Abstract
We have shown previously that when the Escherichia coli chromosomal lacZ gene is put under the control of an extended Shine-Dalgarno (SD) sequence (10 or 6 nucleotides in length), the translation efficiency can be highly variable, depending on the presence of AU-rich targets for ribosomal protein S1 in the mRNA leader. Here, the same strains have been used to examine the question of how strong ribosome binding to extended SD sequences affects the stability of lacZ mRNAs translated with different efficiencies. The steady-state concentration of the lacZ transcripts has been found to vary over a broad range, directly correlating with translation efficiency but not with the SD duplex stability. The observed strain-to-strain variations in lacZ mRNA level became far less marked in the presence of the rne-1 mutation, which partially inactivates RNase E. Together, the results show that (i) an SD sequence, even one that is very long, cannot stabilize the lacZ mRNA in E. coli if translation is inefficient; (ii) inefficiently translated lacZ transcripts are sensitive to RNase E; and (iii) AU-rich elements inserted upstream of a long SD sequence enhance translation and stabilize mRNA, despite the fact that they constitute potential RNase E sites. These data strongly support the idea that the lacZ mRNA in E. coli can be stabilized only by translating, and not by stalling, ribosomes.
As a rule, prokaryotic mRNAs are unstable, with half-lives in the minute range. In Escherichia coli, pathways of mRNA decay are complex and include an initial rate-limiting endonucleolytic cleavage (generally mediated by RNase E) followed by 3′→5′ exonucleolytic degradation of the cleavage products (reviewed in references 6, 10, 19, and 34). For several mRNAs, decay pathways have been studied in detail, allowing molecular models to be formulated (4-6, 11, 13, 28). Several lines of evidence suggest that translating ribosomes usually protect the mRNA from endonucleolytic attacks (10). Much of the available information has been derived from experiments with lacZ translational fusions. In this case, mRNA translation and degradation are intimately correlated. In particular, mutations in the ribosome binding site (RBS) which reduce translation initiation efficiency, and hence efficiency of the overall translation, accelerate mRNA degradation (14, 22, 38). However, despite all efforts, the mechanism of the lacZ mRNA decay still remains poorly characterized.
Although in many cases translation efficiency positively correlates with stability of the lacZ mRNA in E. coli, the question of whether merely the ribosome binding to the RBS is able to stabilize the whole transcript remains equivocal. On one hand, it has been suggested that a ribosome bound to a strong Shine-Dalgarno (SD) sequence, but not translation per se, could be the main stability factor for the lacZ mRNA (37). This would imply that E. coli mRNAs can be protected from degradation by the same mechanism as in bacilli, where ribosome stalling within 5′ untranslated regions can protect kilobases of the downstream mRNA (1, 7). In contrast, other data (15) show that a lacZ mRNA which carries an extended SD sequence but no initiation codon, i.e., which can bind a ribosome but cannot be translated, is not more stable than its counterpart lacking an SD sequence. Both mRNAs are very sensitive to RNase E, a key player in mRNA degradation in E. coli (25, 27), suggesting that the ribosome tightly bound to a strong SD does not represent a substantial barrier for this enzyme (10, 15).
Here, we further address this question with a standard system in which the lacZ mRNA is translated normally. Earlier, we created specialized E. coli strains in which the lacZ transcripts synthesized from the chromosomal lac promoter carry extended SD sequences (6 to 10 nucleotides [nt] in length), ensuring strong ribosome binding but not necessarily a high translation yield. When AU-rich sequences serving as targets for ribosomal protein S1 were inserted upstream of the SD sequence, the translation yield was increased significantly (18). Here, we show that the steady-state level and the half-life of the lacZ mRNA in these strains change in parallel with the β-galactosidase activity, thus indicating a positive correlation between translation efficiency and lacZ mRNA stability. The data obtained strongly support the idea that only translating ribosomes can protect the lacZ mRNA from rapid decay in E. coli.
MATERIALS AND METHODS
Bacterial strains.
E. coli strains AKSD10H (formerly IBCENS-SD1/2 [17]), AKSD10, AKSD6, BASD10, CompBASD10, MutBASD10, BASD6, and CompBASD6 are derivatives of ENSO (formerly HfrG6Δ12 [9]) and were described previously (17, 18). The ams/rne-1(Ts) mutation (25, 27) was P1 transduced into these strains by selecting for tetracycline resistance and thermosensitive growth.
