Abstract
Methods to label cell populations selectively or to modify their gene expression are critical tools in the study of developmental or physiological processes in vivo. A variety of approaches have been applied to the zebrafish model, capitalizing on Tol2 transposition to generate transgenic lines with high efficiency. Here we describe the adoption of the Q system of Neurospora crassa, which includes the QF transcription factor and the upstream activating sequence (QUAS) to which it binds. These components function as a bipartite regulatory system similar to that of yeast Gal4/UAS, producing robust expression in transient assays of zebrafish embryos injected with plasmids and in stable transgenic lines. An important advantage, however, is that QUAS-regulated transgenes appear far less susceptible to transcriptional silencing even after seven generations. This chapter describes some of the Q system reagents that have been developed for zebrafish, as well as the use of the QF transcription factor for isolation of tissue-specific driver lines from gene/enhancer trap screens. Additional strategies successfully implemented in invertebrate models, such as a truncated QF transcription factor (QF2) or the reassembly of a split QF, are also discussed. The provided information, and available Gateway-based vectors, should enable those working with the zebrafish model to implement the Q system with minimal effort or to use it in combination with Gal4, Cre, or other regulatory systems for further refinement of transcriptional control.
1. BACKGROUND
Transgenic approaches to manipulate gene expression in spatially or temporally restricted ways have been instrumental in the study of biological processes. The transparency of zebrafish embryos and larvae make them particularly well suited for selective labeling of cell populations and observing their behavior over time, for assessing the effects of genetic perturbations, for optogenetic methods to alter neural activity, and for monitoring of genetically encoded biosensors. In these and other strategies to explore cellular function in vivo, versatility is an important property of the transgenic tools being generated. One way to increase flexibility and maximize utility is with a binary system, in which the expression of a collection of reporter or effector genes is activated by a transcription factor that is itself placed under the control of different cell type—specific promoters/enhancers.
The use of bipartite transcriptional regulatory systems in zebrafish was first accomplished in the laboratory of Jose Campos-Ortega with promoters driving near ubiquitous expression of the gene encoding the yeast Gal4 transcription factor. Five copies of an optimized upstream activating sequence (UAS) where Gal4 binds, placed upstream of the minimal E1b promoter of adenovirus and a synthetic start site from the carp β-actin promoter, controlled transcription of sequences encoding the intracellular domain of the zebrafish Notch1 protein fused with six myc tags (Scheer & Campos-Ortega, 1999). Expression was visualized in embryos derived from matings between the driver and responder transgenic lines by anti-myc antibody labeling. This early demonstration showed that the Gal4/UAS system could function in zebrafish embryos, albeit at low levels of expression as detection of the UAS-regulated gene product required signal amplification by immunolabeling.
Expression levels were significantly increased through the adoption of methods to overexpress randomly targeted genes in a Drosophila insertional mutagenesis screen (Rorth, 1996). Widespread and intense labeling from the green fluorescent protein (GFP) was achieved in zebrafish embryos using a self-reporting vector that contained the transcriptional activation domain (AD) of the VP16 protein of Herpes simplex virus fused to the Gal4 DNA binding domain and 14 UAS (14X) binding sites in a tandem array upstream of the GFP gene (Koster & Fraser, 2001). Injection of circular or linearized plasmids in transient assays yielded robust GFP labeling in a variety of zebrafish embryonic tissues depending on the promoter driving Gal4-VP16. Transgenic lines were not established on account of lethality, presumably due to “squelching” of factors required for the transcription of endogenous genes (Koster & Fraser, 2001). Moreover, the injected DNA likely integrated into the genome in high copy number as complex concatemers that would be targets for transcriptional silencing.
The discovery and application of Tol2 transposition (Kawakami, Shima, & Kawakami, 2000; Urasaki, Morvan, & Kawakami, 2006) circumvented the problem of high copy number and lethality due to overexpression. Several groups modified Gal4-VP16 constructs with the addition of Tol2 arms (eg, Asakawa & Kawakami, 2008; Davison et al., 2007; Scott et al., 2007). When injected with Tol2 transposase into 1-cell stage embryos, integration into the zebrafish genome by transposition results in single copy insertions, survival of the resultant embryos and germ-line propagation of transgene. Construction of gene/enhancer trap vectors for Tol2 transposition has yielded numerous Gal4 driver lines from screens for tissue-specific patterns of expression (eg, Distel, Wullimann, & Koster, 2009; Kawakami et al., 2010; Marquart et al., 2015; Otsuna et al., 2015; Takeuchi et al., 2015) as well as generated new insertional mutations (Balciuniene & Balciunas, 2013).
