Abstract
Anaerobic biodegradation of toluene and ethylbenzene is of environmental concern and biochemical interest due to toxicity and novel reactions, respectively. The denitrifying strain EbN1 is unique in anaerobically degrading both alkylbenzenes via different pathways which converge at benzoyl coenzyme A. The organization of genes involved in both pathways was only recently determined for strain EbN1. In the present study, global expression analysis (DNA microarray and proteomics) indicated involvement of several thus-far-unknown proteins in the degradation of both alkylbenzenes. For example, orf68 and orf57, framing the ebd operon, are implicated in ethylbenzene degradation, and the ebA1932 and ebA1936 genes, located 7.2 kb upstream of the bbs operon, are implicated in toluene degradation. In addition, expression studies were now possible on the level of the complete pathways. Growth experiments demonstrated that degradative capacities for toluene and ethylbenzene could be simultaneously induced, regardless of the substrate used for adaptation. Regulation was studied at the RNA (real-time reverse transcription-PCR and DNA microarray) and protein (two-dimensional-difference gel electrophoresis) level by using cells adapted to anaerobic growth with benzoate, toluene, ethylbenzene, or a mixture of toluene and ethylbenzene. Expression of the two toluene-related operons (bss and bbs) was specifically induced in toluene-adapted cells. In contrast, genes involved in anaerobic ethylbenzene degradation were induced in ethylbenzene- and toluene-adapted cells, suggesting that toluene may act as a gratuitous inducer. In agreement with the predicted sequential regulation of the ethylbenzene pathway, Ebd proteins (encoding subunits of ethylbenzene dehydrogenase) were formed in ethylbenzene- but not in acetophenone-adapted cells, while Apc proteins (subunits of predicted acetophenone carboxylase) were formed under both conditions.
Alkylbenzenes are abundant constituents of crude oil and fuels and are widely used solvents and starting compounds in chemical synthesis. Among them, benzene, toluene, ethylbenzene, and xylenes have attracted particular attention due to their toxicity. Environmental pollution by crude oil or fuels often affects (leads to) anoxic systems such as groundwater aquifers, leading to the demand for knowledge about bacterial degradative capacities under anoxic conditions. In recent years, numerous pure cultures of novel bacteria which degrade alkylbenzenes completely to CO2 in the absence of molecular oxygen were isolated (for an overview, see references 22, 47, 52, and 53). Most of these bacteria are denitrifying isolates belonging to a recently recognized phylogenetic group clustering with the known genera Azoarcus and Thauera within the Betaproteobacteria. Due to the remarkable degradative capacities of its members, this group is currently described as a new genus, and the Azoarcus-like strain EbN1 is described as a new species therein (R. Rabus et al., unpublished data).
Biodegradation of alkylbenzenes has to overcome their exceptional chemical stability. Aerobic bacteria achieve this by employing highly reactive oxygen species in monooxygenase- or dioxygenase-catalyzed reactions. An understanding of the fundamentally different reactions used by anaerobic bacteria was obtained only in recent years (for an overview, see references 22, 47, 52, and 53).
Benzylsuccinate synthase catalyzes the initial formation of benzylsuccinate from toluene and fumarate (Fig. 1A). The glycyl radical of the active enzyme is generated by a bssD-encoded activase (1, 3, 10, 15, 26, 30, 32). Benzylsuccinate is further degraded to the common aromatic intermediate benzoyl coenzyme A (benzoyl-CoA) via a modified β-oxidation pathway (33-35). In denitrifying bacteria, anaerobic degradation of ethylbenzene (Fig. 1B) is initiated by the molybdenum-containing ethylbenzene dehydrogenase forming (S)-1-phenylethanol (2, 24, 25, 28, 29, 38, 39, 42). The maturation of this periplasmic enzyme is presumably mediated by the ebdD product. Subsequent conversion to benzoyl-CoA is proposed to involve a carboxylation reaction and thiolytic removal of an acetyl-CoA moiety. Further degradation of the common intermediate benzoyl-CoA begins with a reductive dearomatization catalyzed by benzoyl-CoA reductase (4) and proceeds via hydrolytic ring cleavage and β-oxidation to acetyl-CoA units. The latter units are then terminally oxidized to CO2 (4, 21).
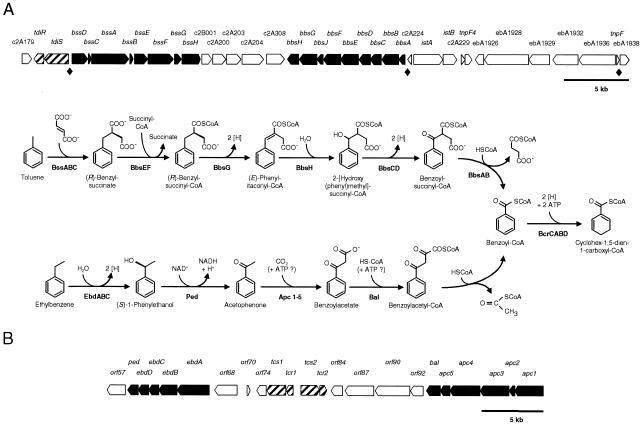
FIG. 1.