To change the initiation codon in front of the lacZ gene in AKSD6 from a strong AUG to a weak UUG, a two-step PCR technique was used. In the first step, two PCR fragments were obtained with pSD6 (18) as a template and two pairs of primers, UPlac-SD6UUG_comp and SD6UUG_for-DSlac. UPlac covers positions −64 to −41 with respect to the start point of lac operon transcription, and DSlac is complementary to positions +57 to + 82 of the lacZ mRNA (+1 is the A in the lacZ start codon). SD6UUG_for is 5′-CCAAAGGAGAAAAACTTGAAACAAAGC, and SD6UUG_comp is complementary to this sequence. In the second step, the two PCR products were mixed and amplified in the presence of UPlac and DSlac. Finally, the resulting fragment was treated with BamHI and HindIII and then cloned in pEMBLΔ46 to generate pSD6UUG. The corresponding ENSO strain SD6UUG was obtained by homologous recombination.
Cell growth and β-galactosidase assays.
Unless otherwise stated, cells were grown at 37°C in Luria-Bertani medium in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside) (0.2 mM) and harvested in the exponential phase (A600 of ca. 0.4 to 0.6). β-Galactosidase activities in clarified cell lysates were measured as described previously (18) and expressed in nanomoles of o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolyzed per minute per milligram of total soluble protein. In experiments with the rne-1 mutants, cells were grown at 30°C to and A600 of ca. 0.3 in LB medium supplemented with IPTG (0.2 mM) and tetracycline (10 μg/ml) and then transferred to 42°C for 30 min. Control cultures were grown continuously at 30°C.
RNA isolation and Northern blotting.
To quantify the steady-state level of lacZ mRNA, aliquots of the cultures used for β-galactosidase measurements were collected, and total RNA was isolated essentially as described previously (12). RNA samples (10 μg) were separated on 1.6% agarose-formaldehyde gels and vacuum blotted onto nylon membranes (21). To visualize the lacZ transcripts, the membrane was probed with the 1.8-kb HincII fragment of lacZ uniformly labeled with 32P (internal probe). After washing, the blots were reprobed with the 5′-labeled oligonucleotide GATCCCAAATTGTTATCCGCTCACAATT, complementary to the first 28 nt common to all lacZ transcripts (5′ probe), and with the 5′-labeled oligonucleotide AAGGTTAAGCCTCACGGTTC, complementary to the 3′ region of 23S rRNA. Hybridization signals specific for 23S rRNA served as an internal standard to correct lane-to-lane variations in RNA loading. All procedures were carried out as described by Lopez et al. (21). Hybridization signals were quantified with a Fuji FLA3000 Imager.
To evaluate the half-lives of lacZ mRNAs, transcription initiation in early-log-phase cultures (optical density at 600 nm of 0.5) was arrested by addition of rifampin to 400 μg/ml. Aliquots were drawn from cultures at serial time points (0, 1, 2, 4, 6, and 8 min) after rifampin addition for analysis of RNA decay. RNA isolation and Northern blot analysis were performed as described above.
RESULTS AND DISCUSSION
Strong SD sequences do not necessarily ensure a high translation yield.
Earlier, we created a series of specialized strains in which expression of the chromosomal lacZ gene is controlled at the transcriptional level by the Plac promoter-operator region and at the translational level by artificial ribosome binding sites varying in the length of the SD sequence (from 6 to 10 nt) and in the nucleotide context downstream (17) or upstream (18) of the SD sequence. The longest, 10-nt SD sequence was found to be the least efficient in driving protein synthesis despite the fact that it tightly binds ribosomes in vitro (18). The β-galactosidase activity in this case was several times lower than that in the presence of the 6-nt SD sequence, indicating that excessive stability of the SD duplex may inhibit translation, presumably by interfering with the RBS clearance necessary for the next initiation event. In all cases, insertions of AU-rich targets for protein S1 upstream of the SD sequence significantly increased the translation yield, with the enhancing effect being most pronounced for the 10-nt SD sequence. Thus, these constructs can be very useful for elucidating how the length of the SD sequence, on one hand, and the efficiency of translation, on the other, affect the stability of the lacZ mRNAs in E. coli.
