2016). In this Sodalis-like endosymbiont, pseudogenes indicated the recent loss of this pathway. It is unclear if the retention of thiamin synthesis in Puchtella has any biological relevance. Red colobus monkeys feed on a variety of resources, including plants (leaves and flowers), lichens, invertebrates, and soil (Struhsaker 2010). However, young leaves constituted the majority of their diet (Struhsaker 2010). It is possible that this leaf heavy diet is low in thiamin and the lice are also deprived of this essential vitamin. Therefore, their parasitic louse may require thiamin from its endosymbionts, while human and chimpanzee lice parasitize hosts with more complex diets that contain thiamin. Hence their endosymbionts of human lice scavenge thiamin rather than synthesizing thiamin (Boyd et al. 2014).
We find that human, chimpanzee, and gorilla louse endosymbionts (Riesia species) carry the small plasmid needed for vitamin B5 synthesis as part of their genomes (table 2 and fig. 5). However, the sister taxon, Puchtella, endosymbiont from the red colobus monkey, does not. Instead, the genes for vitamin B5 synthesis are located on its primary chromosome, a configuration more typical of Eubacteria. It seems likely that the plasmid arose through genome re-organization in the common ancestor of hominid louse endosymbionts. When comparing the genomes of Riesia and Puchtella, there appears to have been movement of genomic regions (fig. 3). This reorganization may have facilitated the origins of the plasmid. The lack of the plasmid in the red colobus monkey endosymbionts, Puchtella, also suggests that this chromosomal structure is not essential for louse endosymbionts. A recent study of an endosymbiont of another mammalian sucking louse (Boyd et al. 2016) also found this pathway was not plasmid based, but has a similar arrangement to Puchtella. Therefore, movement of these genes into close physical proximity on the genome is not essential; however, it may be advantageous, because it has been conserved through multiple rounds of speciation events.
In many ways, this system appears analogous to the movement of amino acid synthesis genes onto plasmids in the endosymbiont of aphids (Buchnera; Baumann et al. 1999; Wernegreen and Moran 2001). Here the rate-limiting steps in tryptophan and leucine synthesis are encoded on plasmids and these plasmids have been maintained over evolutionary time scales (Rouhbakhsh et al. 1996; Wernegreen and Moran 2001). Wernegreen and Moran (2001) noted there was a slight increase in the substitution rate in genes on plasmids in Buchnera. We compared the pairwise distance between genes on the plasmid and in the linear chromosome of Riesia and only see slight greater pairwise distance in plasmid-based genes (supplementary table S2, Supplementary Material online). It seems likely that the plasmid-based genes in louse endosymbionts are not experiencing a significantly higher substitution rate.
Our results are a departure from previous findings and support monophyly of human and primate louse endosymbionts instead of endosymbiont replacement. Previous studies have found that Riesia species were derived from an Arsenophonus-like bacteria ∼ 20–25 Ma (Allen et al. 2009; Novakova et al. 2009). Unlike Riesia, which have genomes ranging from 0.53 to 0.58 Mb in size, Arsenophonus endosymbionts possess larger genomes. Sequenced genomes include the 3.67 Mb genome of A. nasoinae (Wilkes et al. 2010) and 2.95 Mb genome of Arsenophonus sp. (Xue et al. 2014). This would have suggested there was a rapid reduction of the Riesia genome shortly after it colonized lice. Instead endosymbionts of lice might be more ancient and have undergone more long-term genome reductions. This system is biologically interesting, because louse, and hence endosymbiont evolution, are uniquely tied to primate and human evolution. Therefore, shifts in primate biology might impact endosymbiont genome evolution and loss of metabolic functionality (e.g., loss or retention of thiamin synthesis). Phylogenomics and methodological advances will inevitably provide additional insights into this group.
