Abstract
Neisseria gonorrhoeae (the gonococcus) is an obligate human pathogen and the causative agent of the disease gonorrhea. The gonococcal pilus undergoes antigenic variation through high-frequency recombination events between unexpressed pilS silent copies and the pilin expression locus pilE. The machinery involved in pilin antigenic variation identified to date is composed primarily of genes involved in homologous recombination. However, a number of characteristics of antigenic variation suggest that one or more recombinases, in addition to the homologous recombination machinery, may be involved in mediating sequence changes at pilE. Previous work has identified several genes in the gonococcus with significant identity to the pilin inversion gene (piv) from Moraxella species and transposases of the IS110 family of insertion elements. These genes were candidates for a recombinase system involved in pilin antigenic variation. We have named these genes irg for invertase-related gene family. In this work, we characterize these genes and demonstrate that the irg genes do not complement for Moraxella lacunata Piv invertase or IS492 MooV transposase activities. Moreover, by inactivation of all eight gene copies and overexpression of one gene copy, we conclusively show that these recombinases are not involved in gonococcal pilin variation, DNA transformation, or DNA repair. We propose that the irg genes encode transposases for two different IS110-related elements given the names ISNgo2 and ISNgo3. ISNgo2 is located at multiple loci on the chromosome of N. gonorrhoeae, and ISNgo3 is found in single and duplicate copies in the N. gonorrhoeae and Neisseria meningitidis genomes, respectively.
The ability of Neisseria gonorrhoeae to infect its human host is due to a variety of virulence factors. One of the factors, the gonococcal type IV pilus, is made up of monomers of pilin encoded by the pilE gene and is responsible for the primary attachment of N. gonorrhoeae to the host epithelium (12, 35). In addition to mediating attachment, the gonococcal pilus undergoes high-frequency antigenic variation (9, 10, 26, 37). This variation is believed to be responsible for the ability of N. gonorrhoeae to evade the immune system and cause reinfection in people previously exposed to the organism (10, 36, 47).
Gonococcal pilus variation is mediated mainly by high-frequency, nonreciprocal, homologous recombination reactions between silent copies of pilin information and the pilin expression locus pilE. Nineteen individual copies of silent pilin information are separated into six loci (pilS) throughout the chromosome of strain FA1090 (23). Recombination between pilE and pilS silent copies occurs at short regions of identity and results in alterations in the coding sequence of the pilE gene (9, 10, 26, 36). The multiple donor silent copies, in conjunction with the frequent regions where crossovers between the recombining genes can occur, contribute to the large repertoire of possible expressed pilin monomers and hence antigenically distinct pili that a single strain of the gonococcus (Gc) can produce. Most of the factors known to be required for antigenic variation at pilE are part of the cell's homologous recombination machinery. This includes recA (13), recX (34), recO, recQ (18), and recJ (32). All of these gene products also play a role in DNA repair. In addition to genes involved in homologous recombination, a potential link to cell division has been demonstrated with the reported requirement of the cell division-associated gene rdgC in pilin variation (17). All homologous recombination processes have been shown to increase under iron-limited conditions in N. gonorrhoeae (31), underscoring the possibility that a global regulatory system controlling recombination exists in this organism.
A number of characteristics of the recombination reactions responsible for pilin variation cannot be explained by homologous recombination reactions alone. First, the regions of identity between recombining segments have been shown to be as short as 7 bp (11). Second, the nonreciprocal nature of the recombination reaction, which is reflected in the exchange of DNA sequences between pilE and pilS copies resulting only in gene conversion at pilE, is not consistent with a mechanism mediated solely by generalized recombination functions (11). Finally, the 3′ region of all pilin loci contains sequences reported to be similar to defined site-specific recombinase binding sites (43). Taken together, these observations support the hypothesis that one or more recombinases act in cooperation with the homologous recombination machinery to mediate gonococcal pilin antigenic variation.
Moraxella lacunata and Moraxella bovis are ocular pathogens of humans and cattle, respectively. These members of the family Neisseriaceae elaborate type IV pili which are phase or antigenically variable. Previous work has demonstrated that antigenic and phase variation of the M. bovis and M. lacunata pili is mediated exclusively by a site-specific recombinase known as Piv, for pilin inversion protein (15, 16). The PivMB and PivML proteins belong to a novel family of DNA recombinases known as the Piv/MooV family; the family is unusual because the related recombinases include both site-specific recombinases and DNA transposases (40). Many of the recombinases of this family mediate transposition of insertion sequence (IS) elements from the IS110 family, including MooV, the transposase encoded by IS492. IS110-related elements are atypical insertion sequences because they lack terminal inverted repeat sequences and because they do not appear to generate target site duplications (4). Structure-function studies and molecular modeling with the defining members of this recombinase family, Piv and MooV, have suggested that all members of this family use a similar mechanism for recombination (40; C. Carpenter, D. Perkins-Balding, and A. C. Karls, unpublished data). A gene predicted to encode a protein with weak identity (18%) and similarity (34%) to Piv was identified in N. gonorrhoeae in a screen for proteins capable of inverting Moraxella pilin DNA in Escherichia coli. This gene was named gcr for gonococcal recombinase, and a loss-of-function mutant revealed that this gene product is dispensable for pilin antigenic variation (24). More recently, Southern blot analysis has revealed the presence of multiple genes encoding proteins with significant identity to Piv and MooV in the chromosome of numerous strains of N. gonorrhoeae but not the commensal bacteria Neisseria subflava and Neisseria lactamica (3). The gonococcus is not the only member of Neisseria species with Piv/MooV homologues, as three potential members of the Piv/MooV family, PivNM-1A, PivNM-1B, and PivNM-2, have been identified in the human pathogen Neisseria meningitidis strain MC58 genome sequence (38). Like N. gonorrhoeae, N. meningitidis is capable of pilin antigenic variation (39).
In this work, we identify eight Piv/MooV homologues encoded in the genome of N. gonorrhoeae strain FA1090 and confirm that they are found only within the pathogenic Neisseria. We named these genes invertase-related genes 1 through 8 (irg1-8) based on their similarity to the Piv proteins. We demonstrate that at least some of these genes are expressed; however, a mutant strain inactivated for all eight genes and a strain that overexpresses one irg gene still exhibit wild-type levels of pilin antigenic variation, DNA transformation, and DNA repair. While biochemical assays for complementation of Piv invertase and MooV transposase reactions did not prove the recombinase activities of Irg proteins, comparative analyses of the DNA flanking the coding sequences support the hypothesis that the Piv homologues are transposases for two new members of the IS110 family of insertion sequences.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli One Shot TOP10 competent cells (Invitrogen) or DH5α [F− endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF) U1691 φ80 dlacZΔM15] and HMS174(DE3) [recA1 hsdR Rifr galλ(DE3)] were grown in Luria-Bertani broth or agar at 37°C and used to propagate plasmids. Solid medium contained 15 g of agar per liter. FA1090 and VD300 (a derivative of strain MS11) are gonococcal laboratory strains. The remaining gonococcal strains used in this study were collected directly from patients presenting to an urban sexually transmitted disease clinic with gonorrhea. Chromosomal DNA preparations from N. gonorrhoeae laboratory strains FA19, RD5, UU1, and F62; clinical isolates 1084, 1918, 1349, 1402, 1384; N. meningitidis strains NMB and GA0929 (clinical isolate, group Y); and commensal strains Neisseria cineria and N. lactamica were graciously provided to A.C.K. by William Shafer. Gonococcal strains were grown on Gc medium base (Difco) plus Kellogg supplements (GCB) [22.2 mM glucose, 0.68 mM glutamine, 0.45 mM cocarboxylase, 1.23 mM Fe(NO3)3 (all from Sigma)] (12) at 37°C in 5% CO2. FA1090 recA6 strains were created by transformation of FA1090 with plasmid DNA carrying an isopropyl-β-d-thiogalactopyranoside (IPTG)-regulatable gonococcal recA allele, recA6, which allows control of recA and subsequent control of recA-dependent recombination and repair processes, including pilin antigenic variation (27). An IPTG (Diagnostic Chemicals Limited) concentration of 1 mM in the medium allows for maximal induction of recA transcription and restoration of transformation competence to near-wild-type levels (27). Antibiotics were added at the following concentrations for E. coli: chloramphenicol, 30 mg/liter; erythromycin, 250 mg/liter; kanamycin, 40 mg/liter; ampicillin, 100 mg/liter; and tetracycline, 12.5 mg/liter. For N. gonorrhoeae, antibiotic concentrations were as follows: chloramphenicol, 2 mg/liter, erythromycin, 1 mg/liter, kanamycin, 50 or 250 mg/liter, and nalidixic acid, 2 mg/liter.