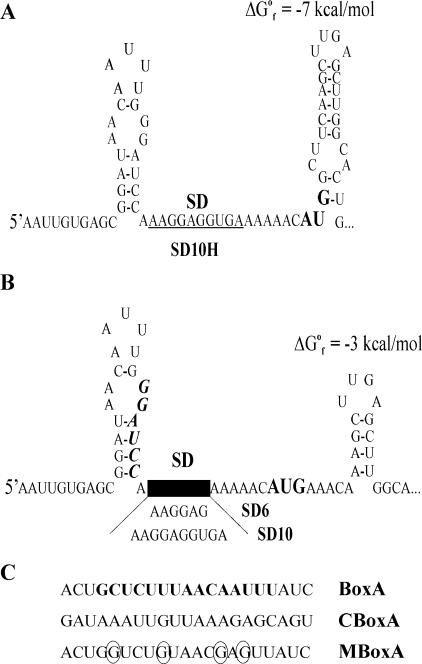
Structure predictions suggest that in all constructs the extended SD sequences are single stranded and hence are accessible for ribosome binding (Fig. 1). In SD10H, the AUG initiation codon is sequestered in a local secondary structure (Fig. 1A), which might explain the lowest translatability of the corresponding lacZ mRNA. In all other constructs, the AUG is single stranded due to the alteration of the second and third codons (GCU→AAA and UCA→CAA, respectively). The significant increase (about fourfold) in translation activity in SD10 compared to SD10H (17) can most likely be attributed to the removal of the inhibitory secondary structure in the vicinity of the start codon. However, a positive effect of the AAA triplet itself cannot be excluded either, since the AAA was shown to be optimal as a second codon (30). The BoxA-SD constructs bear the transcriptional antiterminator BoxA from the rrnB operon inserted upstream of the SD sequence. This AU-rich element has been shown to be a target for protein S1 (18, 24). In CBoxA-SD, the BoxA insert is changed to a complementary sequence retaining a comparable affinity for S1. MBoxA-SD carries four mutations in BoxA reducing this affinity (Fig. 1C). Presumably due to the increased affinity of mRNA leaders for S1, both BoxA and CBoxA act as translational enhancers (18). Consistent with this interpretation, the replacement of BoxA by MBoxA reduces this enhancing effect.
FIG. 1.
Structures of the 5′ regions of chromosomally encoded lacZ mRNAs bearing extended SD sequences. (A) SD10H, construct carrying the start AUG codon (boldface) partly involved in a stable stem-loop structure. The 10-nt SD sequence is underlined. (B) SD10 and SD6 constructs, comprising a single-stranded AUG codon (boldface). The position of the BamHI site used to insert the AU-rich S1 targets is shown in boldface italic. (C) Inserts upstream of the SD sequence which serve as S1 targets (18): BoxA, rrnB transcriptional antiterminator BoxA (boldface) with natural flanking regions; CBoxA, an insert complementary to BoxA; MBoxA, a mutated variant of BoxA with a lower affinity to S1 (mutated nucleotides are circled).
Correlation between translation efficiency and lacZ mRNA level during exponential growth.
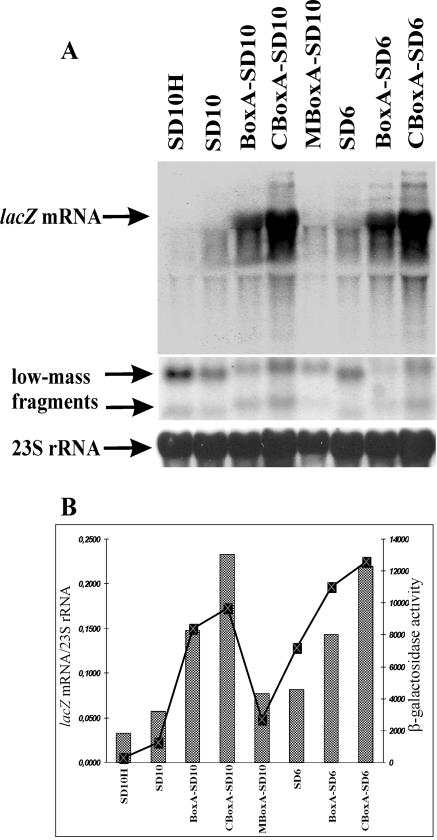
The effects of variations in SD sequence length or S1 targets on the steady-state concentration of the full-length lacZ mRNA were then examined. This concentration was measured on Northern blots with a lacZ internal probe (Fig. 2A). The intensity of the lacZ signals was further normalized to that of 23S rRNA in the same lane. These relative intensities varied markedly with the RBS used (Fig. 2B). The SD10H construct, which is the weakest in translation, showed the faintest full-length lacZ mRNA signal (Fig. 2). For the SD10 and SD6 transcripts, the full-length signals were also weaker than those for their more efficiently translated counterparts bearing AU-rich S1 targets, BoxA and CBoxA (Fig. 2). Moreover, mutations within BoxA that decreased its affinity for S1 (18) decreased not only the translation yield but also the level of the full-length lacZ mRNA (MBoxA-SD10 in Fig. 2). Together, the steady-state levels of the full-length lacZ transcripts clearly show positive correlation with the β-galactosidase activities in the same series of strains (Fig. 2).