Owing to its widespread use in the Drosophila model, many zebrafish researchers have used the Gal4/UAS system to manipulate gene expression in cells or tissues of interest, but with varying results. Both the Gal4-VP16 and UAS components have been revised to maximize expression levels while reducing toxicity (Akitake, Macurak, Halpern, & Goll, 2011; Asakawa & Kawakami, 2008; Distel et al., 2009). However, a persistent problem has been the loss of expression from transgenes in which transcription is under the control of a multicopy UAS. The UAS contains essential CpG dinucleotides for Gal4 binding, which also makes it a preferred site for DNA methylation and, hence, transcriptional silencing (Goll, Anderson, Stainier, Spradling, & Halpern, 2009). Although UAS-regulated transgenes may be robustly expressed initially, in subsequent generations, expression often becomes variable in individuals from the same clutch, and, in extreme cases, fully extinguished (Akitake et al., 2011; Goll et al., 2009; Pang, Wang, Zhu, & Sun, 2015). Transgenic lines have sometimes been maintained by selecting embryos or larvae that show more intense or complete labeling patterns, but this only is possible when a transgene contains fluorescent or visible markers that can be readily scored. Alternatively, constructs have been reinjected and lines rederived from new transgenic founders.
To potentially combat transgene silencing and expand the repertoire of transgenic tools, another binary system derived from Neurospora crassa was adapted for Tol2 transposition in zebrafish (Subedi et al., 2014). The qa gene cluster was originally identified from the discovery of Neurospora mutants that failed to metabolize quinic acid as a carbon source, and its regulatory genes were later determined (refer to Giles et al., 1985). Components of this cluster (named the Q system) were shown to activate transcription in Drosophila embryonic and adult stages at a higher level than the Gal4/UAS system and to operate in cultured mammalian cells (Potter, Tasic, Russler, Liang, & Luo, 2010) and, subsequently, in nematodes (Wei, Potter, Luo, & Shen, 2012). This chapter reviews how the Q system has been applied to zebrafish transgenesis, as well as for gene/enhancer trap screens to recover new tissue-specific driver lines. The wild-type AB laboratory strain (Walker, 1999) was used for all of the described experiments and to generate new transgenic lines.
2. COMPONENTS OF THE Q TRANSCRIPTIONAL REGULATORY SYSTEM
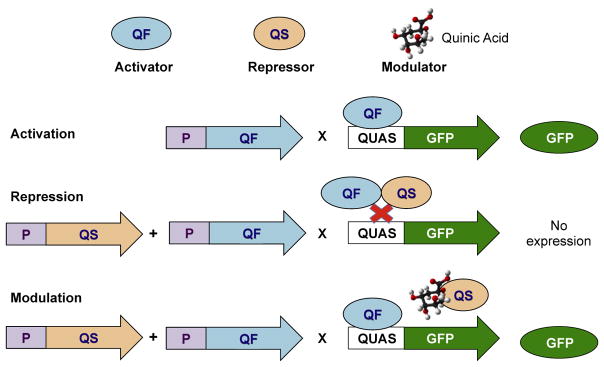
The key features of the Q system are depicted in Fig. 1. The QF transcription factor is an 816 amino acid protein comprised of structurally distinct regions: a DNA binding domain (DBD), a middle region of unknown function (DM), and a transcriptional AD. QF binds to QUAS sites to induce transcription of adjacent genes. A repressor protein, QS, functions to inhibit QF binding, while exposure to quinic acid relieves this inhibition and restores QF-mediated transcriptional activation.
FIGURE 1.