DNA and RNA manipulations and analyses.
Standard procedures were performed as described previously by Sambrook et al. (25). Plasmid DNA was isolated from strains of E. coli by using QIAGEN (Valencia, Calif.) plasmid kits. Enzymes were used according to manufacturers' directions (Promega Corp. and New England Biolabs [NEB]). E. coli strains were transformed by electroporation using the Gene Pulser II electroporation system (Bio-Rad Laboratories) according to the manufacturer's specifications. For Southern blot analysis, DNA was transferred to a Magnagraph nylon membrane according to the manufacturer's specifications (Micro Separations Inc.). Sequencing reactions were performed by using the Big-Dye Terminator Cycle Sequencing kit (Perkin-Elmer Corp.), and sequencing products were separated with an ABI Model 377 automated DNA sequencer. DNA sequence analysis was performed by using Lasergene software (DNASTAR, Inc.) and VectorNTI software (Informax, Inc.). PCR fragments were purified by using a Qiaquick PCR purification kit (QIAGEN). The BLAST program (1) was used to search the National Center for Biotechnology Information (NCBI) nonredundant database. pilE sequences were determined by amplifying pilE from the chromosome with primers PILRBS (5′-GGCTTTCCCCTTTCAATTAGGAG-3′) and SP3A (5′-CCGGAACGGACGACCCCG-3′) (29) using Taq polymerase and sequencing the resulting PCR product with CONST-F2 (5′-TACCAAGACTACACCGCCCG-3′) (29). RNA isolation was performed by using the RNeasy Mini protocol for isolation of total RNA from bacteria (QIAGEN). DNase treatment was performed by using OnColumn DNase digestion with the RNase-Free DNase set (QIAGEN). A260 and A280 levels were taken with a spectrophotometer to assess purity. RNA was run in a 0.8% agarose gel to assess purity, and 28S rRNA bands were quantified with ImageQuant software (Molecular Dynamics, Piscataway, N.J.). RNA folding analysis was performed by using a derivative of the Vienna Folding package from the DNASTAR package. cDNA synthesis was performed as described previously by Serkin and Seifert (30). The primer used for the synthesis of cDNA was LCirgrev (5′-GACCGGCAATACTGCGC-3′). Real-time PCR was performed with a LightCycler instrument (Roche Diagnostics, Indianapolis, Ind.) using a fluorescence resonance energy transfer (FRET) detection method (45, 46). FRET requires the use of two probes: the first (donor) probe is labeled with a fluorophore at the 3′ end, and the second (acceptor) probe is labeled with a fluorophore at the 5′ end. When both probes properly anneal to target DNA so that they are in close proximity, excitation of the donor results in emission, which in turn excites the acceptor. In this manner, the ratio of acceptor fluorescence to donor fluorescence is measured. Hybridization probes were created in accordance with previously published guidelines (7). The distance between the donor and acceptor probes was 1 bp. The probes used were LCHybirg1 (5′-CGTAGTGAACCCGCTGAAAATAAGCAAGTATGC-3′) and LCHybirg2 (5′-GAAAGCAGGTTCAAGCGAACCAAAACAG-3′) for the irg genes and LC HybrecA 1 (5′-CGGGCGCATCGTCGAAATCTTCGG-3′) and LC HybrecA 2 (5′-CCCGAATCCTCCGGCAAAACCACA-3′) for recA. All donor probes were labeled with fluorescein isothiocyanate at the 3′ end, while all acceptor probes were labeled at the 5′ end with LightCycler-Red 640-N-hydroxysuccinimide ester. Acceptor probes also had phosphate groups attached to the 3′ end to prevent probe extension (IT Biochem). Primer and probe melting temperatures were calculated by using the freeware program TM Utility, version 1.3 (Idaho Technology [http://www.idahotech.com/downloads_up/Tmutility_form.htm]). Mixtures in hybridization experiments contained 2 μl of LightCycler DNA Master Hybridization Probes mix (Roche Diagnostics), 3.2 μl of MgCl2 (for a final concentration of 5 mM), 2 μl of each primer (0.5 mM), 2 μl of each hybridization probe (0.2 mM), 2 μl of template, and PCR-grade sterile water for a final volume of 20 μl. Fluorescence was acquired at the end of every extension step during the PCR. Primers used for PCR were LCirg (5′-CGTAGTGAACCCGCTGAAAATAAGCAAGTATGC-3′) and LCirgrev (5′-GACCGGCAATACTGCGC-3′).
Identification, cloning, and inactivation of the irg copies.
The Gonococcal Sequencing Project database (23) was used to identify the individual irg genes in the chromosome of N. gonorrhoeae strain FA1090. BLASTN analysis of the complete gonococcal genome was performed by using the pivNG sequence (3). Sequences greater than 54 bp with E values of 10−56 or less were concluded to be Piv homologues and hence irg copies. This BLASTN analysis did not produce any hits longer than 100 bp besides the eight irg genes.