Materials and Methods
Sample Collection
Endosymbionts are intracellular and obligate associates of insects. To sequence the genomes of different louse endosymbionts, we first collected lice from humans and primates. This included lice from red colobus monkeys, chimpanzees, mountain gorillas, and humans (table 1). Chimpanzees have structured populations with limited gene flow (Becquet et al. 2007). Human head lice have divergent mitochondrial haplotypes (Ashfaq et al. 2015). Therefore, we expected divergent strains of endosymbionts and identified likely candidate lice using the mitochondrial cytochrome oxidase subunit1 (COI; supplementary methods, Supplementary Material online). Because mitochondria and endosymbionts are both maternally inherited, mitochondria were treated as proxies for identifying divergent endosymbiont groups. Each endosymbiont used in sequencing was assigned a strain name based on host association and collection site (supplementary methods, Supplementary Material online).
DNA Extraction and Sequencing
Total gDNA was extracted from chimpanzee, gorilla, and human pubic lice using the Zymo Genomic DNA-Tissue MicroPrep kit following the manufacturer’s instructions except that lice were manually macerated prior to incubation with proteinase-K. Total gDNA from red colobus monkey lice was extracted using a phenol–chloroform method following Boyd et al. (2014). DNA extracts from the Netherlands human head lice were provided by Ascunce (see Ascunce et al. 2013 for collection methods).
gDNA from each louse was sonicated using the Covaris M220 instrument to an average fragment size of 300–450 bp (actual range was 200–600 bp). The sheared gDNA was prepared for next-generation sequencing using TruSeq DNAseq or Kapa Library preparation kits. The resulting library was sequenced on one-half lane of Illumina HiSeq2000 or 2500 using the TruSeq SBS sequencing kit v.1-2 for 161 cycles. All samples were sequenced paired-end, with 100 or 160 bp reads. Fastq files were produced using Casava v.1.8.2. Sequence data for the Uganda chimpanzee louse were obtained from the Genbank Short Read Archive (accession SRX390495; see Boyd et al. 2014 and Johnson et al. 2014 for complete sequencing methods).
Genome Assembly
The gDNA libraries contained sequence data from the lice, any nonendosymbiont bacteria, and the endosymbionts. We used a combination of approaches to isolate the endosymbiont data and assemble their genomes. This included reference based and de novo assembly methods. To reconstruct the human louse endosymbiont genomes, we used a reference-guided approach. We first aligned Illumina paired-end reads to the Riesia pediculicola str. USDA genome sequence (gi295698239 and gi292493920) using bowtie2 v.2.1.0 (local option, default sensitivity, results exported as SAM file; Langmead and Salzberg 2012). Resulting SAM files were converted to BAM and sorted using SAMtools v.0.1.19 view and sort functions (Li et al. 2009). We then visualized the sorted BAM files in Geneious v.7.1.7 (www.geneious.com) and searched for evidence of large insertions or deletions (indels) and to check for assembly errors. To make an accurate assembly, it was essential to check for indels as it was suspected that these strains may have diverged more than 1 Ma, ample time for indels to have arisen. No evidence of indels was found in human louse endosymbionts from Cambodia and Netherlands. These endosymbionts came from lice that had the same COI haplotype as the source for the reference genome. However, we did find evidence for indels in the human louse endosymbiont from Honduras when compared with the USDA genome (table 1). This Honduras sample came from a louse with a different COI haplotype. Therefore, to obtain a more accurate assembly of this divergent strain, we used the aTRAM software (Johnson et al. 2013; Allen et al. 2015) to build small-targeted de novo contigs around potential indels. The resulting aTRAM contigs were aligned to the str. USDA genome and the genome sequence was modified to include insertions and remove larger deletions. This modified sequence served as a reference and we aligned reads to it using bowite2 (end-to-end, default sensitivity, results exported as a SAM file). A consensus genome sequence was then called using vcfutils.pl (script distributed as part of BCF/samtools package) from all final BAM files using this modified reference.
To reconstruct the genomes of chimpanzee louse endosymbionts, we download the Riesia pediculschaeffi str. PTSU genome assembly (gi746672782) and the Illumina read library used in that assembly (SRX390495). The original str. PTSU genome consisted of five genomic scaffolds that made up the primary linear chromosome. To improve this genome build, we used the aTRAM software to build small contigs that overlapped the end existing str. PTSU contigs. We then aligned the new aTRAM contigs to the existing contigs to assemble the primary chromosome into one continuous genome sequence. Next, we aligned the Illumina paired-end reads to the corrected genome sequence using Bowtie2 (end-to-end, default sensitivity, results exported as SAM file). The data were converted to a BAM file and sorted using samtools and the genome consensus sequence called using vcfutils.pl. This corrected genome sequence served as the reference for a reference guided assembly of the Riesia pediculschaeffi str. PTSK genome. Illumina paired-end reads were mapped to the updated PTSU genome sequence using bowtie2 (end-to-end, default sensitivity, results exported as SAM file). The SAM file was converted to a BAM file and sorted using samtools and the genome consensus sequence called using vcfutils.