Schematic of the Q system. GFP, green fluorescent protein; QUAS, Q upstream activating sequence. (See color plate)
2.1 QF AND QF DERIVATIVES
To assess whether QF would be active in zebrafish embryos, the entire QF coding sequence was initially placed under the control of either the Xenopus laevis elongation factor 1-alpha (EF1α) or zebrafish ubiquitin B (ubb) promoter, promoters that are known to drive high levels of expression throughout the zebrafish embryo (Amsterdam, Lin, & Hopkins, 1995; Mosimann et al., 2011). Constructs were produced in Tol2 vectors as previously described (Subedi et al., 2014) and coinjected in one to two cell stage embryos along with a QUAS:GFP Tol2 plasmid (see Section 2.2) and RNA encoding the Tol2 transposase (25 ng/μL). Robust fluorescent labeling was observed in diverse tissues of the injected embryos after 1 day post fertilization (dpf), validating the use of the Q system in transient expression assays. Lethality was measured and the optimal concentration for the driver and reporter plasmids was determined (1–10 ng yielded a survival rate of ranging from 88% to 95%). Following these encouraging findings, Tol2 vectors with tissue-specific promoters (eg, mnx1, fabp1a, and ins) driving QF expression were assembled for the generation of stable transgenic lines (Subedi et al., 2014). Transgenic QF reporters have also been produced to label cells activated by Notch or retinoic acid (RA) signaling using tp1 (Parsons et al., 2009; Subedi et al., 2014) and RA response elements (Huang et al., 2014), respectively. To facilitate the inclusion of promoters or regulatory sequences of interest, a PCR fragment containing the QF gene and the SV40 termination sequence was inserted as a middle entry (ME) clone in the Gateway Recombination Cloning system (Invitrogen, Thermo Fisher Scientific Inc.). In this modular system, any promoter in a 5′ entry clone can be recombined along with the pME-QF clone and a 3′ reporter gene into a Gateway destination vector reengineered for Tol2 transposition (refer to Kwan et al., 2007).
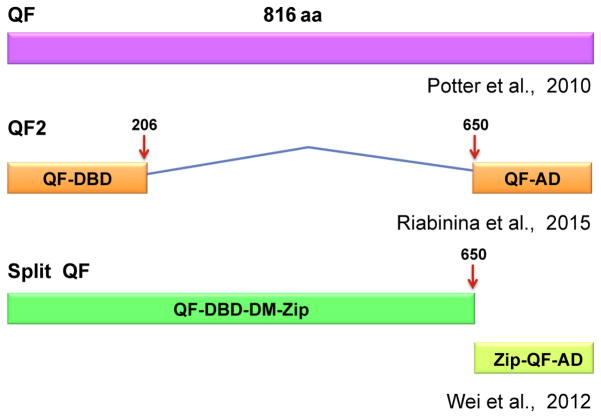
A drawback to QF is that, as with ectopic Gal4-VP16 activity, widespread overexpression of this transcriptional activator appears to be toxic to developing embryos. It has therefore been challenging to recover zebrafish transgenic founders with broadly expressed promoters driving QF. To circumvent this problem in Drosophila, Riabinina et al. (2015) identified the region of the QF protein responsible for toxicity and devised other versions of the protein that would significantly reduce it. By testing a number of QF variants, they determined that lethality corresponded with the presence of the middle region that, fortunately, was dispensable for QF function. One of the resultant truncated and codon-optimized products, QF2 (Fig. 2), fulfilled the criteria of promoting high levels of gene expression, being suppressible by QS and nontoxic when activated throughout the tissues of larval or adult Drosophila (Riabinina et al., 2015).
FIGURE 2.
The QF transcription factor and modifications.
In preliminary experiments to test the efficacy of QF2 in zebrafish, a 1061 bp fragment (including the terminal 58 bp of the synaptobrevin (syb) promoter and a translation initiation sequence 5′ and 28 bp of the Hsp70 termination site 3′) was amplified from the pattB-synaptobrevin-7-QFBDAD-hsp70 plasmid developed for Drosophila (Riabinina et al., 2015), and introduced as a Gateway ME clone. This clone was recombined with a Gateway 5′ entry clone bearing the ubb promoter (Mosimann et al., 2011) into a Tol2 destination vector, and the resulting plasmid coinjected into one to two cell embryos together with QUAS:GFP Tol2 plasmid and RNA encoding Tol2 transposase. By 1 dpf, robust labeling was observed. Moreover, QF2 appeared to be less toxic than in parallel tests with a comparable range of QF concentrations (Monge and Halpern, unpublished results). Thus, QF2 is functional in zebrafish, and its smaller size may be a significant advantage for gene/enhancer trap vectors or for integration into genes of interest by Crispr/Cas9 genome editing.
A further modification of QF is the division of the coding sequence into two parts that can each be expressed under the control of a different promoter for the purpose of intersectional gene expression (Fig. 2). The split QF system is based on split Gal4, whereby sequences containing the DNA binding and transcriptional ADs are separated and fused with heterodimeric leucine zipper sequences (Luan, Peabody, Vinson, & White, 2006). With transcription of each module regulated by different promoters, the two peptides are synthesized only in those cells in which both promoters are active and, upon binding through the leucine zipper, assemble into a functional transcription factor. This method was recently applied to zebrafish by splitting an optimized version of Gal4 and demonstrating its reassembly by activation of UAS reporters (Almeida & Lyons, 2015). In a similar fashion, split QF components were designed through fusion of sequences for leucine zipper heterodimers to those encoding the QF DNA binding and ADs. In transgenic worms, fluorescent labeling was observed in a single neuron, in which expression from a promoter driving the DBD construct overlapped with that of another promoter driving the AD construct (Wei et al., 2012). Although a powerful technique to restrict gene expression, a downside to the split QF approach is that the activity of the reconstituted protein, as measured by fluorescence intensity, was only 42% of that of the intact transcription factor and not all of expected cells were labeled (Wei et al., 2012).
