Abstract
Bacteriophage φYeO3-12 is a T7/T3-related lytic phage that naturally infects Yersinia enterocolitica serotype O:3 strains by using the lipopolysaccharide O polysaccharide (O antigen) as its receptor. The phage genome is a 39,600-bp-long linear, double-stranded DNA molecule that contains 58 genes. The roles of many of the genes are currently unknown. To identify nonessential genes, the isolated phage DNA was subjected to MuA transposase-catalyzed in vitro transposon insertion mutagenesis with a lacZ′ gene-containing reporter transposon. Following electroporation into Escherichia coli DH10B and subsequent infection of E. coli JM109/pAY100, a strain that expresses the Y. enterocolitica O:3 O antigen on its surface, mutant phage clones were identified by their β-galactosidase activity, manifested as a blue color on indicator plates. Transposon insertions were mapped in a total of 11 genes located in the early and middle regions of the phage genome. All of the mutants had efficiencies of plating (EOPs) and fitnesses identical to those of the wild-type phage when grown on E. coli JM109/pAY100. However, certain mutants exhibited altered phenotypes when grown on Y. enterocolitica O:3. Transposon insertions in genes 0.3 to 0.7 decreased the EOP on Y. enterocolitica O:3, while the corresponding deletions did not, suggesting that the low EOP was not caused by inactivation of the genes per se. Instead, it was shown that in these mutants the low EOP was due to the delayed expression of gene 1, coding for RNA polymerase. On the other hand, inactivation of gene 1.3 or 3.5 by either transposon insertion or deletion decreased phage fitness when grown on Y. enterocolitica. These results indicate that φYeO3-12 has adapted to utilize Y. enterocolitica as its host and that these adaptations include the products of genes 1.3 and 3.5, DNA ligase and lysozyme, respectively.
Yersinia enterocolitica is a gram-negative bacterial species that belongs to the family Enterobacteriaceae. This species includes over 30 known serotypes, a number of which are human pathogens. While the most common pathogenic serotypes in Europe, Canada, Japan, and South Africa are O:3 and O:9, in the United States serotype O:8 is more prevalent. Y. enterocolitica is widely distributed in nature, with swine being the major reservoir of the pathogenic strains (6).
φYeO3-12 is a lytic bacteriophage that infects Y. enterocolitica serotype O:3 strains (28, 30). The receptor of the phage is Y. enterocolitica O:3 O antigen, and the phage is able to infect and proliferate in Escherichia coli strains that express the Y. enterocolitica O:3 O-antigen gene cluster (2). The genome of φYeO3-12 is a 39,600-bp-long linear, double-stranded DNA molecule, whose nucleotide sequence has been completely determined (GenBank accession no. AJ251805). The genome harbors 58 putative genes that all are transcribed from the same DNA strand (30). φYeO3-12 belongs to the T7 group of lytic phages, with E. coli phage T3 being its closest relative. The genome organizations of T7, T3, and φYeO3-12 are colinear, with early, middle, and late regions, and the overall identity between the φYeO3-12 and T3 genomes is 84%. Based on similarity to T7 and T3 genes, probable functions of many φYeO3-12 genes have been predicted (25, 29). However, several apparent genes, represented by open reading frames, still remain without obvious role. Here, we used efficient MuA transposase-catalyzed in vitro transposon insertion mutagenesis (13, 38) to further study which genes are nonessential for the phage and, furthermore, whether different genes are needed for the phage to proliferate on Y. enterocolitica and on E. coli.
(Part of this work has been published in the Proceedings of the 8th International Symposium on Yersinia [20].)
MATERIALS AND METHODS
Bacterial strains, DNA, phages, and media.
The bacterial strains, plasmids, and transposon used in this work are listed in Table 1, and bacteriophages are listed in Table 2. For convenience, Y. enterocolitica strain 6471/76-c is called YeO3-c. Purification of the phage has been described elsewhere (28). Tryptic soy broth medium (Oxoid) was used for bacterial liquid cultures. Soft-agar medium additionally included 0.4% (wt/vol) agar (Biokar Diagnostics), and it was supplemented with ampicillin (133 or 440 μg/ml), IPTG (isopropyl-β-d-thiogalactopyranoside) (1.33 mM), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (0.067%) when required. Luria agar (32) was used as a solid medium for bacteria, and lambda agar (tryptone, 10 g/liter; NaCl, 2.5 g/liter; agar, 15 g/liter) was used for phage plates. Plates were supplemented with ampicillin (100 or 150 μg/ml) or chloramphenicol (30 μg/ml) when required. Phages were grown at room temperature (RT).
TABLE 1.
Bacterial strains, plasmids, and transposon
| Strain, plasmids, or transposon | Genotype or relevant features | Source or reference |
|---|---|---|
| Bacteria | ||
| Y. enterolitica O:3 6471/76-c | Plasmid-cured host for φYeO3-12 | 35 |
| E. coli | ||
| JM109 | F′ traD36 proA+B+lacIq Δ(lacZ)ΔM15/Δ(lac-proAB) glnV44e14−gyrA96 recA1 relA1 endA1 thi hsdR17 | 39 |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 Δ(ara leu)7697 galU galK λ−rpsL nupG λ−tonA | Life Technologies |
| C600 | thi thr leuB tonA lacY supE | 3 |
| HB101 | F− Δ(gpt-proA)62 leuB6 glnV ara-14 galK2 lacY1 Δ(mcr-mrr) rpsL20 (Strr) xyl-5 mtl-1 recA13 | 7 |
| S. sonnei IJ286 | D2 371-48 | Ian Molineux |
| S. enterica serovar Typhimurium | ||
| His515 | Δ(his-rfb) | 26 |
| TV-163 | rfaL | 5, 37 |
| Plasmids | ||
| pAY100 | ampR; O-antigen gene cluster of Ye O:3 cloned in pBR322 | 2 |
| pTM100 | Clmr Tetr; mobilizable cloning vector | 24 |
| pBAD33oT | Clmr; oriT from pTM100 cloned into pBAD33 | C. I. Pacot-Hiriart et al., unpublished data |
| pRK2013 | Kmr; helper plasmid for conjugation | 9 |
| pBAD33oT-gl.3 | Clmr; φYeO3-12 gene 1.3 cloned into pBAD33oT | This study |
| pEcligA | Clmr; E. coli C600 ligA cloned into pTM100 | This study |
| pg3.5long | Clmr; φYeO3-12 gene 3.5 cloned into pBAD33oT | This study |
| pprg3.5 | Clmr; φYeO3-12 promoter 2.5 and gene 3.5 cloned into pBAD33oT | This study |
| pCSS810 | Clmr Kmr; lucFF in E. coli-Bacillus subtilis shuttle vector | 22 |
| pMP300 | Clmr; reporter plasmid, a pTM100 derivative; the lucFF gene from pCSS810 was cloned between φYeO3-12 genes 9 and 10 downstream of the φ10 promoter | This study |
| Transposon LacZ′-Mu(NotI) | Promoterless lacZ′, NotI sites close to transposon ends | 38 |
TABLE 2.
