Abstract
DNA repair is essential for life, yet its efficiency declines with age for reasons that are unclear. Numerous proteins possess Nudix homology domains (NHDs) that have no known function. We show that NHDs are NAD+ (oxidized form of nicotinamide adenine dinucleotide) binding domains that regulate protein-protein interactions. The binding of NAD+ to the NHD domain of DBC1 (deleted in breast cancer 1) prevents it from inhibiting PARP1 [poly(adenosine diphosphate–ribose) polymerase], a critical DNA repair protein. As mice age and NAD+ concentrations decline, DBC1 is increasingly bound to PARP1, causing DNA damage to accumulate, a process rapidly reversed by restoring the abundance of NAD+. Thus, NAD+ directly regulates protein-protein interactions, the modulation of which may protect against cancer, radiation, and aging.
The oxidized form of nicotinamide adenine dinucleotide (NAD+) is critical for redox reactions and as a substrate for signaling by the PARPs [poly(adenosine diphosphate–ribose) polymerases] and the sirtuins (SIRT1 to SIRT7) in the regulation of DNA repair, energy metabolism, cell survival, and circadian rhythms, among other functions (1–3). Raising NAD+ concentrations or directly activating the sirtuins delays aging in yeast, flies, and mice (4–6). Increased amounts of sirtuin and PARP1 activity are also associated with improved health and longevity in humans (4, 7). Thus, understanding how cells modulate NAD+ and PARP1 activity may shed light on new therapies for major diseases, such as diabetes and cancer, and possibly also the aging process (3, 8).
Whether NAD+ has a third role in cells as a direct regulator of protein-protein interactions is a matter of speculation (9). The protein DBC1 (deleted in breast cancer 1) is one of the most abundant yet enigmatic proteins in mammals (10, 11), with a conserved domain similar to Nudix hydrolases that hydrolyze nucleoside diphosphates but lacking catalytic activity due to the absence of key catalytic residues (9, 12, 13).
DBC1 is known to inhibit SIRT1 (14), so we tested whether DBC1 might also inhibit PARP1 as a way to coregulate these two major NAD+-responsive pathways. In human embryonic kidney (HEK) 293T cells, a SIRT1-independent interaction between DBC1 and PARP1 was detected (Fig. 1A and fig. S1, A and B). The PARP1 inhibitors PJ-34 and 3-aminobenzamide (3-AB) had no effect on the interaction (fig. S1C), nor did overexpression of the adenosine diphosphate (ADP)–ribose hydrolase MACROD1 (MACRO domain containing 1) (fig. S1D) or the PARP1 catalytic mutant, PARP1-E988K (where E988K denotes Glu988→Lys988) (fig. S1E). Thus, PARP1-DBC1 binding is independent of PARP1 catalytic activity.
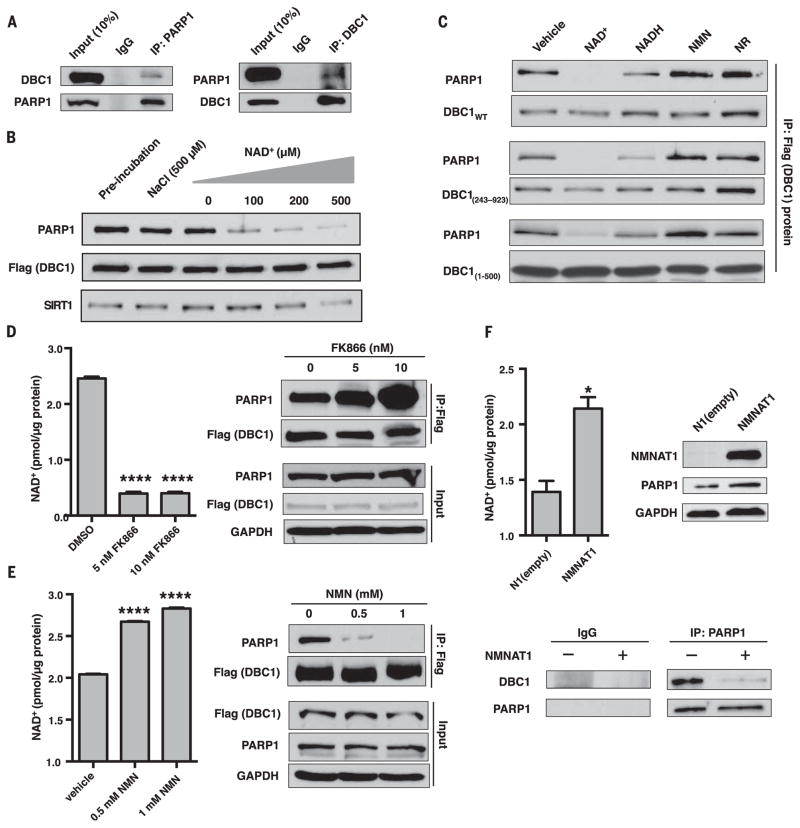
Fig. 1. Regulation of the PARP1-DBC1 interaction by NAD+.