The functionality of split QF in zebrafish has, so far, been verified in transient expression assays. Sequences corresponding to the CZipper(−)::AD(QF) (678 bp) and the NLS-BD-DM(QF)::NZipper(+) (2118 bp) regions were amplified, respectively, from plasmids XW52 and XW54 of Wei et al. (2012) to produce ME Gateway clones. The ME clones were individually recombined with the ubb promoter 5′ entry clone and an SV40 polyA tail 3′ entry clone into a Tol2 destination vector (Kwan et al., 2007). The two destination vectors were co-injected with QUAS:GFP plasmid and Tol2 RNA into one to two cell stage zebrafish embryos or, alternatively, co-injected into QUAS:GFP transgenic embryos (see Section 2.2). All groups were scored for the presence of fluorescently labeled cells a day later. Relative to sibling embryos injected with a ubb:QF construct that produces widespread GFP labeling, those expressing the split QF components displayed far fewer, brightly labeled cells, scattered throughout a variety of tissues (Monge and Halpern, unpublished results). GFP labeled cells were not detected when the AD or BD construct was injected alone. The split QF ME clones were also assembled into Gateway destination vectors with a 5′ entry clone containing the promoter of the Xenopus neural beta tubulin gene (XlTubb) (gift of Paul Krieg, University of Arizona). In QUAS:GFP transgenic embryos, coinjected with the two vectors, GFP positive cells were only recovered in the central nervous system (CNS) where the XlTubb promoter selectively functions (Ghosh and Halpern, unpublished results). Thus, reassembly of split QF components can be successfully accomplished in zebrafish tissues, however, in stable lines, optimal activation of QUAS-regulated genes will likely require strong promoters driving expression of the separate DBD and AD modules.
2.2 Q UPSTREAM ACTIVATING SEQUENCE
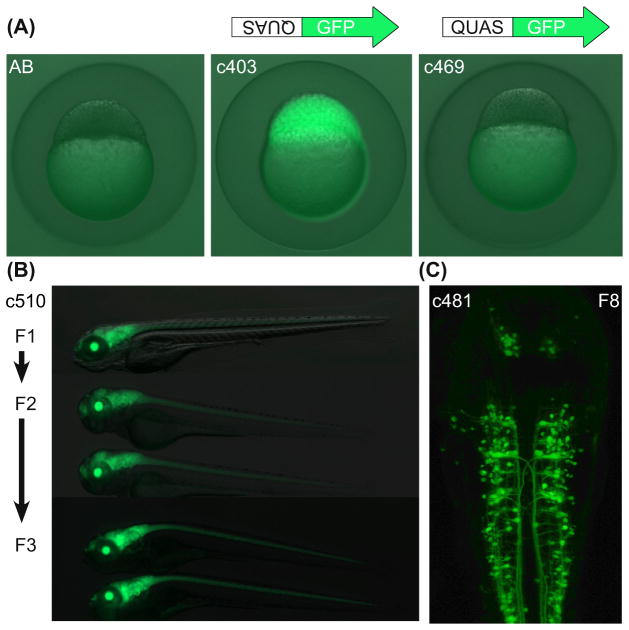
The QUAS is a conserved 16 bp sequence that is present at multiple sites and in varying numbers of tandem copies within the qa gene cluster (Giles et al., 1985). In the first plasmid produced (Subedi et al., 2014), a restriction fragment containing the five copy QUAS array was obtained from pQUAST (Potter et al., 2010) and cloned upstream of the carp β-actin minimal promoter and GFP coding sequence in a Tol2 plasmid in reverse orientation relative to the original construct used in Drosophila. As described earlier, coinjection into one to two cell embryos of the reversed QUAS:GFP Tol2 vector and plasmids containing ubiquitous promoters controlling QF results in robust mosaic labeling in many tissues. A stable transgenic line was established, Tg(QUASR:GFP)c403, that gives consistently high levels of GFP labeling. Fluorescence from this transgenic reporter does not vary when tested with the same QF driver over multiple generations and high levels of activation have persisted for over seven generations (Fig. 3). These results contradict the assertion that QUAS-regulated transgenes will be transcriptionally silenced similar to those regulated by a multicopy UAS (Suli, Guler, Raible, & Kimelman, 2014). Tg(QUASR:GFP)c403 can also be maintained as a homozygous line.