Bacteriophage φYeO3-12 and its derivatives
| Phage | Descriptiona | Source or reference(s) |
|---|---|---|
| φYeO3-12 | Wild type isolated from sewage | 2, 28, 30 |
| ΔPK | φYeO3-12 with deletion in gene 0.7 | 28, 30 |
| φ::lacZ1 | lacZ′ insertion in gene 5.5; 16237 (TTAAA) | This study |
| φ::lacZ2 | lacZ′ insertion in gene 0.45; 1747 (CAGGG) | This study |
| φ::lacZ3 | lacZ′ insertion in gene 5.5; 16269 (GACAG) | This study |
| φ::lacZ4 | lacZ′ insertion in gene 1.3; 7830 (CCGCG) | This study |
| φ::lacZ5 | lacZ′ insertion in gene 1.1; 6475 (ATGGC) | This study |
| φ::lacZ6 | lacZ′ insertion in gene 1.3; 7680 (φ::lacZ61) (ATGAC) and gene 3.7; 11300 (φ::lacZ62) (AGTGC) | This study |
| φ::lacZ7 | lacZ′ insertion in gene 4.5; 13523 (φ::lacZ71) (AGAAG) and gene 5.5-5.7; 16529 (φ::lacZ72) (TACCT) | This study |
| φ::lacZ8 | lacZ′ insertion in gene 1.3; 7799 (ACATC) | This study |
| φ::lacZ10 | lacZ′ insertion in gene 3.5; 11206 (AACTG) | This study |
| φ::lacZ11 | lacZ′ insertion in gene 0.7; 2772 (GACGC) | This study |
| φ::lacZ12 | lacZ′ insertion in gene 1.6; 8623 (TACTG) | This study |
| φ::lacZ13 | lacZ′ insertion in gene 4.3; 13322 (AAACA) | This study |
| φ::lacZ14 | lacZ′ insertion in gene 0,45; 1682 (ATTGG) | This study |
| φ::lacZ15 | lacZ′ insertion in gene 0.7; 2423 (AGGCT) | This study |
| φ::lacZ16 | lacZ′ insertion in gene 3.5; 11134 (ACCAT) | This study |
| φ::lacZ17 | lacZ′ insertion in gene 1.1; 6528 (GCGGT) | This study |
| φ::lacZ18 | lacZ′ insertion at nt position 888 and deletion of nt 888 to 2449 (genes 0.3 to 0.7) | This study |
| φΔ888-2449 | Deletion of nt 888 to 2449 (genes 0.3 to 0.7) | This study |
| φΔ1681-1741 | Deletion of nt 1681 to 1741 (in gene 0.45) | This study |
| φΔ7801-7823 | Deletion of nt 7801 to 7823 (in gene 1.3) | This study |
| φΔ11141-11200 | Deletion of nt 11141 to 11200 (in gene 3.5) | This study |
For transposon insertion mutants, the interrupted gene and the first base pair after the 5-bp duplication are indicated. The duplicated sequence is shown in parentheses. Deletion mutants are named by indicating the base pairs that are deleted. nt, nucleotides.
DNA techniques.
Phage DNA was extracted as previously described for bacteriophage lambda (32). Small-scale isolations were done with the Wizard Lambda Preps DNA purification system (Promega), where the protocol was modified by including extra steps to break the phage particles with proteinase K-sodium dodecyl sulfate-EDTA treatment and phenol-chloroform extractions (32) prior to adding the DNA purification resin. Standard DNA techniques were performed as described previously (32), and enzymes were used as recommended by the suppliers. Transposon LacZ′-Mu(NotI) was isolated from its carrier plasmid by BglII digestion as described previously (38). Primers used for sequencing and construction of plasmids are described in Table 3.
TABLE 3.
Oligonucleotides used in this work
| Oligonucleotide | Sequencea | Comments and usage |
|---|---|---|
| Lpsn-1 | ATTTCACACCGCATATGGTGCACT | Sequencing of lacZ′ insertion sites |
| Lpsn-2 | CAGGCATGCAAGCTTGGCGT | Sequencing of lacZ′ insertion sites |
| g1.3For1 | GGGGTACCCCAATGAGGAACAACCGTA | Cloning of gene 1.3, KpnI site |
| g1.3Rev1 | GCTCTAGACTAACTTATACTTTCTCTTGTG | Cloning of gene 1.3, XbaI site |
| EcligAFor | GGGGTACCGCATTGATGGTGCGATATGG | Cloning of E. coli ligA gene |
| EcligARev | GCTCTAGACCATCTCAGCTACCCAGCA | Cloning of E. coli ligA gene |
| Gene3.5For | ATGGCTAAAGTTCAATTCAAACC | Cloning of gene 3.5 into pg3.5long |
| MP38BACK | ATCGCTAGGCCAACACG | Cloning of gene 3.5 into pg3.5long |
| G3.5For3 | CGGGATCCCGAAAGCCTAATTACCCTCACTAAAGGGAACA ACCCAATCCTAAGAAAGGAGTAAAGAAAAATGGC | φYeO3-12 promoter φ2.5, cloning of gene 3.5 into pprg3.5 |
| G3.5Rev1 | GCTCTAGAGTATCACCCTCGGTCAGATG | XbaI site, cloning of gene 3.5 into pprg3.5 |
| Gene9for | TCCCCCCGGGCTGAATCTAATGCAGACG | XmaI site, construction of pMP300 |
| Gene10back | TCCCCCCGGGTACCAACCGCAGAGCGG | XmaI site, construction of pMP300 |
| SK-8 | CATGTTGAATCTCCTTATGTTAA | Construction of pMP300 |
| SK-7 | TTAACATAAGGAGATTCAACATGAGGGGATCCGAAGACGC | 5′ end complementary to SK-8, construction of pMP300 |
| SK-5 | TTAACCCTCACTAAAGGGAG | Construction of pMP300 |
| SK-3 | CTCCCTTTAGTGAGGGTTAATTACAATTTGGACTTTCCGCC | 5′ end complementary to SK-5, construction of pMP300 |
| pFK1INBACK2 | AGCCAGCAGCTTAGCGGC | RT-PCR |
| pFK1INFOR | CGTAACTAAACGTTCGGTC | RT-PCR |
Sequence in 5′ to 3′ orientation. Flanking restriction sites are shown in boldface; promoter sequence is underlined.
In vitro DNA transposition reaction, electroporation of the reaction products, and plating in the presence of an indicator strain.