Anaerobic toluene and ethylbenzene degradation in strain EbN1 proceed via different reaction sequences to the first common intermediate, benzoyl-CoA. (A) Pathway of anaerobic toluene degradation (modified from references 4 and 33). Enzyme names of indicated gene products (shown in boldface type) (31) are as follows: BssABC, benzylsuccinate synthase; BbsEF, succinyl-CoA:(R)-benzylsuccinate CoA-transferase; BbsG, (R)-benzylsuccinyl-CoA dehydrogenase; BbsH, phenylitaconyl-CoA hydratase; BbsCD, 2-[hydroxy(phenyl)methyl]-succinyl-CoA dehydrogenase; BbsAB, benzoylsuccinyl-CoA thiolase. (B) Pathway of anaerobic ethylbenzene degradation (modified from references 2, 28, 38, and 39). Enzyme names of indicated gene products (shown in boldface type) (42) are as follows: EbdABC, ethylbenzene dehydrogenase; Ped, (S)-1-phenylethanol dehydrogenase; Apc1-5, acetophenone carboxylase; Bal, benzoylacetate CoA-ligase. Anaerobic degradation of benzoyl-CoA is initiated by benzoyl-CoA reductase (BcrCABD) and then further oxidized via reductive ring cleavage to carbon dioxide (not shown; 4). Reducing equivalents ([H]) are used for the reduction of nitrate to dinitrogen. A scale model for the organization of the involved genes is displayed for both pathways. Annotations for ebA1926 through ebA1938 genes are provided in Table 4, while those for all other genes have already been described elsewhere (31, 42). ♦, consensus sequence TTA(A/G)GTGTTCGCACCAATTG in the promoter regions of bssD, bbsA, and the ebA1936 gene is located 118, 68, and 450 bp upstream of the respective translational starts.
Strain EbN1 is unique among known alkylbenzene degraders for its capacity to anaerobically degrade toluene as well as ethylbenzene. These alkylbenzenes are utilized not only when supplied as pure substances (39) but also directly from crude oil (40). Strain EbN1 employs the above-described pathways, which were previously suggested to be regulated by their respective substrates (6). The complete genetic blueprints for both pathways in strain EbN1 include three related two-component regulatory systems (31, 42). Only recently was the complete genome sequence of strain EbN1 determined (42a).
Regulation of anaerobic hydrocarbon degradation is to date only poorly understood. Initial studies were concerned with the bss operon of anaerobic toluene degradation (8, 23). The genomic reconstruction of anaerobic toluene and ethylbenzene metabolism in strain EbN1 (31, 42) allowed for the first time an investigation of regulation of anaerobic hydrocarbon degradation on the level of the complete pathways. Physiological adaptation experiments were combined with global expression profiling (DNA microarrays and proteomics) to pursue the following lines of research: (i) the influence of adaptation substrates on induction of pathways, (ii) simultaneous induction and activity of both pathways, (iii) differing modes of regulation for the two pathways, (iv) specificities of the predicted sensors for their respective substrates, and (v) the search for additional gene products not previously correlated with the two pathways.
MATERIALS AND METHODS
Media and cultivation.
The denitrifying bacterium strain EbN1 was cultivated under nitrate-reducing conditions as previously described (39). Substrates with low solubility in water were provided as dilutions in 2,2,4,4,6,8,8-heptamethylnonan (HMN) rather than added directly to the medium. The used chemicals were of analytical grade, and purity of toluene and ethylbenzene was confirmed by gas chromatographic analysis.
Mass cultivation was performed to supply sufficient cell material for RNA and protein analysis. Substrate-adapted cells (see below) were used for inoculation. To reduce slime formation, a phosphate-buffered mineral medium supplemented with NaCl (1 g/liter) was used (51). For DNA microarray and two-dimensional-difference gel electrophoresis (2D DIGE) experiments, strain EbN1 was grown with the following substrate concentrations (vol/vol) in HMN: 1% toluene, 2.5% ethylbenzene, a mixture of 0.5% toluene and 1.25% ethylbenzene, and 1% acetophenone. For real-time reverse transcription (RT)-PCR experiments, cultures with the following substrate concentrations (vol/vol) in HMN were used: 1% toluene, 2% ethylbenzene, and a mixture of 1% toluene and 2% ethylbenzene. The concentration of benzoate was 4 mM. Cultivation was carried out in 500-ml bottles containing 400 ml of medium, 20 ml of carrier phase, and 80 ml of headspace. Up to 20 parallel cultures for each substrate condition were harvested at an optical density at 660 nm of around 0.2, corresponding to the mid-exponential-growth phase, as described previously (6). Cells were washed twice with 100 mM Tris-HCl (pH 7.5) containing 5 mM MgCl2, and the obtained pellets were immediately frozen in liquid nitrogen and stored at −80°C.
Physiological adaptation experiments.
Cells were adapted to growth with toluene, ethylbenzene, a mixture of both alkylbenzenes, or benzoate over at least five passages. These four differently adapted subcultures served as inoculum for subsequent cultivation under three different substrate conditions: toluene (0.3%, vol/vol), ethylbenzene (0.3%, vol/vol), and a mixture of both alkylbenzenes (each 0.15%, vol/vol). Chemical analysis of alkylbenzenes (see below) required reduced substrate concentrations in HMN. Controls lacked either organic substrate or inoculum. During incubation, samples were withdrawn from the carrier phase (0.2 ml) and the aqueous phase (1 ml) at intervals of about 3 h. To maintain anoxic conditions during sampling, N2-flushed sterile syringes were used. Samples from the carrier phase were stored in Teflon-sealed 0.7-ml flange bottles (WiCom, Heppenheim, Germany) and subsequently analyzed by gas chromatography to determine substrate consumption (see below). Samples from the aqueous phase were directly used to monitor growth by measuring the optical density at 660 nm (UV-mini 1240; Shimadzu, Duisburg, Germany). The end of incubation was indicated by the depletion of the electron acceptor nitrate and intermediately produced nitrite. This process was monitored with Merckoquant test strips (Merck, Darmstadt, Germany). Four parallels (based on two independent inoculum cultures) were carried out for each tested substrate condition. Parallel cultures yielded highly similar time courses of growth and alkylbenzene utilization.
Chemical analysis.
