Abstract
Recombinational repair-dependent mutants identify ways to avoid chromosomal lesions. Starting with a recBC(Ts) strain of Escherichia coli, we looked for mutants unable to grow at 42°C in conditions that inactivate the RecBCD(Ts) enzyme. We isolated insertions in ackA and pta, which comprise a two-gene operon responsible for the acetate↔acetyl coenzyme A interconversion. Using precise deletions of either ackA or pta, we showed that either mutation makes E. coli cells dependent on RecA or RecBCD enzymes at high temperature, suggesting dependence on recombinational repair rather than on the RecBCD-catalyzed linear DNA degradation. Complete inhibition of growth of pta/ackA rec mutants was observed only in the presence of nearby growing cells, indicating cross-inhibition. pta rec mutants were sensitive to products of the mixed-acid fermentation of pyruvate, yet none of these substances inhibited growth of the double mutants in low-millimolar concentrations. pta, but not ackA, mutants also depend on late recombinational repair functions RuvABC or RecG. pta/ackA recF mutants are viable, suggesting, together with the inviability of pta/ackA recBC mutants, that chromosomal lesions due to the pta/ackA defect are of the double-strand-break type. We have isolated three insertional suppressors that allow slow growth of pta recBC(Ts) cells under nonpermissive conditions; all three are in or near genes with unknown functions. Although they do not form colonies, ackA rec and pta rec mutants are not killed under the nonpermissive conditions, exemplifying a case of synthetic inhibition rather than synthetic lethality.
The term “synthetic lethals” was introduced by Dobzhansky to describe the relationship between natural alleles in wild-type populations of Drosophila pseudoobscura that, having no phenotype individually, are lethal when combined pairwise in the same organism (11). The contemporary analysis of synthetic lethals involves finding secondary mutations inviable in combination with a query mutation (35). Although originally perceived as a way to reveal metabolic redundancies, by identifying the activities that are necessary for survival in the absence of a particular function, analysis of synthetic lethals exposes the contribution of frequently unrelated and mechanistically different metabolic branches towards the same common goal (28, 29). Because of inviability of the double mutant, identification of a synthetically lethal combination facilitates subsequent selection of suppressors, either of inactivation type or multicopy suppressors (3). Identification of suppressors in genes with known functions immediately suggests potential mechanisms of the original synthetic lethality (5, 19). Thus, identification of inviable combinations with a particular mutation is a potentially powerful tool for unraveling the cellular consequences of the corresponding metabolic defect. However, analysis of synthetic lethals, so popular for the yeast Saccharomyces cerevisiae, is used relatively infrequently in bacteria (4).
Chromosomal lesions are local DNA lesions with genomic consequences, because they block chromosomal replication and/or segregation, thus inactivating not only individual genes, in which they happened to occur, but the whole genetic system of the organism (22). Mechanistically, chromosomal lesions comprise DNA lesions that affect both DNA strands of a duplex opposite each other, the so-called two-strand lesions, the best-known examples of which are double-strand DNA breaks and interstrand cross-links (23). To fix chromosomal lesions, all organisms employ recombinational repair (36, 39) as well as some other DNA repair pathways like translesion DNA synthesis (14) and nonhomologous end joining (41). In eubacteria, recombinational repair is the major way to mend chromosomal lesions (23).
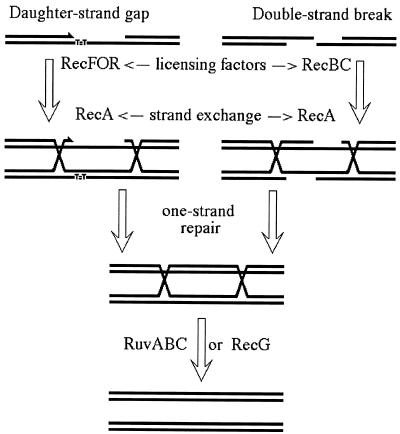
In Escherichia coli, two separate pathways for the repair of chromosomal lesions exist (23) (Fig. 1). The central step in both pathways is homologous strand exchange between damaged and intact duplexes which is catalyzed by RecA (9). However, the RecA protein itself is incompetent to initiate repair because it cannot recognize chromosomal lesions in their original form. Repair of unfillable single-strand gaps is initiated by the RecFOR complex of activities (34, 43), whereas repair of double-strand breaks is initiated by the RecBCD enzyme (20). Both activities target RecA to the corresponding chromosomal lesions and license RecA to displace single-stranded DNA (ssDNA)-binding protein from ssDNA, which is always associated with chromosomal lesions. The RecA-promoted strand exchange generates a region of hybrid DNA (in which the original chromosomal lesion can be repaired) bracketed by Holliday junctions. Removal of the spent RecA filaments and resolution of Holliday junctions require either RuvABC resolvasome or RecG helicase, the two alternative clean-up teams of recombinational repair (23) (Fig. 1).
FIG. 1.
A scheme of recombinational repair in E. coli. RecFOR activities license RecA polymerization of daughter-strand gaps, whereas the RecBC enzyme does the same for double-strand breaks. After the RecA-catalyzed homologous strand exchange enables one-strand repair (excision repair) to fix the irregularities in the individual DNA strands, RuvABC or RecG activities remove the spent RecA filaments and Holliday junctions from the repair intermediate, freeing the participating chromosomes.
Interestingly, although recombinational repair-deficient mutants in E. coli cannot mend chromosomal lesions, such mutants are nevertheless viable (7, 33), suggesting the existence of pathways acting to avoid chromosomal lesions. Inactivation of such an avoidance pathway should increase the frequency of chromosomal lesions, making the mutant cells dependent on recombinational repair. Thus, by characterizing rec-dependent mutants, it should be possible to identify the metabolic pathways that contribute to the generation of chromosomal lesions as well as the activities that neutralize this “metabolic poisoning” (5, 19, 38). Here we describe the isolation and characterization of two E. coli mutants which are dependent on the RecBCD enzyme.
MATERIALS AND METHODS
Media, growth conditions, and general methods.
Cells were grown in Luria-Bertani (LB) broth (10 g of tryptone/liter, 5 g yeast extract (YE)/liter, 5 g of NaCl/liter [pH 7.2]) or on LB plates (15 g of agar per liter of LB broth). When necessary, media were supplemented with antibiotics: 50 μg of kanamycin/ml, 10 μg of chloramphenicol/ml, or 10 μg of tetracycline/ml. To determine viability, rapidly growing cultures were serially diluted 10-fold at each step in 1% NaCl, and 10 μl of every dilution was spotted onto plates and developed under permissive as well as nonpermissive conditions. To detect the effect of cell density, if any, we routinely spotted all dilutions. DNA was kept in TE buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA).
Bacterial strains.