FIGURE 3. QUAS:GFP transgenic reporters.
(A) In the absence of QF, fluorescence in blastula stage embryos derived from Tg(QUASR: GFP) mothers correlates with the orientation of the 5 copy Q upstream activating sequence (QUAS) array relative to the GFP transcriptional start site. Maternal expression is abolished in the c469 line. (Image courtesy of Estela Monge). (B) Labeling from the Tg(QUASR:GFP)c403 transgene regulated by the ET(QF)c510 driver is undiminished in 5-day-old larvae over three generations. (C) In the eighth generation of Tg(QUASR:GFP)c403 progeny, green fluorescent protein (GFP) is robustly expressed at 48 hpf in a subset of neurons in which the ET(QF)c481 driver is active. (See color plate)
Image courtesy of Jean Michael Chanchu. The chromosomal locations of the c510 and c481 transgenic insertions have not yet been determined.
An unexpected finding was that the embryonic progeny of Tg(QUASR:GFP)c403 mothers show faint GFP labeling from the 1-cell stage to several days, even in the absence of QF activity. Because early embryos are weakly fluorescent, irrespective of their genotype, it is most likely due to activation of the QUAS in the maternal germ line and deposition of GFP. Accordingly, when the Tg(QUASR:GFP)c403 reporter is transmitted from the paternal genome, basal expression from the transgene is not observed (Subedi et al., 2014). To determine whether reversal of the 5X QUAS relative to the GFP transcription start site was the basis for the maternal expression, a Gateway 5′ entry clone was constructed with the QUAS sequences in the original orientation (Potter et al., 2010) and recombined into a Tol2 destination vector. To expedite the identification of transgenic founders and long-term maintenance of the QUAS reporter line in the absence of QF, an additional marker for the lens was supplied with a 3′ entry clone consisting of the zebrafish β-crystallin b1 (crybb1) promoter driving an enhanced cyan fluorescent protein (CFP) gene (a gift of M. Parsons, Johns Hopkins University). The Tg(QUAS:GFP)c469 line, established from this vector, has been bred for over five generations. Embryos derived from either heterozygous or homozygous c469 mothers are brightly labeled upon QF activation but do not show any evidence of basal fluorescence in the absence of a QF driver (Fig. 3), indicating that the QUAS transgene is not constitutively expressed. Therefore, all QUAS reporter and effector lines have been subsequently produced with the QUAS array in the same orientation as in Tg(QUAS:GFP)c469. Screening for CFP labeling of the lens also permits identification of larvae bearing QUAS-regulated transgenes or propagation of QUAS responder lines independent of QF.
2.3 QS AND QUINIC ACID
QS, the QF repressor, was assessed for its ability to reduce or eliminate GFP labeling in Tg(QUASR:GFP)c403 embryos coinjected with Tol2 plasmids containing the EF1α or ubb ubiquitous promoters driving either QF or QF2 expression (Monge and Halpern, unpublished results; Subedi et al., 2014). The QS gene was also placed under the control of the same two strong promoters in Tol2 vectors and those plasmids were injected at a higher concentration (50 ng/μL) relative to the QF plasmids (3.2 ng/μL). In all experiments, QF and QF2-dependent GFP labeling was significantly reduced in the presence of either QS construct and unaffected in the absence of QS. Fluorescence intensity measurements recorded from individual Tg(QUASR:GFP)c403 embryos at 1 dpf corroborated that, on average, the level of GFP labeling decreased by approximately 50% in those that had received the QS plasmid. Repression of QF activity by QS proteins synthesized from transcription of integrated transgenes has not yet been accomplished and will require careful calibration of the relative concentrations of both proteins. Until this is achieved, it will be difficult to assay the ability of quinic acid to alleviate QS-mediated repression of QF or QF2. Since a saturated quinic acid solution (~300 mg/mL) is added as a supplement to fly food vials (Potter & Luo, 2011) or to nematode growth medium (Wei et al., 2012), it is not possible to extrapolate the equivalent amount to administer to zebrafish embryos in system water. In a dose-response test, embryonic and larval zebrafish were found to survive concentrations of quinic acid equal to or lower than 0.3 mg/mL for up to 5 days of exposure. Higher doses produced morphological abnormalities and lethality. Whether this baseline of quinic acid treatment will be adequate to disrupt the QS—QF interaction remains to be determined.