Concentrations of toluene and ethylbenzene in samples from the carrier phase were determined by gas chromatography. The system consisted of a gas chromatograph equipped with a flame ionization detector (Perkin-Elmer Autosystems, Rodgau-Jügesheim, Germany) and an OPTIMA-5 column (0.25 μm by 50 m; Macherey-Nagel, Düren, Germany). Retention times were 4.6 min (toluene) and 7.1 min (ethylbenzene). The dynamic range was between 0.1 and 10 mM in both cases. Undiluted samples (1 μl) of the carrier phase were injected. Separation was achieved with hydrogen as the carrier gas at a flow rate of 120 ml min−1, a split of 1:70, and the following temperature gradient: injection port temperature, 250°C; column temperature, 40°C for 2 min, ramping at 4°C min−1, 60°C for 0.1 min, ramping at 20°C min−1, and 320°C for 10 min; flame detector temperature, 350°C.
Preparation of mRNA.
Total RNA was prepared from substrate-adapted cells by using the hot-phenol method described previously by Oelmüller et al. (36). To consider biological variations, RNA was prepared from two independent cultures for each substrate condition. After treatment with RNase-free DNase (MBI Fermentas, St. Leon Roth, Germany), the removal of DNA was confirmed by the absence of target genes which are amplifiable by PCR, RNA quality was controlled as described previously (31), and RNA aliquots were further purified with RNeasy Mini purification columns (QIAGEN, Hilden, Germany).
Real-time RT-PCR.
Gene-specific primers (Table 1) were designed with the MacVektor program (Accelrys, Munich, Germany). The antisense primers were used for reverse transcription with H minus M-MLuV reverse transcriptase (MBI Fermentas) applied according to the manufacturer's instructions and using 2 μg of total RNA. A 40-cycle real-time PCR was performed on a 25-μl scale by using a SYBR Green ready mix (Eurogentec, Seraing, Belgium) and standard PCR conditions. For each reaction, 2 μl of the individual RT reactions served as a template. Primer pairs complementary to the bcrC, bssA, and ebdA sequences were used to amplify gene-specific products (Table 1). Specificity of accumulated products was verified by melting curve analysis and sequencing. Relative expression levels were calculated, and PCR efficiencies were determined as described previously by Pfaffl (reference 37, see equation 3 therein) and Ramakers et al. (43). Concurrent substrate concentrations and time points of cell harvesting (see above) as well as highly similar PCR efficiencies (around 1.9) for the three target genes under the different growth conditions underline the significance of determined relative expression levels (see Fig. 3). For each tested RNA preparation, at least three independent real-time RT-PCR experiments were conducted.
TABLE 1.
Primer sequences for genes of anaerobic alkylbenzene degradation in denitrifying strain EbN1
| Primera | Target gene | Sequence (5′→3′) | Product length (nt)b |
|---|---|---|---|
| bcrC 537F | bcrC | GCTGAAGAAAGTGCTCGC | 459 |
| bcrC 995R | bcrC | ATACGGATACGGAAGGGG | |
| bssA 983F | bssA | AGAAGGAAGATTCGCTGC | 195 |
| bssA 1177R | bssA | CCAAGGTCAGGATGAAGAG | |
| ebdA 2433F | ebdA | TGCCCAGTTCTACCTTGAC | 496 |
| ebdA 2928R | ebdA | TGCTTTCTTGSTGCTTSCC |
Antisense primers (R) were used for reverse transcription. Primer pairs (F and R) were used for PCR amplification.
nt, nucleotides.
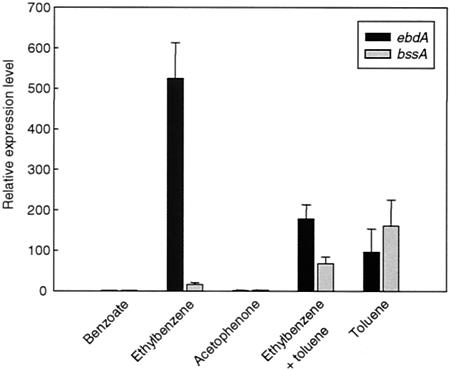
FIG. 3.
Expression levels of bssA and ebdA in substrate-adapted cells of strain EbN1. bssA and ebdA encode the catalytic subunits of benzylsuccinate synthase and ethylbenzene dehydrogenase, respectively. bcrC was used as the reference gene, and benzoate-adapted cells were used as the reference state. Substrates used for adaptation are indicated. Relative expression levels were determined with real-time RT-PCR.
Probe amplification (DNA microarray).
PCR products were amplified from genomic DNA fragments as described previously (42). Suitable gene-specific primers (average length, 23 bp; average melting point temperature, 60 to 63°C) were designed by using the PRIDE plug-in (20) from the Staden program package (48). Following amplification, 25 μl of PCR products was concentrated by isopropanol precipitation. The pellets were dried at room temperature, resuspended overnight in 15 μl of spotting buffer (3× SSC [1× SSC is 0.15 M NaCl, 10 mM sodium citrate], 1.5 M betaine [13]) on a rotary shaker and subsequently transferred to 384-well polystyrene spotting plates (Genetix, New Milton, United Kingdom). Size and concentration of the PCR products were verified by agarose gel electrophoresis and spectrophotometric measurements.
DNA microarray fabrication.
Spotting was performed with a modified Genetix Qarray spotting robot by using TeleChem (Sunnyvale, Calif.) CMP4 split pins and TeleChem SuperAmine slides. After incubation for 12 h at 20°C and 55% relative humidity, the slides were snap dried on a heating plate (200°C) for 30 s. DNA was linked to the slide surface by UV irradiation with a Stratagene UV-Stratalinker (twice at 1,200 μJ). The slide surface was subsequently blocked as described previously (13). Slides were stored at room temperature and protected from light and humidity until further use.
cDNA synthesis (DNA microarray).