Bacterial strains were E. coli K-12 derivatives (Table 1). pta and ackA deletion-replacement alleles were confirmed by PCR. recA(Ts), recBC(Ts), ΔrecA, ΔrecBCD, ΔrecF, recG, and ΔruvABC mutations were confirmed by their characteristic UV sensitivities. Gene replacements with either chloramphenicol or kanamycin resistance markers were constructed according to the method described by Datsenko and Wanner (10). The sequences of the primers used in the construction of the replacement alleles are shown in Table 2.
TABLE 1.
Strains
| Strain | Backgrounda | DNA metabolism | Source, derivation, or reference |
|---|---|---|---|
| Previous studies | |||
| AB1157 | AB1157 | Rec+ | 1 |
| AK4 | AB1157 | Δ(srlR-recA)306::Tn10 | This laboratory |
| AM3 | AB1157 | ΔrecF20::cat | 33 |
| AM547 | AB1157 | Δ(ruvA-C)65 | 26 |
| DH5α::pir | pir+ | 32 | |
| JB1 | AB1157 | ΔrecBCD3::kan | 33 |
| JC9239 | AB1157 | recF143 | 17 |
| JC9941 | AB1157 | recA200(Ts) | A. J. Clark |
| L-56 | AB1157 | recA200(Ts) ΔmltB1085::cat | 19 |
| N2057 | AB1157 | ruvA60::Tn10 | 26 |
| N4452 | AB1157 | ΔrecG265::cam | 31 |
| SK129 | AB1157 | recB270(Ts) recC271(Ts) | 21 |
| TNM1072 | AB1157 | recG263::kan | 26 |
| This study | |||
| JB17 | AB1157 | Δpta51::cat | Deletion-replacement (pKD46) |
| JS1 | AB1157 | Δpta51::cat recB270(Ts) recC271(Ts) | SK129 × P1 JB17 |
| JS2 | AB1157 | Δpta51::cat recA200(Ts) | JC9941 × P1 JB17 |
| JS4 | AB1157 | Δpta52::kan | Deletion-replacement |
| JS5 | AB1157 | Δpta52::kan recB270(Ts) recC271(Ts) | SK129 × P1 JS4 |
| JS6 | AB1157 | Δpta52::kan recA200(Ts) | JC994 × P1 JS4 |
| JS7 | AB1157 | Δpta52::kan Δ(ruvA-C)65 | AM547 × P1 JS4 |
| JS8 | AB1157 | Δpta52::kan recF143 | JC9239 × P1 JS4 |
| JS9 | AB1157 | Δpta52::kan ΔrecG265::cat | N4452 × P1 JS4 |
| JS12 | AB1157 | Δpta52::kan Δ(srlR-recA)306::Tn10 | JS4 × P1 AK4 (pEAK2) |
| JS13 | AB1157 | Δpta51::cat ΔrecBCD3::kan | JB17 × P1 JB1 (pK134) |
| IYS1 | AB1157 | Δpta51::cat recBC(Ts) | SK129 × P1 JB17 |
| IYS2 | AB1157 | Δpta51::cat recA200(Ts) | JC9941 × P1 JB17 |
| IYS3 | AB1157 | Δpta52::kan | AB1157 × P1 JS4 |
| IYS4 | AB1157 | ΔackA45::kan | Deletion-replacement (pKD46) |
| IYS5 | AB1157 | ΔackA46 | IYS4 kan removal (pCP20) |
| IYS6 | AB1157 | ΔackA45::kan recBC(Ts) | SK129 × P1 IYS4 |
| IYS7 | AB1157 | ΔackA45::kan recA200(Ts) | JC9941 × P1 IYS4 |
| IYS9 | AB1157 | ΔackA45::kan ΔrecA304 | JC10287 (pBEU14) × P1 IYS4 |
| IYS11 | AB1157 | ΔackA45::kan ΔrecF20::cat | AM3 × P1 IYS4 |
| IYS14 | AB1157 | ΔackA45::kan ruvA60::Tn10 | IYS4 × P1 N2057 |
| IYS15 | AB1157 | ΔackA46 recG263::kan | IYS5 × P1 TNM1072 |
Complete genotypes of backgrounds are as follows: AB1157, F− λ− rac− thi-1 hisG4 Δ(gpt-proA)62 argE3 thr-1 leuB6 kdgK51 rfbD1 araC14 lacY1 galK2 xylA5 mtl-1 tsx-33 supE44(glnV44) rpsL31 (strR); DH5α::pir, F′ λ::pir endA1 hsdR17(r− m+) glnV44 thi-1 recA1 relA1 Δ(lacIZYA-argF) U169 deoR(φ80-ΔlacZM15) gyrA(Na1R).
TABLE 2.
Sequences of primers used for ackA and pta gene replacement
| Purpose | Name | Sequencea |
|---|---|---|
| pta deletion | pta-for | CGAGCCGCCTGACTGCCTGATTTCACACCGCCAGCTCAGCGTGTAGGCTGGAGCTGCTTC |
| pta deletion | pta-rev | GTATGCAAAGTGGGATGGCGCAATTCATTGATGCAGCGCAGCATATGAATATCCTCCTTAG |
| pta verification | ptaF2 | CTGGCTGCACGTTTCGGC |
| pta verification | ptaR2 | GTGATTGCGGACATAGCGC |
| ackA deletion | ACK1 | CTGACGTTTTTTTAGCCACGTATCAATTATAGGTACTTCCTGTAGGCTGGAGCTGCTTCG |
| ackA deletion | ACK2 | TGATGAAACCAGATTTGCCGAAACGTGCAGCCAGGTTGCCATATGAATATCCTCCTTAG |
| ackA verification | VP-ack-1 | CATGCGCTACGCTCTATGGC |
| ackA verification | VP-ack-2 | GGCAGTCAGGCGGCTCGC |
The nucleotides complementary to the pKD3/4 templates are underlined.
Quantification of CFU.
A fresh culture grown overnight at 28°C was serially diluted 10-fold up to 10−5 by using 1% NaCl. Five microliters of each dilution was spotted onto an LB plate in the presence of cross-inhibiting streaks of AB1157. After incubating the plate at 45°C for 16 to 24 h, individual spots at a 10−1 dilution were punched out, shaken at 28°C in 1 ml of 1% NaCl for 1 h to disperse the cells, serially diluted, spotted onto LB plates, and grown at 28°C for 16 to 24 h. Individual colonies were then counted to determine the number of CFU in the original spot grown at 45°C. The values were not normalized to the titer of the original culture that was grown overnight since we found that the titers of the cultures grown overnight were almost the same (±10%), independent of the genotype of the strain.
Pulsed-field gel electrophoresis.