(A) Endogenous DBC1 and PARP1 interact. IgG, immunoglobulin G; IP, immunoprecipitation. (B) NAD+ dissociates the PARP1-DBC1 interaction. (C) Effects of NAD+ and structurally related molecules on the PARP1-DBC1 interaction. Flag-DBC1 was incubated with molecules (200 μM) for 1 hour and then probed for PARP1. NMN, nicotinamide mononucleotide; NR, nicotinamide riboside; WT, wild type. (D to F) The PARP1-DBC1 interaction after treatment with (D) FK866 or (E) NMN for 24 hours or (F) in cells overexpressing NMNAT1, an NAD+ salvage pathway gene to raise NAD+. DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Error bars indicate mean ± SEM. (D and E) One-way analysis of variance (ANOVA) and Sidak’s post-hoc correction. (F) Unpaired two-tailed t test. *P < 0.05, ****P < 0.0001.
The PARP1-DBC1 complex was abrogated by NAD+ in a concentration-dependent manner, whereas the SIRT1-DBC1 interaction was unaffected within physiological ranges of NAD+ (15), except at 500 μM (Fig. 1B). This effect was surprisingly specific: 200 μM of nicotinamide mononucleotide (NMN), nicotinamide riboside (NR), adenosine, adenosine triphosphate, and ADP-ribose, or 500 μM of nicotinamide and its structural analog 3-AB (2 mM), had no effect on the PARP1-DBC1 complex, and NADH (the reduced form of NAD+) (200 μM) or adenine (200 μM) were less effective than NAD+ (Fig. 1C and fig. S2, A to C).
DBC1 mutants lacking regions outside the NHD (DBC11-500 and DBC1243-923) behaved similarly to full length DBC1 (Fig. 1C), whereas MACROD1, poly(ADP-ribose) glycohydrolase (PARG) (fig. S2D), or PARP1 inhibitors (fig. S1C) had no effect. PARP1-E988K behaved similarly to the wild type (fig. S1E and fig. S2E). Carba-nicotinamide adenine dinucleotide, a nonreactive PARP1 substrate, abrogated the complex (fig. S2F). Thus, disruption of the PARP1-DBC1 complex by NAD+ does not require NAD+ cleavage or a covalently attached ADP-ribose.
In HEK 293T cells, FK866, an inhibitor of NAD+ biosynthesis (16), increased the PARP1-DBC1 interaction (Fig. 1D), as did depletion of NAD+ by genotoxic stress (fig. S3A). Interventions that increased NAD+ abundance (5, 17) decreased the PARP1-DBC1 interaction (Fig. 1, E and F, and fig. S3B). The SIRT1-DBC1 interaction was slightly diminished by FK866, possibly through the sequestration of DBC1 by PARP1 (fig. S3C). Together, these data indicate that NAD+ inhibits PARP1-DBC1 complex formation in cells.