Fifteen micrograms of total RNA was reverse transcribed using Cy-labeled dUTP (Perkin-Elmer), SuperScript II RT (Invitrogen, Karlsruhe, Germany), and random nonamer primers as described elsewhere (27). The labeled cDNAs were purified by using Microcon YM-30 spin columns (Millipore, Bedford, Mass.). The quality of cDNA synthesis was verified by agarose gel electrophoresis and fluorescence scanning of the gel with a Fuji FLA-8000 scanner at 532 and 640 nm followed by subsequent staining of the gel with ethidium bromide.
Hybridization procedure (DNA microarray).
Cy3- and Cy5-labeled reference and test cDNAs were pooled and mixed with 2 μl of a DNA mixture consisting of 2.5 μg of herring sperm DNA (Invitrogen)/μl, 25 ng of acetylated bovine serum albumin (Promega, Mannheim, Germany)/μl, and 0.6 μM unlabeled oligonucleotides. This mixture was added to the labeled samples, concentrated with a SpeedVac (volume of 3 to 5 μl), resuspended in 50 μl of DIG Easy Hyb buffer (Roche Diagnostics, Mannheim, Germany), and denatured at 95°C for 3 min prior to transfer to the slide surface. Hybridization was conducted for 16 h at 42°C in a hybridization chamber (Scienion AG, Berlin, Germany). After washing (once with 0.2 × SSC-0.1% [wt/vol] sodium dodecyl sulfate and twice with 0.2 × SSC for 5 min) and drying by centrifugation, the slides were scanned with a GMS 418 microarray scanner (MWG Biotech AG, Ebersberg, Germany). Image processing and grid placement were achieved with the AIDA image analysis software package (Raytest, Straubenhardt, Germany). For each investigated test state (growth with ethylbenzene, toluene, or a mixture of both), a total of 7 slides were hybridized with random primed Cy3- or Cy5-labeled cDNA. Each slide contained four identical blocks. For each gene, three replicate samples were spotted per block. Thus, 84 (7 × 4 × 3) replicates were used for each gene and treatment in the final calculations.
Image analysis (DNA microarray).
The quantification was based on the pixel positions resulting from the grid placement with the AIDA software. Intensities of spots and background were calculated and normalized (49) by applying a median method to three reference genes. For this purpose, bcrC (γ-subunit of benzoyl-CoA reductase), mdh (malate dehydrogenase), and ssb (single-strand binding protein) were selected since they were assumed to be constitutively expressed. The median of this reference set (88 replicates per slide) was determined for each single slide and for the whole data set (49). In the normalized data, all replicates of a single gene were averaged, resulting in the mean value. The ratios of the means between a test state (adaptation to ethylbenzene, toluene, or a mixture of both) and the reference state (adaptation to benzoate) were calculated. Standard error and Welch test indicated significance of differential expression (46). The significance level for the P value was 0.01.
Reproducibility of the hybridization experiments was as follows. When different RNA preparations from the same test state were used for parallel hybridizations, the correlation coefficients (linear scale) of spot intensities ranged between 0.83 and 0.93. Changing the order of CyDye labeling between test and reference states resulted in correlation coefficients closer to the lower limit, while constant CyDye labeling gave values closer to the upper limit. Comparing spot intensities from independent cultures of the same test state gave correlation coefficients in the same range. Correlation coefficients were low (<0.5) when spot intensities representing different test states were compared.
2D DIGE.
Cell disruption with a PlusOne Sample Grinding kit (Amersham Biosciences, Freiburg, Germany) and preparation of protein extracts were carried out as recently reported (17). Protein concentration was determined according to the method described previously by Bradford (5).
Isoelectric focusing (IEF) was performed as described previously (17, 41) by using the IPGphor system and 24-cm IPG (immobilized pH gradient) strips with linear pH gradients of 4 to 7 and 3 to 10 (Amersham Biosciences). To enhance reproducibility and resolution of IEF in the alkaline range of pH 3 to 10, the rehydration buffer contained 1.2% DeStreak reagent (Amersham Biosciences) instead of 0.4% dithiothreitol. The EttanDalt II system (Amersham Biosciences) was used for separation according to molecular weight in 12.5% Duracryl gels (Genomic Solution, Ann Arbor, Mich.) as described previously (17). Low-fluorescence glass plates were used for 2D DIGE.
2D DIGE was essentially carried out as described previously by Gade et al. (17). A total of 200 pmol of CyDye was used to label 50 μg of protein sample. Preelectrophoretic labeling with different fluorescent dyes allows coseparation of up to three samples in a single gel. An individual experiment in the present study contained (per gel) reference state, test state, and internal standard. To achieve statistical confidence, five parallel gels were run per experiment. Protein extracts from benzoate-adapted cultures served as the reference state and were labeled with Cy5. Protein extracts from cultures adapted to anaerobic growth with toluene, ethylbenzene, a mixture of both, or acetophenone represented the test states and were each labeled with Cy3. All four test states were related to the same reference state. All performed experiments contained the same preparation of internal standard, which was composed of equal amounts of reference state and all test states and was labeled with Cy2.
2D DIGE gels were scanned immediately after electrophoresis with a Typhoon 9400 scanner (Amersham Biosciences). Analysis of cropped images was performed with DeCyder software (version 5.0; Amersham Biosciences). Parameters for codetection of spots were as follows: (i) detection of 2,500 spots in pH 4 to 7 gels and of 3,000 spots in pH 3 to 10 gels and (ii) exclusion of signals with a slope of >1, an area of <200, a peak height of <190, and a volume of <60,000. Statistical analysis was based on independent spot maps. Differentially regulated spots were manually controlled and fulfilled the following criteria: an average ratio of <−2.5 or >2.5, a P value of <0.05 by analysis of variance, t test value of <10−4, >45 out of 60 matched gels. The pH 4 to 7 master gel contained 1,724 matched protein spots. Total numbers of substrate-specific regulated protein spots were 215, 182, 239, and 188 up-regulated and 26, 24, 20, and 73 down-regulated protein spots in cells adapted to toluene, ethylbenzene, a mixture of both, and acetophenone, respectively. For better perception, the Cy2 channel was omitted in Fig. 4 and 5. A second set of 2D DIGE experiments with samples from independent mass cultures yielded essentially the same results for each of the four investigated test states (data not shown).
