Summary
In model organisms, comprehensive loss-of-function genetic screens have facilitated the identification of pathway components and regulatory nodes. However, although we now have tools that allow the systematic manipulation of genes in mammalian cells, studies using genome scale tools have focused almost exclusively on a small number of genes among the many identified by such approaches. Here we describe a systematic approach to interrogate 177 genes identified as essential for the proliferation of cancer cell lines exhibiting aberrant β-catenin activity. We show the utility of integrating shRNA and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9-mediated gene editing to assess gene function and then systematically characterized the interactions of these co-dependencies using both proteomic and genetic interaction approaches. We identified new regulators of β-catenin signaling and defined functional networks required for the survival of β-catenin active cancers. In particular, we found that the transcriptional regulator YAP1 regulates chromatin-modifying complexes in β-catenin active cancers. More generally, these studies provide an experimental framework to define signaling networks in mammals.
Introduction
WNT/β-catenin signaling plays key roles in development and tissue homeostasis and is deregulated in colon and other cancers (Clevers and Nusse, 2012). Indeed, APC and β-catenin mutations occur in the majority of colon cancers and suffice to drive colon adenomas. Although many of the major components of the WNT/β-catenin signaling pathway are known, recent work suggests that several different β-catenin complexes operate in colon cancers (Rosenbluh et al., 2014; Schwitalla et al., 2013) and the regulation of such β-catenin-containing complexes remains incompletely understood.
The development of high throughput methods to manipulate gene function provides the means to interrogate the consequences of increasing or decreasing gene expression at genome scale and have been successfully used to identify genes associated with particular genetic contexts, such as cancer cell survival, drug resistance and viral infection (Bernards, 2014; Boehm and Hahn, 2011). In principle, these types of screens should provide global information about the components of signaling networks and how these components function in defined genetic contexts; however, the interpretation of these studies is complicated by the genomic heterogeneity of cancer cell lines and incomplete knowledge of the components and interactions of signaling pathways.
Moreover, to date, we and others have performed genome scale screens and focused our efforts on detailed mechanistic studies of selected, often single, high confidence candidates (Mullenders and Bernards, 2009; Rosenbluh et al., 2012). This approach clearly has value; however, by definition, this tactic ignores the other lower confidence and poorly characterized genes identified in such screens. Interrogating all of the genes identified in such screens with orthogonal and complementary approaches should allow one to identify genes that truly contribute to the phenotypes under study and provide greater biological insights.
To develop an approach that allows one to systematically study lists of genes that emerge from genomic studies, we focused on 177 genes whose expression we found was essential for the proliferation of cancer cell lines harboring aberrantly active β-catenin. We demonstrate the utility of combining RNAi and CRISPR-Cas9 loss-of-function screens for classification of genetic dependencies and use proteomic profiling and genetic interaction mapping to gain insights into the molecular functions of genetic dependencies associated with deregulation of the WNT/β-catenin activity in cancer.
Results
Identification of β-catenin co-dependencies
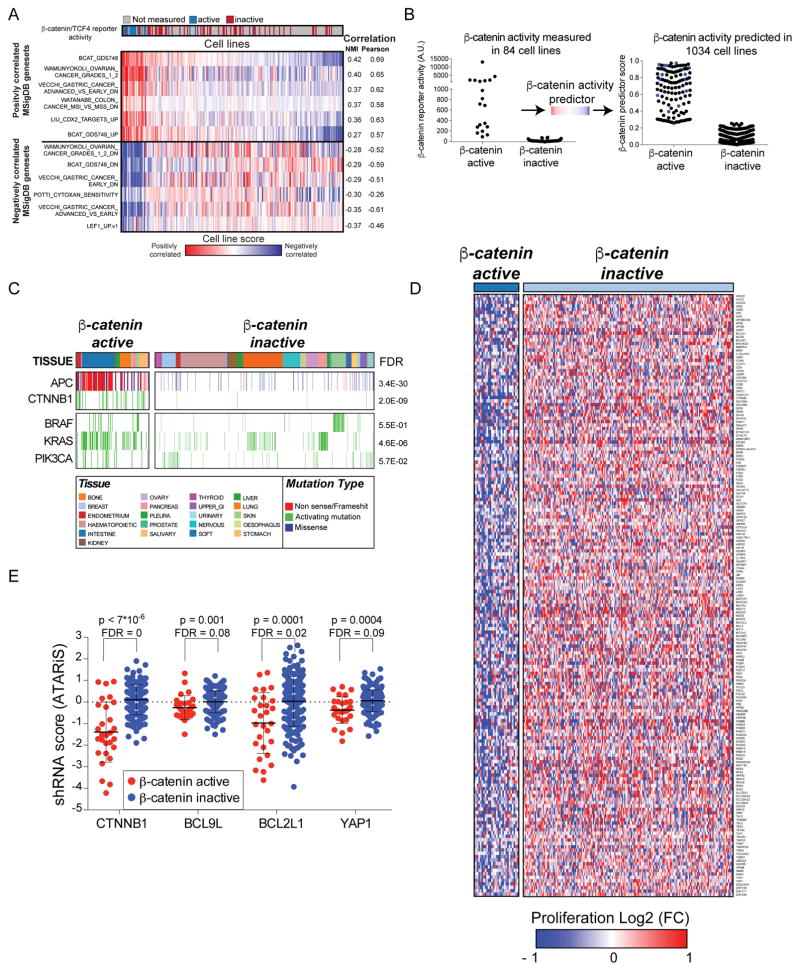
To interrogate cancers with deregulated β-catenin activity, we previously measured endogenous β-catenin activity in 84 cancer cell lines using a β-catenin/TCF4 reporter (Rosenbluh et al., 2012). To generate a β-catenin activity gene expression classifier, we first projected the gene expression profiles of these 84 cell lines (Barretina et al., 2012) onto the Molecular Signatures Database (MSigDB) (Liberzon et al., 2011). Specifically, using single-sample Gene Set Enrichment Analysis (ssGSEA) (Barbie et al., 2009), we calculated a score reflecting the correlation between each cell line and an MSigDB gene set and used this score to compute a vector of 84 enrichment scores for each MSigDB gene set. We next used a mutual information-based metric to find gene sets that share the most information with measured β-catenin reporter activity (Fig. 1A) (Abazeed et al., 2013). In addition, we performed an empirical permutation on the β-catenin/TCF4 reporter values and repeated the matching procedure to generate a null distribution from which nominal p-values and False Discovery Rates (FDR) were computed (Fig. 1A). We found that the BCAT_GDS748_UP, a MSigDB expression signature generated by over expression of β-catenin (GEO dataset GDS748 (Chamorro et al., 2005) (Table S1), most closely matched the measured β-catenin/TCF4 reporter.
Figure 1. Identification of β-catenin co-dependencies.
(A) A mutual information-based metric was used to identify expression signatures from the MsigDB database that correlate with TCF/β-catenin reporter activity assay. (B) Left panel; β-catenin activity measured in 84 cell lines using a β-catenin/TCF4 reporter assay. Right panel; β-catenin activity in 904 cell lines predicted using the β-catenin expression signature. Blue dots represent cell lines with high β-catenin/TCF4 reporter activity. (C) Recurrent somatic mutations and β-catenin activity in 904 cancer cell lines from various lineages. (D) The β-catenin signature was used to classify cells screened in Project Achilles and identified 177 genes as β-catenin co-dependencies. (E) Proliferation changes induced by RNAi suppression of known β-catenin co-dependencies. Cell line permutation analysis was used for calculating FDR and p-values.