To better understand the mechanism by which NAD+ inhibits complex formation, we identified the DBC1 domain necessary for interaction with PARP1. A truncated version of DBC1 lacking an NHD conserved region (DBC1Δ354-396) had impaired PARP1 binding in cells (fig. S4, A to D), whereas a recombinant DBC1 mutant containing the NHD domain (DBC1-NHD, residues 239 to 553) bound to a truncated PARP1 lacking the catalytic domain (PARP1-ΔCAT, residues 1 to 654) (fig. S5, A and B) and was not abrogated by NAD+, indicating that residues outside the minimal NHD domain may be necessary for NAD+ to dissociate the complex.
DBC1 interacted with the BRCT [BRCA1 (breast cancer 1) C-terminal] domain of PARP1 (fig. S6A) (18) but not with the PARP1 catalytic domain or PARP2, which lacks a BRCT domain (fig. S6, B to D). A BRCT-deficient PARP1 had higher activity than the wild type (fig. S6, E to F), together indicating that PARP1-DBC1 is mediated by contacts between the NHD and PARP1-BRCT.
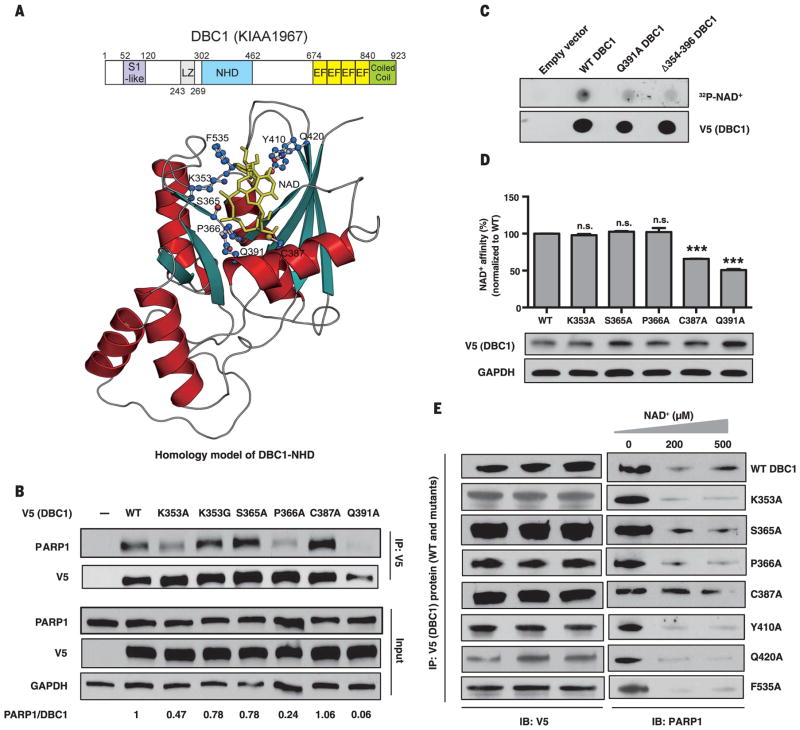
We generated an atomic-resolution homology model for the human DBC1-NHD. Our model is based on five known crystal structures of Nudix domains from other proteins (Fig. 2A, fig. S7, and supplementary materials and methods). NAD+ had the best fit of all riboside nucleotides. Substitutions of the amino acids predicted to alter NAD+ binding inhibited PARP1-DBC1 binding either slightly (K353A and P366A) or substantially (Q391A) (Fig. 2B and fig. S8, A and B). Radio-labeled or biotin-labeled NAD+ directly bound to DBC1 (Fig. 2, C and D) and was competed off with unlabeled NAD+ (fig. S9, A to D). Partial deletion of the NHD (DBC1Δ354-396) or Q391A (DBC1Q391A) reduced NAD+ binding, whereas N-terminal and C-terminal truncations did not (Fig. 2, C to D, and fig. S9E). Mutation of C387, a residue close to Q391 on the same helix (Fig. 2A), also decreased NAD+ binding (Fig. 2D) and reduced the responsiveness of the PARP1-DBC1 complex to NAD+ (Fig. 2E).