The following mutant irg genes were created and introduced into FA1090 recA6 in the order presented to create a strain with all eight irg genes knocked out. irg5, irg6, and irg8 were PCR amplified by using either PivDir (5′-TCGGGATTTACGCCGATTTG-3′) with PivPrimer (5′-ATAAGGGTAATCCCTG-3′) for irg6 or PivFor (5′-CGCCGAAAGGAACGTGTATGCT-3′) with PivRev2 (5′-CGGATTTCAGACGCGGCAAAGCA-3′) for irg5 and irg8 and cloned into the pBlunt vector (Invitrogen). Chloramphenicol and kanamycin resistance cassetts were then inserted into SnaBI and NdeI sites internal to irg5 and irg6 genes, respectively. A kanamycin resistance cassette was inserted into irg8 at the NdeI site. These vectors were then transformed into FA1090 recA6, and the appropriate antibiotic resistance was selected to inactivate the individual irg copy by insertion. irg7 was inactivated by using transposon shuttle mutagenesis (28), resulting in insertional inactivation of irg7 with an erythromycin minitransposon. irg1 and irg4 were inactivated through the creation of in-frame deletions. To create an in-frame deletion in irg1, a 4,590-bp segment of DNA containing irg1 was amplified from the chromosome of N. gonorrhoeae by using primers PivNG3B-For (5′-ACCGCAGCAAACCCTCTCCCTAAC-3′) and PivNG3B-Rev (5′-CGAATACGACATCCGCCCAGAAGA-3′). This fragment was cloned into pBlunt (Invitrogen), and a 357-bp fragment internal to irg1 was removed by double digestion with AgeI and NdeI. The resulting construct was transformed into N. gonorrhoeae, and successful deletions were screened with primer PivNG3BScreen (5′-ATCAGTTTTCGGACCGGTATGCG-3′), which spans the newly formed junction. To create an in-frame deletion in irg4, primer pairs were created to generate products that when ligated together would result in the removal of 568 nucleotides internal to irg4. The primers used to amplify the 3′ portion of irg4 including downstream sequence were PivNG4Right-For (5′-GAATCCGGGACAAGCGTAAGGGG-3′) and PivNG4Right-Rev (5′-CCTGAAAGTCAGTGCCGATGCCG-3′), and the primers used to amplify the 5′ portion of irg4 including upstream sequence were PivNG4Left-For (5′-TGAGTCAATACCTGTCGCAACGCC-3′) and PivNGLeft-Rev (5′-TCCCAAGCTTTTCAACCGGTCCG-3′). The two products ligated together were cloned into pCR-BluntII (Invitrogen) and transformed into N. gonorrhoeae, and successful deletions were screened with primer PivNG4-Screen (5′-GGTTGAAAAGCTTGGGAGAATCCG-3′). irg3 was inactivated by deleting part of the gene and inserting a spectinomycin resistance cassette, pHP45Ω (22). A 2,278-bp segment of DNA which included irg3 was amplified from the FA1090 chromosome by using primers Irg 3 Upstream (5′-TCG TAA TGG GCG GAA AGT CC-3′) and Irg 3 Downstream (5′-CGC TAA TGG GCT TCA GAC GG-3′). The PCR product was cloned into pCR-Blunt II (Invitrogen). A 571-bp fragment internal to irg3 was dropped out by double digestion with AgeI and AflII (NEB), and the vector was treated with T4 DNA polymerase (NEB). pHP45Ω was digested with SmaI (NEB), and the fragment containing ΩSpc was ligated into the blunted pCR-BluntII-irg3 vector. The resulting construct, pCR-Blunt irg3::Spc was then sequenced. FA1090 recA6 irg145678 was transformed with this plasmid, and transformants were selected on Spec45 plates. To create an in-frame deletion in irg2, primer sets were created to generate products that when ligated together would result in the removal of 764 bases internal to irg2, leaving 199 bp of the gene with an intervening 8-bp PacI site. The primers used to amplify the 5′ end of irg2, including upstream sequence, were Irg2UP Uptake, which contains the Gc uptake sequence (5′-TTG CCG TCT GAA GCA ATG AGG GCG GTA CAG G-3′) and Irg2 Pac I, which contains a PacI site (5′-GAT TAA TTA AAT CTA AAC CTT TTG AAT CGT TG-3′). This 835-bp PCR fragment was cloned into pCR-Blunt II (Invitrogen). For the 3′ end of irg2 including downstream sequence, a 1,384-bp fragment was amplified using primers Irg 2 Pac I-1599, containing a PacI site (5′-TTA ATT AAA GCA TTG ATG CGT AAA CTC G-3′), and Irg 2 Downstream (5′-GCA CAT GCT ATT CAA ATC AAG G-3′). This PCR product was also cloned into pCR-Blunt II, and then each plasmid was digested with PacI and PstI (NEB). The fragment containing the 3′ portion of the gene was gel isolated and subcloned into the linear plasmid containing the 5′ portion of irg2. The resulting plasmid was sequenced and transformed into FA1090 recA6 irg1345678. The colonies were screened for the deleted irg2 gene by using a primer that spanned the newly formed junction containing the PacI site, IRG2INFRAME (5′-AGG TTT AGA TTT AAT TAA A-3′). Successful combination of all eight mutations in one strain was confirmed by Southern blot and PCR analysis (data not shown).
To create a strain to be used in overexpression analysis, irg5 was amplified and cloned into the Neisserial Insertional Complementation System vector pGCC6 (32), which allows for IPTG-inducible expression of irg5. This construct was then transformed into FA1090 recA6 at a site in the gonococcal chromosome irrelevant to pilin antigenic variation to create FA1090 recA6 NICS6::irg5.
In vivo inversion and transposition assays.
Expression plasmids were created for two of the irg genes. irg2 (pAG712) and irg7 (pAG711) were amplified with Pfu polymerase (Promega) by using primer pairs 5′-CAGTTCAGCTAGCCGTAACGCCGTAGGATTGG-3′ and 5′-CTCGTCTCGAGCGCCGATTTGTAACGCGATGG-3′ as well as 5′-CAGTTCACATATGAATATAATCGGGCCGGACATC-3′ and 5′-CTCGTCTCGAGTTGATTCAATCGGTGTCTTTCC-3′ introduced into the NheI/XhoI and NdeI/XhoI sites, respectively, of pET21a (Novagen) and expressed from the T7 promoter as His6-tagged proteins in E. coli HMS174(DE3). The pivML (pAG1300) and mooV (pAG921) expression vectors, containing the recombinase genes in pET21a between the NdeI/XhoI sites, have previously been shown to exhibit in vivo inversion and precise excision activities, respectively (21, 41). Recombinase expression was induced with 0.2 mM IPTG, and samples were taken at 2 h postinduction from both induced and uninduced cultures for Western blot analysis as described previously by Tobiason et al. (40). Anti-His6 rabbit antibody (Santa Cruz Biotechnology) was used for the primary antibody.
The in vivo inversion assay was performed essentially as described previously by Tobiason et al. (40). The inversion substrate pAG862 and each recombinase expression vector were introduced into HMS174(DE3), and transformants were grown in Luria broth for 12 h without IPTG or for 5 and 12 h after induction at an optical density at 600 nm of 0.6 with 0.005 mM IPTG. The lower level of induction, compared to that of the cultures used for Western blot analysis, was necessary due to toxicity of high-level expression of the recombinases. Isolated plasmid DNA from these cultures was restricted with KpnI and SalI and electrophoresed on a 1% agarose gel. Southern blot analysis of the inversion-diagnostic restriction fragments utilized probe prepared from pMxL1 (16), which contains the full invertible segment from M. lacunata. The 32P-labeled probe was prepared by PCR as described previously by Perkins-Balding et al. (21). Three primers were used to generate probes for both orientations of the invertible segment: 5′-CGGAATTCGCTTGCAATACTGGCTCTTTAC-3′, 5′-TATGCCCGCTGTCATAGTCAGCT-3′,and 5′-GAGGTGATTTCGCATAATATA-3′.
The in vivo transposition assay was performed as described previously (21), using pAG921, pAG711, and pAG712 to complement the transposition substrate for precise excision and circle junction formation. The transposition substrate pAG713 was constructed by first replacing the BamHI and XhoI segments of pACYC177 with the IS492-containing BamHI/XhoI fragment from pAG949 (21). Sequence between the MfeI sites of mooV was then replaced with the chloramphenicol acetyltransferase gene (cat) from pACYC184 (PCR amplified with Pfu DNA polymerase and the primers 5′-CCTACAATTGACGGAAGATCACTTCGCAGAA-3′ and 5′-GCGCAATTGAGGGCACCAATAACTGCCTT-3′) to generate pAG713. The induction of recombinase expression was done with 0.005 mM IPTG; induction of higher levels of His6-tagged MooV inhibited the excision of IS492 (21).
Kinetic pilus-dependent colony variation assay.