We then used this signature profile as an input for a logistic function that was fitted to the active versus inactive β-catenin profile to generate a simple probabilistic classifier. Using this classifier to predict β-catenin activity in a larger set of 1034 cancer cell lines, we classified 124 of these cell lines as β-catenin active (Fig. 1B,C and Table S2). To further validate this β-catenin activity classification, we used a β-catenin/TCF4 reporter assay in 5 cell lines (3 classified as β-catenin active and 2 as β-catenin inactive) and found high reporter activity (500–2000 fold increase) only in the 3 cell lines predicted as β-catenin active (Fig. S1A). As expected, most cell lines classified as β-catenin active harbored APC or β-catenin mutations (73%); however, 31 β-catenin active cell lines did not harbor mutations in known components of the canonical WNT/β-catenin signaling pathway, suggesting that other genetic or epigenetic mechanisms deregulate β-catenin activity in these cells (Fig. 1C). Indeed, when we used the same approach to predict β-catenin activity in 561 tumors, we found that APC mutations were highly enriched in β-catenin active cancers (FDR = 0, Fig. S1B).
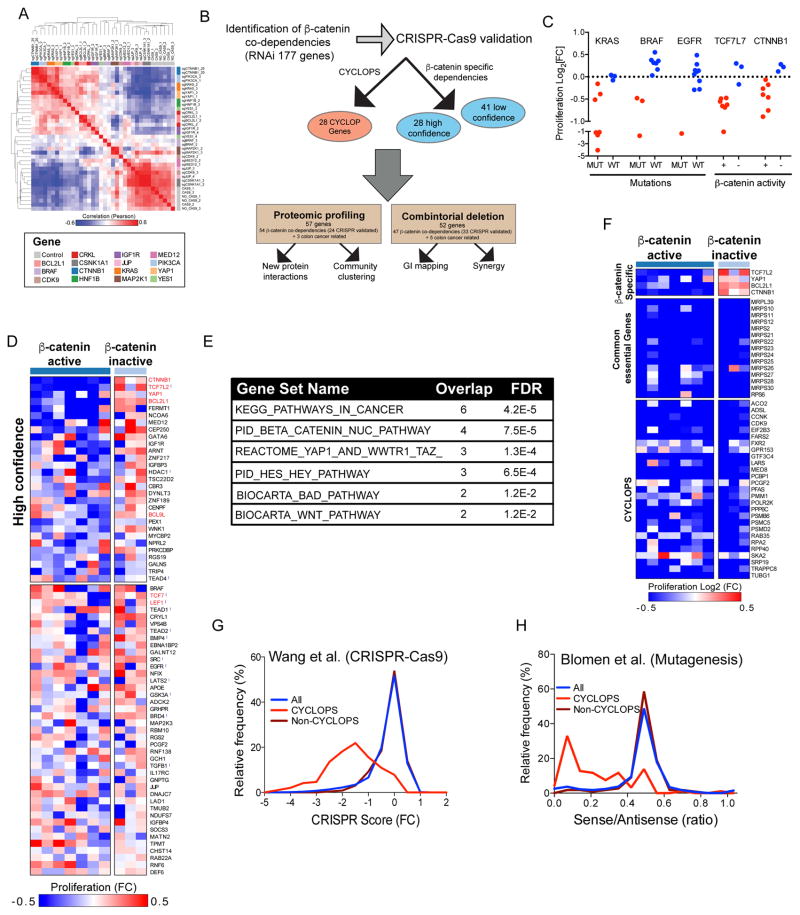
To expand our initial search for genes required for the survival of β-catenin active cell lines, we used the β-catenin expression signature to predict β-catenin activity in 216 cancer cell lines in which we had performed genome scale shRNA screens (Cowley et al., 2014) and found 28 of them to be β-catenin active. shRNA level dependency scores were collapsed to a gene score using a previously described ATARiS algorithm (Shao et al., 2013). To identify genes required for the proliferation of β-catenin active cell lines, we compared the mean gene dependency score in β-catenin active and inactive cell lines and used a cell line permutation analysis to assess statistical significance (Fig. 1D and Table S3). Genes with an FDR<0.25 were then ranked based on the magnitude of difference between classes or using a mutual information based metric (https://www.broadinstitute.org/achilles/resources/paris). For further studies, we selected the top 190 genes scoring by either method. Genes that were not expressed in β-catenin active cell lines (Mean RNAseq RPKM < 1) were excluded. Based on these criteria, we defined 177 candidate β-catenin co-dependencies (Fig. 1D and Table S3). These β-catenin co-dependencies include β-catenin, BCL9L and YAP1, confirming that this approach identified genes known to be required for β-catenin activity (Fig. 1E).
Characterization of dependencies associated with genomic alterations
Among the genes required for the proliferation of β-catenin active cell lines, we expected to find direct and indirect regulators of β-catenin activity as well as dependencies that are associated with genomic alterations that are frequent in β-catenin active cell lines. For example, the oncogenes KRAS, BRAF, PIK3CA and CTNNB1 that scored as β-catenin co-dependencies are also frequently mutated in β-catenin active cancers (Fig. 1C). When we examined the dependency of these genes in non-mutated cell lines, we found that only CTNNB1 was differentially essential for the proliferation of β-catenin active cell lines regardless of its mutational status (Fig. S2A).
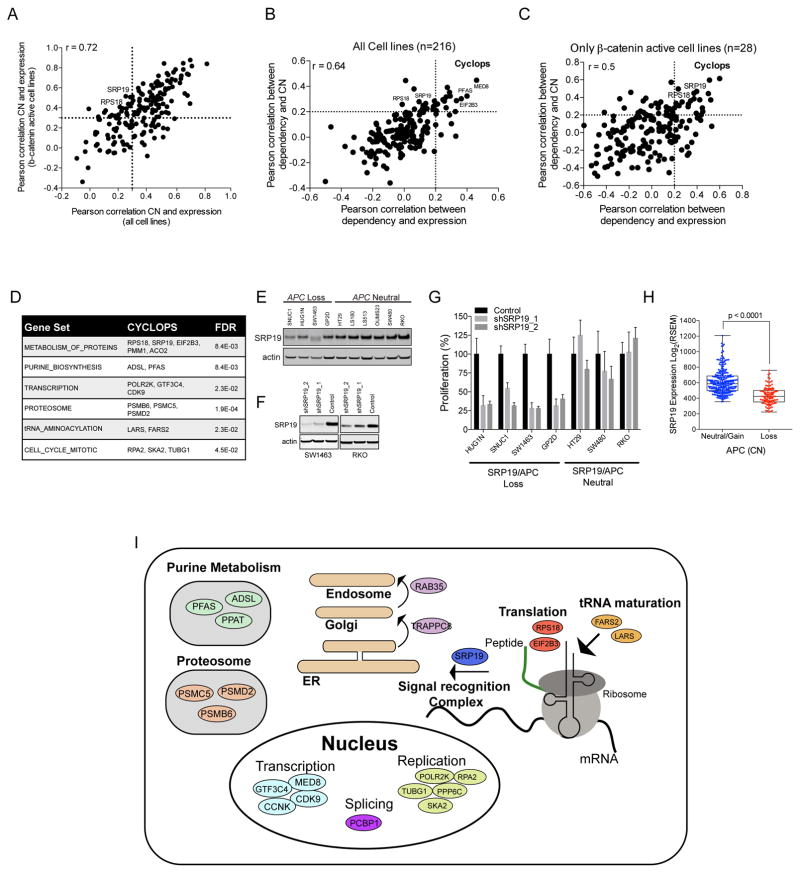
We previously defined a class of genetic dependencies we termed CYCLOPS, where one copy of a cell essential gene is lost due to its close proximity to a tumor suppressor gene (Nijhawan et al., 2012). As a consequence, cancers harboring one CYCLOPS allele express lower levels of the CYCLOPS gene and are highly sensitive to suppression of the remaining CYCLOPS gene (Fig. S2B). When we compared the expression of the 177 β-catenin co-dependencies identified by the RNAi screens to copy number (CN) across all of the cancer cell lines used in the shRNA screen (Fig. 2A), we found 114 genes in which CN and mRNA levels were concordant (Rpearson>0.3). Among these 114 genes, we identified 28 genes that exhibited differential dependency (Rpearson>0.2 for both expression and CN) in cells that harbored low CN and mRNA levels (Fig. 2B,C; Fig. S2C and Table S4) and therefore met the criteria for CYCLOPS. We note that of these 28 genes, 19 of the genes exhibited the CYCLOPS phenotype only in β-catenin active cell lines (Fig. 2C), suggesting that these genes are likely related to gene loss events that occur in β-catenin active cancers. As expected, CYCLOPS genes are enriched with genes required for essential cellular processes (Fig. 2D).