The MuA transposase-catalyzed DNA transposition reaction was performed essentially as described previously (38). Briefly, the reaction mixture (25 μl) contained 1 μg of LacZ′-Mu(NotI) transposon as donor DNA, 1 μg of φYeO3-12 as target DNA, 2.32 μg of MuA transposase, 25 mM Tris-HCl (pH 8.0), 2.5 μg bovine serum albumin, 15% (wt/vol) glycerol, 0.05% (wt/vol) Triton X-100, 100 mM NaCl, and 10 mM MgCl2. The reaction mixture was incubated at 30°C, and 5-μl aliquots were withdrawn at different time points (from 0 to 15 min). An equal volume of 1% sodium dodecyl sulfate was added to the samples, and the incubation was continued at RT for 40 min. Following dilution (1:2) with water, 2-μl aliquots were used for electroporation of competent E. coli DH10B cells (4). The settings for the Bio-Rad Gene Pulser II were 400 Ω, 1.8 kV, and 25 μF. After the pulse, the electroporation mixture was plated with the E. coli JM109/pAY100 indicator strain on soft-agar indicator plates containing ampicillin (440 μg/ml), IPTG (1.33 mM), and X-Gal (0.067%) in the soft-agar layer and ampicillin (150 μg/ml) in the bottom agar.
Identification of mutant phages and determination of transposon insertion sites.
Mutant phage clones were identified on indicator plates by visual inspection of blue color formation at and around the plaques. In general, transposon insertion sites were localized by initial restriction analysis and/or PCR-based analysis followed by sequence determination with transposon-specific primers Lpsn-1 and Lpsn-2 (Table 3). In the cases when transposon-specific primers could not be used, e.g., with mutants with multiple transposon insertions, appropriate phage-specific primers were used instead (not shown). DNA sequencing reactions were performed with the ABI PRISM BigDye Terminator Ready Reaction version 2.0 cycle sequencing kit and analyzed with an ABI 377 DNA analyzer as recommended by the manufacturer. The sequence data were analyzed with the Genetics Computer Group suite of programs (version 10; Accelrys, San Diego, Calif.) and the European Molecular Biology Open Software Suite, version 2.6.0.
Fitness analysis.
Fitness analysis was performed as previously described for T7 (31). Briefly, YeO3-c or JM109/pAY100 cells were grown at RT or at 37°C, respectively, with aeration until the logarithmic growth phase (A600 = 0.3) was reached. A 5-ml aliquot of the culture was infected with 104 to 105 phages and incubated at RT with aeration. Samples were taken at 0 and 45 min and treated with chloroform to release phages, and phage titers were determined. The numerical value for fitness was calculated from the equation F = log2 (Nt/N0)/4t, where Nt and N0 are phage titers at 45 and 0 min, respectively, and t is the incubation time in hours. The fitness value gives the number of doublings of phage numbers per 15 min, which is the typical generation time for T7. Each experiment was repeated at least twice.
Construction of deletion mutants and complementing plasmids.
Deletion mutants φΔ1681-1741, φΔ7801-7823, and φΔ11141-11200 (Table 2) were constructed by deleting sequences between the transposon insertion sites of two mutants having insertions in the same gene: φ::lacZ2 and φ::lacZ14 for φΔ1681-1741, φ::lacZ4 and φ::lacZ8 for φΔ7801-7823, and φ::lacZ10 and φ::lacZ16 for φΔ11141-11200. The left ends of NotI-digested genomes of φ::lacZ14, φ::lacZ8, and φ::lacZ16 were purified from agarose gels and ligated to the calf intestinal alkaline phosphatase-treated right ends of NotI-digested genomes of φ::lacZ2, φ::lacZ4, and φ::lacZ10, respectively. Recombinant genomes were then electroporated into DH10B cells as described above and screened for colorless plaques, which are indicative of transposon loss. Deletion mutant φΔ888-2449 was constructed by digesting the φ::lacZ18 genome with NotI, religating the fragments, eletroporating the religation products, and screening as described above.
Plasmid pBAD33oT-g1.3 (Table 1) was generated by amplifying φYeO3-12 gene 1.3 by PCR (with primers g1.3For1 and g1.3Rev1 [Table 3]), after which the resulting fragment was digested with KpnI and XbaI and ligated into pBAD33oT digested with the same enzymes. For plasmid pEcligA, the ligA gene (GenBank accession no. M30255) of E. coli C600 was amplified by PCR (with primers EcligAFor and EcligARev [Table 3]), phosphorylated, and cloned as a blunt-ended fragment into the EcoRV site of pTM100. Plasmid pg3.5long was constructed by ligating a phosphorylated blunt-ended PCR fragment containing φYeO3-12 gene 3.5 (with primers Gene3.5For and MP38BACK [Table 3]) into the SmaI site of pBAD33oT. To express φYeO3-12 gene 3.5 in trans during the phage infection, plasmid pprg3.5 was constructed: φYeO3-12 gene 3.5 was cloned under the control of the phage φ2.5 promoter by amplifying a PCR fragment containing gene 3.5 preceded by φ2.5 promoter (with primers G3.5For3 and G3.5Rev1 [Table 3]), after which the fragment was phosphorylated, digested with XbaI, and ligated into SmaI-XbaI-digested pBAD33oT. Plasmids were transferred to YeO3-c by triparental conjugation (11) with helper strain HB101/pRK2013.
mRNA analysis.
YeO3-c cells grown to late logarithmic growth phase (A600 = 2.3; i.e., ca. 2 × 109 CFU/ml) were infected with phage at a multiplicity of infection of 3.6 in total of 5 ml of tryptic soy broth. The cultures were incubated at RT, and 1.5-ml samples were collected at 10 and 30 min after the infection. Total RNA was isolated with Bio-Rad Aurum total RNA minikit. The purity of RNA was monitored by PCR, and an extra treatment with RQ1 DNase (Promega) was performed when necessary. For reverse transcriptase PCR (RT-PCR), 3 μg of total RNA was used as the template. The cDNA synthesis was done with Amersham Pharmacia Biotech Ready-To-Go You-Prime First-Strand Beads by using 30 pmol of gene 1-specific primer pFK1INBACK2 (Table 3). The PCR mixtures contained 2 to 16.5 μl of cDNA as the template, gene 1-specific primers pFK1INBACK2 and pFK1INFOR (Table 3), and 1 U of DyNAzyme DNA polymerase (Finnzymes). Otherwise, standard conditions recommended by the enzyme producers were used. The PCR cycling parameters were as follows: 95°C for 2 min; 35 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 40 s; and then 72°C for 2 min.
Construction of luciferase reporter plasmid and luminescence measurements.
The reporter plasmid pMP300 contained the Photinus pyralis luciferase gene (lucFF) under control of φYeO3-12 promoter φ10, surrounded by phage genes 9 and 10. The reporter fragment was made by splicing-by-overlap-extension-PCR (1), using Vent polymerase (New England Biolabs). Initially, three fragments were separately amplified: The first fragment contained φYeO3-12 gene 9 followed by phage promoter φ10 (nucleotides 20572 to 21659). It was made with primers Gene9for and SK-8 (Table 3), using phage DNA as the template. The second fragment contained lucFF and was amplified from plasmid pCSS810 (Table 1) with primers SK-7 and SK-3. The third fragment, containing phage promoter φ10 followed by phage gene 10 (nucleotides 21501 to 22554), was made by using primers SK-5 and Gene10back, and phage DNA as the template. As the last step, the individual PCR fragments containing the overlapping segments were purified, mixed, and combined by splicing-by-overlap-extension-PCR. The final PCR fragment was then digested with XmaI and ligated to pTM100 (Table 1) digested with the same enzyme, resulting in pMP300.