Fig. 2. Binding of the Nudix homology domain (NHD) of DBC1 to NAD+ and PARP1.
(A) Domains and crystallographic-based homology model of the NHD docked with NAD+. S1-like, ribosomal protein S1 OB-fold domain-like; EF, EF-hand; LZ, leucine zipper. Residues predicted to be in the vicinity of bound NAD+ are highlighted. (B) Interaction of V5/His-tagged DBC1 mutants and PARP1. See fig. S8 for additional mutants. (C and D) Direct binding of NAD+ to the DBC1-NHD, assessed using a radiolabeled NAD+ binding assay (C) or a biotin-NAD+ binding assay (D). Error bars indicate mean ± SEM; one-way ANOVA and Sidak’s post-hoc correction. ***P < 0.001; n.s., not significant. (E) Effect of NAD+ on binding of DBC1-NHD mutants to PARP1. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; F, Phe; G, Gly; K, Lys; P, Pro; Q, Gln; S, Ser; and Y, Tyr.
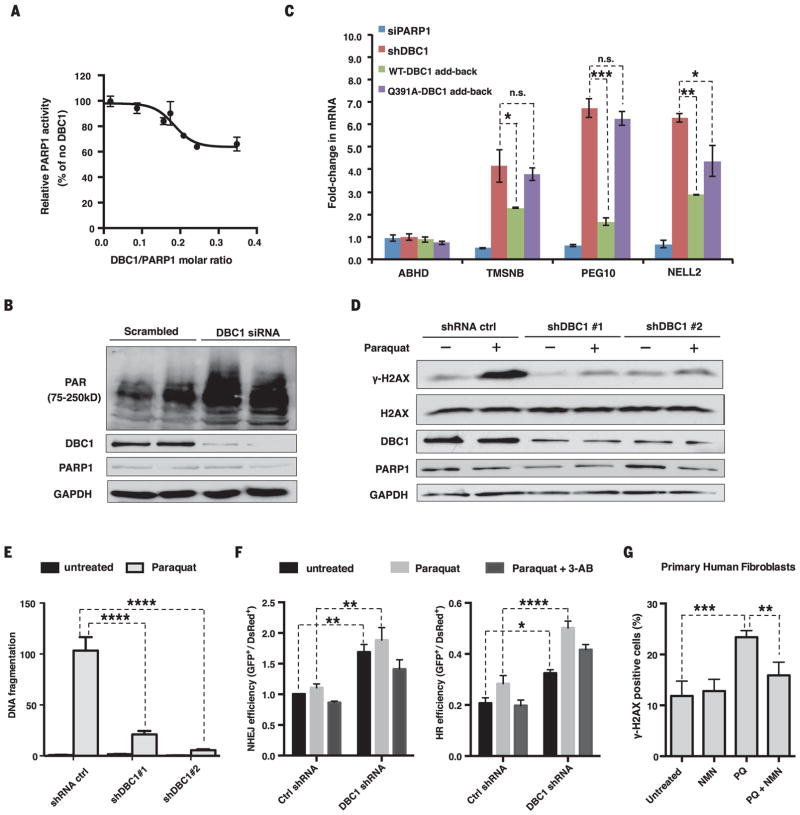
PARP1 activity was inhibited by DBC1 in vitro (Fig. 3A). In cells, knocking down DBC1 increased both PAR [poly(ADP-ribose)] concentrations before and after exposure to paraquat, H2O2, or etoposide (Fig. 3B and fig. S10, A to D) and the abundance of mRNAs positively regulated by PARP1 (Fig. 3C) (19, 20). Reintroduction of wild-type DBC1 reduced PARP1 activity and partially restored gene expression, whereas DBC1Q391A had no effect (Fig. 3C and fig. S10, E and F). Reducing DBC1 lowered the abundance of the phosphorylated form of histone H2AX (γH2AX) (Fig. 3D), reduced DNA fragmentation (Fig. 3E and fig. S11A), increased cell survival after paraquat treatment (fig. S11B), and increased both non-homologous end joining (NHEJ) and homologous recombination (HR) pathways in a PARP1-dependent manner (Fig. 3F and fig. S11C). Similarly, NMN treatment reduced the number of γH2AX foci in paraquat-treated primary human fibroblasts (Fig. 3G and fig. S12). No other major DNA repair proteins appeared to change their interactions with the PARP1-DBC1 complex in the presence of NMN or DNA damage (fig. S13, A and B), though we cannot rule out other interactions. These results are consistent with a model in which binding of NAD+ to the DBC1-NHD regulates the two major pathways of DNA repair.