The kinetic colony phase variation assay (E.V. Sechman and H. S. Seifert, unpublished data) was performed as follows. N. gonorrhoeae strains were streaked for isolated colonies on GCB plates containing 1 mM IPTG and incubated at 37°C at 5% CO2. Isolated colonies were screened at 18, 20, 22, 24, and 26 h for the presence of sectors of variation which are indicative of pilus variation. Scores ranging from 0 to 4 were assigned to colonies based on the number of sectors of variation present per colony. Colonies that contained more than four sectors of variation were given a score of 4. Analysis was performed on at least 20 colonies per screen per experiment, and the assay was performed at least three times per strain.
UV sensitivity assay.
The UV sensitivity of Gc strains was assayed as previously described (14). Gonococci were incubated for 24 h on GCB plates (in the presence of IPTG for recA6 and Neisserial Insertional Complementation System derivatives), collected with a Dacron swab, and suspended in GCB liquid. Serial dilutions were made and exposed to UV radiation at 0 to 80 J/m2 in a Stratagene UV Stratalinker 1800. Relative resistance was expressed as percent survival compared to the number of CFU in the unirradiated culture.
DNA transformation competence assay.
Gonococcal strains were grown on GCB medium overnight and transferred by a Dacron swab into 37°C GCB liquid plus MgCl2 containing uncut pSY6 plasmid DNA (33), which carries a point mutation in the gonococcal gyrB gene that confers nalidixic acid resistance when recombined into the gyrB locus. After a 15-min incubation, unincorporated DNA was degraded with RQ1 DNase I (Promega) for 15 min, and the cells were diluted into GCB, incubated for 4 h, and plated for nalidixic acid-resistant and total CFU.
RESULTS
The genome of N. gonorrhoeae contains eight genes with significant homology to the Moraxella Piv invertases.
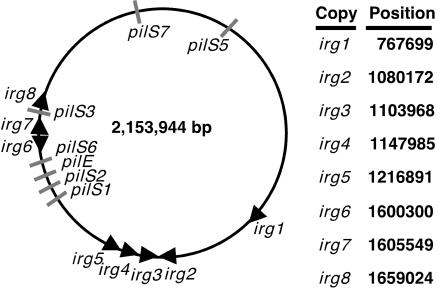
Previously, multiple copies of a gene predicted to encode a protein with significant homology to the pilin invertases PivML and PivMB from Moraxella species were identified in the genome of different strains of N. gonorrhoeae (3). We sought to determine the specific number of irg gene copies in the genome of N. gonorrhoeae strain FA1090. In silico analysis of the unannotated N. gonorrhoeae strain FA1090 genome database (23) revealed the presence of eight copies of genes encoding Piv homologues clustered in three regions of the chromosome (Fig. 1). Each of these genes is predicted to encode a protein with approximately 30% identity and 58% similarity (CLUSTALW alignment over the full amino acid sequences) to the pilin inversion protein from M. bovis (PivMB) and M. lacunata (PivML) (Fig. 2). We named these genes invertase-related genes 1 through 8 (irg1-8) based on their similarity to the Piv proteins and their order on the chromosome of N. gonorrhoeae. They do not show significant sequence similarity to the previously identified Gcr recombinase (23). TBLASTN searches with the Irg proteins yielded only two protein sequences from other bacteria with a very high degree of identity with one of the Irg proteins. PivNM-1A and PivNM-1B from N. meningitidis (MC58 serogroup B and Z2491 serogroup A) exhibit >98% amino acid identity with Irg7, differing at only five residues over the full 318-amino-acid sequence (Fig. 2). PivNM-2 shows only 33.8% identity with Irg7 and 37% identity with the other Irg proteins.
FIG. 1.
Chromosomal location of irg copies in N. gonorrhoeae strain FA1090. A schematic diagram of the N. gonorrhoeae chromosome is shown. pil loci are labeled, and irg copies are denoted by black arrows that point in the predicted direction of transcription. The chromosomal position of the start codon for each irg copy is given for the linear sequence of the N. gonorrhoeae strain FA1090 genome, starting at dnaA (GenBank accession no. AE004969 [23]).
FIG. 2.
Alignment of Irg proteins with Piv and homologues. Conserved residues are boxed in gray. Asterisks are above residues shown to be required for PivML-mediated inversion of Moraxella DNA in E. coli (40). PivNM-1A, PivNM-1B, and PivNM-2 are Piv homologues found in N. meningitidis. MooV is the transposase for IS492 in P. atlantica (21) and IS621 Tnp is the transposase for IS621 in E. coli (5).
The Piv proteins from Moraxella belong to an unusual family of site-specific recombinases and transposases known as the Piv/MooV family. This recombinase family includes transposases of the IS110 family of insertion sequence elements. An alignment of the Irg copies with other members of this family revealed that the predicted Irg proteins have approximately 25% identity and 50% similarity to the transposase MooV from IS492 in Pseudoalteromonas atlantica and approximately 33% identity and 53% similarity to the transposase encoded by the E. coli element IS621 (Fig. 2).
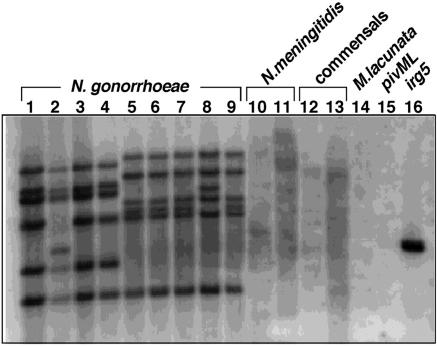
To determine whether strains of N. gonorrhoeae, in addition to FA1090, contain multiple copies of the irg genes, Southern blot analysis was performed on the laboratory strains FA19, UU1, RD5, and F62 as well as five isolates from patients presenting with disseminated gonococcal infections (Fig. 3). The probe, which was generated by PCR from the full irg2 gene, should hybridize to irg1, irg2, irg3, irg4, irg5, irg6, and irg8 but should not hybridize to irg7 or to the divergent N. meningitidis Piv homologues (Fig. 3, lanes 10 and 11). The probe specificity was confirmed by the lack of hybridization with piv sequence on M. lacunata chromosomal DNA or pAG702 (41) (Fig. 3, lanes 14 and 15), while strong hybridization was seen with pAG852, in which irg5 replaces piv on the pAG702 vector (lane 16). This Southern blot analysis revealed the same number of irg gene copies on similarly migrating fragments in both laboratory isolates (Fig. 3, lanes 1 to 4) and minimally passaged clinical isolates (lanes 5 to 9). The only exceptions were laboratory strain UU1 (lane 2) and one of the clinical isolates (lane 8), which each showed one fragment with a different mobility. These shifts in restriction fragment mobility most probably reflect restriction fragment polymorphism but could also represent DNA rearrangements. An additional nine epidemiologically unrelated clinical isolates of N. gonorrhoeae were analyzed by Southern blot and found to exhibit an identical distribution of the irg copies to one another (data not shown). Therefore, the number and chromosomal positions of the irg genes are extremely well conserved within this species. The commensal Neisseria species N. cineria and N. lactamica (Fig. 3, lanes 12 and 13) do not have any DNA sequences that hybridize with the irg probe. Therefore, the irg genes are found only in the pathogenic Neisseria species.
FIG. 3.
Distribution of irg copies in various isolates of N. gonorrhoeae. Southern analysis was performed with chromosomal DNA preparations that were digested with ClaI, electrophoresed on a 0.8% agarose gel, transferred to a nylon membrane, and probed with denatured 32P-labeled PCR product corresponding to the irg2 gene. N. gonorrhoeae strains are as follows: lane 1, FA19; lane 2, UU1; lane 3, RD5; lane 4, F62; lane 5, 1084; lane 6, 1918; lane 7, 1349; lane 8, 1402; lane 9, 1384. N. meningitidis strains are as follows: lane 10, GA0929; lane 11, NMB. Neisseria commensals are in lanes 12 (N. cineria) and 13 (N. lactamica). Negative control lanes are lanes 14 (M. lacunata) and 15 (pivML clone pAG702). The positive control, irg5 clone pAG852, is in lane 16.