Cultures of strains to be tested, which were grown at either 28 or at 45°C, were normalized to an optical density at 600 nm (OD600) of 0.3. Cells from 1.5 ml of the normalized cultures were sedimented by centrifugation and resuspended in 60 μl of TE buffer. After incubation at 37°C for 2 min, 5 μl of 10 mg of proteinase K/ml was added, followed by 65 μl of 1.2% molten agarose in 0.2% sarcosyl-10 mM Tris HCl (pH 8.0)-5 mM EDTA (kept at 70°C). After vortexing and mixing by pipetting, 110 μl of the cell suspension in agarose was pipetted into a plug mold and was allowed to solidify for 2 to 10 min. Each plug was then placed in a small glass tube containing 1 ml of 1% sarcosyl-50 mM Tris-HCl (pH 8.0)-25 mM EDTA and incubated for 1 h at 60°C. Plugs were inserted into 1% agarose gel on 0.5× Tris-borate-EDTA buffer and run in a Gene Navigator unit (Amersham-Pharmacia) with hexagonal electrodes (10°C, 180 V, ramp 3 to 165 s, 20 h).
Insertional mutagenesis.
For Tn5 mutagenesis, 109 cells of SK129 [AB1157 recBC(Ts)] carrying λ cI-expressing plasmid pKB252 (2) were infected with 108 particles of Tn5-carrying λ (MMS704 [Δb221 rex::Tn5 cI857], from Frank Stahl's collection) in a total volume of 150 μl of TM buffer (10 mM Tris HCl [pH 8.0], 10 mM MgSO4), giving a multiplicity of infection of 0.1. After 15 min on ice, injection was effected at 37°C for 5 min, and the adsorption mixture was transferred to 1.5 ml of LB broth and shaken at 28°C for 30 min. The culture was plated onto kanamycin- and tetracycline-supplemented LB plates which were incubated at 28°C for 2 days, and the colonies were screened for the inability to grow at 42°C on LB plates by toothpicking. The kanamycin resistance determinant from the temperature-sensitive candidates was P1 transduced into the recBC(Ts) parent to confirm the associated temperature sensitivity, indicative of dependence on the RecBCD enzyme. The position of the insert was identified by sequencing out of the transposon (using primer 93seq [5′-GCCCAGTCGGCCGCACGATG]) after subcloning the transposon halves with the neighboring chromosomal sequences and selection for the associated kanamycin resistance.
For an alternative insertional mutagenesis, we used pRL27, a plasmid carrying a hyperactive Tn5 transposase gene adjacent to the insertional cassette comprising two inverted Tn5 termini bracketing the kan gene plus the R6K pir-dependent origin (25). The cells of the strain to be mutagenized were electroporated with 10 ng of pRL27 and plated for kanamycin resistance with an immediate selection for the desired phenotype. Once the clones of interest were identified, total DNA from 2 ml of cultures grown overnight were prepared as described previously (24), and 1/20 of it was digested with MluI, ligated onto itself, and electroporated into DH5α pir+ cells with selection for kanamycin resistance. The retrieved plasmids were digested with MluI, and the smallest plasmid was selected for sequencing. The two primers used for sequencing of pRL27 inserts were 27 ori (5′-AACAAGCCAGGGATGTAACG [the ori side of the insert]) and 27 kan (5′-CAGCAACACCTTCTTCACGA [the kan side of the insert]).
Enrichment for mutants inhibited in the absence of RecBC.
The enrichment for mutants inhibited in the absence of RecBC is based on the observation that recombination-dependent mutants, although unable to form colonies when recombinational repair is inactivated, maintain viability for several hours and resume growth once the recombinational repair capacity is restored. Transient growth inhibition due to recombinational repair deficiency allows recombination-dependent mutants to survive treatments with antibiotics, like ampicillin, that kill growing cells. To enrich for recBC-dependent mutants, recBC(Ts) cells were mutagenized (as described above) and plated onto LB agar supplemented with kanamycin to obtain ∼1,000 colonies per plate. The next day, 1 ml of LB broth per plate was used to wash off and combine ∼5,000 colonies. The cell suspension was diluted to 107 cells/ml into 40 ml of LB broth, and the culture was pregrown at 42°C for 1 h to inhibit rec-dependent cells. At this point, ampicillin was added to 100 μg/ml, and incubation at 42°C was continued for 4 more hours, at which point the culture cleared, indicating cell lysis. The surviving cells were collected by centrifugation (15,000 × g, 10 min) and resuspended in 1 ml of LB broth without ampicillin. Fifty microliters of various dilutions of this suspension was spread to single colonies on LB plates supplemented with kanamycin and incubated at 28°C. The next day, 100 colonies were toothpicked to check for the inability to grow at 42°C. One of the multiple temperature-sensitive clones thus obtained (that were assumed to be all siblings) was saved for further analysis.
Sandwich plating.
A 25-ml LB plate was overlaid with a 3.5-ml layer of top agar containing 300 μl of a mid-log-phase AB1157 culture (∼1 × 108 cell/ml) and incubated at 37°C for 4 h. The plate was then overlaid with another 3.5 ml of top agar containing 175 μl of 100× stock of kanamycin (to eventually produce 25 μg of kanamycin/ml per entire ∼36-ml plate). After 2 min, a third 3.5-ml layer of top agar containing 500 μl of the pRL27-electroporated culture of the pta recBC(Ts) mutant was added. The plate was then incubated at 42°C for 36 to 48 h.
RESULTS
Isolation of mutants inhibited in the absence of RecBCD enzyme.
The recBC(Ts) mutants can grow at 42°C as long as they do not suffer chromosomal lesions that require recombinational repair. To find mutants that suffer from chromosomal lesions and that are therefore dependent on recombinational repair, random Tn5 insertions were selected in a recBC(Ts) strain at 28°C (permissive temperature) and screened for the inability to grow at 42°C (nonpermissive temperature), yielding an insertion in pta (Fig. 2). In an enrichment procedure for mutants unable to grow in combination with recBC inactivation at 42°C, a kan (pRL27) insertion into ackA was isolated (Fig. 2). Since the ackA and pta genes form a two-gene operon in E. coli, our isolation of the two mutations as inhibited at 42°C in the absence of RecBCD enzyme activity suggested that the ackA-pta pathway is somehow involved in the avoidance of chromosomal lesions in E. coli.
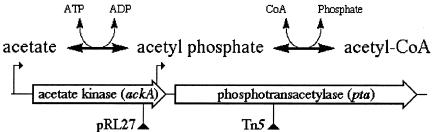
FIG. 2.
A scheme of acetate↔acetyl-CoA conversion and the genes involved. The elements of the ackA-pta operon are drawn to scale. The ackA gene is 1,202 bp, whereas the pta gene is 2,145 bp. The two promoters are shown as small arrows on long vertical stems. The positions of pRL27 and Tn5 inserts are indicated by filled triangles on vertical stems.