Figure 2. Characterization of β-catenin co-dependency CYCLOPS genes.
(A) Correlation between the expression and CN of 177 β-catenin co-dependencies using either the 216 cancer cell lines used in RNAi screen or the subset of β-catenin active cell lines. Genes in the upper right quadrant correlate in both data sets and genes in the upper left quadrant correlate only within β-catenin active cell lines. Comparison of correlation between dependency and expression or CN in (B) all cell lines or (C) β-catenin active cell lines. CYCLOPS genes are found in the upper right quadrant. (D) GSEA of pathways associated with β-catenin activity associated CYCLOPS genes. (E) SRP19 protein levels in APC deleted or WT colon cancer cell lines. (F) SRP19 protein levels measured four days after expression of 2 sgRNAs targeting SRP19. (G) Proliferation of APC WT and loss colon cancer cell lines after deletion of SRP19. (H) SRP19 mRNA levels in 359 colorectal cancer samples segregated by APC CN. A non-parametric T test analysis was used for calculating the p-value. (I) Genes and pathways associated with β-catenin co-dependency CYCLOPS genes.
Of these candidate CYCLOPS genes SRP19, a component of the signal recognition complex (Halic and Beckmann, 2005), is located in close proximity (14kb) to the tumor suppressor APC that is focally deleted in a subset of β-catenin active cancers. We found lower SRP19 protein levels in cell lines harboring APC deletion (Fig. 2E and S2D), and shRNAs targeting SRP19 inhibited the proliferation of cells harboring one SRP19 allele (Fig. 2F,G and S2E). Moreover, SRP19 mRNA levels were lower in colon cancer tumors harboring loss of APC [Fig. 2H, TCGA (Cancer Genome Atlas, 2012)], confirming SRP19 as a CYCLOPS gene in colon cancers that harbor APC loss. In total, of the 177 genes essential for proliferation β-catenin active cell lines, 31 of these genes (28 CYCLOPS and 3 oncogenes) are clearly associated with genomic alterations enriched in β-catenin active cancers (Fig. 2I).
Validation of β-catenin co-dependencies using CRISPR-Cas9-mediated gene editing
Although we used multiple shRNAs and cell lines and employed analytical methods that allowed us to concentrate on on-target shRNAs, RNAi off-target effects confound the interpretation of RNAi screens. To confirm that the candidates that we identified by RNAi were truly related to β-catenin activity, we used CRISPR-Cas9 gene editing as an orthogonal approach to modulate gene expression (Shalem et al., 2014; Wang et al., 2014).
To evaluate the specificity of CRISPR-Cas9 in mediating gene deletion, we measured global expression changes induced by multiple single guide RNAs (sgRNA) targeting 15 β-catenin co-dependent genes in an APC mutated colon cancer cell line (DLD1) in which we had stably expressed Cas9 (Table S5). We found that sgRNAs that target β-catenin or MAP2K1 effectively reduced β-catenin or MAP2K1 protein levels also inhibited β-catenin or MAP2K1 mRNA levels (Fig. S3A,B), suggesting that mRNA levels provided a useful metric to assess whether a particular sgRNA affected its target. Based on these observations, we used mRNA expression of the target gene to select the two most potent sgRNAs for each gene and used unsupervised hierarchical clustering to identify sgRNAs that induced similar expression changes. We found that the global expression changes induced by sgRNAs targeting a particular gene were highly concordant (Fig. 3A), confirming the specificity of CRISPR-Cas9-mediated gene deletion (Shalem et al., 2014; Wang et al., 2014). Furthermore, although some genes (BRAF, MAP2K1, CDK9, JUP, MED12 and CSNK1A1) induced small expression changes, the expression changes induced by sgRNAs targeting known components of the MAPK signaling pathway (MA2K1 and BRAF) were highly similar further demonstrating the high specificity of CRISPR-Cas9-mediated gene deletion (Fig. 3A).
Figure 3. Validation of β-catenin co-dependencies using CRISPR-Cas9-mediated gene editing.
(A) Hierarchical clustering of the two most effective sgRNAs/gene (determined by mRNA levels of the target gene) based on global expression changes induced by CRISPR-Cas9 mediated deletion of 15 different genes in DLD1 cells. (B) Diagram summarizing the strategy used for identification and functional validation of β-catenin co-dependencies. (C) Proliferation changes (mean of two best correlating sgRNAs) following CRISPR-Cas9-mediated deletion of known oncogenes. (D) Heat map of proliferation changes induced by CRISPR-Cas9-mediated depletion of validated β-catenin co-dependencies. The average of the most correlated sgRNAs was used to calculate a gene score. Due to the pooled format of these experiments, non-targeted control sRNAs score at ~0.5. For high confidence β-catenin co-dependencies, we considered genes that were essential in at least 4 β-catenin active cell lines and non-essential in at least two β-catenin inactive cell lines. Known β-catenin co-dependencies are marked in red, and genes that were included in the CRISPR-Cas9 validation screen but did not score as β-catenin co-dependencies by RNAi are marked with
 (E) MsigDB enrichment analysis for pathways associated with high confidence β-catenin co-dependencies. (F) Proliferation changes following CRISPR-Cas9-mediated depletion of known β-catenin co-dependencies (Top), common essential control genes (middle), or CYCLOPS genes (bottom). (G) Distribution of proliferation changes following genome scale CRISPR-Cas9 screens (Wang et al., 2015). (H) Distribution of proliferation changes following mutagenesis of two haploid cell lines (Blomen et al., 2015). A ratio of 0.5 represents genes whose deletion had no effect on proliferation.
(E) MsigDB enrichment analysis for pathways associated with high confidence β-catenin co-dependencies. (F) Proliferation changes following CRISPR-Cas9-mediated depletion of known β-catenin co-dependencies (Top), common essential control genes (middle), or CYCLOPS genes (bottom). (G) Distribution of proliferation changes following genome scale CRISPR-Cas9 screens (Wang et al., 2015). (H) Distribution of proliferation changes following mutagenesis of two haploid cell lines (Blomen et al., 2015). A ratio of 0.5 represents genes whose deletion had no effect on proliferation.