Fig. 3. DBC1 inhibits PARP1 activity and DNA repair.
(A) Inhibition of PARP1 activity by DBC1 purified from HEK 293T cells. (B) PAR [poly(ADP-ribose)] abundance in HEK 293T cells lacking DBC1. siRNA, small interfering RNA. (C) Opposing effects of reintroducing the wild type or DBC1Q391A into MCF-7 cells on mRNA of PARP1-regulated genes. TMSNB, Thymosin beta; PEG10, paternally expressed gene 10; NELL2, neural EGFL-like 2. ABHD2 was a negative control. DBC1 and PARP1 abundance are shown in fig. S10E. sh, short hairpin. (D) γH2AX abundance in DBC1 knockdown cells after paraquat treatment (1 mM, 24 hours). (E) DNA fragmentation after paraquat treatment (0.5 mM, 24 hours) in DBC1 knockdown cells, assessed by a comet assay (>50 cells per group). See fig. S11A. (F) DNA break repair [nonhomologous end joining (NHEJ) and homologous recombination (HR)] in DBC1 knockdown cells treated with paraquat (1 mM) or 3-AB (5 mM). n = 3 biological replicates. (G) Protection of human primary fibroblasts from paraquat (PQ)–induced DNA damage (300 μM paraquat) by NMN (500 μM). 100 ± 20 cells per condition, n = 4 biological replicates (two cell lines, twice for each), 24-hour treatment. See fig. S12. Error bars indicate SEM; one-way ANOVA [(C) and (G)] and two-way ANOVA [(E) and (F)], Sidak’s post-hoc correction. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; n.s., not significant.
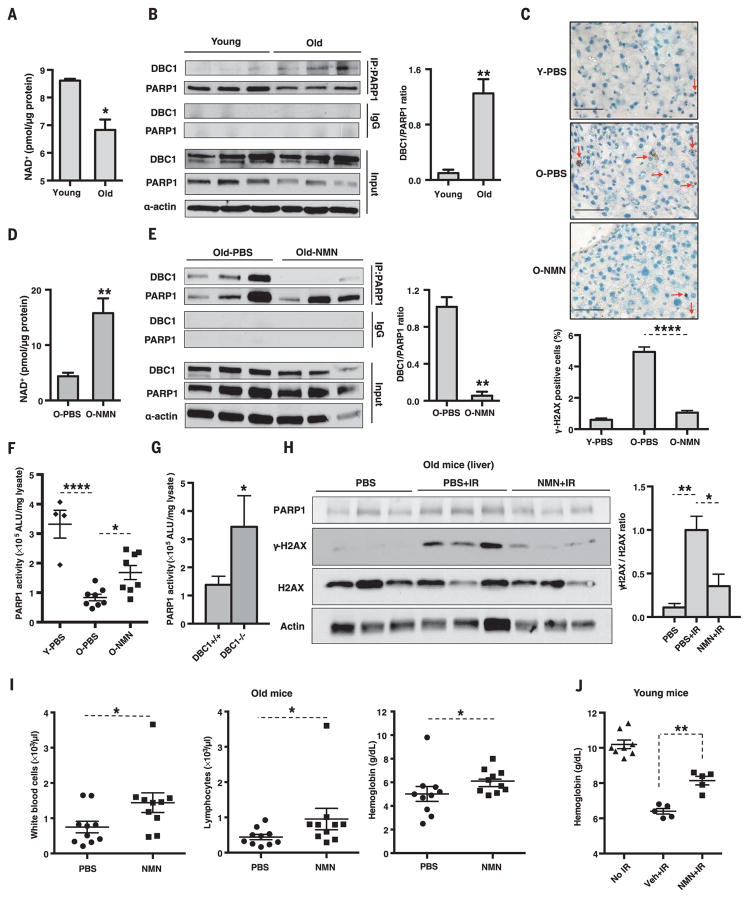
DNA repair declines with age (21), in concert with lower PARP1 activity (7). Our data indicate that a cause may be increased binding of DBC1 to PARP1 as NAD+ levels decline during aging. To test this, we examined the effect of NMN treatment on young and old mice. Hepatic NAD+ concentrations were lower in old mice (Fig. 4A), coincident with a higher amount of the DBC1-PARP1 complex (Fig. 4B) and an increase in γH2AX staining (Fig. 4C and fig. S14). A week of NMN treatment (500 mg/kg per day intraperitoneally) increased hepatic NAD+ concentrations and disrupted the PARP1-DBC1 complex in young (fig. S15, A and B) and old mice (Fig. 4, D and E) and reduced the abundance of γH2AX in old mice (Fig. 4C and fig. S14).
Fig. 4. Increases in the PARP1-DBC1 complex and DNA damage with age are reversed by NMN, a precursor to NAD+.