To measure the light production from the reporter strain YeO3-c/pMP300 during the phage infection, a mid-exponential-growth-phase cell culture (A600 = 0.4) was infected with phage at a multiplicity of infection of 5. The culture was incubated at RT, and light production was monitored at 5-min intervals by mixing 100 μl of the cell suspension with 100 μl of 0.2 mM d-luciferin (pH 5.0) (Labsystems) and measuring the luminescence with a Luminova 1254 luminometer (Bio-Orbit).
RESULTS AND DISCUSSION
Transposon mutagenesis of phage φYeO3-12.
Even though the functions of most of the phage φYeO3-12 genes have been predicted (30), there are still open reading frames in the genome whose roles are not known. In this study, we were able to identify a number of nonessential genes in the phage genome by using a transposon insertion mutagenesis strategy that utilizes an efficient MuA transposase-catalyzed in vitro DNA transposition reaction (13, 38). The features of transposon insertion mutants with altered phenotypes were then confirmed by studying mutants having deletions in the corresponding genes and by complementation analysis.
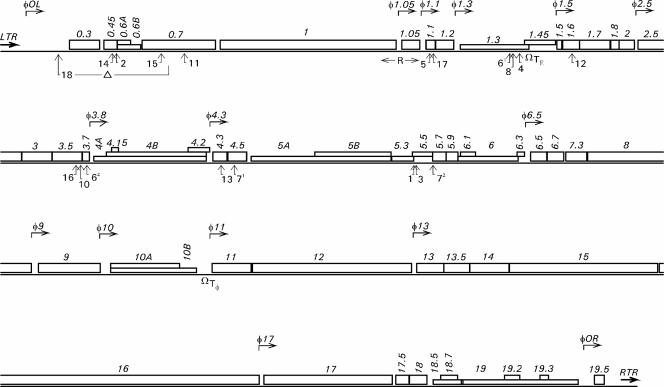
A total of 17 transposon insertion mutants were obtained, most of them from the 5-min time point of the transposition reaction (see Materials and Methods). In 15 mutants there was a single transposon inserted, and two of the mutants harbored two individual transposons (Fig. 1; Table 2). Due to the promoterless transposon and color-based screening method used, only mutants in which lacZ′ was transcribed from a phage promoter were obtained. Insertion sites were determined by sequence analysis, revealing their distribution in a total of 11 genes. All of these genes were located in the early and middle regions of the phage genome: 0.45 (unknown function), 0.7 (coding for protein kinase), 1.1 (unknown function), 1.3 (coding for DNA ligase), 1.6 (unknown function), 3.5 (coding for lysozyme), 3.7 (unknown function) 4.3 (unknown function), 4.5 (coding for homing endonuclease), 5.5 (unknown function), and 5.5 to 5.7 (coding for a predicted fusion protein).
FIG. 1.
Transposon insertion sites in the genome of φYeO3-12. The phage genes are represented by open boxes, and promoters are represented by bent arrows above the genes. LTR and RTR, left and right terminal repeats, respectively; TΕ and Tφ, transcription termination sites for host and phage RNA polymerases, respectively; R, primary origin of replication. The positions of transposon insertions are indicated below the genes with upward-pointing arrows and the number of the corresponding transposon mutant. The range of the deletion in the mutant φ::lacZ18 is also shown.
Thirteen of the insertion mutant clones were stable as judged by the plaque characteristics and restriction enzyme analysis of the genomes. However, four insertions (φ::lacZ3, φ::lacZ16, φ::lacZ71, and φ::lacZ13) generated unstable genomes as indicated by loss of the blue color of the plaques, PCR analysis, and sequencing of some of the reversion mutants that had lost the transposon (data not shown). The unstable mutants were not studied further.
Insertions upstream of gene 1 cause growth defects on Y. enterocolitica O:3.
The early region of φYeO3-12 includes genes 0.3, 0.45, 0.6a, 0.6b, and 0.7 (30). The product of gene 0.3, S-adenosyl-l-methionine hydrolase is 98% identical to T3 gp0.3, which is known to be nonessential (21). Gene 0.45 is unique to φYeO3-12, and nothing is known about its role during phage infection. Genes 0.6a and 0.6b are 34 and 25% identical to their homologues in T7, respectively, but there is no information available about their function (30). The gene 0.7 product, protein kinase, is known to be needed for T7 phage only when grown in suboptimal conditions (16).
Since the in vitro transposon insertion mutagenesis strategy used was based on screening of the mutants in E. coli, it was important to analyze whether the mutants would still grow on Yersinia. To study this, the mutants were plated on YeO3-c and E. coli JM109/pAY100 (Table 4). In general, YeO3-c was a more restrictive host than E. coli, since the plating efficiency was slightly lower for most mutants and even for the wild-type φYeO3-12. However, for mutants harboring the insertion in gene 0.45 (φ::lacZ14 and φ::lacZ2) or gene 0.7 (φ::lacZ15 and φ::lacZ11), the difference in the efficiency of plating (EOP) between YeO3-c and E. coli was clearly more pronounced. Also φ::lacZ18, where the insertion was mapped between the A3 promoter for host RNA polymerase (RNAP) (30) and gene 0.3, had a very low EOP on YeO3-c. For φ::lacZ11 the growth defect on YeO3-c was also evident in fitness analysis (Fig. 2A), since the fitness value was negative after 45 min of infection. The value then became positive when a 90-min infection time was used (not shown). The fitness of φ::lacZ14 on YeO3-c was also slightly lower than that of the wild-type phage.
TABLE 4.
Efficiency of plating compared to that on JM109/pAY100
| Phage | EOP on:
|
|||
|---|---|---|---|---|
| YeO3-c | IJ286/pAY100 | His515/pAY100 | TV-163/pAY100 | |
| φYeO3-12 | 0.6 | 1.1 | 0.9 | 1.0 |
| φ::lacZ14 | 0.2 | 0.9 | 1.3 | 1.5 |
| φ::lacZ2 | 0.1 | NDa | ND | ND |
| φΔ1681-1741 | 1.3 | ND | ND | ND |
| φ::lacZ15 | 0.02 | 1.1 | 1.2 | 1.2 |
| φ::lacZ11 | 0.1 | ND | ND | ND |
| ΔPK | 0.8 | 1.0 | 1.1 | 1.3 |
| φ::lacZ18 | 0.1 | 1.2 | 1.3 | 1.4 |
| φΔ888-2449 | 0.9 | ND | ND | ND |
| φ::lacZ8 | 0.5 | ND | ND | ND |
| φ::lacZ4 | 0.5 | 1.2 | 0.8 | 1.0 |
| φΔ7801-7823 | 0.9 | ND | ND | ND |
| φ::lacZ10 | 0.5 | 1.0 | 0.8 | 1.0 |
| φΔ11141-11200 | 1.0 | ND | ND | ND |
ND, not determined.