Based on these observations, we constructed a lentivirally delivered, pooled sgRNA library targeting all 177-candidate β-catenin co-dependencies (6–10 sgRNAs/gene) as well as 23 genes encoding known components of the WNT/β-catenin signaling pathway and oncogenes recurrently altered in colon cancer (Fig. 3B and Table S6). In addition, we included 146 negative controls (targeting non-human genes) and 105 sgRNAs targeting 19 common essential genes. We introduced this library into 10 Cas9-expressing cancer cell lines (7 β-catenin active and 3 β-catenin inactive) and one cell line that did not express Cas9 and measured the effect of each sgRNA on cell proliferation by calculating the difference in sgRNA abundance at 3 and 28 days post infection (DPI) (Table S7). Replicate experiments were highly concordant (Fig. S3C), and in contrast to what we have observed with RNAi (Cheung et al., 2011), none of the negative control sgRNAs exhibited effects on cell proliferation (Fig. S3D). As expected, sgRNAs targeting known oncogenes specifically inhibited the proliferation of cell lines that harbor mutations in these oncogenes (Fig. 3C and S3E–H). Furthermore, we observed that sgRNAs targeting different sequences in the same gene exhibited highly concordant phenotypes (Fig. S3I,J).
To identify genes required for the proliferation of β-catenin active cell lines, we used the mean of 4 replicate experiments and collapsed sgRNA level proliferation scores to a gene score based on the mean of the two best-correlated sgRNAs (determined by comparing the proliferation changes across 10 cell lines) (Table S7). We defined high confident genetic dependencies specific for β-catenin active cancers by considering genes that were essential in at least 4 β-catenin active cell lines and not essential in at least 2 of the three β-catenin inactive cell lines (log2[fold change]<-0.1) (Fig. 3D). We found 28 high confidence β-catenin co-dependencies including known regulators of β-catenin activity such as TCF7L2, YAP1 and BCL9L. In addition, we found that other known regulators of β-catenin activity such as TCF7 and LEF1 were essential only in 2–3 β-catenin active cell lines suggesting these genes are required for proliferation of β-catenin active cell lines only in specific contexts. High confidence β-catenin co-dependencies were enriched with regulators of β-catenin activity (Fig. 3E) and previously reported β-catenin co-dependencies (Fig. 3D). In addition, we identified 23 genes that have not been previously associated with β-catenin activity.
When we evaluated the consequence of CRISPR-Cas9-mediated deletion of candidate CYCLOPS genes, we found that sgRNAs targeting genes that we identified by RNAi as CYCLOPS genes inhibited proliferation in all cell lines regardless of expression levels (Fig. 3F). We further confirmed these observations in two recently reported independent datasets that used CRISPR-Cas9-mediated gene deletion (Wang et al., 2015) or extensive mutagenesis in human haploid cell lines (Blomen et al., 2015) to identify human cell essential genes. We found that all the genes we identified as CYCLOPS also scored as common essential genes in these studies (Fig. 3G,H and S3K-P). These observations demonstrate the utility of combining RNAi and CRISPR-Cas9-mediated gene deletion to facilitate the identification of CYCLOPS genes associated with β-catenin active cancers.
In total, we identified 28 high confidence β-catenin co-dependencies (25 of which scored in both CRISPR-Cas9 and RNAi experiments) and 41 genes (30 that scored in both CRISPR-Cas9 and RNAi) that scored only in a subset of β-catenin active cell lines. In addition, we defined 28 CYCLOPS genes associated with genomic alterations in β-catenin active cancers. To interrogate how these genes are related to β-catenin activity, we developed a systematic approach that includes proteomic profiling and genetic interaction (GI) mapping (Fig. 3B).
Proteomic characterization of β-catenin co-dependencies
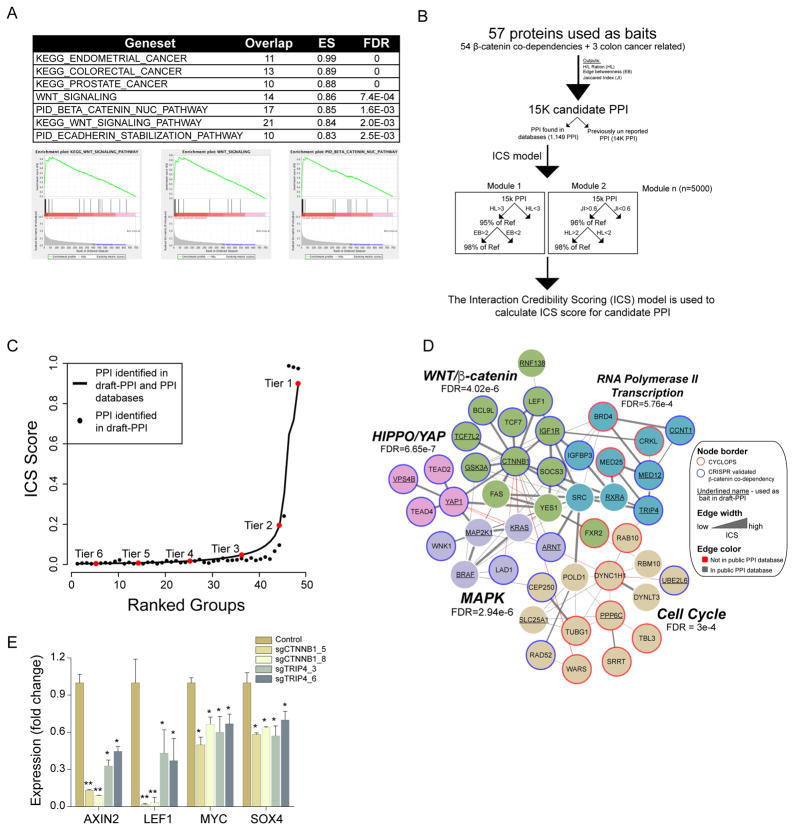
The specificity of affinity-based protein interaction profiling is increased by Stable Isotope Labeling by Amino acids in Cell culture (SILAC). However, large-scale SILAC experiments are currently limited by cost and throughput. Using lower concentrations of input lysate and shorter acquisition times, we developed a medium-throughput proteomic method we termed draft-PPI. Draft-PPI retains high quality SILAC labeling while greatly reducing cost. Based on the availability of expression clones, we selected 57 genes including 24 CRISPR-Cas9 validated β-catenin co-dependencies, 30 β-catenin co-dependencies that scored only in RNAi, and 3 colon cancer related oncogenes (KRAS, PIK3CA and MAP2K1) and used draft-PPI (one replicate/bait) to identify 15,189 candidate protein interactions with these 57 genes (Table S8). Pre-ranked GSEA analysis (based on the heavy/light ratio) of these β-catenin interacting proteins correctly identified components of the WNT signaling pathway (Fig. 4A), demonstrating that draft-PPI identifies known protein interactions.
Figure 4. Proteomic characterization of β-catenin co-dependencies.
(A) Pre-ranked GSEA enrichment analysis based on the heavy/light ration of proteins interacting with β-catenin in draft-PPI. (B) Analytical pipeline used for ICS. (C) Distribution of ICS scores for PPI identified using draft-PPI (black circles). Solid line represents previously reported PPI. The tiers were determined by measuring the curve deflection. Based on the distribution of true positive PPI (solid line) tiers 1–3 were considered as credible PPI. (D) Depiction of the β-catenin co-dependency network. Colors denote distinct communities. FDR was determined using GSEA analysis. (D) Following CRISPR-Cas9-mediated deletion of β-catenin or TRIP4, the expression of known β-catenin target genes was measured using quantitative PCR. A non-parametric T test analysis was used for significance scoring (* indicates p<0.03 and ** indicates p<0.0001).