(A and B) NAD+ concentrations and PARP1-DBC1 interactions in livers of young and old mice (6 versus 22 months, n = 3 per group). (C) γH2AX-positive cells (red arrow) in the livers of young (6 months, Y) and old (30 months, O) mice (n = 3 per group) treated for 7 days with vehicle [phosphate-buffered saline (PBS)] or NMN (500 mg/kg per day intraperitoneally, n = 3 per group). Scale bars, 50 μm. (D and E) NAD+ concentrations and PARP1-DBC1 interactions in the livers of old mice (22 months) treated as in (C). (F) PARP1 activity in young (6 months, n = 4) and old (26 ± 4 months, n = 8 per group) mice. (G) PARP1 activity in 18- to 20-month-old DBC1 knockout mice livers (n = 3 or 4). (H) γH2AX abundance in the livers of 26-month-olds after irradiation (IR) [7.5 grays (Gy), 137CsCl], treated as in (C) (n = 3 per group). (I) Blood counts of 23-month-olds on the 7th or 8th day after irradiation (n = 10 per group). See fig. S18, A and B. (J) Blood metrics of 4-month-olds given a single oral dose of NMN (2000 mg/kg) 1 hour after irradiation (8 Gy, 137CsCl), followed by another 7 days of treatment (2000 mg/kg per day, n = 5 to 8 per group). Veh, vehicle. See fig. S18C. Errors bars indicate SEM; unpaired two-tailed t test [(A), (B), (D), (E), and (G)], one-way ANOVA [(C), (F), and (H)], Sidak’s post-hoc correction, Mann-Whitney U-test [(I) and (J)]. *P < 0.05, **P < 0.01, ****P < 0.0001.
Old mice had lower levels of PARP1 activity (Fig. 4F and fig. S16A) and a decreased response to DNA damage (fig. S16B), and DBC1 knockout mice had increased PARP1 activity (Fig. 4G and fig. S16C). The reduced PARP1 activity in the old mice was restored by NMN (Fig. 4F and fig. S16D) but not if PARP1 activity was inhibited (fig. S16, E and F). Total amounts of poly(ADP-ribosylation) increased with age in the liver (4), possibly due to reduced PARG abundance (fig. S17A). These data indicate that the decline in NAD+ during aging promotes the binding of DBC1 to PARP1, which inhibits PARP1’s ability to mediate DNA repair. Similar studies were conducted on old mice exposed to gamma irradiation. NMN treatment (Fig. 4E) reduced DNA damage (Fig. 4H and fig. S17, B and C) and protected against alterations in white blood cell counts, lymphocytes, and hemoglobin, even when given after irradiation (Fig. 4, I and J, and fig. S18, A to C).