FIG. 2.
Steady-state level and translation efficiency of lacZ mRNA depend on the structure of its translation initiation region but not simply on the length of the SD sequence. Translation initiation regions in front of lacZ are SD10H, SD10, BoxA-SD10, CBoxA-SD10, MBoxA-SD10, SD6, BoxA-SD6, and CBoxA-SD6 (Fig. 1). (A) Northern blot analysis of lacZ transcripts. Total RNA (10 μg) was analyzed on Northern blots. The full-length lacZ mRNA and two low-mass 5′-terminal RNA fragments (indicated by arrows) were revealed with an internal probe and a 5′-probe, respectively (see Materials and Methods). The bottom panel shows hybridization of the same membrane with the 23S rRNA-specific probe used as an internal control. (B) Relative abundances of the lacZ mRNAs (bars) directly correlates with their translation efficiency (graph). Relative abundance was evaluated as a ratio of a hybridization signal from the full-length lacZ mRNA to that from 23S rRNA. β-Galactosidase activities in strains (graph) are expressed in nanomoles of ONPG hydrolyzed per minute per milligram of total soluble cell protein. Averages from three independent assays are shown, with standard deviations not exceeding 15% of magnitude (18).
Strain-to-strain variations in lacZ mRNA level reflect differences in mRNA stability.
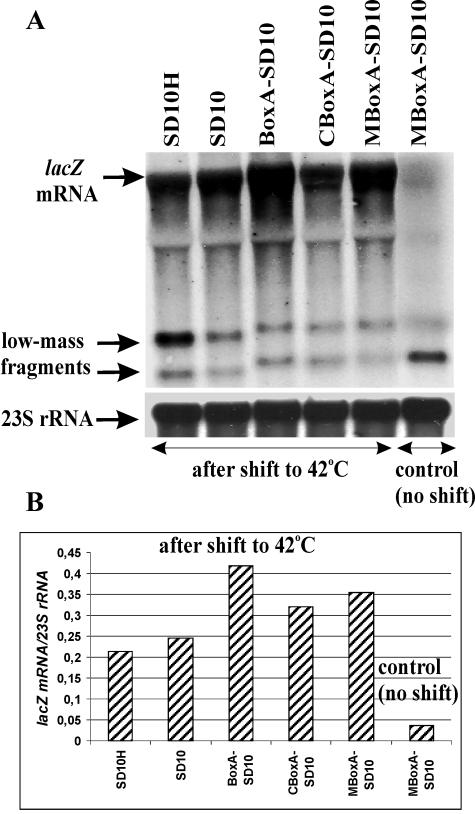
The low abundance of the lacZ mRNA in the case of inefficient translation can be accounted for either by a low stability (an increased accessibility to RNase E) or by transcription polarity (33, 38). To distinguish between these two possibilities, we introduced the ams/rne-1 mutation in our strains by P1 transduction. This mutation results in temperature-sensitive RNase E activity (25, 27). Northern blot analysis of RNA isolated after shifting the cultures to 42°C (see Materials and Methods) revealed that inactivation of RNase E largely reduces the differences in mRNA abundance caused by the different translation efficiencies in rne+ strains (Fig. 3). This result indicates that the strain-to strain variations in the lacZ mRNA level observed in the rne+ background reflect mainly variations in mRNA stability, thus further confirming the major role of RNase E in degrading the lacZ mRNA in E. coli observed earlier with other genetic constructions (14, 15, 38).
FIG. 3.
Northern blot analysis of the lacZ transcripts in strains bearing the rne-1(Ts) allele. Cultures were grown at 30°C to early log phase (A600 of 0.3) and then shifted for 30 min to 42°C to inactivate RNase E before RNA isolation. Control cultures (only that for MBoxA-SD10 is shown) were grown at 30°C without a temperature shift. (A) A representative Northern blot for total RNA (about 10 μg) extracted from rne1 strains (indicated above the lanes). RNA was analyzed as described for Fig. 2. (B) The relative abundances of lacZ transcripts in rne1 strains are higher than that in the rne+ background (see Fig. 2B). Averages from two independent Northern blots are shown, with deviations not exceeding 20% of magnitudes.