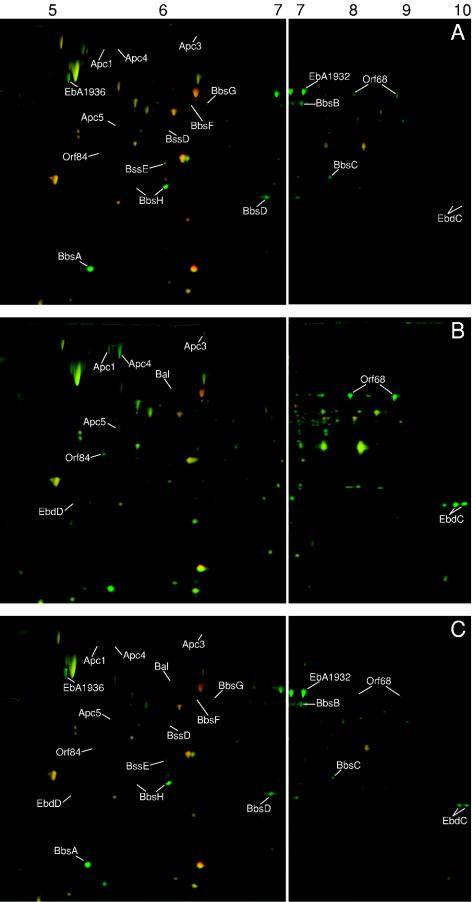
FIG. 4.
Formation of pathway-specific proteins in substrate-adapted cells of strain EbN1 as determined by 2D DIGE. (A) Toluene-adapted cells. (B) Ethylbenzene-adapted cells. (C) Cells adapted to a mixture of toluene and ethylbenzene. Enzyme abbreviations are as described in the legend of Fig. 1. Mass spectrometric identification and relative abundances of marked proteins are shown in Table 3.
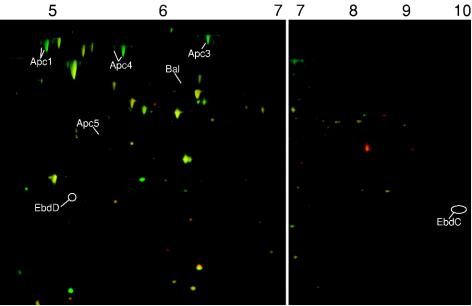
FIG. 5.
Formation of pathway-specific proteins in acetophenone-adapted cells of strain EbN1 as determined by 2D DIGE. Enzyme abbreviations are as described in the legend of Fig. 1. Identification of annotated spots was based on mass spectrometric analyses. For unknown reasons, spot coordinates of Apc proteins deviate from those in alkylbenzene-adapted cells, while about 800 other protein spots could be mapped.
In order to analyze regulated proteins by mass spectrometry, separate preparative (500-μg protein load) gels were run and stained with colloidal Coomassie brilliant blue according to the method described by Doherty et al. (14).
Protein identification by mass spectrometry.
Selected up-regulated protein spots were excised with an automatic excision workstation (PROTEINEER sp; Bruker Daltonics, Bremen, Germany). Tryptic digest of excised proteins and preparation of the matrix-assisted laser desorption ionization target with α-cyano-4-hydroxycinnamic acid as matrix was carried out with a PROTEINEER dp digest and a sample preparation robot (Bruker Daltonics) as recently described (31). Acquisition of mass spectrometry fingerprints was achieved with an Ultraflex tandem time of flight instrument (Bruker Daltonics) as recently described (31). Fingerprint information was submitted to a protein database search by using the MASCOT search engine (Matrix Science, London, United Kingdom). The database contained essentially the protein sequences predicted to be involved in the anaerobic degradation of toluene (31) and ethylbenzene (42) by denitrifying strain EbN1.
Nucleotide sequence accession numbers.
DNA sequences of new genes of strain EbN1 have been deposited in the EMBL database under the accession numbers CR792447 (ebA1926, ebA1928, ebA1929, ebA1932, ebA1936, tnpF, and ebA1938), CR792444 (bcrC), CR792558 (mdh), and CR792561 (ssb).
RESULTS
Regulation of the anaerobic degradation pathways for toluene and ethylbenzene in strain EbN1 (Fig. 1) by their respective substrates was investigated in vivo by measuring substrate consumption in cultures (Fig. 2) and in vitro on RNA (Fig. 3 and Table 2) and by measuring protein levels (Fig. 4 and 5 and Table 3). The common basis for these experiments was cells of strain EbN1 adapted to one of five different substrate conditions. Compounds used for adaptation were, on the one side, the key substrates of the investigated pathways, i.e., toluene and ethylbenzene (supplied as a single substrate or as a mixture) and acetophenone, and, on the other side, benzoate, which represents the first common intermediate (Fig. 1). Benzoate-adapted cells served as the reference state, since the degradative capacities for the other substrates were not induced in them. To allow optimal comparison, global expression studies (DNA microarrays or proteomics) were conducted with the same cell batches.