The irg gene products have the motifs shown to be essential for Piv catalytic activity but do not complement for Piv-mediated inversion or MooV-mediated transposition.
Previous work has identified three amino acid motifs that are essential for Piv inversion of the Moraxella pilin DNA segment in E. coli (40). The amino acids of these required motifs in Piv include G, D, and K at positions 7, 9, and 12; KTD at positions 99 to 101; and the S and G residues at positions 240 and 241 (40). All eight irg gene products have retained these residues necessary for Piv catalytic activity (Fig. 2). We therefore asked whether purified protein could complement a Piv-mediated DNA inversion reaction or a MooV-mediated excision reaction. Irg2 and Irg7 were expressed in E. coli as His6-tagged proteins from the expression vectors pAG712 and pAG711, respectively, and expression was confirmed by Western blot analysis (Fig. 4A). The DNA substrate for the inversion assays in E. coli was pAG862, which encodes the M. lacunata invertible segment with the piv gene inactivated by insertion of a spectinomycin-streptomycin resistance cassette. Southern analysis of the restriction fragments from pAG862 that are diagnostic of inversion of the M. lacunata segment indicated no complementation of Piv activity by His6-tagged Irg2 or Irg7, while His6-tagged PivML mediated the expected level of inversion activity (Fig. 4B). An untagged form of Irg2 expressed from the lac promoter in pAG852 confirmed that the lack of complementation for Piv activity was not due to the His6 tag (data not shown).
FIG. 4.
Assays for complementation of Piv invertase and MooV transposase activities by Irg2 and Irg7 in E. coli. (A) Western blot analysis of His6-tagged MooV (MooV-H6), PivML (Piv-H6), Irg2 (Irg2-H6), and Irg7 (Irg7-H6) expression. Whole-cell lysates from induced (I) and uninduced (U) cultures were assayed on the Western blot as described in Materials and Methods. The proteins range in size from 318 to 322 amino acid residues without the His6 tag; the arrow marks the approximate location of the tagged Irg proteins. The molecular weights (in thousands) of protein standards (S) are indicated. (B) His6-tagged Irg7, Irg2, and the invertase PivML were assayed for invertase activity in HMS174(DE3) by using the Piv inversion substrate (pAG862), which contains the M. lacunata pilin invertible segment. Expression of the recombinases was induced and inversion products were isolated as described in Materials and Methods. Southern blot analysis of the pAG862 KpnI/SalI restriction fragments which are diagnostic for inversion of the pilin segment is shown. Arrows indicate the restriction fragments that are unique to the plasmid substrate that has undergone Piv-mediated inversion of the pilin segment, as illustrated. Tc, tetracycline. (C) His6-tagged MooV, Irg2, Irg7, and PivML were assayed for transposase activity in HMS174(DE3) by using the plasmid substrate pAG713, containing IS492ΔmooV::cat that is defective for excision due to the substitution of cat for most of the mooV transposase gene. Excision of IS492ΔmooV::cat to yield a circular element is detected in a PCR-based assay using primers (arrows) directed outward from the ends of the element which amplify the circlejunction. The 405-bp PCR product is indicated on the agarose gel by the arrow. Lane M contains pBR322/MspI (sizes in base pairs are shown). Ap, ampicillin; Kn, kanamycin.
Complementation of MooV transposase activity by His6-tagged Irg2 and Irg7 in HMS174(DE3) was assayed by using pAG713, which contains the IS492 element with mooV disrupted by cat. Excision of IS492 was detected in a PCR-based assay using outwardly directed primers from the ends of the element which give a product from the excised circular form of IS492 (21). His6-tagged MooV supported excision of IS492ΔmooV::cat to yield the circular form of the element, but neither of the Irg proteins was capable of complementing for MooV-mediated excision of the IS element (Fig. 4C).
The irg genes are not involved in pilin variation or general recombination.
To determine whether the Irg proteins may play a role in a recombination or repair process of the gonococcus, all eight irg genes were inactivated in FA1090 recA6 by using a mixture of antimicrobial resistance cassettes, minitransposons, and in-frame deletions to create strain FA1090 recA6 irg1-8, as confirmed by PCR and Southern blot analyses (data not shown). In addition, to determine whether overexpression of an irg gene alters recombination or repair processes, irg5 was placed under Plac control and introduced into an intergenic region on the chromosome of strain FA1090 recA6, creating FA1090 recA6 NICS6::irg5. The irg mutant and overexpressor strains were generated in a recA6 strain, in which recA expression is controlled by LacI (27), so that initiation of antigenic variation could be regulated. Because different pilE sequences can lead to changes in pilin-dependent phenotypes, including antigenic variation, the pilE genes of these various constructs were sequenced. All genes were found to be identical, ensuring that any observed differences were not caused by different starting pilE sequences.
Reverse transcription (RT)-PCR analysis of FA1090 recA6 total RNA showed that at least one of these irg genes produces a detectable transcript (data not shown). A quantitative RT-PCR assay for irg mRNA levels demonstrated that the overexpressing strain (FA1090 recA6 NICS6::irg5) showed an approximately sevenfold increase in total mRNA compared to that of the parental strain (FA1090 recA6) (Fig. 5). Therefore, the parental strain FA1090 recA6 irg1-8 and FA1090 recA6 NICS6::irg5 were used to determine whether elimination or overexpression of irg affected pilin antigenic variation.
FIG. 5.
Quantitative real-time PCR analysis of irg transcript levels in the overexpressed strain. Transcript levels were measured by using FRET probes in a LightCycler (Roche). Black bars are levels of irg transcripts, and gray bars are levels of recA transcripts included as a control. Total RNA was isolated from FA1090 recA6, in which recA gene expression is inducible by IPTG, and FA1090 recA6 NICS6::irg5, which overexpresses irg5 upon induction with IPTG.
A pilus-dependent colony variation frequency assay (34), which uses changes in colony morphology as a surrogate measure of antigenic variation at pilE, did not reveal any differences in the frequency of pilin variation between FA1090 recA6, FA1090 recA6 irg1-8, and FA1090 recA6 NICS6::irg5 (data not shown). This result was confirmed with a kinetic pilE variation assay that measures pilin-dependent changes in colony morphology over time (Sechman and Seifert, unpublished) (Fig. 6A). These results indicate that the irg copies are not involved in pilin antigenic variation and do not encode the putative recombinases used in this specialized recombination system.
FIG. 6.
Inactivation or overexpression of irg genes does not affect pilin antigenic variation, DNA transformation, or DNA repair. (A) Kinetic variation assay of N. gonorrhoeae measuring the number of sectors of a colony that undergo pilin-dependent colony variation over time. Error bars represent the standard errors of the means of at least four experiments analyzing 20 individual colonies per experiment. (B) DNA transformation competence measuring the introduction of a point mutation in gyrB that confers NAL resistance. Error bars represent the standard deviations from the means for at least four replicate experiments. (C) Relative survival after irradiation with UV light. Error bars represent the standard deviations from the means for four experiments.
To address the possibility that the irg genes encode recombinases involved in DNA transformation or DNA repair promoted by UV light irradiation, transformation assays and UV survival assays were performed on FA1090 recA6, FA1090 recA6 irg1-8, and FA1090 recA6 NICS6::irg5. Neither inactivation nor overexpression of the irg genes altered the frequency of DNA transformation (Fig. 6B) or survival to UV light (Fig. 6C). These results show that the irg genes do not encode recombinases involved in vegetative recombination or repair processes measured by these phenotypic assays.