Premature termination of transcription within the lacZ gene.
Besides the full-length lacZ RNA, two low-molecular-weight species, i.e., a major one ca. 0.42 kb and a minor one ca. 0.25 kb in length, were detected when a lacZ 5′ probe was used (Fig. 2A). The larger fragment is much more abundant in the case of the weakest construct, SD10H, than in other constructs, and its level inversely correlates with translation efficiency in the strains. Regarding the origin of these fragments, two possibilities must be considered: either they are stable intermediates of the RNase E-mediated decay, emerging when 3′→5′ exonucleolytic degradation of the initial cleavage products encounters a stable secondary structure, or they result from transcription polarity, i.e., from premature termination of transcription caused by inefficient translation (33, 38). As a test of this issue, we examined the level of 5′-terminal species in the rne-1 background. While the steady-state level of the full-length SD10H transcript increased notably after inactivation of RNase E (compare Fig. 2A and Fig. 3A), the abundance of the 5′-terminal upper fragment in this case remained the highest among the constructs (Fig. 3A), indicating that this species is not related to the RNase E-mediated decay. Thus, it seems likely that its accumulation occurs due to transcription polarity. Indeed, the sequestration of the start codon in the SD10H transcript (Fig. 1A) should delay formation of the true initiation complex and impede the transition to elongation that must result in the extended spacing between the RNA polymerase and the elongating ribosome, increasing the extent of premature termination (33).
The accumulation of 5′ lacZ fragments has been repeatedly reported (15, 29, 33). Ruteshouser and Richardson (29) have mapped their extremities and analyzed mechanisms for their formation. They have found the 421-nt fragment to originate from transcription termination at the unique intrinsic Rho-independent terminator within the lacZ coding region. The length of our abundant upper fragment (ca. 420 nt [not shown]) is consistent with the position of this Rho-independent termination, suggesting that this fragment represents a transcription abortion product. Interestingly, this unusual terminator is not formed by a local palindromic unit, as is typical for most Rho-independent terminators, but arises from a very stable long-range pairing between positions 1 to 22 and 401 to 421 of the lacZ transcript (29), with the pairing being strong enough to be cleaved by RNase III in vitro (31). As we noticed, these regions correspond to locations of a principal lac operator, O1, and a so-called pseudo-operator, O2, which have high sequence similarity but opposite orientations (26), thus ensuring the stable pairing at the RNA level. Because translating ribosomes behind the RNA polymerase will interfere with such a long-distance interaction, termination at this site must inversely correlate with translation efficiency, which is just what we observe for the upper 5′ fragment (Fig. 2A and 3A).
The origin of the minor fragment (ca. 0.25 kb in length) is less clear. There is a possibility that it originates from Rho-dependent transcription termination typical for the early lacZ sequence (29, 33). However, the same band was observed on the Northern blots when lacZ transcripts were generated by the fast T7 RNA polymerase, which is insensitive to Rho-dependent terminators (15). It therefore seems more likely that this species is a decay intermediate of the larger fragment, arising when the 3′→5′ degradation machinery encounters a stable secondary structure. Formation of a long, stable stem-loop covering positions 215 to 269 of the lacZ transcript has been reported (29, 31), arguing in favor of the latter interpretation.
The abundance of the lacZ mRNA is a measure of its stability in a cell.
The steady-state level of an mRNA in a cell depends on the balance between its rates of synthesis and decay. It is evident that, in our strains, construct-to-construct variations in the lacZ mRNA level reflect primarily variations in its stability. Indeed, in all cases the lacZ mRNA is transcribed from the same lac promoter and bears the same 5′ terminus, so that the transcripts vary only in the structure of the translation initiation region. Furthermore, the large variations in lacZ mRNA abundance visibly decrease when RNase E is inactivated (Fig. 3), indicating that in rne+ cells these variations reflect different susceptibilities to RNase E-mediated cleavages, i.e., different stabilities. An interesting exception is the BoxA-containing mRNA: in rne-1 cells this mRNA is more abundant than those from other constructs. As shown earlier by Vogel and Jensen (36), the presence of the transcription antiterminator BoxA in the lacZ leader elevates the rate of transcription about twofold, which could explain the higher mRNA level in this case. Yet, the overabundance of the BoxA-containing mRNA is not observed in rne+ cells (Fig. 2); moreover, its level is distinctly lower than that of the CBoxA mRNA, although the latter has no antiterminator sequence and therefore should be synthesized at a normal rate. We speculate that the elevated transcription rate can extend a naked region of a nascent transcript between the RNA polymerase and the first elongating ribosome, increasing the risk for this region to be cut by RNase E in rne+ cells. A similar effect, disappearing in the rne background, has been reported for the lacZ transcript generated by the fast T7 RNA polymerase (14).