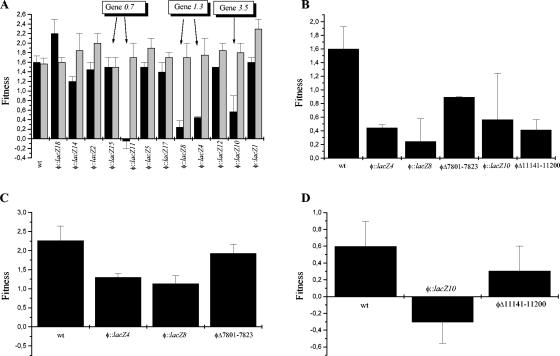
FIG. 2.
Fitness analysis of the transposon mutants. Fitness represents the number of doublings in phage numbers in 15 min. (A) The fitnesses of wild-type (wt) φYeO3-12 and transposon mutants grown on YeO3-c and JM109/pAY100 are shown by black and gray bars, respectively. (B) Fitnesses of mutants having transposon insertions or deletions in genes 1.3 (φ::lacZ4, φ::lacZ8, and φΔ7800-7823) and 3.5 (φ::lacZ4 and φΔ11140-11200) on YeO3-c. (C) Fitnesses of gene 1.3 mutants on YeO3-c/pEcligA. (D) Fitnesses of gene 3.5 mutants on YeO3-c/pprg3.5. Error bars indicate standard deviations.
These growth problems might either be truly Y. enterocolitica specific or be caused by the fact that E. coli JM109 is a laboratory strain developed for molecular biology research and YeO3-c is a virulence plasmid-cured derivative of the wild-type strain (Table 1). To study whether the low EOP is really specific for Yersinia, the EOP for one representative of each group of phage mutants having the insertion in the same gene was measured on Shigella sonnei (IJ286/pAY100) and Salmonella enterica serovar Typhimurium (His515/pAY100 and TV-163/pAY100), representing other enterobacteria. As seen in Table 4, for each transposon mutant studied (φ::lacZ14, φ::lacZ15, and φ::lacZ18), the EOPs on S. sonnei and S. enterica serovar Typhimurium were comparable to that on JM109/pAY100. This demonstrates that the growth defects of these transposon mutants are specific to Y. enterocolitica.
To study whether the low EOP on YeO3-c was due to a loss of the particular gene function or to the insertion of the transposon itself, we constructed φYeO3-12 mutants having out-of-frame nonpolar deletions in the corresponding genes (φΔ1681-1741 for gene 0.45, ΔPK for gene 0.7, and φΔ888-2449, in which genes 0.3, 0.45, and 0.6 and part of gene 0.7 are deleted) (Table 2). All of these deletion mutants were found to plate on YeO3-c as efficiently as on JM109/pAY100 (Table 4), indicating that the low EOPs of the transposon mutants were caused by the transposon insertion rather than by inactivation of the individual genes.
Insertions upstream of gene 1 are polar to gene 1.
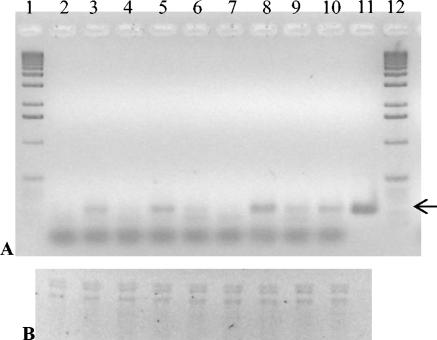
In order to answer the question whether the growth defects of the mutants having transposon insertions in the early region of the phage genome were due to polar effects, the mRNA levels of gene 1, coding for the RNA polymerase, were studied by RT-PCR. As can be seen in Fig. 3, after 10 min of infection there was a clear difference in the gene 1 mRNA levels between the mutants: The insertion mutants φ::lacZ14, φ::lacZ15, and φ::lacZ11 produced almost undetectable amount of gene 1 mRNA, whereas in the deletion mutants φΔ1681-1741, ΔPK, and φΔ888-2449, there was more of the 237-bp gene 1 RT-PCR product than in the wild-type phage. For φ::lacZ18, the amount of RT-PCR product was roughly comparable to that of the wild-type phage. After 30 min of infection, there were more or less similar amounts of gene 1 mRNA in all of the phages (not shown).
FIG. 3.
Transcription of φYeO3-12 gene 1 at 10 min after infection of YeO3-c. (A) The 237-bp RT-PCR products were analyzed on a 1.8% agarose gel and visualized with ethidium bromide. The arrow shows the position of the correct RT-PCR product. (B) RNA samples were analyzed on a 1.5% agarose-MOPS (morpholinepropanesulfonic acid)-formaldehyde gel and visualized with ethidium bromide. Lanes 1 and 12: DNA size standard (1-kb DNA ladder, Invitrogen). Lane 2: Uninfected YeO3-c. Lane 3: wild-type φYeO3-12. Lane 4: φ::lacZ14. Lane 5: φΔ1681-1745. Lane 6: φ::lacZ15. Lane 7: φ::lacZ11. Lane 8: ΔPK. Lane 9: φ::lacZ18. Lane 10: φΔ888-2449. Lane 11: φYeO3-12 DNA control. The whole procedure for detecting mRNA was repeated several times to ensure reproducibility, and a typical result is shown.
The mRNA analysis clearly indicates that the transposon insertions in the early region of the φYeO3-12 genome lead to the delayed expression of the phage RNA polymerase, which is thus the probable cause for the growth problems of these mutants. A possible explanation for the delayed RNAP expression in the insertion mutants is that the approximately 500-bp-longer early genome region of these mutants enters the bacterial cell more slowly (10, 19). This hypothesis is supported by the fact that φ::lacZ18, in which the transposon insertion is followed by a 1.56-kb deletion, produces gene 1 mRNA after 10 min of infection in a quantity roughly similar to that for the wild-type phage. At present it is still unclear, however, whether the slower DNA entry is the (sole) reason for the delayed RNAP expression. It is also not yet understood why these mutants have their growth defects only on Yersinia.
Mutations in the early genes drive the phage infection cycle out of balance.
To be able to study more closely the activity of the phage RNA polymerase in the different mutants, a reporter system was set up. In the reporter plasmid pMP300 (Table 3), the gene coding for P. pyralis luciferase (lucFF) (22) was cloned under the control of φ10, a strong consensus promoter of φYeO3-12 (30), resulting in a system in which lucFF is transcribed by the phage RNA polymerase during infection. Luciferase is a fairly stable enzyme, with a half-life of ca. 2 h (18), and thus the rise in the light production curve was considered an indicator of the phage RNAP activity.