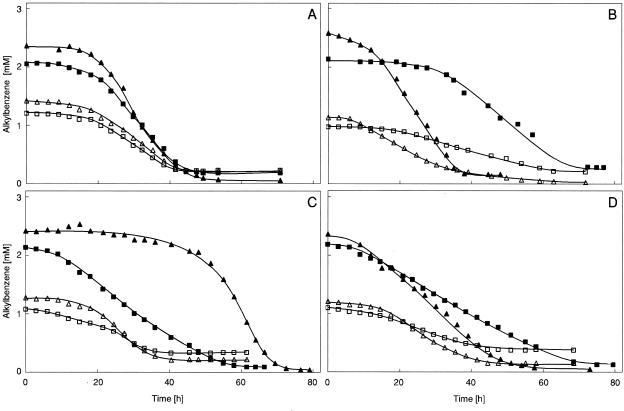
FIG. 2.
Consumption of toluene and ethylbenzene during growth of strain EbN1 under nitrate-reducing conditions. Cultures were inoculated with anaerobic substrate-adapted cells. (A) Benzoate-adapted cells. (B) Toluene-adapted cells. (C) Ethylbenzene-adapted cells. (D) Cells were adapted to a mixture of toluene and ethylbenzene. Symbols: ▴, toluene provided as single substrate; ▪, ethylbenzene provided as single substrate; ▵, toluene provided as mixture with ethylbenzene; □, ethylbenzene provided as mixture with toluene.
TABLE 2.
Average ratios of expression of genes involved in anaerobic alkylbenzene degradation as determined by DNA microarray analysis of substrate-adapted cells of strain EbN1
| Geneb | Avg ratios (±SD) of cells adapted to anaerobic growth witha
|
||
|---|---|---|---|
| Ethylbenzene | Toluene | Ethylbenzene + toluene | |
| Toluene-related genes | |||
| bssD | 1.1 ± 0.1 | 6.4 ± 0.8 | 3.2 ± 0.3 |
| bssC | 1.4 ± 0.1 | 12.8 ± 1.7 | 6.2 ± 0.8 |
| bssA | 1.6 ± 0.2 | 28.8 ± 4.4 | 13.6 ± 1.7 |
| bssB | 1.5 ± 0.2 | 18.1 ± 2.3 | 8.8 ± 0.9 |
| bssE | 1.2 ± 0.1 | 9.2 ± 1.1 | 5.0 ± 0.5 |
| bssF | 1.2 ± 0.2 | 4.6 ± 0.5 | 2.7 ± 0.3 |
| bssG | 1.3 ± 0.1 | 3.7 ± 0.3 | 2.1 ± 0.2 |
| bssH | 1.1 ± 0.1 | 3.5 ± 0.3 | 1.7 ± 0.1 |
| c2B001 | 1.3 ± 0.1 | 3.2 ± 0.3 | 1.5 ± 0.1 |
| c2A200 | 0.8 ± 0.1 | 2.5 ± 0.4 | 1.2 ± 0.2 |
| c2A203 | 1.1 ± 0.1 | 4.8 ± 0.4 | 1.9 ± 0.1 |
| c2A204 | 1.1 ± 0.1 | 3.4 ± 0.3 | 1.5 ± 0.1 |
| c2A308 | 1.1 ± 0.1 | 3.6 ± 0.4 | 1.6 ± 0.2 |
| bbsH | 1.1 ± 0.1 | 11.1 ± 1.4 | 3.9 ± 0.4 |
| bbsG | 1.4 ± 0.2 | 17.4 ± 2.2 | 9.1 ± 0.9 |
| bbsJ | 1.8 ± 0.2 | 13.9 ± 1.8 | 7.5 ± 1.0 |
| bbsF | 1.2 ± 0.1 | 7.8 ± 0.9 | 3.9 ± 0.4 |
| bbsE | 1.4 ± 0.1 | 4.5 ± 0.5 | 2.8 ± 0.2 |
| bbsD | 1.3 ± 0.1 | 4.5 ± 0.5 | 3.8 ± 0.4 |
| bbsC | 1.7 ± 0.3 | 5.2 ± 0.7 | 4.0 ± 0.6 |
| bbsB | 2.0 ± 0.2 | 4.3 ± 0.5 | 3.2 ± 0.3 |
| bbsA | 1.3 ± 0.1 | 2.3 ± 0.2 | 1.7 ± 0.1 |
| Ethylbenzene-related genes | |||
| orf57 | 14.9 ± 1.9 | 5.1 ± 0.6 | 5.8 ± 0.7 |
| ped | 24.5 ± 3.6 | 5.7 ± 0.8 | 5.7 ± 0.8 |
| ebdD | 11.4 ± 1.5 | 5.1 ± 0.7 | 3.6 ± 0.5 |
| ebdC | 21.1 ± 2.2 | 7.8 ± 0.8 | 4.7 ± 0.5 |
| ebdB | 15.8 ± 1.9 | 6.2 ± 0.8 | 4.3 ± 0.5 |
| ebdA | 12.0 ± 1.4 | 5.7 ± 0.6 | 2.6 ± 0.3 |
| orf68 | 22.5 ± 4.1 | 12.0 ± 2.2 | 4.7 ± 0.8 |
| orf84 | 6.8 ± 0.7 | 2.8 ± 0.3 | 3.6 ± 0.3 |
| orf87 | 22.1 ± 2.3 | 7.4 ± 0.8 | 14.3 ± 1.5 |
| orf90 | 15.3 ± 1.8 | 5.0 ± 0.6 | 6.7 ± 0.8 |
| orf92 | 9.1 ± 1.1 | 3.7 ± 0.4 | 5.2 ± 0.5 |
| bal | 16.1 ± 1.9 | 3.8 ± 0.4 | 6.6 ± 0.8 |
| apc5 | 14.1 ± 2.0 | 3.7 ± 0.5 | 6.4 ± 0.7 |
| apc4 | 22.2 ± 2.7 | 4.5 ± 0.5 | 7.6 ± 0.9 |
| apc3 | 12.5 ± 1.6 | 2.7 ± 0.3 | 3.4 ± 0.4 |
| apc2 | 11.8 ± 1.4 | 2.0 ± 0.2 | 2.3 ± 0.3 |
| apc1 | 10.9 ± 1.5 | 2.0 ± 0.3 | 2.3 ± 0.3 |
| Other genes | |||
| orf206 | 1.8 ± 0.2 | 2.3 ± 0.2 | 1.6 ± 0.1 |
| fadB | 1.6 ± 0.2 | 2.4 ± 0.2 | 1.5 ± 0.2 |
| fadA | 2.3 ± 0.2 | 2.7 ± 0.2 | 2.0 ± 0.2 |
| fadE | 1.5 ± 0.2 | 2.1 ± 0.2 | 1.6 ± 0.2 |
Average ratios were calculated with benzoate-grown cells as a reference as described in Materials and Methods. Average ratios are provided with standard deviations.