The ackA gene encodes acetate kinase, an enzyme that catalyses the interconversion of acetate and acetyl-phosphate. The downstream pta gene codes for phosphotransacetylase, an enzyme that catalyzes the interconversion of acetyl-phosphate and acetyl coenzyme A (acetyl-CoA). In effect, the pta-ackA pathway converts acetyl-CoA via acetyl-phosphate into acetate or vice versa, with the direction of the overall reaction dictated by growth conditions (Fig. 2) (6, 12, 37). In a minimal medium with acetate as the sole energy source, the ackA-pta pathway charges acetate on CoA, thus enabling acetate to enter the metabolism. In a rich medium, the pathway reverses (acetyl-CoA is converted to acetate), allowing the cell to dump the excess of acetylation potential in exchange for energy in the form of ATP. We set out to characterize the rec dependence of pta and ackA mutants in E. coli and aimed to identify the type of chromosomal damage involved when the ackA-pta pathway is inactivated.
Chromosomal fragmentation in ackA+ and ackA mutant cells.
Deletion-replacement alleles ΔackA::kan and Δpta::kan with the corresponding open reading frames precisely removed as well as an ΔackA allele with the drug resistance gene removed were constructed. Both ΔackA (data not shown) and Δpta (Fig. 3A) mutations made the original recBC(Ts) strain unable to grow at 42°C, confirming the observations made with Tn5 and pRL27 insertions.
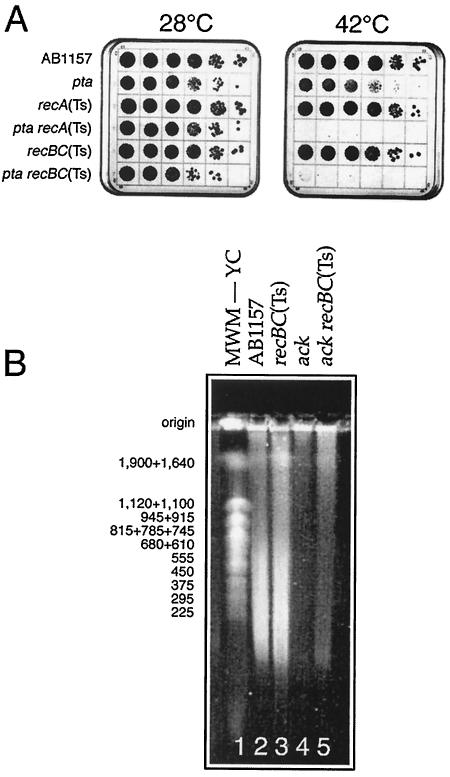
FIG. 3.
Δpta/ΔackA recBCD mutants are inviable and fragment their chromosome. A. pta recA(Ts) and pta recBC(Ts) are inviable at 42°C. Serial 10-fold dilutions of growing cultures were spotted onto plates which were incubated at the indicated temperatures for 36 to 48 h. The strains are as follows: for pta, JB17; for recA(Ts), JC9941; for pta recA(Ts), JS2; for recBC(Ts), SK129; for pta recBC(Ts), JS1. B. Pulsed-field gel of chromosomal DNA isolated from AB1157, recBC(Ts), ΔackA, and ΔackA recBC(Ts). The cells were grown at 45°C to an OD600 of 0.3. MWM—YC, yeast chromosomes serving as molecular weight markers (their length in kilobase pairs is indicated on the left). The strains are as follows: for recBC(Ts), SK129; for ackA, IYS4; for ackA recBC(Ts), IYS6.
In terms of the chromosome, dependence of a mutant on the RecBCD enzyme indicates the formation of double-strand DNA breaks, which translates into chromosomal fragmentation, detectable by pulsed-field gel electrophoresis (5, 19, 30). In pulsed-field gels, intact chromosomal DNA stays in the wells, whereas subchromosomal fragments migrate into the gel, forming a characteristic smear. We carried out pulsed-field gel electrophoresis of DNA isolated from ackA recBC(Ts) double mutants grown at 45°C as well as from the corresponding single mutants and wild-type cells. There was definitely more chromosomal fragmentation in the ackA recBC(Ts) double mutant than in the ackA single mutant (Fig. 3B, compare lanes 4 and 5). However, we have encountered two types of difficulties with quantification of chromosomal fragmentation in ackA recBC(Ts) double mutants: (i) an unexpectedly high level of chromosomal fragmentation in wild-type and recBC(Ts) mutant cells at 45°C (the temperature we had to use to obtain the ackA mutant phenotype [see below]) (Fig. 3B, lanes 2 and 3) and (ii) a much reduced chromosomal fragmentation in ackA mutants compared to that in the ackA+ cells, most likely due to the slow growth of the mutant (Fig. 3B, compare lanes 2 and 4 and lanes 3 and 5). These results suggest that (i) there is chromosomal instability of unknown origin in the wild-type cells at 45°C and (ii) the apparent inhibition of chromosomal fragmentation in recBC(Ts) mutant cells by the ackA mutation is due to the slow rate of DNA replication in ackA mutants. The fragmentation of the chromosome in wild-type cells at 45°C was particularly unexpected and warrants further investigation; however, such an investigation is beyond the scope of this project.
ΔackA and Δpta mutants depend on RecA and RecBC.
In terms of DNA repair, dependence of an E. coli mutant on RecBCD enzyme indicates that the mutant relies on either linear DNA degradation or recombinational repair of DNA nicks or breaks. If the former is true, the mutant is viable in combination with recA inactivation (18, 40); if the latter is the case, the mutant requires RecA as well as RecBCD (5, 16, 19). To see whether ackA and pta mutants depend on DNA degradation capabilities or recombinational repair capabilities of RecBCD, we combined them with the recA(Ts) mutation and found that the resulting strains are inviable at 42°C (Fig. 3A and data not shown), suggesting the dependence of ackA-pta mutants on recombinational repair.
pta and ackA mutants are sensitive to cross-inhibition.