There was no close reference genome available for the gorilla or the human pubic louse endosymbiont genomes; therefore, we took a combined de novo and reference guided approach. First, we trimmed low quality bases from the Illumina 2500 sequence data using Timmomatic v.032 (Bolger et al. 2014) by removing bases with a phred score <30 from the 3’ end of the read for both libraries. After trimming, any reads shorter than 75 bp and their mate pairs were removed. The quality trimmed reads were then de novo assembled into contigs and scaffolds using abyss-pe v.1.5.2 (k = 64). We then used BLASTn v2.2.28+ to identify assembled contigs belonging to the endosymbiont genome (str. USDA genome served as the target for BLASTn). Next, we used LAST v.531 to find the assembled plasmid by aligning the conserved protein coding sequences to the contigs library (str. USDA pPAN plasmid served as the target sequence). As with the str. PTSU genome, we used aTRAM to build targeted small de novo assemblies to join the endosymbiont contig ends. Contigs were aligned in Geneious and a reference sequence called. We then aligned the Illumina paired-end reads to the draft genome sequence using bowtie2 (end-to-end, default sensitivity). We then examined the resulting read alignments in BAM format to identify assembly errors. The final consensus genome sequence was called using vcfutils.pl. The pubic louse endosymbiont genome was expected to be similar to the gorilla louse endosymbiont. Therefore, we aligned the pubic louse endosymbiont reads to the gorilla louse endosymbiont genome using bowtie2 (local, default sensitivity). The genome sequence was called using vcfutils.pl. Overall, sequencing depth for the pubic louse endosymbiont genome was low and we could not identify indels like we did with human head louse endosymbionts. Therefore we skipped this step for this genome.
In all instances where a plasmid was found in de novo or reference guided assemblies, we used read associations to verify the chromosome was circular. We did this by retrieving BAM files from the assemblies and isolating the plasmid-contig and its reads. The BAM was then visualized in Geneious and we manually verified that read pairs supported circularization of the plasmid by overlapping the end of the contig. Finally, each of the Riesia genomes were annotated using the RAST pipeline (Overbeek et al. 2005; Aziz et al. 2008).
The genome of the red colobus monkey louse endosymbiont was largely unknown. Therefore, we followed the same method described by Boyd et al. (2014) to assemble and annotate the original chimpanzee louse endosymbiont genome (Riesia pediculschaeffi str. PTSU) genome when it was unknown. This method used a de novo assembly of all reads into contigs, BLAST was used to identify contigs belonging to the endosymbiont genome (target consisted of a custom database of representative bacterial genomes), the genome was re-assembled in isolation with only reads belonging to the genome, and annotation of the genome in the RAST pipeline.
To compare the annotated genes in louse endosymbionts we used reciprocal best tBLASTx 2.5.0+ to identify shared genes (scripts for filtering blast results obtained from http://goo.gl/csx730). However, this only provides a summary of potential shared genes without regard to functionality of genes. To better understand the metabolic capacity for each genome we used comparison tools in SEED (Overbeek et al. 2005) to compare each endosymbiont genome to that of the genome of the endosymbiont from the human body louse. Total numbers of genes belonging to different functional categories were summarized for each genome and then shared and unique functions were identified using the “compare function” tools within SEED.