TABLE 3.
2D DIGE-determined average ratios of amounts of mass spectrometrically identified proteins of anaerobic alkylbenzene degradation in substrate-adaptated cells of strain EbN1
| Proteina | MS-identificationb
|
Avg ratios in cells adapted to anaerobic growth withe
|
|||
|---|---|---|---|---|---|
| No. of repeatsc | Avg scored | Toluene | Ethylbenzene | Toluene + ethylbenzene | |
| Toluene pathway | |||||
| BssD | 4 | 226 | 9 | 1 | 9 |
| BssE | 4 | 383 | 14 | 1 | 13 |
| BssG | 4 | 163 | ND | ND | ND |
| BbsH | 4 | 278 | 155 | 2 | 138 |
| BbsG | 3 | 176 | 4 | 1 | 3 |
| BbsF | 4 | 180 | 9 | 1 | 7 |
| BbsD | 3 | 280 | 34 | 2 | 27 |
| BbsC | 4 | 149 | 77 | −2 | 65 |
| BbsB | 3 | 117 | 25 | −2 | 19 |
| BbsA | 4 | 231 | 27 | 1 | 24 |
| EbA1932 | 3 | 246 | 54 | 1 | 44 |
| EbA1936 | 3 | 271 | 46 | 1 | 45 |
| Ethylbenzene pathway (upper part) | |||||
| EbdD | 4 | 391 | 3 | 14 | 8 |
| EbdC | 4 | 114 | 40 | 242 | 92 |
| Orf68 | 11 | 222 | 163f | 281f | 63f |
| Ethylbenzene pathway (lower part) | |||||
| Orf84 | 3 | 216 | 6 | 28 | 15 |
| Bal | 3 | 425 | 2 | 4 | 3 |
| Apc5 | 3 | 249 | 5 | 34 | 17 |
| Apc4 | 4 | 466 | 13 | 88 | 32 |
| Apc3 | 4 | 188 | 4 | 13 | 9 |
| Apc1 | 4 | 423 | 3 | 8 | 5 |
Order and names of proteins are as described for the corresponding genes in the legend of Fig. 1.
A MASCOT score of >100 was used as a threshold for positive identification by peptide mass fingerprints.
Regulated proteins were identified under each growth condition at least three times.
Average MASCOT score was calculated from the repetitive identification experiments.
Average ratios were determined with five parallel gels; benzoate-grown cells were used as a reference. For details, see Materials and Methods. ND, not detected in 2D DIGE gels although detected in Coomassie- and silver-stained gels. Possibly, the presence of only one lysine residue in BssG rendered this protein unsuitable for labeling with CyDyes under the applied conditions.
Protein species that are separated as two spots with different pI and similar Mr on the 2D DIGE gel.
Differential alkylbenzene consumption during growth.
Four differently adapted types of cultures of strain EbN1 were studied. In each case, cultures were shifted to either toluene, ethylbenzene, or a mixture of both alkylbenzenes as the sole source of organic carbon. Due to their toxicity and low water solubility, the alkylbenzenes were added as a solution in an inert carrier phase. During growth, consumption of the alkylbenzenes was monitored in samples withdrawn from the carrier phase. Results are shown in Fig. 2.
When cultures were adapted to benzoate utilization, consumption of toluene and ethylbenzene occurred only after prolonged incubation (Fig. 2A). Regardless of whether the alkylbenzenes were added as single substrates or as a mixture, similar induction times of less than 15 h were observed. Interestingly, degradative capacities for both alkylbenzenes were induced simultaneously when provided as a mixture. Thus, neither of the alkylbenzenes repressed the onset of the degradation of the other alkylbenzene.
Toluene-adapted cultures (Fig. 2B) utilized toluene, as expected, almost instantly. In contrast, consumption of ethylbenzene required an even longer induction time in toluene-adapted cells than that observed in case of the benzoate-adapted cells. When cells were supplied with a mixture of both alkylbenzenes, toluene utilization again set in early. During the course of toluene consumption, degradation of ethylbenzene had already started. This finding indicated that ongoing toluene degradation does not repress induction of the capacity to degrade ethylbenzene.
In the case of ethylbenzene-adapted cultures (Fig. 2C), an analogous pattern of substrate utilization was observed. Ethylbenzene was consumed rather rapidly whether supplied as a single substance or as a mixture with toluene. The latter was utilized only after a long induction time but then also while ethylbenzene was still consumed.
Time courses of toluene and ethylbenzene utilization were similar when cells were adapted to their mixture (Fig. 2D).
Monitoring bssA and ebdA expression with real-time RT-PCR.
As a first step towards the transcriptional analysis of the pathway regulation, a real-time RT-PCR approach was established to monitor expression of the bssA and ebdA genes (Fig. 3). Both genes are particularly relevant since they encode the catalytic subunits of benzylsuccinate synthase and ethylbenzene dehydrogenase, respectively, the initial enzymes of the two pathways. Gene expression was investigated in cells adapted to benzoate, toluene, ethylbenzene, the mixture of both alkylbenzenes, and acetophenone. Benzoate-adapted cells served as the reference state. bcrC was selected as the reference gene since it codes for the γ-subunit of benzoyl-CoA reductase, the first common enzyme after convergence of the two pathways. In agreement with the in vivo experiments (Fig. 2A), only basal expression of bssA and ebdA was observed in benzoate-adapted cells.