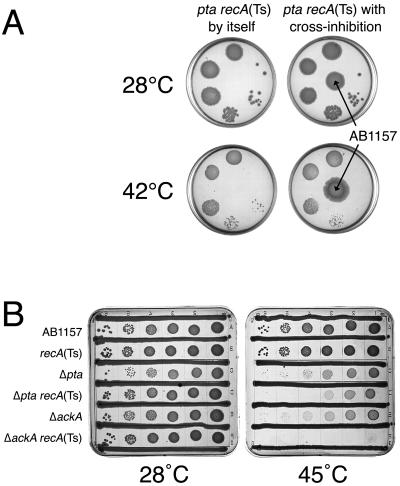
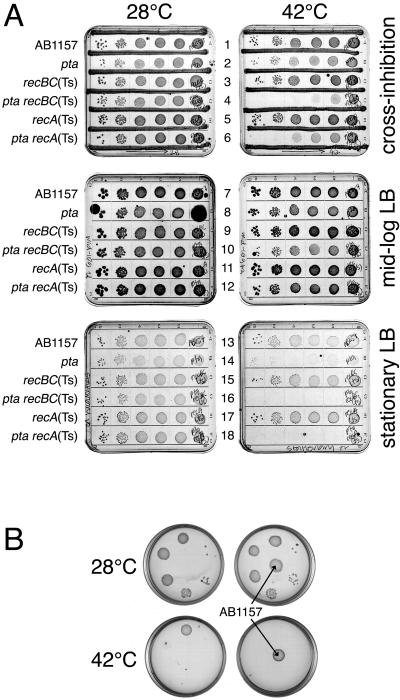
Remarkably, when we spot tested ackA rec (data not shown) or pta rec (Fig. 4A) cells at 42°C or even at 45°C on individual LB plates rather than on a common plate with other strains, the double mutants were able to form small colonies, suggesting that the inability of the same strains to form colonies on the original plates was due to cross-inhibition from the growing neighbors (which can be seen on the lower-right plate shown in Fig. 4A). To eliminate possible inconsistencies due to this cross-inhibition by neighbors, from then on, we scored ackA rec and pta rec double-mutant phenotypes on plates with cross-inhibiting streaks of AB1157, which made the mutant phenotype unequivocal (Fig. 4B).
FIG. 4.
pta/ackA rec mutants are inviable at temperatures of 42°C and higher in the presence of growing cells. Serial 10-fold dilutions of growing cultures were spotted onto plates which were incubated at the indicated temperatures. Where present, cross-inhibiting spots or streaks were those of AB1157. A. pta recA(Ts) is viable at 42°C when spotted onto a plate without other strains present. Incubation at both temperatures was done for 36 h. Note cross-inhibition from the AB1157 spot (the bottom-right plate). The strain is JS2. B. A typical cross-inhibition plate. Incubation was done for 36 h at 28°C but for only 24 h at 45°C. The strains are as follows: for recA(Ts), JC9941; for Δpta, JS4; for Δpta recA(Ts), JS6; for ΔackA, IYS4; for ΔackA recA(Ts), IYS7.
pta/ackA rec mutants are sensitive to medium modification.
The cross-inhibition of pta/ackA rec mutants could be due to either exhaustion of critical medium components or, alternatively, excretion of inhibitory waste substances by nearby growing cells. When we grew various strains on “spent” LB agar (AB1157 was grown to saturation, the cells were removed, and the medium was made solid by the addition of agar), all strains grew (slowly) at 28°C; at 42°C, the pta single mutant grew extremely poorly, while pta rec double mutants failed to grow completely (Fig. 5A, rows 13 to 18). When we grew the same strains on partially spent (“mid-log” LB) medium (obtained by growing AB1157 in LB medium until an OD600 of 0.5 was reached, removing cells, and making a solid medium out of such a “preconditioned” LB medium), the effect was opposite: at 42°C, the double mutants grew almost as well as single mutants and actually grew better than they did on the regular LB agar with cross-inhibition (Fig. 5A, compare rows 7 to 12 with rows 1 to 6), as if pta mutants were helped by the products excreted by mid-log-phase cells.
FIG. 5.
pta recA(Ts) and pta recBC(Ts) cells cannot grow at 42°C on spent medium or on medium containing only yeast extract. Serial 10-fold dilutions of growing cultures were spotted onto plates which were incubated at the indicated temperatures for 36 to 48 h. Where present, cross-inhibiting spots or streaks were those of AB1157. A. pta mutants grow better on a mid-log medium but cannot grow on a spent medium. Stationary LB, a sterilized supernatant of a saturated AB1157 culture solidified with 1.5% agar; mid-log LB, a sterilized supernatant of a growing AB1157 culture (OD600 of 0.5) solidified with 1.5% agar. The strains are as follows: for pta, IYS3; for recBC(Ts), SK129; for pta recBC(Ts), IYS1; for recA(Ts), JC9941; for pta recA(Ts), IYS2. B. Yeast extract medium does not support growth of a pta recA(Ts) mutant at 42°C. The medium contains (per liter) 2.5 g of YE, 5 g of NaCl, and 15 g of agar (pH 7.2). The strain is JS6.
We also found that of the two major components of LB agar, tryptone enhanced the growth of the double mutants (data not shown), while YE inhibited it. This inhibition was so strong that on the medium composed of only 0.5× YE (without tryptone), a pta recA(Ts) mutant would not grow at 42°C even in the absence of cross-inhibition (Fig. 5B, lower-left plate). At the same time, all other strains tested, including single pta mutants, had no problem growing on this yeast extract medium (data not shown). We conclude that ackA rec and pta rec mutants at 42°C are sensitive to products accumulated in stationary-phase cultures and some components of the yeast extract. Alternatively, the double mutants may require some substances in tryptone that are exhausted by other cells growing nearby.
pta rec mutants are sensitive to products of mixed-acid fermentation of pyruvate.
When grown aerobically in rich medium, E. coli turns pyruvate exclusively into acetyl-CoA and then uses the pta-ackA pathway to dump the excess acetate, which is excreted and then reused as the cells enter stationary phase (8, 12). To test the idea that cross-inhibition is due to pta rec double mutant sensitivity to acetate excreted by their pta+/ackA+ neighbors, we plated pta recA(Ts) and pta single-mutant strains at 42°C on LB plates supplemented with increasing concentrations of acetate. We found that the double mutant could not grow in the presence of acetate in excess of 50 to 60 mM, whereas the single mutants grew normally in the presence of 150 mM acetate (Table 3). Still, the inhibiting concentration of acetate was at least 10 times higher than the maximal concentration of acetate that accumulated in cultures of rapidly growing cells (8, 45). Moreover, the pta/ackA rec double-mutant strains were inhibited equally well by growing pta/ackA single-mutant neighbors (data not shown), ruling out acetate as the inhibitor.
TABLE 3.
Concentrations of various metabolites that completely inhibit growth of pta recA(Ts) mutants at 42°C
| Metabolite | Concn that inhibits growth completely (mM)a
|
|
|---|---|---|
| pta recA(Ts) | ptab | |
| (Sodium) acetate | 50 | >150 |
| (Ammonium) acetate | 60 | >150 |
| Ammonium chloride | 60 | >150 |
| Ethanol | 3.0% | >5.0% |
| Lactic acid | 0.05% | >0.1% |
| (Sodium) formate | 85 | 120 |
| (Sodium) pyruvate | 60 | >150 |
| (Sodium) succinate | 80 | >150 |
Total concentration per plate in mM or in percent (where indicated). Concentrations that still fail to inhibit pta single mutants are shown for comparison.
Wild-type cells and recA(Ts) mutants were also not inhibited at these concentrations (in excess of 150 mM for salts, 5.0% for ethanol, and 0.1% for lactic acid).