Recognizing the variability introduced by the lack of replicates and shorter acquisition time in draft-PPI, we developed a method for protein interaction credibility scoring (ICS)(Data S1). For each of the 15,189 PPI identified in draft-PPI, we computed three predictors: (1) Heavy/light ratio, (2) Jaccard similarity coefficient (Jaccard, 1912); and (3) Edge betweenness centrality (Girvan and Newman, 2002). As true positive protein interactions, we considered 1,149 that were identified in draft-PPI and are also reported as high confidence protein interactions in publicly available PPI databases (Lage et al., 2007). We found that using each of these predictors we correctly identified only a subset of the true positive PPI (Fig. S3A–C), suggesting that none of predictors alone was suitable for this application. As such, we developed ICS using the Random Forest (RF) binary response classifier (Breiman, 2001), which uses the true positive PPI as a response and computes 5000 different combinations (modules) of these classifiers that correctly enrich true positive PPI. The ICS score is calculated using the number of modules in which a draft-PPI protein interaction scores together with the true positives (Fig. 4B). We trained the RF model on 70% of the data and used the area under the ROC curve (AUC) on the remaining 30% of the dataset to calculate classification power. The ICS showed high AUC (AUC: 96.9 confidence intervals (95.7%, 98.1%) (Fig. S4E), and 5-fold cross validation showed similar AUC estimates (Fig. S4F). Furthermore, ICS correctly identified the majority of true positive PPI (Fig. S3D), demonstrating the reliability of this approach.
To identify credible draft-PPI interactions, we used the distribution of ICS in PPI found in publicly available databases and a change-point analysis (e.g. defilation from curve) to group draft-PPI protein interactions into six tiers (Fig. 4C). Since the majority of true positive protein interactions were in tiers 1–3 (92%), we defined tiers 1–3 as credible PPI. This approach identified well-characterized relationships such as interactions between β-catenin and TCF7L2 (Poy et al., 2001) or YAP1 and TEAD transcription factors (Vassilev et al., 2001), as well as more recently reported observations between YAP1 and β-catenin (Heallen et al., 2011; Rosenbluh et al., 2012). In total, using draft-PPI and ICS, we identified 3639 high-credibility PPI including 2589 previously uncharacterized PPI (Table S8).
To gain mechanistic insights into the molecular processes regulated by β-catenin co-dependencies, we combined credible interactions from draft-PPI with protein interactions from literature-curated databases (Lage et al., 2007) and used direct physical interactions between β-catenin co-dependencies to identify relationships among these candidates. Using community-based network clustering constructed with the number of interacting partners and the interaction credibility (Clauset et al., 2004), we found 5 distinct protein communities (Fig. 4D). GSEA enrichment analysis revealed these communities were enriched with components of known signaling networks. Specifically, we found communities containing components of the MAPK, WNT, HIPPO signaling networks as well as genes involved in transcription and cell cycle control. As expected, the majority of CYCLOPS genes (10 out of 11 CYCLOPS) were clustered into two separate communities that contain cell essential genes demonstrating that CYCLOPS genes are a functionally distinct class of β-catenin co-dependencies. However, we found 1 CYCLOPS gene (FXR2) in a community containing known components of the WNT/β-catenin signaling pathway suggesting that FXR2 is a regulator of β-catenin activity. In addition, we found three communities that were enriched with components of the WNT, HIPPO and MAPK signaling pathways demonstrating these pathways are required for the proliferation of β-catenin active cancers.
In addition to these known interactions, we found potential new roles and interactions for other β-catenin co-dependent genes. For example, we found TRIP4, a known regulator of NF-κB signaling (Jung et al., 2002), in a community that contains known regulators of RNA polymerase II transcription (MED12, MED25, CCNT1 and BRD4), suggesting a role for TRIP4 in regulation of β-catenin transcription activity. To test this hypothesis, we measured the expression of β-catenin target genes following CRISPR-Cas9-mediated depletion of TRIP4. Using two TRIP4 targeting sgRNAs, we found that TRIP4 deletion inhibited the expression of known β-catenin target genes (Fig. 4E). Furthermore, depletion of TRIP4 did not lead to global suppression of gene expression (Fig. S4G) suggesting that in β-catenin active cell lines TRIP4 is a specific regulator of β-catenin activity. Taken together, PPI mapping allowed us to classify the products of many β-catenin co-dependent genes into distinct functional communities and suggest that three known signaling pathways (WNT, HIPPO, MAPK) are β-catenin co-dependencies.
Genetic interaction mapping
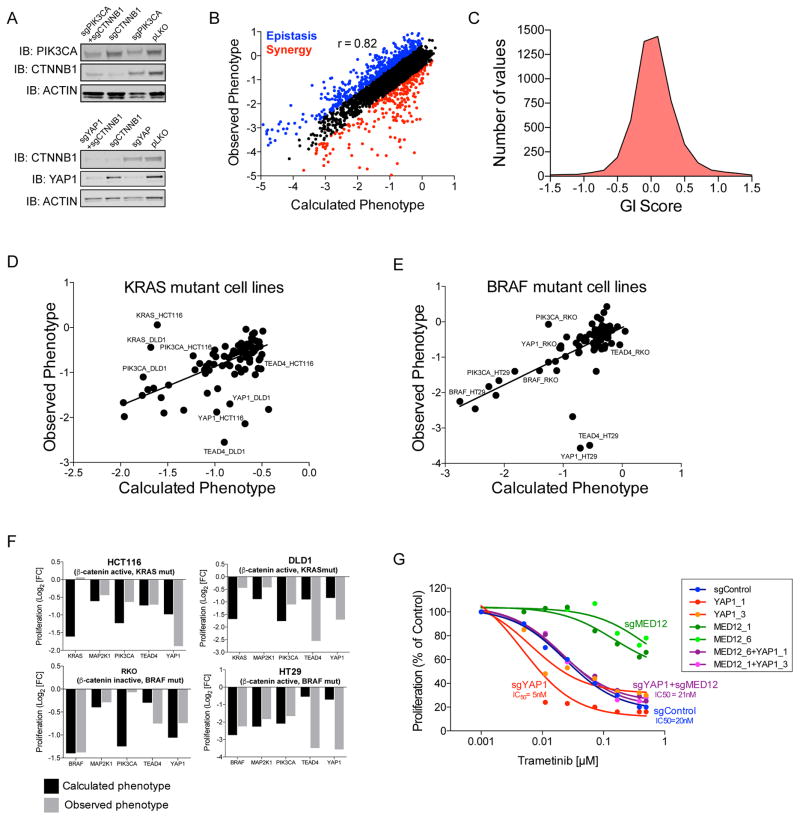
To complement our proteomic profiling approach, we performed genetic interaction (GI) mapping by assessing the consequences of co-deleting every combination of two β-catenin co-dependencies using a modified sgRNA lentiviral vector that carries 2 sgRNA targeting different genes (Fig. S5A). We found that individual gene knockout efficiency was not affected by co-expression of 2 sgRNAs targeting distinct genes (Fig. 5A). We selected 52 genes including 33 CRISPR-Cas9 validated β-catenin co-dependencies, 14 genes that scored as β-catenin codependences in the RNAi screens, and 5 genes commonly altered in colon cancer (KRAS, BRAF PIK3CA, MAP2K1 and SMAD4) and constructed a double sgRNA library encoding all pairwise combinations of 115 sgRNAs (2 sgRNAs/gene + 11 negative controls, 13,225 constructs). We introduced this library into 4 cancer cell lines (DLD1, HCT116, HT29 and RKO) and measured the abundance of each sgRNA after 28 days (Table S9). Replicate experiments were highly correlated, and expression of negative control sgRNA combinations did not induce substantial changes in cell proliferation (Fig. S5B,C). To assess the effects on proliferation induced by single gene knockouts with the same vector, we included 11 negative controls (sgRNAs targeting non-human genes) and measured the phenotypes of deletion combinations that included a negative control sgRNA and gene targeting sgRNA. We found that the introduction of vectors encoding a control and gene-specific sgRNAs combination induced nearly identical changes in proliferation as observed when we expressed a single sgRNA (Fig. S5D).
Figure 5. Combinatorial deletion of β-catenin co-dependencies.