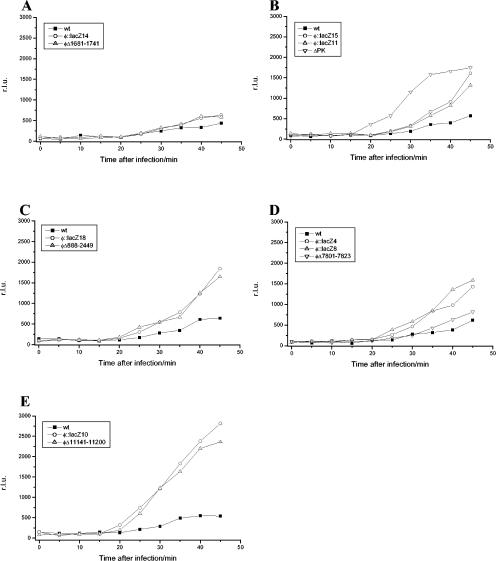
Figure 4 shows the kinetics of light production after infection of the reporter strain with wild-type φYeO3-12 and the different phage mutants. For the wild-type phage, the light production began ca. 25 to 30 min after infection and increased to ca. 35 min. A plateau phase then followed after 35 to 40 min, indicating that no more luciferase was synthesized. The remarkable finding in this assay was that all of the mutant phages produced more light than the wild-type phage. For the gene 0.45 mutants φ::lacZ14 and φΔ1681-1741 (Fig. 4A), the difference between the wild-type phage and the mutants was small but reproducible. The gene 0.7 mutants differed clearly from the wild type (Fig. 4B): the light production of the deletion mutant ΔPK began earlier and reached a higher level, but the timing of the plateau phase was comparable to that for the wild-type. The insertion mutants φ::lacZ15 and φ::lacZ11, however, began to produce light at the same time as the wild-type, but the light production was higher and there was no plateau phase. This was also true for the mutants in which the whole early region of the phage genome was deleted, φ::lacZ18 and φΔ888-2449 (Fig. 4C).
FIG. 4.
Light production after phage infection of the luciferase reporter strain YeO3-c/pMP300. Shown are the light productions by phage mutants having an insertion or deletion in gene 0.45 (A), protein kinase mutants (B), mutants having a 1.56-kb deletion in the early region of the phage genome (C), DNA ligase mutants (D), and lysozyme mutants (E). The light production of each mutant group was measured three times, and representative curves are shown. wt, wild type; r.l.u., relative light units.
The luminescence reporter system did not detect the delay in RNAP expression that was obvious in the RT-PCR analysis, since by the time the reporter system could measure the RNAP activity (ca. 20 min after infection) (Fig. 4), the short delay was no longer evident. Instead, the differences in the light production were due to the missing contribution of the gene product(s) of the interrupted gene(s). The increased luciferase activity in the mutant phage-infected bacteria most likely reflects either (i) the availability of more template for RNAP (i.e., less efficient degradation of pMP300 DNA by phage DNases), (ii) a longer half-life of the lucFF transcripts due to less efficient (phage) RNase activity, or (iii) increased activity of the phage RNAP due to the loss of regulation. Thus, the interrupted gene products could play a role directly or indirectly in the coordinated shift of nucleic acid metabolism during the phage infection.
The gene 0.45 mutants, φ::lacZ14 and φΔ1681-1741, produced slightly more light than the wild-type phage. So far nothing is known about the function of gp0.45, but the present result suggests that it may have a small role in the regulation of the phage growth cycle. gp0.7 (protein kinase) of phage T7 is known to have many functions, one of which is participation in the shutoff of host transcription (15, 23). This is in accordance with the finding that all of the mutants having deficient gp0.7 (φ::lacZ15, φ::lacZ11, ΔPK, φ::lacZ18, and φΔ888-2449) produced more light than the wild-type phage, indicating increased RNAP expression. The deletion mutant ΔPK differed clearly, with the strongest light production, from the other gene 0.7 mutants, probably due to its more rapid growth under laboratory conditions (28).
Genes 1.3 and 3.5, coding for DNA ligase and lysozyme, respectively, are needed for the phage to propagate efficiently on Y. enterocolitica O:3.
Middle region genes 1.1, 1.6, and 5.5 could be interrupted with a transposon without an obvious change in the phage phenotype. However, integration of the transposon into middle region genes 1.3 and 3.5 clearly altered the phage growth characteristics. When plated on a lawn of YeO3-c, the plaque sizes of φ::lacZ4 and φ::lacZ8 (insertions in gene 1.3) and φ::lacZ10 (insertion in gene 3.5) were significantly smaller than that of the wild type (Fig. 5 and data not shown). To study the growth rate of the mutants more thoroughly, a fitness analysis was conducted. As illustrated in Fig. 2A, the fitnesses of the wild-type phage and all of the studied mutants were relatively constant on JM109/pAY100. In contrast, on YeO3-c the fitness varied considerably. In accordance with the plaque sizes, the above-mentioned insertion mutants had significantly lower fitness than the wild-type φYeO3-12. The mutant φ::lacZ10 was shown to be somewhat unstable, having a reversion rate of ca. 10% per generation (data not shown), which explains the relatively large standard deviation obtained in the fitness analysis for this mutant (Fig. 2A and B).
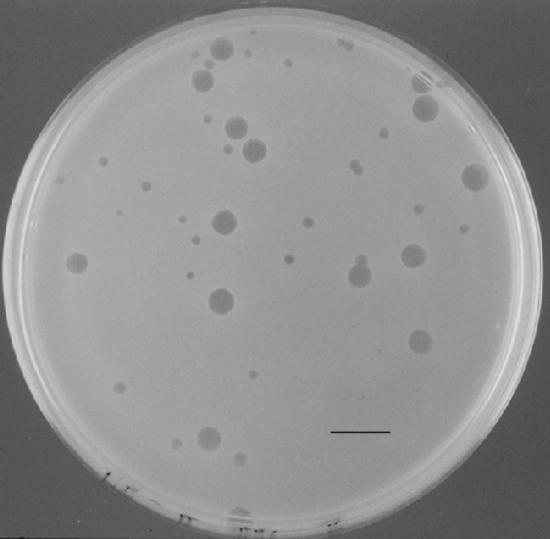
FIG. 5.
Plaques of φ::lacZ4 after a 45-min infection of YeO3-c/pBAD33oT-g1.3 plated on YeO3-c cells. Small plaques are φ::lacZ4, and large plaques are φ::lacZ4 in which gene 1.3 has been corrected by recombination between the phage genome and the plasmid. Bar, 1 cm.
To study whether the slow-growth phenotypes of φ::lacZ4, φ::lacZ8, and φ::lacZ10 were caused by the inactivation of their respective genes, corresponding deletion mutants were constructed: φΔ7801-7823, which has a short out-of-frame deletion in gene 1.3, and φΔ11141-11200, which has an out-of-frame deletion in gene 3.5 (Table 2). As can be seen in Fig. 2B, all of the mutants having either insertions or deletions in these genes had clearly lower fitness than the wild-type phage, indicating that inactivation of genes coding for DNA ligase or lysozyme were enough to cause the growth defect of φYeO3-12 on Y. enterocolitica.
Mutations in genes 1.3 and 3.5 can be reversed by homologous recombination with wild-type genes.