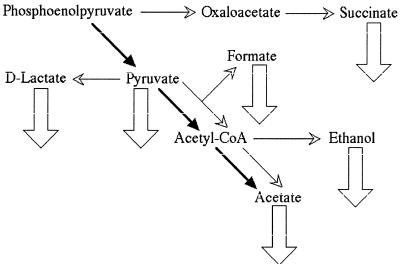
In an attempt to find the substance excreted by the cells growing on LB plates which would inhibit pta rec double mutants in low concentrations, we considered a possibility that cells growing in colonies experience a microaerobic environment, which we thought might lead to the activation of mixed-acid fermentation of pyruvate, resulting in excretion of not only acetate but also lactate, ethanol, formate, succinate, and pyruvate itself (Fig. 6) (45). We found that wild-type cells as well as single pta and recA(Ts) mutants can grow on LB plates supplemented with high concentrations of these various metabolites (Table 3). In contrast, pta recA(Ts) mutants are completely inhibited by lower concentrations of all these metabolites, suggesting that cross-inhibition could indeed be due to excretion of these products into the medium surrounding growing cells. However, there was not a single metabolite among those we tested that would inhibit the double pta recA(Ts) mutant in submillimolar or even low-millimolar concentrations (expected during cross-inhibition). Moreover, we found that even ammonium chloride inhibits pta recA(Ts) mutants in similar concentrations (Table 3), suggesting that these mutants may generally be sensitive to salts in concentrations that are not inhibitory to pta or rec single mutants.
FIG. 6.
Metabolic neighbors of the pta-ackA pathway during anaerobic growth in a rich medium. The aerobic overflow pathway (phosphoenolpyruvate→pyruvate→acetyl-CoA→acetate) is indicated by boldface arrows. The anaerobic mixed-acid fermentation is shown by thin arrows with open arrowheads. Excreted products are marked by thick downward open arrows. Only the key substances are shown.
ackA rec and pta rec double mutants are synthetically inhibited rather than synthetically lethal.
The inability to form colonies under nonpermissive conditions (inviability) may represent either death of the cells (lethality) or inhibition without actual loss of viability. The two alternatives can be distinguished by first subjecting cells to nonpermissive conditions, making sure they do not form colonies, and then shifting the cells to permissive conditions to detect decreases in colony-forming ability. Tests like these, conducted previously with dut-1 rec and tdk rec mutants (E. A. Kouzminova and A. K. Kuzminov, unpublished data), demonstrated quantitative killing of the double mutants under the nonpermissive conditions. However, when we returned pta rec or ackA rec mutants to permissive conditions, not only did they form colonies, but also the titer of the CFU was the same as that in cultures incubated throughout under permissive conditions (data not shown). Thus, pta rec and ackA rec mutants showed “synthetic inhibition” rather than synthetic lethality.
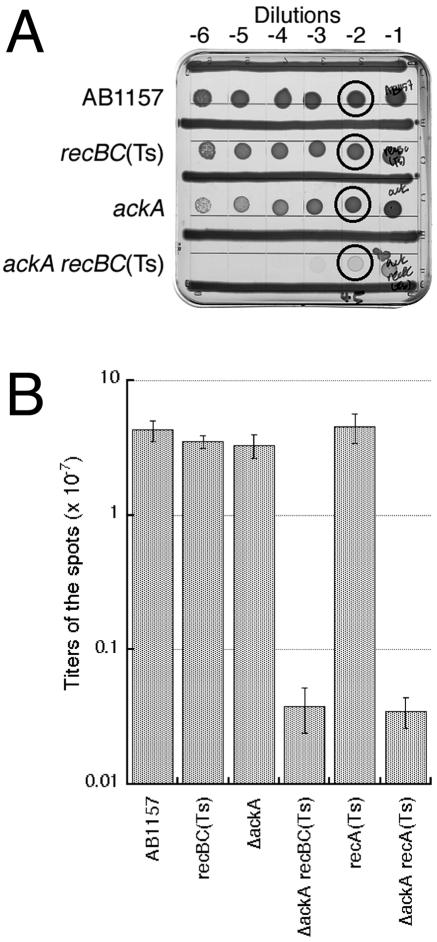
To quantify this inhibition, we spotted serial dilutions of saturated cultures, grew the spots under inhibitory conditions, and determined the titer of the CFU in the “−2” dilution spot at the permissive conditions (Fig. 7A). The titers of single rec or ackA mutant spots determined by this procedure were similar to the titer of spots of wild-type cells (Fig. 7B). However, the titer of ackA rec mutant spots was down by 2 orders of magnitude. Thus, although ackA rec and pta rec mutant cells do not lose viability in the nonpermissive conditions, they in effect form 100-times-smaller colonies, which makes them almost invisible. We suggest that ackA rec and pta rec combinations be called synthetically inhibited rather than synthetically lethal.
FIG. 7.
Quantification of ackA recBC(Ts) and ackA recA(Ts) inhibition relative to wild-type or single mutant strains. A. A representative plate to show the spots that were taken out for the analysis (circled). The strains are as follows: for recBC(Ts), SK129; for ackA, IYS4; for ackA recBC(Ts), IYS6. B. The titers of CFU. The data are averages of two to five independent determinations ± standard errors.
pta mutants depend on RecG and RuvABC but not RecF.
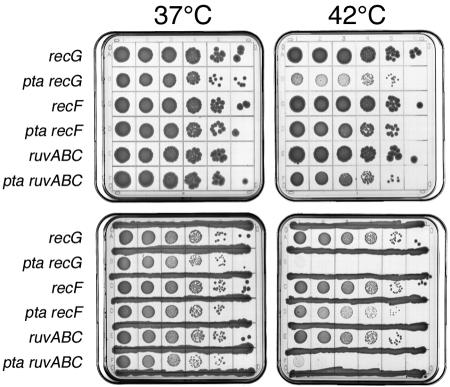
As pointed out above, recombinational repair of chromosomal lesions has three phases (Fig. 1). First, the targeting or licensing activities of RecBCD and RecFOR recognize specific chromosomal lesions and target the RecA protein to these lesions, licensing it to displace ssDNA-binding protein from the lesion-associated ssDNA. Second, RecA filament catalyzes homologous pairing and strand exchange between the affected and the intact DNA duplexes, forming hybrid duplexes between Holliday junctions. Third, after the now one-strand DNA lesions have been repaired in the hybrid duplexes, RuvABC or RecG activities remove spent RecA filaments and resolve the DNA junctions, restoring functional chromosomes. Our demonstration of the RecA and RecBC dependence of pta and ackA mutants prompted us to see if other recombinational repair activities are required when the acetyl-CoA↔acetate pathway is inactivated. We found that ackA and pta mutants do not require RecF, either with or without cross-inhibition (Fig. 8 and data not shown), suggesting that the chromosomal lesions in these mutants do not include daughter-strand gaps. On the other hand, we found that pta mutants depend on both RecG and RuvABC (with cross-inhibition at 42°C) (Fig. 8). This result is unexpected because the late phase of recombinational repair is believed to be catalyzed by either activity, so RecA-dependent mutants usually show dependence on both late activities but could survive if one of them is still active. In contrast to the Δpta mutant, our ΔackA mutant was viable in combination with either recG or ruvABC mutations (data not shown), which was the only difference in phenotypes between pta and ackA mutants detected in this work.