These data show that NAD+ has a third function in cells: to directly regulate protein-protein interactions. We speculate that this mechanism evolved to allow a cell to adapt to fluctuations in NAD+ abundance without degrading it and, in the case of DBC1, to serve as a negative-feedback loop to prevent PARP1 from depleting NAD+ down to lethal levels during DNA damage (15) (fig. S19). The data also provide an explanation for why DBC1 mutations are associated with cancers (22) and indicate that assessing DBC1 status in tumors will help inform ongoing clinical trials of PARP1 inhibitors for treating cancer (23). Although the reason NAD+ declines with age is unclear, this work provides a plausible explanation for why DNA repair capacity declines as we age (24), pointing to NAD+ replenishment as a means of reducing the side effects of chemotherapy, protecting against radiation exposure, and slowing the natural decline in DNA repair capacity during aging.
Supplementary Material
Acknowledgments
D.A.S. was supported by the Glenn Foundation for Medical Research, American Federation for Aging Research, E. Schulak, and grants from the National Institute on Aging and the NIH; D.Z. and L.A. by the National Library of Medicine/NIH intramural program; and C.S. by Deutsche Forschungsgemeinschaft (STE1701/15). Z.L. was supported by grants CA130996 and CA203561 from the National Cancer Institute and J.L. by the Sinclair Harvard Fund. We thank J.-H. Yang, A. Gomes, J. Pfahler, M. Pannek, S. Armour, K. Chwalek, M. Cooney, W. L. Kraus, S.-I. Imai, E. Chini, R. London, M. Cuneo, W. Bohr, J. Mitchell, C. Hine, and Y. Lu. This paper is dedicated to Diana Sinclair, who bravely survived cancer for two decades. D.A.S. is an inventor on patent applications held by The University of New South Wales that cover the use of NAD precursors to modulate DNA repair, fertility, and blood flow, and by Harvard Medical School for the treatment of diseases of aging and mitochondrial disorders. J.L. and D.A.S. are inventors on a patent application held by Harvard Medical School that covers the modulation of Nudix hydrolase domain proteins by small molecules. M.S.B. was a paid consultant to OvaScience. D.A.S. is an unpaid consultant, board member, inventor on patent applications, and holds equity in companies developing NAD precursor–based medicines (EdenRoc, Liberty Biosecurity, Metrobiotech, and Jumpstart Fertility) and is a paid consultant and inventor on a patent application licensed to OvaScience for improving in vitro fertilization.
Footnotes
REFERENCES AND NOTES
- 1.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonkowski MS, Sinclair DA. Nat Rev Mol Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai S, Guarente L. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouchiroud L, et al. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshino J, Mills KF, Yoon MJ, Imai S. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhullar KS, Hubbard BP. Biochim Biophys Acta. 2015;1852:1209–1218. doi: 10.1016/j.bbadis.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Grube K, Bürkle A. Proc Natl Acad Sci USA. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigis MC, Sinclair DA. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anantharaman V, Aravind L. Cell Cycle. 2008;7:1467–1472. doi: 10.4161/cc.7.10.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armour SM, et al. Mol Cell Biol. 2013;33:1487–1502. doi: 10.1128/MCB.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mildvan AS, et al. Arch Biochem Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Gagné JP, et al. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JE, Chen J, Lou Z. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, et al. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasmann M, Schemainda I. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 17.Berger F, Lau C, Dahlmann M, Ziegler M. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 18.Bork P, et al. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 19.Zhang T, et al. J Biol Chem. 2012;287:12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnakumar R, Kraus WL. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbunova V, Seluanov A, Mao Z, Hine C. Nucleic Acids Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamaguchi M, et al. Proc Natl Acad Sci USA. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Audeh MW, et al. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 24.Lombard DB, et al. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.