3. GENE/ENHANCER TRAPPING USING THE Q SYSTEM
In a pilot screen, the QF transcription factor was used effectively for gene/enhancer trapping with a focus on isolating driver lines specific for the CNS. A gene trap vector, p(GT-QF), similar to the Gal4 gene trap of Davison et al. (2007), was generated through recombination of a rabbit β-globin splice acceptor, the QF transcription factor, and SV40 polyA termination sequences into a Tol2 destination vector (Kwan et al., 2007). A related plasmid, p(GT-QF2), was produced with the QF2 ME clone. In contrast to the Gal4 gene trap vector, a QUAS-regulated reporter gene was provided in trans, rather than included within the same construct. To test the effectiveness of p(GT-QF) and p(GT-QF2), the Tol2 vectors were separately injected into 1-cell stage Tg(QUAS:GFP) embryos, which were screened for GFP expression from 1 to 5 days later and raised to adulthood. Transgenic founders were identified by screening for patterns of GFP labeling in their F1 progeny, which were raised and outcrossed to establish QF driver lines. GFP labeled cells were confined to the CNS (Fig. 3B and C) in approximately 20% of the QF driver lines derived from 33 identified founders. Cloning and sequencing of the Tol2 QF insertion sites is necessary to confirm that they correspond to gene trap events, as prior work indicated that some Gal4 drivers map outside of coding sequences and are likely regulated by nearby enhancers (Davison et al., 2007). In contrast to p(GT-QF), the QF2 Tol2 gene trap vector induced basal GFP labeling in the midline of the brain and spinal cord, which confounded the identification of founders. We are currently testing whether this is due to residual syb promoter sequences amplified along with the QF2 gene. However, the preliminary screens support gene/enhancer trapping as a means of obtaining QF driver lines in cell types of interest at a frequency similar to that of Gal4-based vectors.
4. FUTURE PROSPECTS
For any transcriptional regulatory system to be adopted by the zebrafish research community, reagents and transgenic lines must be available, easy to apply, and validated in vivo. We are in the process of generating new QUAS lines that will be useful tools for a variety of applications, including fluorescent reporters to label subcellular structures, effectors for cell-autonomous ablation, and optogenetic modulators and genetically encoded calcium indicators of neural activity. A limitation to any bipartite system in zebrafish is the paucity of identified tissue or cell type—specific promoters to drive expression of QF or other regulatory genes. This can, to some extent, be overcome by expanding the collection of tissue-specific driver lines isolated from gene/enhancer trap screens. Exciting advances in Crispr/Cas9 technology will enable the creation of driver lines in a more targeted manner. In pilot studies, QF2 was integrated into specific gene loci following the strategy of Kimura, Hisano, Kawahara, and Higashijima (2014), and transgenic embryos were recovered that activate transcription of QUAS reporters in patterns recapitulating endogenous gene expression (Choi and Halpern, unpublished results). Even so, to perform experiments on selective populations of cells, particularly for functional studies of subsets of neurons in specific regions of the zebrafish brain, will require additional refinements.
The split QF approach, designed for intersectional gene expression, is a promising direction but whether it will provide sufficient levels of activity in single copy transgenic lines remains to be demonstrated. Alternative strategies involve combining the Q system with other binary regulatory systems. QF/QUAS and Gal4/UAS have been shown to operate independently in the fly (Potter et al., 2010) and zebrafish (Subedi et al., 2014). However, to achieve the sophistication of methods commonly used in Drosophila, such as mosaic analysis with a repressible cell marker (MARCM) or coupled MARCM that capitalizes on both Gal4 and QF (refer to Potter & Luo, 2011), will necessitate optimization of their respective repressors to confine the spatial or temporal extent of their activity. Suppression of Gal4 by transgene supplied Gal80 has been demonstrated in zebra-fish embryos (Faucherre & Lopez-Schier, 2011; Fujimoto, Gaynes, Brimley, Chien, & Bonkowsky, 2011), but does not work for modified versions of the protein that have the native transcriptional AD replaced by another. While the QS repressor inhibits QF and QF2 activation of QUAS-regulated transgenes when provided from injected plasmids, appropriate levels still must be attained in transgenic lines. This should be possible using strong promoters to drive QS expression or by developing lines that have multiple copies of QS transgenes.