Analyses of the sequences flanking the Irg coding sequences.
As members of the Piv/MooV family, the Irg proteins are likely to function as site-specific recombinases or as transposases for IS elements belonging to the IS110 family. Because our results indicate that the Irg proteins are not generalized recombinases, we investigated the possibility that the irg genes and their flanking sequences define IS elements. An unusual feature of most IS110-related elements is the absence of both target site duplication and terminal inverted repeats (4), making it difficult to define the ends of possible elements. Alignments of chromosomal sequences upstream and downstream of the irg genes, performed with the MegAlign tools of LaserGene Navigator, revealed that four (irg2, irg3, irg5, and irg6) of the eight irg genes are adjacent to long repeated sequences, encoding putative filamentous phage proteins (Fig. 7A). Preference for insertion next to repeated sequences or site-specific insertion has been reported for other members of the IS110 family (2, 21). The alignment of the flanking sequences for the irg genes reveals 14 bp of identity immediately upstream of the predicted start codons for irg2, irg3, irg4, irg5, irg6 (irg2-6), and irg8 and 133 bp of near-identity (1-bp difference in the irg8 flanking sequence) in the sequence downstream of the stop codons for irg1-6 and irg8 (Fig. 7B). The Irg proteins encoded by irg2-6 and irg8 are >99.3% identical in their primary amino acid sequence and, based on the homology in the flanking DNA sequence, are probably the transposases for an IS element belonging to the IS110 family. The name attributed to this putative element is ISNgo2 (http://www-is.biotoul.fr/IS.html). The termini of ISNgo2 cannot be definitively assigned until movement of the element has been characterized. However, because the putative ISNgo2 element containing irg2 is inserted in the repeated prophage sequence in the opposite orientation of the irg3-, irg5-, and irg6-associated ISNgo2 copies, the conserved phage sequence helps to delimit the left- and right-end sequences of the element (Fig. 7B and C). As illustrated in Fig. 7C, the 5′-ATA of the conserved left-end sequence appears to be part of the prophage target sequence and not part of ISNgo2. This definition of the left end makes ISNgo2 1,107 bp long with very short terminal inverted repeats of two base pairs (5′-AG). Possible target site duplications are not evident for five of the six copies, indicating that like other IS110-related elements, insertion of the element does not generate a duplication of target sequence. Examination of the conserved left-end sequence for a potential ribosome binding site for irg identified the sequence AGGG, ending 7 bp before the irg start codon. This sequence can base pair with the 3′ end of N. gonorrhoeae 16S RNA (8) but includes one GU base pair (Fig. 7B). An additional AU base pair is contributed by the flanking sequence.
FIG. 7.
Sequences flanking the irg copies and definition of ISNgo2 and ISNgo3. (A) The relative order and orientation of the irg genes are shown with flanking genes; gaps are indicated by //. The irg1 and irg8 genes are inserted into sequences predicted to encode a conserved hypothetical protein and a nitroreductase, respectively (dark gray arrows). Putative prophage genes zot, rstA, and tspB (light gray arrows) are described in Discussion; this repeated sequence is nearly identical over 6,874 bp upstream of irg3 and irg5, 4,836 bp downstream of irg2, and 3,319 bp upstream of irg6. Potential prophage sequence associated with irg7 differs from the sequence associated with other irg genes but still encodes a filamentous phage structural protein with a Zot domain (zot2). Parentheses indicate truncated genes or sequences. hrpA is a helicase. (B) Thirty-six base pairs of sequence 5′ to the start codon and 59 bp of sequence (with a 100-bp gap of identical sequence) 3′ to the stop codon of irg1-6 and irg8 are grouped together and aligned. Thirty-seven base pairs of sequence upstream from the start codon and 59 bp of sequence (with a 119-bp gap in which 3 bp of irg7-associated sequence differs from those of pivNM-1A and pivNM-1B) downstream of the stop codon of irg7, pivNM-1A, and pivNM-1B are aligned. The completely conserved base pairs are highlighted in gray; the proposed left and right ends for ISNgo2 and ISNgo3 are boxed with a black line. Interaction of the 3′ end of the N. gonorrhoeae 16S RNA with the proposed ribosome binding sites for the irg genes of ISNgo2 and ISNgo3 is shown below the aligned sequences. dRS3 repeat sequence is underlined, and the flanking prophage sequences are italicized. (C) The putative IS elements are shown with flanking sequences to illustrate their insertion relative to the repeated prophage sequences (italicized). The irg genes of ISNgo2 are black boxes, the left ends are white boxes, and the right ends are horizontally hatched boxes. The ISNgo3 elements, with the irg and pivNM genes as gray boxes and the left and right ends as white and diagonally hatched boxes, respectively, are inserted in the same orientation relative to prophage sequence (see above).
The irg1 nucleotide sequence exhibits 98% identity with irg2-6 and irg8 and has the conserved 133-bp sequence downstream of the stop codon. However, there are an additional 39 bp after the start codon of irg1, and the sequence upstream of irg1 diverges significantly from the flanking sequences for the other irg genes (Fig. 7B). Therefore, it is likely that irg1 encoded the transposase for an ISNgo2 element but that a deletion or insertion event altered the left end of the element and the coding sequence for the amino terminus of Irg1.
BLASTN searches of bacterial sequences in the NCBI database using each of the irg genes and their flanking sequences as query sequences revealed pivNM-1A and pivNM-1B and their surrounding sequences, including 1,874 bp of upstream and 155 bp of downstream sequence, as nearly identical to irg7. There were no other DNA sequences that showed significant homology to the other irg genes and their flanking sequences at the DNA level. As described above, while all the Irg proteins possess approximately the same level of homology with the Piv/MooV family proteins (∼20 to 30% identity), Irg2-6 and Irg8 all share >99.3% amino acid identity with each other but only 40% identity with Irg7 (Fig. 2). Also, the sequence flanking irg7 is not homologous to that of irg2-6 and irg8. Alignment of irg7, pivNM-1A, and pivNM-1B and their flanking sequences predicts a putative 1,124-bp IS110-related element, designated ISNgo3 (http://www-is.biotoul.fr/IS.html) (Fig. 7B). The left terminus of this element is uncertain but is currently defined by homology with the left end of the ISNgo2 element. There are no terminal inverted repeats and no apparent target site duplications.
Comparison of the sequences flanking the putative elements reveals that both ISNgo2 and ISNgo3 may prefer repeated elements as target sites or have a preferred target sequence. Four of the six putative ISNgo2 elements (containing irg genes irg2, irg3, irg5, and irg6) are inserted at exactly the same site in the repeated prophage sequence (Fig. 7). One of these elements (irg5) is inserted between the prophage and type dRS3 repeat sequences (42) (Fig. 7B). The irg8-associated element is inserted into a homologue of the putative nitroreductase genes from N. meningitidis strains MC58 and Z2491 (GenBank accession numbers NC_003112 and NC_003116). Based on the N. meningitidis sequence for the uninterrupted nitroreductase gene, the ISNgo2 element is immediately adjacent to a palindromic 34-bp sequence, 5′-TATATTGCGCCCCTTCTTAGGGACGCAATATATA-3′, that is present in the nitroreductase gene found in N. gonorrhoeae. Interestingly, the irg4-associated ISNgo2 copy is inserted in the same orientation relative to a sequence that is identical to 27 bp of this 34-bp palindromic sequence which is located within a truncated copy of the IS1016 transposase from N. meningitidis MC58 (Fig. 7A). The putative ISNgo3 elements are found adjacent to identical repeated prophage sequences in both N. gonorrhoeae and N. meningitidis (Fig. 7B and C).