Long SD sequences cannot stabilize the lacZ mRNA in E. coli.
The main conclusion reached here is that the lacZ mRNA stability depends mainly on the efficiency of its translation (Fig. 2 and 4) but not on the strength of the potential SD duplex, as was proposed earlier by Wagner et al. (37). Indeed, in the absence of translational enhancers, the 6-nt SD sequence provides higher stability of the lacZ mRNA than the 10-nt SD sequence (SD6 and SD10 constructs in Fig. 2), which correlates directly with translation efficiency but inversely with SD duplex stability. Moreover, had the strong ribosome binding to the long SD sequence been sufficient to protect the full-length lacZ RNA, the transcripts with the same SD sequence length would show similar (or, at least, comparable) stability. This is clearly not the case in our experiments, where the lacZ mRNA abundance varies widely for the same 10- or 6-nt SD sequence. These variations parallel those of β-galactosidase expression (Fig. 2B), as expected if mRNA stability strongly depends upon translation efficiency. We therefore conclude that only translating, not stalling, ribosomes can protect the lacZ transcript in E. coli, a conclusion at variance with that of Wagner et al. (37). Those authors have probed the stability of the untranslated lacZ mRNA by using a primer extension technique to monitor the disappearance of the 5′-terminal region after a rifampin block. However, this technique should also detect the abundant 5′-terminal fragments resulting from premature transcription termination in the lacZ gene (see above), which does not decay like a full-length mRNA and which might be stabilized by an SD sequence-bound ribosome. We show here that the lower the translatability of the lacZ mRNA in a strain, the higher the accumulation of the 5′-terminal lacZ fragments.
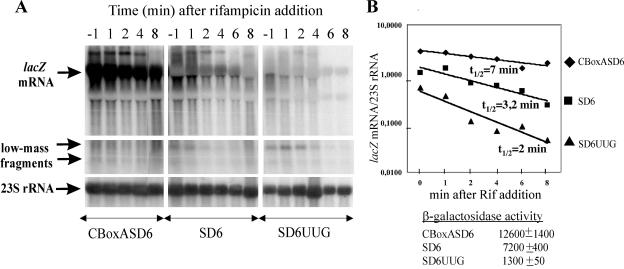
FIG. 4.
Stabilization of lacZ mRNA by efficient translation. (A) Northern blot analysis of total RNA (10 μg) extracted from CBoxASD6, SD6, and SD6UUG strains at various times (above each lane) after addition of rifampin (400 μg per ml). RNA was analyzed on Northern blots as described for Fig. 2. (B) Decay of lacZ mRNAs bearing the same 6-nt SD sequence depends on their translation efficiency. A plot of the lacZ mRNA chemical stability versus time is shown. The β-galactosidase level in exponentially growing cells (in nanomoles of ONPG hydrolyzed per minute per milligram of total protein) is indicated below the graph. The relative chemical stability of lacZ mRNA at each time point after the rifampin block was evaluated as a ratio of the hybridization signal from the full-length lacZ mRNA to that from 23S rRNA. A logarithmic scale was used because of the large range covered by the data. Assuming exponential decay kinetics, the lacZ mRNA chemical half-lives were estimated to be 6.9, 3.18, and 2 min for CBoxASD6, SD6, and SD6UUG, respectively.
To strengthen our conclusion that for the same SD sequence length the mRNA stability strongly depends on translation efficiency, we created an additional strain in which a strong AUG initiator codon downstream of an optimal SD6 was mutated to a weak UUG codon. This mutation caused more than a fivefold decrease in the β-galactosidase activity (1,300 ± 50 U for SD6UUG versus 7,200 ± 400 U for SD6AUG). As a result, we obtained the following range of translation efficiencies for the same 6-nt SD sequence: CBoxASD6/SD6/SD6UUG ≈ 10:5:1. A comparison of decay rates for the lacZ transcripts in this set of strains after a rifampin block (Fig. 4) clearly shows that both abundances and half-lives of full-length lacZ mRNAs parallel their translational yields.