Cre recombinase-based technology has also been successfully used in zebrafish in concert with Gal4 to restrict gene expression to more limited numbers of cells. For example, stochastic Cre-mediated activation of Gal4-VP16 permitted labeling of single tectal neurons and their dendritic and axonal processes in the brains of larval zebrafish (Sato, Hamaoka, Aizawa, Hosoya, & Okamoto, 2007). Techniques to induce Cre activity conditionally in zebrafish (eg, Hans, Kaslin, Freudenreich, & Brand, 2009; Hesselson, Anderson, Beinat, & Stainier, 2009) provide further ways of regulating QF alleles disrupted by loxP-stop-loxP cassettes, in subsets of cells at desired developmental stages.
The initial investigation into the Q system indicates that it will be a valuable addition to the increasing genetic toolkit for transgenic approaches in zebrafish. As more QF drivers and QUAS responders are developed and shared, this alternative regulatory system, and the potential for coupling it with other methods, is certain to become widely exploited.
Acknowledgments
We thank Jean Michael Chanchu, Tyler Harvey, Ayse Keskus, Michelle Macurak, and Estela Monge for their expert technical assistance in developing reagents and providing results described in this chapter. Abigyna Subedi and Mary Goll were instrumental in initiating this project and Chris Potter has generously provided plasmids and advice. We are grateful to Shannon Fisher, Koichi Kawakami, Kristen Kwan, Michael Parsons, and Kang Shen for sharing plasmids. The zebrafish Q project is supported by the National Institute of Child Health and Human Development (4R01HD078220).
References
- Akitake CM, Macurak M, Halpern ME, Goll MG. Transgenerational analysis of transcriptional silencing in zebrafish. Developmental Biology. 2011;352(2):191–201. doi: 10.1016/j.ydbio.2011.01.002. http://dx.doi.org/10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RG, Lyons DA. Intersectional gene expression in zebrafish using the split KalTA4 system. Zebrafish. 2015;12(6):377–386. doi: 10.1089/zeb.2015.1086. http://dx.doi.org/10.1089/zeb.2015.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Lin S, Hopkins N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Developmental Biology. 1995;171(1):123–129. doi: 10.1006/dbio.1995.1265. http://dx.doi.org/10.1006/dbio.1995.1265. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Development, Growth & Differentiation. 2008;50(6):391–399. doi: 10.1111/j.1440-169X.2008.01044.x. http://dx.doi.org/10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- Balciuniene J, Balciunas D. Gene trapping using gal4 in zebrafish. Journal of Visualized Experiments: JoVE [Electronic Resource] 2013;79:e50113. doi: 10.3791/50113. http://dx.doi.org/10.3791/50113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, … Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Developmental Biology. 2007;304(2):811–824. doi: 10.1016/j.ydbio.2007.01.033. http://dx.doi.org/10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13365–13370. doi: 10.1073/pnas.0903060106. http://dx.doi.org/10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre A, Lopez-Schier H. Delaying Gal4-driven gene expression in the zebra-fish with morpholinos and Gal80. PLoS One. 2011;6(1):e16587. doi: 10.1371/journal.pone.0016587. http://dx.doi.org/10.1371/journal.pone.0016587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto E, Gaynes B, Brimley CJ, Chien CB, Bonkowsky JL. Gal80 intersectional regulation of cell-type specific expression in vertebrates. Developmental Dynamics: an Official Publication of the American Association of Anatomists. 2011;240(10):2324–2334. doi: 10.1002/dvdy.22734. http://dx.doi.org/10.1002/dvdy.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles NH, Case ME, Baum J, Geever R, Huiet L, Patel V, Tyler B. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiological Reviews. 1985;49(3):338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics. 2009;182(3):747–755. doi: 10.1534/genetics.109.102079. http://dx.doi.org/10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS One. 2009;4(2):e4640. doi: 10.1371/journal.pone.0004640. http://dx.doi.org/10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(35):14896–14901. doi: 10.1073/pnas.0906348106. http://dx.doi.org/10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Wang G, Delaspre F, del Vitery MC, Beer RL, Parsons MJ. Retinoic acid plays an evolutionarily conserved and biphasic role in pancreas development. Developmental Biology. 2014;394(1):83–93. doi: 10.1016/j.ydbio.2014.07.021. http://dx.doi.org/10.1016/j.ydbio.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Abe G, Asada T, Asakawa K, Fukuda R, Ito A, … Yoshida M. zTrap: zebrafish gene trap and enhancer trap database. BMC Developmental Biology. 