(A) Protein levels following single or combined deletion of β-catenin and PIK3CA (top panel) or β-catenin and YAP1 (bottom panel). (B) Calculated and observed proliferation changes following introduction of double sgRNAs. As synergistic GI, we considered GI<-0.5 (red) and GI>0.5 were considered as epistatic (blue). (C) Distribution of GI scores. (D–E) Calculated and observed proliferation changes following introduction of double sgRNAs targeting MED12 and the indicated gene in (D) KRAS mutant cell lines (DLD1 and HCT116) or (E) BRAF mutant cell lines (HT29 and RKO). The observed and expected phenotypes were used to generate a linear regression line. (F) The observed and calculated proliferation changes following combined deletion of MED12 with the indicated genes. (G) Following CRISPR-Cas9 mediated deletion of MED12, YAP1 or a combination of MED12 and YAP1, HT29 cells were treated with trametinib. Cell titer glo (CTG) was used to evaluate cell proliferation 5 days post treatment.
To evaluate GI between β-catenin co-dependencies, we calculated the anticipated phenotype for every pair of double deletions. Using the sum of proliferation changes induced by single gene knockouts, we found that a simple linear model predicted the proliferation phenotype induced by ~93% of deletion combinations, indicating that the majority of double deletion combinations result in an additive phenotype (Fig. 5B). As expected, we found that combinations containing 2 sgRNAs targeting the same gene did not induce a larger effect (Fig. S5E), demonstrating that each of the sgRNA used in these studies was highly effective by itself in deleting their target genes. Corroborating this finding, we confirmed that introduction of 2 sgRNAs targeting the same gene failed to decrease β-catenin and YAP1 protein levels further than single sgRNAs targeting these genes in HT29 cells (Fig. S5F,G). Based on these observations, we concluded that we could use this approach to identify interactions between deleting pairs of genes.
Specifically, we also noted that 375 (7%) of the combinations led to phenotypes that were greater than or less than the predicted additive phenotype (-0.5 > GI > 0.5, Fig. 5B). For example, we found that co-deleting MED12 with KRAS, BRAF or PIK3CA in cell lines harboring mutations in these oncogenes attenuated the proliferation defects observed when depleting each of the oncogenes alone (Fig. 5D–F). This observation corroborates prior work that showed that suppression of MED12 attenuated the effects of inhibiting these oncogenic pathways (Huang et al., 2012). Unexpectedly, we found that combined deletion of MED12 and YAP1 or MED12 and TEAD4 induced a synergetic anti-proliferation effect (Fig. 5F), suggesting that YAP1 modulates MED12. Indeed, suppression of MED12 has been reported to attenuate cancer cell drug response by induction of an EMT-like phenotype (Huang et al., 2012), and suppression of YAP1 was shown to enhance the effect of MEK inhibitors in cancer cells (Lin et al., 2015). Consistent with these observations, we found that sensitivity of HT29 cells to a MEK inhibitor (trametinib) was attenuated by CRISPR-Cas9 mediated deletion of MED12 and enhanced by deletion of YAP1 (Fig. 5G and S5H). Furthermore, sensitivity to trametinib in MED12 deleted HT29 cells was restored by the additional deletion of YAP1 (Fig. 5G and S5I). These observations demonstrate that YAP1 modules the activity of MED12, and more broadly that GI mapping allowed us to identify interactions among β-catenin co-dependent genes.
Creating a β-catenin co-dependency GI map
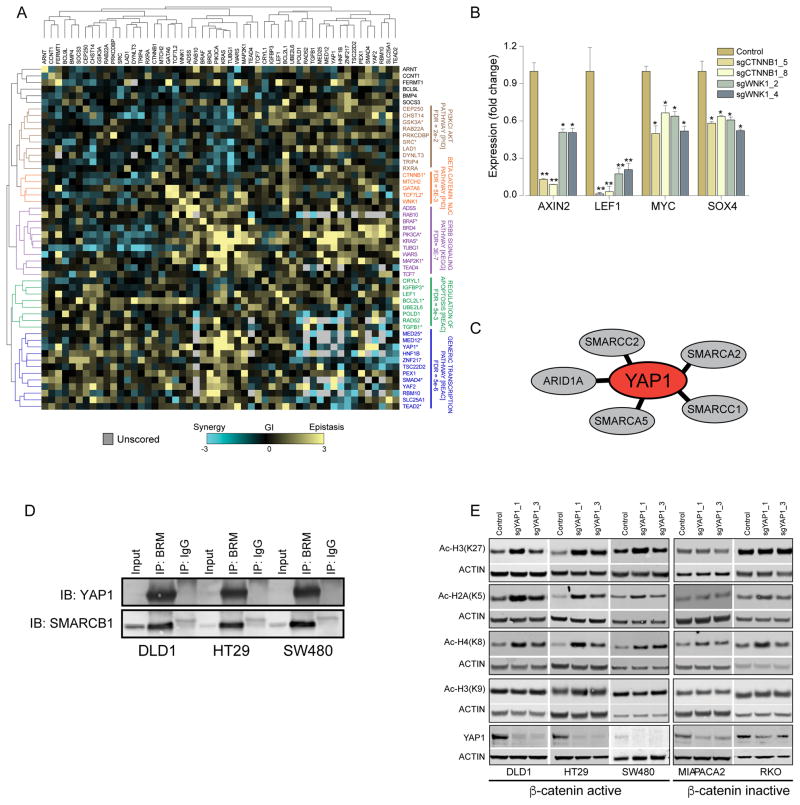
Systematic GI mapping studies in model organisms have demonstrated that functionally related genes exhibit similar GI patterns, which facilitates the identification of mechanistically related genes (Collins et al., 2007). To evaluate the consequence of pairwise deletions, we assigned a GI score to every pair of double deletions that reflects the deviation of the observed proliferation phenotype from the calculated phenotype (Supplementary Table 9). We labeled deviations that induced decreased proliferation beyond the sum of two sgRNA as ‘synergistic’, and pairwise deletions that exhibited a smaller than predicted proliferation defect as ‘epistatic’ (Fig. 5B). Using the median GI score derived from three β-catenin active cell lines and Spearman rank hierarchal clustering, we constructed a β-catenin co-dependency GI map (Fig. 6A). GSEA enrichment analysis showed clusters containing genes in well-defined pathways such as the MAPK and WNT/β-catenin signaling pathway.
Figure 6. β-catenin co-dependency GI-map.
(A) The median GI-score in three β-catenin active cell lines was used for Spearman correlation-based hierarchal clustering of β-catenin co-dependencies. (B) Expression of β-catenin target genes was evaluated 8 DPI after expression of WNK1 or β-catenin-specific sgRNAs. A non-parametric T test analysis was used to calculate p-values (* p<0.03, ** p<0.0001). (C) Components of chromatin modifying complexes that interact with YAP1. (D) Binding of YAP1 to BRM measured in three colon cancer cell lines by BRM immunoprecipitation. (E) Effects of CRISPR-Cas9-mediated deletion of YAP1 in β-catenin active and inactive colon cancer cell lines on acetylated histones H3k27, H2AK5, H4K8 and H3K9.