Our conclusion that in the absence of efficient translation the SD sequence-bound ribosome cannot stabilize the full-length lacZ transcript, which was supported earlier by another source of evidence (15), implies that the lacZ mRNA decay in E. coli does not follow the same pathway as in Bacillus subtilis. In B. subtilis, which has no RNase E homologue (7), the degradation of the mRNAs is thought to be controlled by an as-yet-unidentified 5′-dependent scanning nuclease that cannot bypass a stalled ribosome (1, 7). In contrast, the E. coli RNase E, which also binds preferentially to accessible 5′ ends (6), is able either to skip over a ribosome to reach internal cleavage sites or to access these sites directly in a 5′-independent manner (2, 7, 10). In this case, only protection of internal cleavage sites by translating ribosomes can prevent a rapid decay.
Stabilizing and destabilizing effects of AU-rich elements.
Our observation that AU-rich S1 targets within the lacZ mRNA leader enhance both translation efficiency and the mRNA stability deserves further comment. Protein S1, an essential component of the E. coli translational machinery (32), is necessary for mRNA recognition during translation initiation (3, 18, 35). S1 possesses a broad sequence specificity and recognizes single-stranded AU-rich motifs that serve as translational enhancers when located within 5′-untranslated mRNA leaders (18, 35). Meanwhile, AU-rich single-stranded regions have been shown to serve as targets for RNase E (2, 16, 20, 23). At first glance, the similar RNA binding specificities of S1 and RNase E is not surprising, because RNase E comprises a so-called S1 domain that is essential for its RNA binding properties, catalytic activity, and feedback regulation (8). The stabilizing effect of AU-rich S1 binding sites on the lacZ mRNA observed here suggests that S1 bound to ribosomes wins the competition with RNase E during translation initiation in exponentially growing cells. On the other hand, when known RNase E cleavage sites (called ectopic sites, also AU rich) were introduced in the rpsT leader sequence, marked destabilization of the rpsT transcripts was observed even in the presence of a 5′ stem-loop protector, suggesting the effectiveness of a 5′-independent RNase E cleavage in this case (2). Thus, it appears that the single-stranded, AU-rich S1 targets serving as stabilizing elements due to their ability to enhance translation (this study) differ in some yet-unknown features from the single-stranded, AU-rich destabilizing elements (ectopic RNase E sites [2]). Further studies are needed to gain more insight into this intriguing mRNA stabilization-destabilization problem.
Acknowledgments
This work was supported by a 2002 FEBS Summer Fellowship to A.V.K., by grants from ARC (no. 4633) and MENRT to M.D., and by RFBR grant 03-04-49131 and the Programme for Basic Research (MCB) of the Presidium of RAS to I.V.B.
We are grateful to Isabelle Iost and Jean Guillerez for their helpful advice on Northern blot analysis.
REFERENCES
- 1.Agaisse, H., and D. Lereclus. 1996. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20:633-643. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. E., and G. A. Mackie. 2003. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Mol. Microbiol. 47:75-88. [DOI] [PubMed] [Google Scholar]
- 3.Boni, I. V., D. M. Isaeva, M. L. Musychenko, and N. V. Tzareva. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, F., J. Le Derout, and P. Régnier. 1998. Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J. 17:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coburn, G. A., and G. A. Mackie. 1998. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 279:1061-1074. [DOI] [PubMed] [Google Scholar]
- 6.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 7.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diwa, A. A., X. Jiang, M. Schapira, and J. G. Belasco. 2002. Two distinct regions on the surface of an RNA-binding domain are crucial for RNase E function. Mol. Microbiol. 46:959-969. [DOI] [PubMed] [Google Scholar]
- 9.Dreyfus, M. 1988. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 204:79-94. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfus, M., and S. A. Joyce. 2002. The interplay between translation and mRNA decay in prokaryotes: a discussion on current paradigms, p. 165-183. In J. Lapointe and L. Brakier-Gingras (ed.), Translational mechanisms. Kluwer Academic/ Plenum Publishers, New York, N.Y.