To study whether the slow-growth phenotype of phage mutants having defective DNA ligase or lysozyme could be complemented with the corresponding wild-type genes, homologous recombination was used to replace the mutated genes in the phage genome. Since successful complementation of phage development in trans would be unlikely due to the requirement for correct timing and the expression level of the transcomplementing gene, we used the recombination rate as an indicator of complementation. To this end, genes 1.3 and 3.5 were cloned into pBAD33oT to yield plasmids pBAD33oT-g1.3 and pg3.5long, respectively (Table 1). In these constructs, the genes were cloned under the control of the tightly regulated PBAD promoter (12) and were not expressed in the complementation analysis. Phage mutants having inactive DNA ligase (φ::lacZ4, φ::lacZ8, and φΔ7801-7823) or lysozyme (φ::lacZ10 and φΔ11141-11200) were then used to infect YeO3-c/pBAD3oT-g1.3 or YeO3-c/pg3.5long, respectively. If the slow-growth phenotype was due to inactivated gene 1.3 or 3.5 and not to any outside mutation, then double homologous recombination should result in the wild-type phenotype. The rate of reversion to wild-type phage was demonstrated by observing the plaque sizes and by PCR analysis. Already after 45 min of infection, 10 to 20% of the mutants had been reverted to wild type (Fig. 5). In the case of DNA ligase mutants, after 3 h of infection, 100% of the recovered plaques had reverted to wild type (data not shown). For lysozyme mutants, after 3 h of infection, three of five and two of six plaques studied for the insertion and deletion mutants, respectively, showed the wild-type pattern. The mutations were thus complemented by the corresponding genes, which confirms that the growth defects were caused by the loss of gene 1.3 or 3.5 and not by a mutation elsewhere in the phage genome.
E. coli LigA and phage lysozyme expressed in trans can partially complement the growth defects of DNA ligase and lysozyme mutants, respectively.
It is known that E. coli DNA ligase can complement bacteriophage T7 DNA ligase, even though gp1.3 mutants do have a reduced burst size. The T7 DNA ligase becomes essential if the phage infects a ligase-deficient strain (31; I. J. Molineux, personal communication). Since Y. enterocolitica DNA ligase did not seem to complement φYeO3-12 gp1.3, we wanted to study whether expression of E. coli DNA ligase in YeO3-c could complement the slow-growth phenotype of mutants φ::lacZ4, φ::lacZ8, and φΔ7801-7823. Figure 2C shows the fitnesses of these mutants on YeO3-c/pEcligA, a strain that constitutively expresses E. coli LigA. Compared to fitnesses of the same mutants on YeO3-c (Fig. 2B), it is evident that the expression of E. coli LigA aided the growth of the mutant phages. In particular, the deletion mutant φΔ7801-7823 grew on YeO3-c/pEcligA virtually as well as the wild-type φYeO3-12.
To study whether lysozyme expression could facilitate the growth of φ::lacZ10 and φΔ11141-11200 on YeO3-c, gene 3.5 was cloned into plasmid pBAD33oT under the control of the phage φ2.5 promoter (Table 1). In this system, gene 3.5 will be transcribed by the φYeO3-12 RNAP after infection of the strain by the phage, which minimizes the deleterious effects of lysozyme on the bacterial cell. As illustrated in Fig. 2D, the expression of gp3.5 severely impeded the growth of φYeO3-12, and this effect was even more pronounced with φ::lacZ10. For this mutant, fitness was negative in the normal 45 min and even in a 90-min assay (data not shown). However, the fitness of the deletion mutant φΔ11141-11200 on YeO3-c/pprg3.5 approached that of the wild-type phage (Fig. 2D), implying that the lysozyme expressed in trans could complement the growth defect of the gene 3.5 deletion mutant.
Inactivation of lysozyme results in a loss of RNAP regulation.
The effects of inactivation of DNA ligase and lysozyme on phage RNA polymerase activity were also studied by use of the above-mentioned luminescence reporter system. As can be seen in Fig. 4D, all of the mutants having defective DNA ligase produced more light than the wild-type phage. For the deletion mutant φΔ7801-7823, however, the increase was not as apparent as for the transposon insertion mutants, perhaps indicating that the transposon insertion itself and not the loss of ligase activity was the main reason for the increased luminescence. In contrast, phage mutants having either an insertion or a deletion in gene 3.5 showed faster and more intense luciferase activity than any other mutants studied (Fig. 4E). This demonstrates that there is a clear defect in the RNAP regulation in the mutants having deficient lysozyme.
Functions of DNA ligase and lysozyme.
During the growth cycle of E. coli bacteriophage T7, DNA ligase is needed to seal the nicks that occur during phage DNA replication (25). The results described above indicate that φYeO3-12 DNA ligase is needed for efficient propagation of the phage on YeO3-c but not on E. coli. This is in contradiction to the case for T7, where gp1.3 defects can be complemented by the host DNA ligase (see above). It is known that NAD+-dependent DNA ligases (LigA) are essential for growth of bacteria and are very conserved (36). We identified the ligA gene from the recently finished genomic sequence of Y. enterocolitica O:8 (sequence data produced by the Yersinia enterocolitica Sequencing Group at the Sanger Institute [www.sanger.ac.uk/Projects/Y_enterocolitica]), and its deduced protein sequence shows 79% identity with that of E. coli LigA (GenBank accession no. M30255). Therefore, it could be assumed that there are no significant differences between the functions of the Y. enterocolitica and E. coli ligases. The T7 and φYeO3-12 DNA ligases also are very similar, with the deduced protein sequences having 73.5% identity (30). At present, we do not know why φYeO3-12 gp1.3 mutants seem to be fully complemented by E. coli but not by Y. enterocolitica DNA ligase, at least at physiological concentrations.
Bacteriophage T7 lysozyme is known to have important regulatory roles during the phage infection cycle. It binds to T7 RNA polymerase by forming a 1:1 complex with it and regulates transcription by inhibiting initiation from class II (middle) promoters to a larger degree than class III (late) promoters, thereby contributing to the switch from middle to late gene expression. T7 lysozyme-RNAP interaction also stimulates replication, maturation, and packaging of the phage DNA (17, 40). In addition, gp3.5 is an enzyme having amidase activity, the probable role for which is to release phage from cell debris after the lysis of host cells. Lysozyme is important but not essential for T7, even though gp3.5 mutants do have lysis defects (25). The structure of T7 gp3.5 is known, and the amino acid residues responsible for different activities have been determined (8). Since φYeO3-12 and T7 lysozymes share 91.4% identity, it is feasible to assume that the two enzymes have similar structures. In the deletion mutant φΔ11141-11200, the amino acid residue that Cheng et al. (8) found to be required for T7 lysozyme amidase activity was deleted, and it thus seems likely that this mutant has lost the amidase activity. Since the luciferase reporter assay (Fig. 4E) obviously indicates the loss of RNAP regulation in the lysozyme mutants, it is evident that they have also lost the RNAP binding activity.