Phylogenetic Database Construction
We searched for Enterobacteriaceae with fully sequenced genomes and selected taxa that were representative of the class along with an outgroup taxa from neighboring class, Pasteurellaceae. We then downloaded all protein coding genes as DNA sequences. To find potential orthologs in downloaded gene sequences, we found bidirectional best hits between the genes. First, tBLASTx v.2.2.28+ was used to find hits between genes in all possible pair-wise comparisons of all taxa (default parameters, outformat -6). Second, BLAST hits were filtered to find only the bidirectional best hits between every possible taxa set (scripts for filtering blast results obtained from http://goo.gl/csx730). Interpreting bidirectional best hits as edges in a network, we grouped genes into separate interconnected networks. Network analysis was done using Cytoscape v.3.2.1 (Shannon et al. 2003), using the “network analysis” and “find interconnected subnetwork” tools. Node lists for all subnetworks that possessed between 45 and 10 nodes were exported from Cytoscape with 195 total subnets recovered. To remove paralogs, we then removed any taxa that were represented by two or more nodes in a network. Any subnets with fewer than ten nodes after paralog removal were then removed leaving 177 subnets that are herein treated as groups of orthologous protein-coding genes. Networks containing genes found on the Riesia plasmid were manually annotated to ensure accuracy. To do this, we aligned based on best hits to the Riesia genes, checking accepted hits by nucleotide and translated alignments. This provided us with a set of genes that were equivocal bidirectional best-BLAST hits in all taxa that our evidence suggests are single copy orthologs.
Only those louse endosymbiont genomes with de novo annotations (human body louse and red colobus monkey louse endosymbiont genomes) were included in the network analysis to find orthologs. This was done because some genomes were only partially assembled. Therefore, if included, we may have excluded otherwise good networks missing taxa due to assembly error leading to missing genes. After candidate orthologs were identified above in the human louse endosymbiont, we used the ortholog set from the human louse endosymbiont to annotate other hominid louse endosymbiont genome using Exonerate v.2.2.0 (Slater and Birney 2005). This allowed us to simultaneously annotate and capture ortholog sequences from these genomes. This software searched the genome, finding potential matches to translated genes from the human body louse endosymbiont genome. The predicted genes were then translated in Geneious using translation table 11. The genes were then examined for pseudogenes or annotation errors. Genes with premature stop codons were removed. Thus each of the 177 gene networks could be treated as groups of orthologous single copy orthologs.
Species Tree Estimation
All 177 single copy orthologs identified in the previous section were used in determining a potential species tree for louse endosymbionts and representative Enterobacteriaceae. Sequences for each gene were translated in Geneious using bacterial translation table 11. Amino acid sequences were then aligned using UPP v.2.0 (Nguyen et al. 2015) and the aligned sequences back translated to nucleotide sequences. Base composition in the first, second, and third positions were plotted. The plot revealed that the %GC content of the third codon position was higher in endosymbiont species when compared with first and second codon positions (supplementary fig. S1, Supplementary Material online). This bias would violate the model assumptions of the GTR + gamma substitution model used in ML tree reconstruction below, therefore the third codon positions were omitted from the gene alignments.
We estimated species trees using 1) two coalescent approaches that estimate species trees from gene trees and 2) a concatenation method where a ML tree is estimated from single data matrix of concatenated sequences. For the coalescent approach we first built individual gene trees for all 177 ortholog sets. Each gene tree was estimated using RAxML (v.8.1.3; Stamatakis 2014) under a GTR + gamma model. The species tree was then estimated from these gene trees using ASTRAL (v.4.9.7; Mirarab et al. 2014a; Mirarab and Warnow 2015) and ASTRID (v.1.4; Vachaspati and Warnow 2015). Support for branches in either tree was computed using local posterior probabilities based on gene tree quartet frequencies (Sayyari and Mirarab 2016).
For the concatenation approach, all aligned orthologs were concatenated into a single matrix. In order to account for substitution rate heterogeneity between genes, we performed a partitioned ML analysis by first estimating the GTR + gamma model parameters for each codon position of each gene. We then performed a principal coordinates analysis of the rate parameters and clustered the codon positions using k-means clustering. We found that seven clusters explained most of the variation in the data. The alignment was partitioned according to the clusters and RaxML was used to find the ML tree using a GTR + gamma model, with an initial starting tree based upon a ML tree estimated using FastTree-2 under GTR + cat (v 2.1.7; Price et al. 2010). Support for branches in the concatenation tree was based on 100 bootstrap replicate trees.