DISCUSSION
Previous work by Carrick et al. (3) indicated that a gene encoding a protein with significant homology to the site-specific recombinase Piv, which controls type IV pilin antigenic and phase variations in M. bovis and M. lacunata, respectively, is found in multiple copies on the N. gonorrhoeae chromosome. Using in silico analysis of the fully sequenced N. gonorrhoeae genome and additional Southern blot analyses, we have demonstrated that there are eight invertase-related genes (irg1-8) clustered in three separate regions of the N. gonorrhoeae chromosome. Comparison of Irg1-8 with each other and with other members of the Piv/MooV family of DNA recombinases showed that Irg2-6 and Irg8 are nearly identical, while Irg7 diverged significantly from the other Irg proteins. However, all eight Irg proteins have retained the residues that have been shown to be required for Piv catalytic activity in an E. coli model system for the inversion reaction. These residues include the putative DED motif that is analogous to the DDE catalytic motif of the retroviral integrases and most IS-transposon element transposases (40). This conservation of the Piv catalytic residues, as well as the significant degree of homology with the Piv-related recombinases, suggested that the irg genes have the potential to encode recombinases.
At least one of the irg copies is actively transcribed in N. gonorrhoeae, as demonstrated by RT-PCR experiments using a primer that would detect any of the irg2-6 or irg8 gene transcripts. Possible recombinase activities for irg2 and the divergent irg7 gene products were examined in vivo (in E. coli) using plasmid-based systems for Piv-mediated site-specific inversion of the M. lacunata pilin segment and for MooV-mediated excision of IS492. Neither of the two homologues could complement Piv or MooV activities in these systems. While there are a number of possible reasons why complementation was not seen, it is most likely due to differences in DNA binding or cleavage site specificity rather than catalytic activity; Piv and MooV cannot complement for each other, as well (41). There is very little similarity between the Piv-binding sites within the inversion region of M. lacunata and the flanking sequences of the irg genes (data not shown), and the sites for MooV binding and DNA cleavage on IS492 have not yet been determined. To determine if the irg genes code for recombinases involved in antigenic variation at pilE, the eight irg copies were inactivated by using a combination of molecular techniques. To further this analysis, a strain was created which overexpressed irg5. Neither overexpression of irg5 nor inactivation of the eight irg copies affects the frequency of antigenic variation at pilE. We conclude that the hypothesis that the irg genes are recombinases involved in pilin antigenic variation is incorrect. Additionally, the irg genes do not code for recombinases involved in DNA transformation or DNA repair.
Although an involvement in site-specific recombination reactions elsewhere in the gonococcus cannot be ruled out, it is most likely that the irg genes encode transposases for IS110-related insertion sequences in the chromosome of N. gonorrhoeae. In support of this hypothesis, all the irg gene products exhibit significant homology to the transposases of the IS110 family of insertion elements. In addition, a high degree of amino acid sequence identity (>99%) is seen among the irg gene products proposed to act as the transposase for ISNgo2 and among the gene products of irg7, pivNM-1A, and pivNM1-B, which are the predicted transposases for ISNgo3. As would be predicted for an IS element, the sequences flanking the proposed transposases for ISNgo2 are identical in each copy, with the exception of only a 1-bp difference found in the right-end sequence of the irg8-associated copy. Nearly the same level of nucleotide sequence identity is seen for the predicted left and right ends of the three ISNgo3 copies. There are no conserved features that define the ends or target sites of IS110-related elements; in general, there are limited or no terminal inverted repeats, and the element does not create target site duplications. Consistent with belonging to the IS110 family, the putative ISNgo2 element has 2-bp terminal inverted repeats and ISNgo3 has no terminal inverted repeats; in addition, neither element appears to generate target site duplications. While there is no common nucleotide sequence for the termini of all the IS110-related elements, there are subgroups to the family in which the IS elements share nucleotide motifs at or near the termini that may be involved in transposase recognition and cleavage (20). We found some nucleotide sequence identity at or near the ends of IS492 with the proposed termini of the ISNgo2 element. For example, 5′-AGGNNAATC-3′ is at the left terminus of ISNgo2 and 19 bp from the left end of IS492. Also, 5′-TAGTNTCT-3′ is at the right terminus of ISNgo2 and the juncture of the right end of IS492 and the 5-bp target sequence. The significance of homology between the ends of IS492 and ISNgo2 will not be evident until we have further characterized the features and functionality of the ISNgo2 element.
We cannot conclude that ISNgo2 and ISNgo3 are active IS elements, since we have not observed movement. Carrick et al. (3) reported that the mobility of irg-hybridizing restriction fragments changes during the passage of strain MS11. We have attempted to repeat this observation with both MS11 and FA1090. After 2 to 3 weeks of daily serial passage, neither strain showed a change in the mobilities of irg-hybridizing fragments (data not shown). However, the association of the irg genes with repeated sequences is a feature of the N. meningitidis IS elements IS1016, IS1106, and IS1655, which are preferentially inserted in or near repeat DNA elements (19). In addition, it has recently been reported that the IS110-related elements IS621 and IS1594 target repetitive extragenic palindromic-like repeated sequences in E. coli and Anabaena sp. strain PCC7120, respectively (5).
ISNgo2 and ISNgo3 are the only new IS elements assigned to the N. gonorrhoeae genome since the discovery of the Correia elements (6). Interestingly, four of the six ISNgo2 copies and the one copy of ISNgo3 are inserted adjacent to degenerate prophage sequences that are found repeated on the N. gonorrhoeae and N. meningitidis chromosomes (23). Based on BLASTX analysis, these ISNgo-associated prophage sequences are predicted to encode a filamentous phage structural protein that has a Zot domain (zonular occludens toxin [zot]), a putative surface antigen (tspB), and a filamentous phage replication protein (rstA). Zot was characterized from the filamentous phage CTXφ of pathogenic Vibrio cholerae and has the unusual activity of reversibly altering intestinal epithelial tight junctions, allowing macromolecules to pass through mucosal barriers (8, 44). The tspB-like gene exhibits approximately 80% identity with tspB from N. meningitidis which is reported in NCBI (accession number NP_284510) and EMBL (accession number O87783) databases to encode a potent T-cell- and B-cell-stimulating Neisseria-specific antigen. The role, if any, of these prophage-encoded proteins in the pathogenesis of N. gonorrhoeae is unknown.
We are certain from the inactivation and overexpression data that the irg genes have no role in pilin variation, thereby disproving the hypothesis that this gene family has a direct role in this immune evasion mechanism. However, the presence of ISNgo2 and ISNgo3 in the pathogenic Neisseria, but not the commensal Neisseria, and their association with potential prophage-encoded virulence factors suggest that they could be associated with virulence properties. The observation that N. meningitidis has copies of the ISNgo3 element, but not ISNgo2, is suggestive of the evolutionary history, although it is not predictive. Although several scenarios could explain the acquisition of these elements in the pathogenic Neisseria, our preferred model is that ISNgo3 was acquired as part of the bacteriophage genome by a progenitor of the pathogenic Neisseria after it diverged from the commensal species and that ISNgo2 was acquired separately by the progenitor of N. gonorrhoeae after it diverged from N. meningitidis. The ISNgo2 element inserted into preferred target sites associated with the now repeated, degenerate prophage sequence. The possible selective force for duplication of prophage sequences could have been increased expression of the Zot, TspB, or RstA protein, if these phage-associated proteins play a role in gonococcal pathogenesis. Regardless of the evolutionary pathway to the contemporary organisms, these sequences could be used as diagnostic markers for the pathogenic Neisseria.