2010;10:105. doi: 10.1186/1471-213X-10-105. http://dx.doi.org/10.1186/1471-213X-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(21):11403–11408. doi: 10.1073/pnas.97.21.11403. http://dx.doi.org/10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Scientific Reports. 2014;4:6545. doi: 10.1038/srep06545. http://dx.doi.org/10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Developmental Biology. 2001;233(2):329–346. doi: 10.1006/dbio.2001.0242. http://dx.doi.org/10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, … Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental Dynamics: an Official Publication of the American Association of Anatomists. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. http://dx.doi.org/10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52(3):425–436. doi: 10.1016/j.neuron.2006.08.028. http://dx.doi.org/10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart GD, Tabor KM, Brown M, Strykowski JL, Varshney GK, LaFave MC, … Burgess HA. A 3D searchable database of transgenic zebra-fish Gal4 and Cre lines for functional neuroanatomy studies. Frontiers in Neural Circuits. 2015;9:78. doi: 10.3389/fncir.2015.00078. http://dx.doi.org/10.3389/fncir.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138(1):169–177. doi: 10.1242/dev.059345. http://dx.doi.org/10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuna H, Hutcheson DA, Duncan RN, McPherson AD, Scoresby AN, Gaynes BF, … Dorsky RI. High-resolution analysis of central nervous system expression patterns in zebrafish Gal4 enhancer-trap lines. Developmental Dynamics: an Official Publication of the American Association of Anatomists. 2015;244(6):785–796. doi: 10.1002/dvdy.24260. http://dx.doi.org/10.1002/dvdy.24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang SC, Wang HP, Zhu ZY, Sun YH. Transcriptional activity and DNA methylation dynamics of the gal4/UAS system in zebrafish. Marine Biotechnology (New York, NY) 2015;17(5):593–603. doi: 10.1007/s10126-015-9641-0. http://dx.doi.org/10.1007/s10126-015-9641-0. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of Development. 2009;126(10):898–912. doi: 10.1016/j.mod.2009.07.002. http://dx.doi.org/10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Luo L. Using the Q system in Drosophila melanogaster. Nature Protocols Other Titles: Protocols. 2011;6(8):1105–1120. doi: 10.1038/nprot.2011.347. http://dx.doi.org/10.1038/nprot.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141(3):536–548. doi: 10.1016/j.cell.2010.02.025. http://dx.doi.org/10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. Improved and expanded Q-system reagents for genetic manipulations. Nature Methods. 2015;12(3):219–222. doi: 10.1038/nmeth.3250. http://dx.doi.org/10.1038/nmeth.3250, 215 p. following 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(22):12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Hamaoka T, Aizawa H, Hosoya T, Okamoto H. Genetic single-cell mosaic analysis implicates ephrinB2 reverse signaling in projections from the posterior tectum to the hindbrain in zebrafish. Journal of Neuroscience: the Official Journal of the Society for Neuroscience. 2007;27(20):5271–5279. doi: 10.1523/JNEUROSCI.0883-07.2007. http://dx.doi.org/10.1523/JNEURO-SCI.0883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mechanisms of Development. 1999;80(2):153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, … Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nature Methods. 2007;4(4):323–326. doi: 10.1038/nmeth1033. http://dx.doi.org/10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Subedi A, Macurak M, Gee ST, Monge E, Goll MG, Potter CJ, … Halpern ME. Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods. 2014;66(3):433–440. doi: 10.1016/j.ymeth.2013.06.012. http://dx.doi.org/10.1016/j.ymeth.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suli A, Guler AD, Raible DW, Kimelman D. A targeted gene expression system using the tryptophan repressor in zebrafish shows no silencing in subsequent generations. Development. 2014;141(5):1167–1174. doi: 10.1242/dev.100057. http://dx.doi.org/10.1242/dev.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Matsuda K, Yamaguchi S, Asakawa K, Miyasaka N, Lal P, … Hibi M. Establishment of Gal4 transgenic zebrafish lines for analysis of development of cerebellar neural circuitry. Developmental Biology. 2015;397(1):1–17. doi: 10.1016/j.ydbio.2014.09.030. http://dx.doi.org/10.1016/j.ydbio.2014.09.030. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174(2):639–649. doi: 10.1534/genetics.106.060244. http://dx.doi.org/10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. Haploid screens and gamma-ray mutagenesis. Methods in Cell Biology. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- Wei X, Potter CJ, Luo L, Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nature Methods. 2012;9(4):391–395. doi: 10.1038/nmeth.1929. http://dx.doi.org/10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]