- 11.Goodrich, A. F., and D. A. Steege. 1999. Roles of polyadenylation and nucleolytic cleavage in the filamentous phage mRNA processing and decay pathways in Escherichia coli. RNA 5:972-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillerez, J., M. Gazeau, and M. Dreyfus. 1991. In the Escherichia coli lacZ gene the spacing between the translating ribosomes is insensitive to the efficiency of translation initiation. Nucleic Acids Res. 19:6743-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajnsdorf, E., and P. Regnier. 1999. E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol. 288:1033-1043. [DOI] [PubMed] [Google Scholar]
- 14.Iost, I., and M. Dreyfus. 1995. The stability of Escherichia coli lacZ mRNA depends upon simultaneity of its synthesis and translation. EMBO J. 14:3252-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joyce, S. A., and M. Dreyfus. 1998. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Biol. 282:241-254. [DOI] [PubMed] [Google Scholar]
- 16.Kaberdin, V. R. 2002. Probing the substrate specificity of Escherichia coli RNase E in a novel oligonucleotide-based assay. Nucleic Acids Res. 31:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komarova, A. V., L. S. Tchufistova, E. V. Supina, and I. V. Boni. 2001. Extensive complementarity of the Shine-Dalgarno region and 3′-end of 16S rRNA is inefficient for translation in vivo. Russian J. Bioorg. Chem. 27:248-255. [DOI] [PubMed] [Google Scholar]
- 18.Komarova, A. V., L. S. Tchufistova, E. V. Supina, and I. V. Boni. 2002. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8:1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushner, S. R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin-Chao, S., T.-T. Wong, K. J. McDowal, and S. N. Cohen. 1994. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 269:10797-10803. [PubMed] [Google Scholar]
- 21.Lopez, P. J., I. Iost, and M. Dreyfus. 1994. The use of a tRNA as a transcriptional reporter: the T7 late promoter is extremely efficient in Escherichia coli but its transcripts are poorly expressed. Nucleic Acids Res. 22:1186-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick, J. R., J. M. Zengel, and L. Lindahl. 1994. Correlation of translation efficiency with the decay of lacZ mRNA in Escherichia coli. J. Mol. Biol. 239:608-622. [DOI] [PubMed] [Google Scholar]
- 23.McDowel, K. J., S. Lin-Chao, and S. N. Cohen. 1994. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269:10790-10796. [PubMed] [Google Scholar]
- 24.Mogridge, J., and J. Greenblatt. 1998. Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J. Bacteriol. 180:2248-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mudd, E. A., H. M. Krisch, and C. F. Higgins. 1990. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol. Microbiol. 4:2127-2135. [DOI] [PubMed] [Google Scholar]
- 26.Oehler, S., E. R. Eismann, H. Krämer, and B. Müller-Hill. 1990. The three operators of the lac-operon cooperate in repression. The EMBO J. 9:973-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono, M., and M. Kuwano. 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129:343-357. [DOI] [PubMed] [Google Scholar]
- 28.Régnier, P., and E. Hajnsdorf. 1991. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′-stabilizing stem and loop structure. J. Mol. Biol. 217:283-292. [DOI] [PubMed] [Google Scholar]
- 29.Ruteshouser, E. C., and J. P. Richardson. 1989. Identification and characterization of transcription termination sites in the Escherichia coli lacZ gene. J. Mol. Biol. 208:23-43. [DOI] [PubMed] [Google Scholar]
- 30.Sato, T., M. Terabe, H. Watanabe, T. Gojobori, C. Hori-Takemoto, and K. Miura. 2001. Codon and base biases after initiation codon of the open reading frames in the Escherichia coli genome and their influence on translation efficiency. J. Biochem. (Tokyo) 129:851-860. [DOI] [PubMed] [Google Scholar]
- 31.Shen, V., F. Imamoto, and D. Schlessinger. 1982. RNase III cleavage of Escherichia coli β-galactosidase and tryptophan operon mRNA. J. Bacteriol. 150:1489-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sørensen, M. A., J. Fricke, and S. Pedersen. 1998. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli. J. Mol. Biol. 280:561-569. [DOI] [PubMed] [Google Scholar]
- 33.Stanssens, P., E. Remaut, and W. Fiers. 1986. Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell 44:711-718. [DOI] [PubMed] [Google Scholar]
- 34.Steege, D. A. 2000. Emerging features of mRNA decay in bacteria. RNA 6:1079-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzareva, N. V., V. I. Makhno, and I. V. Boni. 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337:189-194. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, U., and K. F. Jensen. 1995. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J. Biol. Chem. 270:18335-18340. [DOI] [PubMed] [Google Scholar]
- 37.Wagner, L. A., R. F. Gesteland, T. J. Dayhuff, and R. B. Weiss. 1994. An efficient Shine-Dalgarno sequence but not translation is necessary for lacZ mRNA stability in Escherichia coli. J. Bacteriol. 176:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarchuk, O., N. Jacques, J. Guillerez, and M. Dreyfus. 1992. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J. Mol. Biol. 226:581-596. [DOI] [PubMed] [Google Scholar]