In this study, φYeO3-12 with defective lysozyme had lower fitness on YeO3-c than wild-type phage. On the other hand, overexpression of gp3.5 was also harmful for the phage. This can be explained by two mechanisms. First, the number of gp3.5 molecules expressed from the medium-copy-number plasmid was larger than that in normal phage infection. Considering the inhibitory activity of gp3.5 on transcription initiation, an excess amount of this enzyme would unquestionably drive the finely tuned regulation of the phage life cycle out of balance. Second, multiple copies of the phage promoter φ2.5 were introduced into the cell with plasmid pprg3.5, which resulted in a competition for the phage RNA polymerase and decreased transcription from the phage genome.
It is widely appreciated that the major factor determining the host specificity of bacteriophages is the ability of the phages to specifically recognize their host cells during adsorption (14, 33, 34). In tailed phages the adhesins are located in the tail fibers. Other factors may also contribute to adaptation of a phage to a certain host; however, these factors are still poorly understood. The host specificity of φYeO3-12 depends mainly on the tail fiber protein, gp17, as replacement of T3 gene 17 with the φYeO3-12 homologue was sufficient to turn T3 into a yersiniophage (27). In this study we found that φYeO3-12 genes 1.3 and 3.5 were required for the phage to be able to proliferate efficiently on its normal host, YeO3-c, but were not needed on E. coli. Therefore, DNA ligase and lysozyme might be considered to be factors important for adaptation of φYeO3-12 to grow on Y. enterocolitica serotype O:3.
Acknowledgments
We thank Ian Molineux for critical reading of the manuscript and Jukka Karhu for technical assistance.
We thank the Academy of Finland, the European Commission (project QRLT-1999-00780), and the Finnish National Technology Agency (TEKES) for financial support.
REFERENCES
- 1.Adams, M. R. 1989. Miscellaneous labour and materials saving methods, p. 239-254. In M. R. Adams and C. F. A. Hope (ed.), Rapid methods in food microbiology, vol. 26. Elsevier Science Publishers B.V., Amsterdam, The Netherlands. [Google Scholar]
- 2.Al-Hendy, A., P. Toivanen, and M. Skurnik. 1991. Expression cloning of the Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 10:47-59. [DOI] [PubMed] [Google Scholar]
- 3.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Beckmann, I., T. V. Subbaiah, and B. A. D. Stocker. 1964. Rough mutants of Salmonella typhimurium. II. Serological and chemical investigations. Nature 201:1299-1301. [DOI] [PubMed] [Google Scholar]
- 6.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, X., X. Zhang, J. W. Pflugrath, and F. W. Studier. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 91:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García, L. R., and I. J. Molineux. 1996. Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J. Bacteriol. 178:6921-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.). 1984. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haapa, S., S. Taira, E. Heikkinen, and H. Savilahti. 1999. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 27:2777-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haggård-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hesselbach, B. A., and D. Nakada. 1977. “Host shutoff” function of bacteriophage T7: involvement of T7 gene 2 and gene 0.7 in the inactivation of Escherichia coli RNA polymerase. J. Virol. 24:736-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch-Kauffmann, M., P. Herrlich, H. Ponta, and M. Schweiger. 1975. Helper function of T7 protein kinase in virus propagation. Nature 255:508-510. [DOI] [PubMed] [Google Scholar]
- 17.Huang, J., J. Villemain, R. Padilla, and R. Sousa. 1999. Mechanisms by which T7 lysozyme specifically regulates T7 RNA polymerase during different phases of transcription. J. Mol. Biol. 293:457-475. [DOI] [PubMed] [Google Scholar]
- 18.Ignowski, J. M., and D. V. Schaffer. 2004. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol. Bioeng. 86:827-834. [DOI] [PubMed] [Google Scholar]
- 19.Kemp, P., M. Gupta, and I. J. Molineux. 2004. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 53:1251-1265. [DOI] [PubMed] [Google Scholar]
- 20.Kiljunen, S., H. Vilen, H. Savilahti, and M. Skurnik. 2003. Transposon mutagenesis of the phage φYeO3-12. Adv. Exp. Med. Biol. 529:245-248. [DOI] [PubMed] [Google Scholar]
- 21.Krüger, D. H., W. Presber, S. Hansen, and H. A. Rosenthal. 1975. Biological functions of the bacteriophage T3 SAMase gene. J. Virol. 16:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lampinen, J., L. Koivisto, M. Wahlsten, P. Mäntsälä, and M. Karp. 1992. Expression of luciferase genes from different origins in Bacillus subtilis. Mol. Gen. Genet. 232:498-504. [DOI] [PubMed] [Google Scholar]
- 23.Marchand, I., A. W. Nicholson, and M. Dreyfus. 2001. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol. Microbiol. 42:767-776. [DOI] [PubMed] [Google Scholar]
- 24.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molineux, I. J. 1999. The T7 family of bacteriophages, p. 2495-2507. In T. E. Creighton (ed.), Encyclopedia of molecular biology. John Wiley and Co., New York, N.Y.
- 26.Nikaido, H., M. Levinthal, K. Nikaido, and K. Nakane. 1967. Extended deletions in the histidine-rough-B region of the Salmonella chromosome. Proc. Natl. Acad. Sci. USA 57:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pajunen, M. 2001. Molecular analysis of Yersinia enterocolitica serotype O:3 -specific bacteriophage φYeO3-12. Ph.D. thesis. University of Turku, Turku, Finland.
- 28.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage φYeO3-12 specific for Yersinia enterocolitica serotype O:3 is related to coliphages T3 and T7. J. Bacteriol. 182:5114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pajunen, M. I., M. R. Elizondo, M. Skurnik, J. Kieleczawa, and I. J. Molineux. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 319:1115-1132. [DOI] [PubMed] [Google Scholar]
- 30.Pajunen, M. I., S. J. Kiljunen, M. E.-L. Söderholm, and M. Skurnik. 2001. Complete genomic sequence of the lytic bacteriophage φYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 183:1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rokyta, D., M. R. Badgett, I. J. Molineux, and J. J. Bull. 2002. Experimental genomic evolution: extensive compensation for loss of DNA ligase activity in a virus. Mol. Biol. Evol. 19:230-238. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Scholl, D., S. Adhya, and C. R. Merril. 2002. Bacteriophage SP6 is closely related to phages K1-5, K5, and K1E but encodes a tail protein very similar to that of the distantly related P22. J. Bacteriol. 184:2833-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56:355-363. [DOI] [PubMed] [Google Scholar]
- 36.Sriskanda, V., and S. Shuman. 2002. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+. J. Biol. Chem. 277:9695-9700. [DOI] [PubMed] [Google Scholar]
- 37.Subbaiah, T. V., and B. A. D. Stocker. 1964. Rough mutants of Salmonella typhimurium. I. Genetics. Nature 201:1298-1299. [DOI] [PubMed] [Google Scholar]
- 38.Vilen, H., J.-M. Aalto, A. Kassinen, L. Paulin, and H. Savilahti. 2003. A direct transposon insertion tool for modification and functional analysis of viral genomes. J. Virol. 77:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, X., and F. W. Studier. 1997. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J. Mol. Biol. 269:10-27. [DOI] [PubMed] [Google Scholar]