Acknowledgments
This work was supported by Public Health Service grants R01-AI33493, R01-AI44239 (H.S.S.), and R01-GM049794 (A.C.K.). E.P.S. was partially supported by the Public Health Service training grant T32 GM08061.
We acknowledge the Gonococcal Genome Sequencing Project, B. A. Roe, L. Song, S. P. Lin, X. Yuan, S. Clifton, T. Ducey, L. Lewis,and D. W. Dyer at the University of Oklahoma (supported by USPHS/NIH grant number AI38399), particularly for assistance in assigning the irg open reading frames. We thank William Shafer for supplying the neisserial DNA. We appreciate the assistance of Deborah Tobiason, Amanda Ozin, and Jonathan Kim in cloning the irg genes and analyses of irg recombinase activity. We thank Kimberly Kline, Eric Sechman, and Brian Higgins for critical reading of the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett, D. H., M. E. Wright, and M. Silverman. 1988. Variable expression of extracellular polysaccharide in the marine bacterium Pseudomonas atlantica is controlled by genome rearrangement. Proc. Natl. Acad. Sci. USA 85:3923-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrick, C. S., J. A. Fyfe, and J. K. Davies. 1998. Neisseria gonorrhoeae contains multiple copies of a gene that may encode a site-specific recombinase and is associated with DNA rearrangements. Gene 220:21-29. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 5.Choi, S., S. Ohta, and E. Ohtsubo. 2003. A novel IS element, IS621, of the IS110/IS492 family transposes to a specific site in repetitive extragenic palindromic sequences in Escherichia coli. J. Bacteriol. 185:4891-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correia, F. F., S. Inouye, and M. Inouye. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194-12198. [PubMed] [Google Scholar]
- 7.Eckert, C., O. Landt, T. Taube, K. Seeger, B. Beyermann, J. Proba, and G. Henze. 2000. Potential of LightCycler technology for quantification of minimal residual disease in childhood acute lymphoblastic leukemia. Leukemia 14:316-323. [DOI] [PubMed] [Google Scholar]
- 8.Fasano, A., B. Baudry, D. W. Pumplin, S. S. Wasserman, B. D. Tall, J. M. Ketley, and J. B. Kaper. 1991. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc. Natl. Acad. Sci. USA 88:5242-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas, R., and T. F. Meyer. 1986. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44:107-115. [DOI] [PubMed] [Google Scholar]
- 10.Hagblom, P., E. Segal, E. Billyard, and M. So. 1985. Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315:156-158. [DOI] [PubMed] [Google Scholar]
- 11.Howell-Adams, B., and H. S. Seifert. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146-1158. [DOI] [PubMed] [Google Scholar]
- 12.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koomey, J. M., and S. Falkow. 1987. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J. Bacteriol. 169:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koomey, M., E. C. Gotschlich, K. Robbins, S. Bergstrom, and J. Swanson. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenich, A. G., and A. C. Glasgow. 1994. Amino acid sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J. Bacteriol. 176:4160-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrs, C. F., F. W. Rozsa, M. Hackel, S. P. Stevens, and A. C. Glasgow. 1990. Identification, cloning, and sequencing of piv, a new gene involved in inverting the pilin genes of Moraxella lacunata. J. Bacteriol. 172:4370-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehr, I. J., C. D. Long, C. D. Serkin, and H. S. Seifert. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 20.Partridge, S. R., and R. M. Hall. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185:6371-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins-Balding, D., G. Duval-Valentin, and A. C. Glasgow. 1999. Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol. 181:4937-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 23.Roe, B. A. 1997. Gonococcal Genome Sequencing Project. University of Oklahoma Health Sciences Center, Oklahoma City, Okla. http://www.genome.ou.edu/gono.html.
- 24.Rozsa, F. W., T. F. Meyer, and M. Fussenegger. 1997. Inversion of Moraxella lacunata type 4 pilin gene sequences by a Neisseria gonorrhoeae site-specific recombinase. J. Bacteriol. 179:2382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Segal, E., P. Hagblom, H. S. Seifert, and M. So. 1986. Antigenic variation of gonococcal pilus involves assembly of separated silent gene segments. Proc. Natl. Acad. Sci. USA 83:2177-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seifert, H. S. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215-220. [DOI] [PubMed] [Google Scholar]
- 28.Seifert, H. S., R. S. Ajioka, D. Paruchuri, F. Heffron, and M. So. 1990. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J. Bacteriol. 172:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seifert, H. S., C. J. Wright, A. E. Jerse, M. S. Cohen, and J. G. Cannon. 1994. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J. Clin. Investig. 93:2744-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serkin, C. D., and H. S. Seifert. 1998. Frequency of pilin antigenic variation in Neisseria gonorrhoeae. J. Bacteriol. 180:1955-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serkin, C. D., and H. S. Seifert. 2000. Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol. Microbiol. 37:1075-1086. [DOI] [PubMed] [Google Scholar]
- 32.Skaar, E. P., M. P. Lazio, and H. S. Seifert. 2002. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J. Bacteriol. 184:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein, D. C. 1991. Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can. J. Microbiol. 37:345-349. [DOI] [PubMed] [Google Scholar]
- 34.Stohl, E. A., and H. S. Seifert. 2001. The recX gene potentiates homologous recombination in Neisseria gonorrhoeae. Mol. Microbiol. 40:1301-1310. [DOI] [PubMed] [Google Scholar]
- 35.Swanson, J. 1973. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J. Exp. Med. 137:571-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson, J., K. Robbins, O. Barrera, D. Corwin, J. Boslego, J. Ciak, M. Blake, and J. M. Koomey. 1987. Gonococcal pilin variants in experimental gonorrhea. J. Exp. Med. 165:1344-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson, J., K. Robbins, O. Barrera, and J. M. Koomey. 1987. Gene conversion variations generate structurally distinct pilin polypeptides in Neisseria gonorrhoeae. J. Exp. Med. 165:1016-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 39.Tinsley, C. R., and J. E. Heckels. 1986. Variation in the expression of pili and outer membrane protein by Neisseria meningitidis during the course of meningococcal infection. J. Gen. Microbiol. 132:2483-2490. [DOI] [PubMed] [Google Scholar]
- 40.Tobiason, D. M., J. M. Buchner, W. H. Thiel, K. M. Gernert, and A. C. Karls. 2001. Conserved amino acid motifs from the novel Piv/MooV family of transposases and site-specific recombinases are required for catalysis of DNA inversion by Piv. Mol. Microbiol. 39:641-651. [DOI] [PubMed] [Google Scholar]
- 41.Tobiason, D. M., A. G. Lenich, and A. C. Glasgow. 1999. Multiple DNA binding activities of the novel site-specific recombinase, Piv, from Moraxella lacunata. J. Biol. Chem. 274:9698-9706. [DOI] [PubMed] [Google Scholar]
- 42.van der Ende, A., C. T. Hopman, and J. Dankert. 1999. Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect. Immun. 67:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wainwright, L. A., J. V. Frangipane, and H. S. Seifert. 1997. Analysis of protein binding to the Sma/Cla DNA repeat in pathogenic Neisseriae. Nucleic Acids Res. 25:1362-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 45.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-131, 134-138. [DOI] [PubMed] [Google Scholar]
- 46.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]
- 47.Zak, K., J. L. Diaz, D. Jackson, and J. E. Heckels. 1984. Antigenic variation during infection with Neisseria gonorrhoeae: detection of antibodies to surface proteins in sera of patients with gonorrhea. J. Infect. Dis. 149:166-174. [DOI] [PubMed] [Google Scholar]