FIG. 8.
pta mutants are viable in combination with recF inactivation but cannot tolerate recG or ruvABC inactivation at 42°C. Serial 10-fold dilutions of growing cultures were spotted onto plates which were incubated at the indicated temperatures for 36 to 48 h. Where present, cross-inhibiting spots or streaks were those of AB1157. The strains are as follows: for recG, N4452; for pta recG, JS9; for recF, JC9239; for pta recF, JS8; for ruvABC, AM547; for pta ruvABC, JS7.
Curiously, although the recG and ruv mutations we used were complete inactivations and the double mutants with pta were inviable at 42°C, they grew normally at 37°C (Fig. 8), suggesting a temperature-sensitive nature of the pta (and, by analogy, ackA) defect. To verify this suspicion, we attempted to build ΔrecA pta/ackA and ΔrecBCD pta strains at low temperature, and we indeed recovered the double mutants. The mutants were fully viable at 28 or even 37°C but, as expected, were unable to form colonies at 42 or 45°C, especially with cross-inhibition (data not shown). We conclude that pta/ackA strains have problems with chromosomes only at high temperature, especially when there are growing cells nearby.
Suppressors of pta rec synthetic inhibition.
Although AB1157 is a standard wild-type strain for DNA metabolism studies of E. coli, it was derived through several mutagenesis rounds (1) and may therefore contain uncharacterized metabolic defects which modify behaviors of other mutations. Indeed, when we introduced ΔackA or Δpta mutants into the less-manipulated MG1655 strain (1), we observed only moderate inhibition in recA or recBCD mutant variants (data not shown), suggesting that the pta rec and ackA rec inhibition is specific for the AB1157 background. However, since we were interested in the mechanisms of Rec dependence, we could still study them in the AB1157 background, where the inhibition of ackA rec and pta rec double mutants is strong.
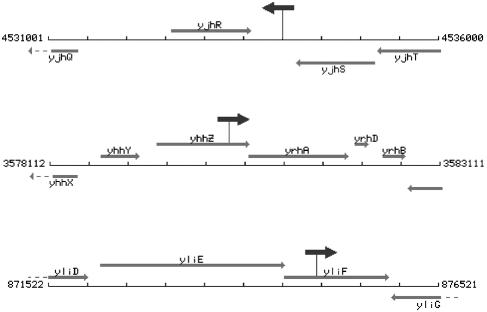
To gain insights into the mechanism of inviability of pta rec mutants, we employed pRL27 mutagenesis to isolate inactivational suppressors which allow pta rec mutants to grow under nonpermissive conditions. For this, we carried out the insertional mutagenesis in the pta recBC(Ts) mutants and plated the mutagenized cells with cross-inhibition at 42°C (see Materials and Methods). The chromosomal inserts from the colonies that grew under these nonpermissive conditions were P1 transduced into the parental pta recBC(Ts) strain and confirmed to suppress its synthetically lethal phenotype. This way, we isolated three weak suppressors, allowing slow growth of the resulting triple mutants (data not shown). We identified positions of inserts in these suppressors by sequencing (Fig. 9). One insert was found in the 583-bp intergenic region between the convergent yjhR and yjhS genes, the second insert was found to inactivate the yhhZ gene, and the third insert inactivated the yliF gene; all genes are currently annotated as “function unknown.” The results of the suppressor analysis are considered in Discussion.
FIG. 9.
Position and orientation of pRL27 inserts relative to the chromosomal regions in the suppressor mutants. The vicinity maps of the disrupted gene with exact chromosomal coordinates were generated with the Colibri server (http://genolist.pasteur.fr/Colibri/genome.cgi). Open reading frames and their direction are shown by thin gray arrows identified by gene names. The positions of pRL27 inserts are shown by short arrows on sticks, with the orientation of these arrows indicating the orientation of the kan gene of the insert.
DISCUSSION
In a screen for mutants dependent on the RecBCD enzyme in E. coli, we tested random Tn5 and pRL27 insertions in the AB1157 recBC(Ts) mutant for the inability to grow at 42°C. We isolated two recBC-dependent mutations which inactivated ackA and pta, the genes that form a two-gene operon responsible for the interconversion of acetate and acetyl-CoA. Subsequent experiments with precise deletion-replacement alleles showed that (i) complete inhibition of ackA rec and pta rec mutants is observed only when they share the same plate with growing E. coli strains (cross-inhibition); (ii) the same effect is emulated by medium modification (yeast extract only, end products of the neighboring metabolic branches); (iii) both ackA and pta mutants fail to grow in combination with recA and recBC mutations but show no effect when combined with a recF inactivation; (iv) the failure of ackA rec and pta rec mutants to form colonies under nonpermissive conditions is due to inhibition (extremely slow growth) rather than lethality (loss of colony-forming potential); (v) this synthetic inhibition is more pronounced in the AB1157 background, perhaps due to an uncharacterized additional mutation; and (vi) in contrast to ackA mutants, pta mutants are also inviable in combination with ruvABC and recG mutations, suggesting a possible role for acetyl phosphate at late stages of recombinational repair.
The dependence of a mutant on recombinational repair is generally interpreted as an increased level of chromosomal lesions caused by the mutation (5, 19), but the mechanisms leading to chromosomal lesions in pta and ackA mutants are currently obscure. One (less likely) possibility is that the opposite defects in production of acetyl phosphate (too much acetyl-phosphate in ackA mutants and too little acetyl-phosphate in pta mutants), which is a known signaling metabolite (27, 42, 44), cause chromosomal lesions via disregulated signaling. This explanation predicts that the synthetic inhibition of pta/ackA rec double mutants will be relieved by inactivation of the target mechanisms of acetyl phosphate signaling. One of the isolated suppressors of the pta recBC synthetic inhibition inactivates yliF, which codes for a GGDEF family protein of unknown function; the members of this family are widespread in eubacterial genomes and are often found in association with two-component response regulators. The enzymatic reaction catalyzed by these proteins is nucleotide cyclization (13).