Origins of a Small Plasmid
We further investigate the phylogeny of a small plasmid found in many louse endosymbionts to determine if it was congruent with the species tree. First we built an ML phylogeny of the plasmid based a concatenation of four genes present on the plasmid and their orthologs in other bacterial species. This tree produced a topology incongruent with any of our estimated species trees with regard to louse endosymbionts, however, this tree was based on only four genes (2,176 sites total). This limited sampling of genes might bias the estimated tree topology.
To explore how decreasing gene sampling may have affected this tree, we compared each gene tree with the presumed species tree (the concatenation tree was selected as the species tree for this comparison because both gene trees and the concatenation tree used bootstraps as a measure of bipartition support). For the gene trees, used the ML gene trees described previously (see section on Coalescent species tree estimation). Each gene tree was examined if to determine if all 38 taxa were present. If one or more taxa were missing in the gene tree, these missing species were trimmed from the species tree for that comparison. By doing this we are comparing two trees with the same tips. Next we collapsed unsupported bipartitions in both the gene and species trees. Bootstrap values were used to identify unsupported bipartitions. Finally, we determined the fraction of supported bipartitions in the gene tree that were in conflict with the species tree using scripts described in Mirarab et al. (2014b). The process of collapsing unsupported bipartitions and comparing the trees was done three times using 50, 75, and 95% bootstraps as cutoffs for unsupported bipartitions.
The resulting gene tree–species tree comparisons yielded 0–1 score of similarity for every gene at differing levels of bootstrap support. Summary statistics for this gene tree–species tree similarity data were calculated in R v.3.1.2. We then isolated scores for gene found on the plasmid and the primary chromosome. This allowed us to compare gene tree–species tree variation by chromosome (i.e., plasmid V primary chromosome) using a Welch two-sampled t-test (test done in R v.3.1.2). This test was selected to determine if gene tree variation was significantly different than gene tree variation on the primary chromosome.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Marina Ascunce (University of Florida) and Katie Shepard (Shepard Institute) for providing lice used in this study and two anonymous reviewers for their comments on this manuscript. This work was supported by the National Science Foundation (DEB1310824 to B.M.B. & D.L.R.; DEB0845392 and DEB0717165 to D.L.R.; DEB0612938, DEB1239788, and DEB1342604 to K.P.J; and ABI-1458652 to T.W.). This research is part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1053575. XSEDE computational resources were provided through XSEDE grant ASC160042 to N.N.
References
- Alikhan NF, Petty NK, Ben Zakour NL, Beastson SA.. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparison. BMC Genomics 12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JM, Reed DL, Perotti MA, Braig HR.. 2007. Evolutionary relationships of “Candidatus Riesia spp.,” endosymbiotic Enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol. 73:1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JM, Light JE, Perotti MA, Braig HR, Reed DL.. 2009. Mutational meltdown in primary endosymbionts: selection limits Muller’s Ratchet. PloS ONE 4:e4969.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JM, Huang DI, Cronk QC, Johnson KP.. 2015. aTRAM-automated target restricted assembly methods: a fast method for assembling loci across divergent taxa from next-generation sequence data. BMC Bioinf. 16:98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JM, Burleigh JG, Light JE, Reed DL.. 2016. Effects of 16S rDNA sampling on estimates of the number of endosymbiont lineages in sucking lice. PeerJ 4:e2187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JM, Boyd B, Nguyen N, Vachaspati P, Warnow T, Huang DI, Gero P, Bell KC, Cronk QCB, Mugisha L, et al. 2017. Phylogenomics from whole genome sequences using aTRAM. Syst Biol. Doi:10.1093/systbio/syw105. [DOI] [PubMed] [Google Scholar]
- Anderson JO, Anderson SG.. 2000. A century of typhus, lice and Rickettsia. Res Microbiol. 29:888–911. [DOI] [PubMed] [Google Scholar]