In addition to these known pathways, we found a GI cluster that contained known regulators of β-catenin activity (CTNNB1, GATA6 and TCF7L2) and WNK1. Other known components of the WNT/β-catenin signaling pathway such as LEF1 and TCF7 that were essential only in SW480 and LS513 cell lines (Fig. 3D) did not cluster with these known regulators of β-catenin activity suggesting them as context specific regulators of β-catenin activity. WNK1 is a serine/threonine kinase that regulates blood pressure by controlling transport of sodium and chloride ions, and germ-line line mutations in WNK1 have been implicated in a rare form of hypertension (Alessi et al., 2014). WNK1 scored as a high confidence β-catenin co-dependency in both the CRISPR-Cas9 and RNAi screens (Fig. 3D), and we found that WNK1 was connected to MAPK21 in the PPI map (Fig. 4D). To further evaluate the role of WNK1 in regulation of β-catenin activity, we measured the expression of β-catenin target genes following CRISPR-Cas9-mediated deletion of WNK1 and found that β-catenin target genes were down regulated following WNK1 depletion (Fig. 6B), implicating WNK1 as a regulator of β-catenin activity. Further supporting these observations, WNK1 was recently reported to act as a regulator of β-catenin activity during Drosophila development (Serysheva et al., 2013; Swarup et al., 2015).
GI and proteomic analysis suggest a role for YAP1 in regulation of chromatin state
In the β-catenin co-dependency GI map, we found the chromatin modifier ZNF217, a member of the HDAC Co-Rest complex (You et al., 2001), exhibited similar patterns to YAF2, a component of the non canonical PRC2 chromatin remodeling complex (Di Croce and Helin, 2013), and to transcriptional regulators including YAP1 and TEAD2 (Fig. 6A), suggesting a mechanistic relationship among these genes. When we examined the β-catenin co-dependency PPI map, we found physical interactions between 5 members of the SWI/SNF chromatin-remodeling complex and YAP1 (Fig. 6C and Table S8). Moreover, we confirmed that YAP1 interacts with SMARCA2 (BRM), the core component of the SWI/SNF complex, in three β-catenin active colon cancer cell lines (Fig. 6D).
To evaluate the role of YAP1 in regulation of chromatin state, we suppressed YAP1 in β-catenin active or β-catenin inactive cell lines. In the draft-PPI, we identified physical interactions between ZNF217 and components of the Co-Rest HDAC complex (Fig. S6A), and deletion of ZNF217 led to increased acetylation of histone H3K27 irrespective of β-catenin activity (Fig. S6B). Similar to what we observed when we suppressed ZNF217, deletion of YAP1 led to increased acetylation of histone H3K27 in β-catenin active cell lines (Fig. 6E). However, unlike ZNF217, acetylation of histone H3K27 was not affected by CRISPR-Cas9-mediated depletion of YAP1 in two β-catenin inactive cell lines (Fig. 6E). Furthermore, we found that acetylation of histone H2AK5 and H4K8 but not H3K9 was increased following CRISPR-Cas9 mediated deletion of YAP1 in β-catenin active cell lines, demonstrating a histone site specific function for YAP1 (Fig. 6E and S6C). Since these histone acetylation marks have been linked to transcriptional activation (Verdin and Ott, 2015), these observations suggest a function for YAP1 in suppressing gene expression in β-catenin active colon cancers. Together, these observations suggest that YAP1 regulates transcription in part through specific interactions with complexes that modify chromatin in β-catenin active cancers.
Discussion
Genome scale functional genomic approaches provide global information regarding the consequence of manipulating gene expression. A major bottleneck in the interpretation of such high throughput genetic screens is the lack of systematic approaches to parse the function and interactions of genes identified in these screens. To date, most mammalian studies involving high throughput screens identify a single gene or interaction, which is studied in some detail, but providing little information about the other genes identified in these large scale experiments.
Here we developed a systematic approach that enabled us to validate and map the relationships among genes that we identified as β-catenin co-dependencies. Specifically, using a combination of RNAi- and CRISPR-Cas9-mediated loss-of-function screens, we defined a set of 28 high confidence genes that were required for the proliferation of the majority of the β-catenin active cancer cell lines studied as well as 41 lower confidence genes that were essential in subsets of β-catenin active cell lines. As such, these studies allowed us to identify new high confidence genetic dependencies associated with deregulation of β-catenin activity, while also defining genes, including several well defined components of the WNT/β-catenin singling pathway (TCF7 and LEF1), that were essential only in a subset of β-catenin active cell lines. In addition, we characterized β-catenin co-dependencies by whether they were related to genomic alterations often found in β-catenin active cancers (CYCLOPS), identified new regulators of β-catenin transcription (TRIP4, WNK1), and found YAP1 interacts with components of the SWI/SNF complex and that suppression of YAP1 upregulates acetylation of histones H3K27, H4K8 and H2AK5 in β-catenin active but not β-catenin inactive cells. Although focused on β-catenin co-dependent genes, these studies demonstrate how the use of multiple approaches to interrogate the function of genes identified in a set of genome scale RNAi screens provided information to allowed us to classify the genes by function, context or pathway.
As others have reported (Shalem et al., 2014; Wang et al., 2014), we found that CRISPR-Cas9-mediated gene editing results in fewer off-target effects than what we observed with RNAi. We found high concordance in the expression and proliferation phenotypes induced by different sgRNAs targeting a similar gene. In addition, we demonstrated the utility of comparing RNAi and CRISPR-Cas9 loss-of-function proliferation screens as an approach to classify genetic dependencies associated with copy number loss (CYCLOPS). Specifically, we found that CYCLOPS genes scored as differentially essential by RNAi and broadly essential following CRISPR-Cas9 mediated gene deletion. These findings suggest that CRISPR-Cas9 and RNAi provide complementary means to perform loss-of-function analyses of gene function. Of the 28 β-catenin co-dependency CYCLOPS genes, we found that SRP19, a component of the signal recognition particle (Halic and Beckmann, 2005), is a CYCLOPS gene associated with loss of APC.
Affinity-based proteomic profiling provides valuable insights into protein function however; large-scale proteomic approaches are limited by cost and throughput. Here we developed draft-PPI that allowed us to profile protein-protein interactions for 57 genes. Although SILAC labeling increases the specificity of affinity-based proteomic studies, experimental variability and non-specific binding are a major obstacle in the interpretation of proteomic studies. In the case of draft PPI, the lack of replicates and short acquisition times adds further noise that requires consideration in interpreting this data. Since most available PPI scoring methods use replicate experiments to address robustness of data (Pu et al., 2015), we developed an analytical approach that uses true positive protein interactions to create a predictive model and assigns a credibility score for each protein interaction identified in draft-PPI. In the top three tiers, we found that 1054 of the 1149 known positives (92%) scored, suggesting that this approach provides a reasonable approach to identifying potentially physiological interactions. Using this approach, we found 3644 high-credibility PPI including 2590 previously uncharacterized PPI.
Using community-based clustering of direct PPI between β-catenin co-dependencies, we identified 5 functionally distinct communities. Two of these communities were enriched with genes that regulate cell essential processes (cell cycle and transcription) and contain 10 of the 11 CYCLOPS genes. In addition, we found three communities that were enriched proteins associated with specific signaling pathways (WNT, HIPPO, MAPK). Based on these observations, we conclude that three major pathways are associated with the survival of β-catenin active cancers.
As a complementary approach for functional annotation of β-catenin co-dependencies, we used CRISPR-Cas9 to evaluate the consequence of pairwise deletions of 52 genes including 47 β-catenin co-dependences and 5 genes commonly altered in colon cancer. In model organisms, combinatorial knockout experiments have been successfully used for GI mapping enabling dissection of complex molecular processes. In particular, by mapping the patterns of double knockout mutants, GI mapping has been used to find genes required for protein folding (Jonikas et al., 2009) and chromosome biology (Collins et al., 2007) in yeast. Although several studies using RNAi-based GI mapping have identified genes and molecular pathways associated with Ricin resistance (Bassik et al., 2013), chromatin regulation (Roguev et al., 2013) and signaling networks (Horn et al., 2011), RNAi off target effects limits this approach to phenotypes with strong signal/noise ratios such as drug resistance or require the use of complex high content image-based phenotypic screens. Here we show that the high specificity of CRISPR-Cas9 enabled us to perform a pooled format GI mapping in mammalian cells using proliferation as a phenotypic readout. Although further mechanistic studies are needed to fully understand these relationships, this approach provides proof-of-principle evidence that these studies can now be performed in mammalian cells.