Another explanation for the synthetic inhibition of pta rec and ackA rec mutants is that the inability of pta and ackA mutants to dump the excess of acetyl-CoA, which is a source of acetyl groups for numerous acetylation enzymes, causes chromosomal lesions via spurious acetylation of DNA-processing enzymes. This explanation predicts that suppressors of pta/ackA rec synthetic inhibition should inactivate acetyltransferases that catalyze this acetylation. Another insertional suppressor of the pta recBC inviability that we found in yhhZ is just downstream of yhhY, which is a putative acetyltransferase of the GNAT family. If the YhhY protein is somehow inactive in our yhhZ mutant, this would be consistent with the idea that the excess of acetyl-CoA causes spurious acetylation.
Yet another explanation for the synthetic inhibition of pta/ackA rec mutants is that the observed upregulation of the tricarboxylic acid cycle in pta/ackA mutants (44) leads to an increase in reactive oxygen species, elevating the background DNA modification. This explanation predicts that suppressors of pta/ackA rec synthetic inhibition may slow down the tricarboxylic acid cycle or inactivate DNA repair activities that remove modified DNA bases, thus interfering with DNA replication. The third suppressor insert landed in the intergenic region between yjhR and yjhS. yjhR, a gene likely to be affected by the insertion due to the antisense effect of the kanamycin resistance gene expression, codes for a putative DNA helicase of superfamily I, to which RecB and RecD helicases (components of the RecBCD enzyme) also belong (15). Thus, yjhR suppressor is compatible with the DNA modification or repair explanation. On the other hand, yjhS codes for a conserved hypothetical protein which is highly related (e-102) to a protein from a cryptic prophage, CP-933P of E. coli O157:H7 EDL933. Interestingly, among the distant homologs of YjhS is a probable acetyl xylan esterase, AxeA, with a possible function in polysaccharide degradation (2e-04) and, with a very low score (8.0), a putative N-acetyltransferase. In summary, the results of our suppressor analysis are intriguing and suggestive but at this point do not allow us to clearly distinguish among the three possible reasons (spurious acetylation of proteins, signaling defect, or DNA modification or repair) why ackA/pta rec double mutants are synthetically inhibited.
Delineating the phenomena of synthetic lethality and synthetic inhibition proved to be the main crux of this work. Conventional tools used in previous studies of recA synthetic lethals, like quantification of chromosomal fragmentation by pulsed-field gel electrophoresis and suppressor analysis (5, 19, 38), are harder to implement in the case of ackA and pta mutants. Finding better conditions will allow us to isolate more suppressors, while biochemical characterization of the proteins inactivated by suppressor mutations should provide insights into the mechanisms involved in conditional recombinational repair dependence of pta and ackA mutants.
Acknowledgments
We are indebted to Jill S. Bradshaw for isolation of the original pta::Tn5 insertion mutant and for construction of the initial Δpta::cam allele and to Paula Welander for isolation of the original ackA::pRL27 mutant. We thank Alvin John Clark, Ichizo Kobayashi, Richard Kolodner, Sydney Kushner, Robert G. Lloyd, William Metcalf, and Benedicte Michel for providing strains and plasmids and Elena Kouzminova for her help, encouragement, and discussion of results as well as for critical reading of the manuscript.
This work was supported by grant MCB-0196020 from the National Science Foundation.
REFERENCES
- 1.Bachmann, B. J. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 1190-1219. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 2.Backman, K., M. Ptashne, and W. Gilbert. 1976. Construction of plasmids carrying the cI gene of bacteriophage λ. Proc. Natl. Acad. Sci. USA 73:4174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt, T. G., and P. A. De Boer. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52:1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradshaw, J. S., and A. Kuzminov. 2003. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol. Microbiol. 48:1711-1725. [DOI] [PubMed] [Google Scholar]
- 6.Brown, T. D. K., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 7.Capaldo, F. N., G. Ramsey, and S. D. Barbour. 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 118:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, D. E., S. Shin, J. S. Rhee, and J. G. Pan. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 181:6656-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. M. 2001. Historical overview: searching for replication help in all the rec places. Proc. Natl. Acad. Sci. USA 98:8173-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobzhansky, T. 1946. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics 31: 269-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.el-Mansi, E. M., and W. H. Holms. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 135:2875-2883. [DOI] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, M. F. 2000. Coping with replication ‘train wrecks’ in Escherichia coli using Pol V, Pol II and RecA proteins. Trends Biochem. Sci. 25:189-195. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitman, J., T. Ivanenko, and A. Kiss. 1999. DNA nicks inflicted by restriction endonucleases are repaired by a RecA- and RecB-dependent pathway in Escherichia coli. Mol. Microbiol. 33:1141-1151. [DOI] [PubMed] [Google Scholar]
- 17.Horii, Z.-I., and A. J. Clark. 1973. Genetic analysis of the RecF pathway of genetic recombination in Escherichia coli K-12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 18.Itaya, M., and R. J. Crouch. 1991. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol. Gen. Genet. 277:424-432. [DOI] [PubMed] [Google Scholar]
- 19.Kouzminova, E. A., and A. Kuzminov. 2004. Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol. Microbiol. 51:1279-1295. [DOI] [PubMed] [Google Scholar]
- 20.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 21.Kushner, S. R. 1974. In vivo studies of temperature-sensitive recB and recC mutants. J. Bacteriol. 120:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzminov, A. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 98:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 26.Mandal, T. N., A. A. Mahdi, G. J. Sharples, and R. G. Lloyd. 1993. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J. Bacteriol. 175:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrill, B. J., and C. Holm. 1998. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics 148:611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrill, B. J., and C. Holm. 1999. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defect of mec1 mutants. Genetics 153:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel, B., G. D. Recchia, M. Penel-Colin, S. D. Ehrlich, and D. J. Sherratt. 2000. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol. Microbiol. 37:180-191. [DOI] [PubMed] [Google Scholar]
- 32.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda, A., and A. Kuzminov. 2003. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 163:1255-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 35.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277-286. [DOI] [PubMed] [Google Scholar]
- 36.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose, I. A., M. Grunberg-Manago, S. R. Korey, and S. Ochoa. 1954. Enzymatic phosphorylation of acetate. J. Biol. Chem. 211:737-756. [PubMed] [Google Scholar]
- 38.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509:49-78. [DOI] [PubMed] [Google Scholar]
- 40.Uzest, M., S. D. Ehrlich, and B. Michel. 1995. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 17:1177-1188. [DOI] [PubMed] [Google Scholar]
- 41.Valerie, K., and L. F. Povirk. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22:5792-5812. [DOI] [PubMed] [Google Scholar]
- 42.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon. p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 43.Webb, B. L., M. M. Cox, and R. B. Inman. 1997. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91:347-356. [DOI] [PubMed] [Google Scholar]
- 44.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 45.Xu, B., M. Jahic, G. Blomsten, and S. O. Enfors. 1999. Glucose overflow metabolism and mixed-acid fermentation in aerobic large-scale fed-batch processes with Escherichia coli. Appl. Microbiol. Biotechnol. 51:564-571. [DOI] [PubMed] [Google Scholar]