- Aschner M, Ries E.. 1933. Das verhalten der kleiderlaus bei ausschaltung iher symbioten. Z Morphol Okol Tiere 26:529–590. [Google Scholar]
- Ascunce MS, Toups MA, Kassu G, Fane J, Scholl K, Reed DL.. 2013. Nuclear genetic diversity in human lice (Pediculus humanus) reveals continental differences and high inbreeding among worldwide populations. PloS One 8:e57619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfaq M, Prosser S, Masood M, Ratnasingham S, Hebert PDN.. 2015. High diversification in the head louse, Pediculus humanus (Pediculidae: Phthiraptera). Sci Rep. 5:14188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet C, Patterson N, Stone AC, Przeworski M, Reich D.. 2007. Genetic structure of chimpanzee populations. PloS Genet. 3:e66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Ann Rev Microbiol 59:155–189. [DOI] [PubMed] [Google Scholar]
- Baumann L, Baumann P, Moran NA, Sandstrom J, Thao ML.. 1999. Genetic characterization of plasmids containing genes encoding enzymes of leucine biosynthesis in endosymbionts (Buchnera) of aphids. J Mol Evol. 48:77–85. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, Allen JM, de Crecy-Lagard V, Reed DL.. 2014. Genome sequence of Candidatus Riesia pediculischaeffi, endosymbiont of chimpanzee lice, and genomic comparison of recently acquired endosymbionts from human and chimpanzee lice. G3 (Bethesda) 11:2189–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, Allen JM, Koga R, Fukatsu T, Sweet AD, Johnson KP, Reed DL.. 2016. Two bacteria, Sodalis and Rickettsia, associated with the seal louse Proechinophthirus flutucs (Phthiraptera: Anoplura). Appl Environ Microbiol. 82:3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, Reed DL.. 2012. Taxonomy of lice and their endosymbiont bacteria in the post-genomic era. Clin Microbiol Infect. 18:324–331. [DOI] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S.. 2010. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 8:218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1965. Endosymbiosis of Animals with Plant Microorganisms. New York: Interscience. [Google Scholar]
- Clark MA, Moran NA, Baumann P, Wernegreen JJ.. 2000. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution 54:517–525. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequences with rearrangements. Genome Res. 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durden LA, Musser GG.. 1994. The sucking lice (Insect, Anoplura) of the world: a taxonomic checklist with records of mammalian hosts and geographical distributions. Bull Am Mus Nat Hist 128: [Google Scholar]
- Eberle MW, Mclean DL.. 1982. Initiation and orientation of the symbiote migration in the human body louse Pediculus humans L. J Insect Physiol. 28:417–422. [Google Scholar]
- Eberle MW, Mclean DL.. 1983. Observation of symbiote migration in human body lice with scanning and transmission electron microscopy. Can J Microbiol. 29:755–762. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatability methods will be positively misleading. Syst Zool. 27:401–410. [Google Scholar]
- Fukatsu T, Hosokawa T, Koga R, Nikoh N, Kato T, Haymam S, Takefushi H, Tanak I.. 2009. Intestinal endocellular symbiotic bacterium of the macaque louse Pedicinus obtusus: distinct endosymbiont origins in anthropoid primate lice and the old world monkey louse. Appl Environ Microbiol. 73:6660–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Koga R, Smith WA, Tanaka K, Nikoh N, Sasaki-Fukatsu K, Yoshizawa K, Cale C, Clayton DH.. 2007. Bacterial endosymbionts of the slender pigeon louse, Columbicola columbae, allied to the endosymbiont of grain weevils and tsetse flies. Appl Environ Microbiol. 73:6660–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes MJ, Latorre A, Lamelas A, Moya A.. 2010. Genomics of intracellular symbionts in insects. Int J Med Microbiol. 300:271–278. [DOI] [PubMed] [Google Scholar]
- Herbeck JT, Degnan PH, Wernegreen JJ.. 2005. Nonhomogenous model of sequence evolution indicates independent origins of primary endosymbionts within the Enterobacteriales (γ-proteobacteria). Mol Biol Evol 22:520–532. [DOI] [PubMed] [Google Scholar]
- Hooke R. 1665. Micrographia: or some physiological description of minute bodies made by magnifying glasses with observations and inquiries thereupon. London, UK: Council of the Royal Society of London for Improving Natural Knowledge. [Google Scholar]