The β-catenin co-dependency PPI and GI maps allowed us to identify interactions among genes that are required for β-catenin active cell proliferation. Although due to the size of this dataset we were limited to comparing these analyses by inspection, we found a previously uncharacterized role for YAP1 in regulation of chromatin state. Specifically, we found that YAP1 interacts with components of the SWI/SNF chromatin-remodeling complex and that histone acetylation is up regulated by deletion of YAP1 in β-catenin active colon cancers. Although further studies are necessary to fully elucidate the roles of these communities in regulating β-catenin activity in normal and malignant tissues, these studies expand our understanding of how β-catenin function is regulated. More generally, although here focused in genes identified in genome scale loss of function screens, the genetic and proteomic approaches described here illustrates a general framework to illuminate the cellular pathways and networks among genes identified by genomic, functional or in silico analyses.
Experimental procedures
β-catenin activity reporter assay
Cell lines were generated by infection with a lentiviral β-catenin/TCF4 reporter (Fuerer and Nusse, 2010). Following puromycin selection (2 μg/ml) 50,000 cells were plated on a 96 well plate and 24 hr later luciferase activity was measured using the Luc-Screen detection kit (Applied Biosystems).
CRISPR-Cas9 loss-of-function screen
Lentiviral particles containing the pooled sgRNA library were transduced at low MOI (4 replicates/cell line) into 10 Cas9-expressing cell lines (DLD1, LS411N, LS513, HCT116, SW480, HT29, GP2D, RKO, MIAPACA2, NCI-H1975). Each sgRNA was stably integrated into at least 600 cells. In addition, parental DLD1 cells that did not express Cas9 were also infected with the same library. DNA extracted at 3 or 28 d post infection was used for massively parallel sequencing as described (Shalem et al., 2014).
Combinatorial CRISPR-Cas9 loss of function screen
To minimize recombination events between repetitive sequences, we inserted a variant U6 promoter downstream of the U6 promoter in pXRP003 (Supplemental Fig. 4A). To generate a pooled double sgRNA library, 115 sgRNAs were individually PCR amplified together with the S. Pyogenes tracer sequence and inserted into position 1 (AgeI/EcoRI restriction sites). Vectors containing an sgRNA in position 1 were pooled and digested with BsmBI (Thermo Scientific) and a pool of the same 115 sgRNAs was ligated into the BsmBI cloning sites (exactly as described above for single sgRNA pooled library). Following ligation the library was electroporated into Stbl4 cells (Life Technologies) grown at 30°C. Lentiviral particles containing the combinatorial CRISPR-Cas9 library were transduced (3 replicates/cell line) into 4 Cas9-expressing cell lines (DLD1, HCT116, HT29 and RKO). Following puromycin selection (2 μg/ml), genomic DNA (5μg) extracted at 3 or 28 DPI was PCR amplified using NEBNext® High-Fidelity 2X PCR Master Mix (New England Biolabs) and F/RPCR1_2sg primers (Supplementary Table 10). This mixture (5μl) was used for a second PCR amplification using barcoded-staggered primers (Supplementary Table 10). Paired end Illumina sequencing was used for sequencing of double sgRNAs. The forward and reverse sequences were aligned to the original sgRNA sequences using Bowtie I suite (Langmead et al., 2009).
Analysis of double CRISPR-Cas9 screen
sgRNA combinations with less then 50 reads at t=0 were discarded. Read counts from sgRNA combinations were combined into one combination score and normalized using equation 1: Y = (Read count + 1)/(total read count) normalized read counts were then normalized to control containing sgRNA combinations using equation 2: Z = Log2((Ycombo/YControl)*1*106). The fold change of every combination was calculated using equation 3: FCcombo = Zcombo_t=0 − Z combo_t=28. For calculating the genetic interaction score we used a previously reported S score that takes into account both consistency and magnitude (Collins et al., 2006) S = (FCExperimental − FCCalculated)/sqrt(SVar/NExperimental + SVar/NCalculated) where SVar = (varExperimental × (NExperimental − 1) + varCalculated × (NCalculated − 1))/(NExperimental + NCalculated − 2).
Supplementary Material
Acknowledgments
We would like to thank Chet Birger for help with analysis of the double CRISPR-Cas9 screen. This work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation in Mexico and was funded in part by US NIH grant U01 CA176058 (W.C.H.).
Footnotes
Author contributions
J.R, W.C.H, J.S.B, K.L, J.J., D.E.R, J.G.D conceived and designed the project. J.R., Y.S., R.O., F.P., E.H., L.F. performed experiments. J.R., J.M., P.T., I.T. performed the analysis. J.R. and W.C.H. wrote the paper.
Literature Cited
- Abazeed ME, Adams DJ, Hurov KE, Tamayo P, Creighton CJ, Sonkin D, Giacomelli AO, Du C, Fries DF, Wong KK, et al. Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res. 2013;73:6289–6298. doi: 10.1158/0008-5472.CAN-13-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, Zhang J, Khanna A, Hochdorfer T, Shang Y, Kahle KT. The WNK-SPAK/OSR1 pathway: master regulator of cation-chloride cotransporters. Sci Signal. 2014;7:re3. doi: 10.1126/scisignal.2005365. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R. Finding effective cancer therapies through loss of function genetic screens. Curr Opin Genet Dev. 2014;24:23–29. doi: 10.1016/j.gde.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet. 2011;12:487–498. doi: 10.1038/nrg3013. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, East-Seletsky A, Ali LD, Gerath WF, Pantel SE, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci U S A. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, Axelsson E, Huber W, Boutros M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C, Prahallad A, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. The distribution of the flora in the alpine zone. New Phytologist. 1912;11:37–50. [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DJ, Sung HS, Goo YW, Lee HM, Park OK, Jung SY, Lim J, Kim HJ, Lee SK, Kim TS, et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol Cell Biol. 2002;22:5203–5211. doi: 10.1128/MCB.22.14.5203-5211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, Asthana S, Neel D, Yan JJ, Lu X, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28:4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy F, Lepourcelet M, Shivdasani RA, Eck MJ. Structure of a human Tcf4-beta-catenin complex. Nat Struct Biol. 2001;8:1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- Pu S, Vlasblom J, Turinsky A, Marcon E, Phanse S, Trimble SS, Olsen J, Greenblatt J, Emili A, Wodak SJ. Extracting high confidence protein interactions from affinity purification data: at the crossroads. J Proteomics. 2015;118:63–80. doi: 10.1016/j.jprot.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, Krogan NJ. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Wang X, Hahn WC. Genomic insights into WNT/beta-catenin signaling. Trends Pharmacol Sci. 2014;35:103–109. doi: 10.1016/j.tips.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Serysheva E, Berhane H, Grumolato L, Demir K, Balmer S, Bodak M, Boutros M, Aaronson S, Mlodzik M, Jenny A. Wnk kinases are positive regulators of canonical Wnt/beta-catenin signalling. EMBO Rep. 2013;14:718–725. doi: 10.1038/embor.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, Schumacher SE, Zack TI, Beroukhim R, Garraway LA, et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res. 2013;23:665–678. doi: 10.1101/gr.143586.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S, Pradhan-Sundd T, Verheyen EM. Genome-wide identification of phospho-regulators of Wnt signaling in Drosophila. Development. 2015;142:1502–1515. doi: 10.1242/dev.116715. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.