- Hypsa V, Krizek J.. 2007. Molecular evidence for polyphyletic origins of the primary symbionts of sucking lice (Phthiraptera, Anoplura). Microb Ecol. 54:242–251. [DOI] [PubMed] [Google Scholar]
- Johnson KP, Walden KO, Robertson HM.. 2013. Next generation phylogenomics using a target restricted assembly method. Mol Phylo Evol. 66:417–422. [DOI] [PubMed] [Google Scholar]
- Johnson KP, Allen JM, Olds BP, Mugisha L, Reed DL, Paige KN, Pittendrigh BR.. 2014. Rates of genomic divergence in humans, chimpanzees and their lice. Proc Biol Sci. 281:20132174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Haas BJ, Sun w, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, et al. 2010. Genome sequence of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci. 10003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light JE, Reed DL.. 2009. Multigene analysis of phylogenetic relationships and divergence times of primate sucking lice (Phthiraptera: Anoplura). Mol Phylo Evol. 50:376–390. [DOI] [PubMed] [Google Scholar]
- Light JE, Smith VS, Allen JM, Durden LA, Reed DL.. 2010. Evolutionary history of mammalian sucking lice (Phthiraptera: Anoplura). BMC Evol Biol. 10:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez MJ, Neef A, Pereto J, Patino-Navarrete R, Latorre A, Moya A.. 2009. Evolutionary convergence and nitrogen metabolism in Blattabacerium strain Bge, primary endosymbiont of cockroach Blattella germanica. PloS Genet. 5:e1000721.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marin A, Oceguera-Figueroa A, Latorre A, Jimenez-Garcia LF, Moya A.. 2015. Solving a bloody mess: b-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthoropods and the leech Haementeria officinalis. Genome Biol Evol. 7:2871–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10:13–26. [DOI] [PubMed] [Google Scholar]
- Mirarab S, Raez R, Simmerman T, Swenson MS, Warnow T.. 2014a. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30:i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Bayzid SM, Boussau B, Warnow T.. 2014b. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 346:1250463.. [DOI] [PubMed] [Google Scholar]
- Mirarab S, Warnow T.. 2015. ASTRAL-II coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31:i44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. [DOI] [PubMed] [Google Scholar]
- Moran NA, Bennett GM.. 2014. The tiniest tiny genomes. Annu Rev Micobiol. 68:195–215. [DOI] [PubMed] [Google Scholar]
- Novakova E, Hypsa V, Moran NA.. 2009. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 9:143.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen ND, Mirarab S, Kumar K, Warnow T.. 2015. Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 16:124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwaards R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti MA, Allen JM, Reed DL, Braig HR.. 2007. Host-symbiont interaction of the primary endosymbiont of human head and body lice. FASEB J. 21:1058–1066. [DOI] [PubMed] [Google Scholar]
- Perotti MA, Kirkness EF, Reed DL, Braig HR.. 2009. Endosymbionts of lice In: Bourtzis K, Miller TA, editors. Insect Symbiosis. V3. Boca Raton (FL: ): CRC Press, p. 205–219. [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PloS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta O. 1955. Eperimentelle untersuchungen uber die bedeutung der symbiose der kleiderlaus Pediculus vestimenti Burm. Z Parasitenkd 17:1. [DOI] [PubMed] [Google Scholar]
- Raoult D, Roux V.. 1999. The body louse as a vector of reemerging human diseases. Clin Infect Dis. 29:888–911. [DOI] [PubMed] [Google Scholar]
- Raoult D, Dutour L, Jankauskas R, Fournier PE, Ardagna Y, Drancourt M, Signoli M, La VD, Macia Y, Adoudharam G.. 2006. Evidence for louse-transmitted diseases in soldiers of Napoleon’s Grand Army in Vinius. J Infect Dis. 193:112–120. [DOI] [PubMed] [Google Scholar]
- Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH.. 2004. Genetic analysis of lice supports direct contact between modern and archaic humans. PloS Biol. 2:e340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DL, Light JE, Allen JM, Kirchman JJ.. 2007. Pair of lice lost or parasites regained: the evolutionary history of anthropoid primate lice. BMC Biol. 5:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries E. 1931. Die symbiose der lause und federlinge. Z Morphol Okol Tiere 20:233–367. [Google Scholar]
- Ries E. 1932. Die prozesse der eibildung und des eiwachstums bei Pediculiden und Mallophagen. Z Zellforsch Mikrosk Anat 16:314–388. [Google Scholar]
- Ries E. 1933. Endosymbiose und parasitismus. Z Parasitenkunde 6:339–349. [Google Scholar]
- Ries E. 1935. Uber den sinn der erblichen insektensymbiose. Naturwissenschaften 23:744–749. [Google Scholar]
- Ries E, van Weel PB.. 1934. Die eibildung der kleiderlaus, untersucht an lebenden, vital gefarbten und fixierten praparaten. 20:565–618. [Google Scholar]
- Rouhbakhsh D, Lai CY, von Dohlen CD, Clark MA, Baumann P, Moran NA, Voetilin DJ.. 1996. The tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the aphidae. J Mol Evol. 42:414–421. [DOI] [PubMed] [Google Scholar]
- Sach JL, Skophammer RG, Bansal N, Stajich JE.. 2014. Evolutionary origins and diversification of proteobacterial mutualists. Proc R Soc B 281:20132146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sach JL, Skophammer RG, Regus JU.. 2011. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci. 108:10800–10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Fukatsu K, Koga R, Nikoh N, Yoshizawa K, Kasai S, Mihara M, Kobayashi M, Tomita T, Fukatsu K.. 2006. Symbiotic bacteria associated with stomach discs of human lice. Appl Environ Microbiol. 72:7349–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer C, Stackebrandt E, Gadau J, Holldobler B, Gross R.. 2000. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol. 50:1877–1886. [DOI] [PubMed] [Google Scholar]
- Sayyari E, Mirarab S.. 2016. Fast-coalescent-based computation of local branch support from quartet frequencies. Mol Biol Evol. 33:1654–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Want JT, Ramage D, Amin N, Schwikowski B, Ideker T.. 2003. Cytoscape: a software environment for integrated model of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WA, Oakeson KF, Johnson KP, Reed DL, Carter T, Smith KL, Koga R, Fukatsu T, Clayton DH, Dale C.. 2013. Phylogenetic analysis of symbionts in feather-feeding lice of the genus Columbicola: evidence for repeated symbiont replacements. BMC Evol Biol. 13:109.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RaxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhsaker TT. 2010. The Red Colobus monkeys: variation in demography, behavior, and ecology of endangered species. Oxford: Oxford University Press. [Google Scholar]
- Summer DK. 1996. The biology of plasmids. Oxford: Blackwell Publishing Ltd. [Google Scholar]
- Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P.. 2000. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 66:2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MA, Kitchen A, Light JE, Reed DL.. 2011. Origins of clothing lice indicates early clothing use by anatomically modern humans in Africa. Mol Biol Evol. 28:29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachaspati P, Warnow T.. 2015. ASTRID: accurate species trees from internode distances. BMC Genomics 16:S3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallari DS, Rock CO.. 1985. Pantothenate transport in Escherichia ecoli. J Bacteriol. 162:1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasenor T, Brom S, Davalos A, Lozano L, Romero D, Garcia-de los Santos A.. 2011. Housekeeping genes essential for pantothenate biosynthesis are plasmid-encoded in Rhizobium etli and Rhizobium leguminosarum. BMC Microbiol. 11:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuven JT, McCutcheon JP.. 2011. An AT mutational bias in the tiny GC-rich endosymbiont genome of Hodgkinia. Genome Biol Evol. 4:24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Moran NA.. 2001. Vertical transmission of biosynthetic plasmids in aphid endosymbionts (Buchnera). J Bacteriol. 183:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes TE, Darby AC, Choi JH, Colbourne JK, Werren JH, Hurst GD.. 2010. The draft genome sequence of Arsenophonus nasoniae, son-killer of Nasonia vitripennis, reveals genes associated with virulence and symbiosis. Insect Mol Biol. 1:59–73. [DOI] [PubMed] [Google Scholar]
- Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW.. 2010. Phylogeny of gammaproteobacteria. J Bacteriol. 102:2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Duncan RP.. 2015. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci. 112:10255–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Zhou X, Zhang CX, Yu LL, Fan HW, Wang Z, Xu HJ, Xi Y, Zhu ZR, Zhou WW, et al. 2014. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15:521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.