In ethylbenzene-adapted cells, expression of ebdA was about 550-fold up-regulated, while expression of bssA was only marginal. Apparently, the predicted sensor for toluene (TdiS) is not responsive to ethylbenzene. In contrast, cells adapted to growth with toluene displayed an up-regulation of ebdA and bssA expression of about 100-fold. This unexpected finding may suggest a relaxed specificity of the predicted ethylbenzene sensor (Tcs2), also recognizing toluene.
In agreement with the in vivo behavior of cells adapted to growth with the alkylbenzene mixture (Fig. 2D), ebdA and bssA were about 180- and 70-fold up-regulated under these growth conditions. In each case, this was about threefold lower than observed with cells adapted to toluene or ethylbenzene as a single substrate. While the rationale behind this observation is not understood, it cannot be attributed to differing substrate concentrations, since these concentrations were always the same for the individual alkylbenzene when supplied as a single substrate or as a mixture.
In acetophenone-adapted cells, the expression of bssA and ebdA was as low as that in benzoate-adapted cells, indicating that neither of the two predicted alkylbenzene sensors (TdiS and Tcs2) recognizes acetophenone. Moreover, this result agrees with the previous hypothesis of a sequential regulation of the ethylbenzene pathway (42).
Transcriptional profiling with DNA microarray.
Following expression analysis of single genes (bssA and ebdA) by means of real-time RT-PCR, a DNA microarray was designed for transcriptional profiling of both complete pathways. The DNA microarray contained probes for all genes predicted to be involved in anaerobic toluene (31) and ethylbenzene (42) degradation. In addition, neighboring genes, mostly coding for proteins of unknown function, were also represented on the DNA microarray. Labeled cDNA for hybridization experiments was generated from total RNA prepared from substrate-adapted cells. Results from the DNA microarray analysis are summarized in Table 2.
Transcriptional profiling of ethylbenzene-adapted cells revealed a specific up-regulation of all genes previously predicted to be involved in anaerobic ethylbenzene degradation (42). The highest average ratios were observed for ebdC (21.1-fold; encoding the γ-subunit of ethylbenzene dehydrogenase), ped [24.5-fold; encoding (S)-1-phenylethanol dehydrogenase], and apc4 (22.2-fold; encoding a subunit of acetophenone carboxylase). The previous suggestion of bal encoding benzoylacetate CoA-ligase was supported by its specific 16.1-fold up-regulation; biochemical characterization will provide final proof. Interestingly, orf92, orf90, orf87, and orf84, which are located directly downstream of the apc/bal gene cluster, were also specifically up-regulated (9.1-, 15.3-, 22.1-, and 6.8-fold, respectively). An unexpected finding was the strong up-regulation of orf68 and orf57 (22.5- and 14.9-fold, respectively). These open reading frames frame the ebd operon, and their products are of unknown function (42). As observed during the real-time RT-PCR experiment with bssA (see above), genes involved in anaerobic toluene degradation (31) were not up-regulated in ethylbenzene-adapted cells. The 17 selected toluene-related genes displayed average ratios ranging between 1.1 and 2.0 (median at 1.4). These results further corroborated the aforementioned specificity of the suggested toluene sensor (TdiS) and also demonstrated the suitability of the used DNA microarray design to study differential regulation of the two pathways.
When cultures of strain EbN1 were adapted to anaerobic growth with toluene, all 17 genes assigned to the bss and bbs operons were up-regulated, in agreement with the growth physiology of the investigated cells. The highest expression levels were observed for bssA, bssB, and bbsG (28.8-, 18.1-, and 17.4-fold, respectively). Several proteins of unknown function are encoded directly downstream of the bss operon by the c2B001, c2A200, c2A203, c2A204, and c2A308 genes. These genes appeared to be up-regulated during anaerobic growth with toluene (average ratios of 3.2-, 2.5-, 4.8-, 3.4-, and 3.6-fold, respectively). Specificity of expression is confirmed by only marginal expression during anaerobic growth with ethylbenzene (average ratios of around 1.1). Previous genomic reconstruction of the toluene pathway in strain EbN1 also indicated the presence of the bbsJ gene within the bbs operon. Only fragments of this gene are detectable in the bbs operon of well-studied Thauera aromatica strain K172 (31). The bbsJ gene was specifically up-regulated by 13.6-fold in toluene-grown cells, while the expression level in ethylbenzene-grown cells was only 1.8. In analogy to the above-described results from the real-time RT-PCR experiment, genes involved in the ethylbenzene pathway were also up-regulated. However, expression levels for ebd and apc genes were two- to threefold and four- to fivefold lower, respectively, than those for ethylbenzene-adapted cells.
Genes for both pathways were clearly up-regulated in cells adapted to simultaneous utilization of ethylbenzene and toluene. In both cases, the determined expression levels were about three- to fourfold lower than those of cells grown with the individual alkylbenzene, in agreement with the results from the real-time RT-PCR experiment.
In the present study, the average ratios for most highly up-regulated genes ranged between 20 and 30. These values are considerably lower than relative expression levels determined with the real-time RT-PCR approach. This result agrees with the observations from several laboratories that DNA microarray analyses generate about 10-fold-lower expression levels than other generally applied methods for RNA analysis (7).
Analysis of relative protein abundances with 2D DIGE.
Soluble proteins extracted from the differently adapted cells were electrophoretically separated in a pH range from 3 to 10. Expanding the IEF to alkaline pH was required since some of the predicted proteins displayed a theoretical isoelectric point in this range (e.g., EbdC with a pI of 9.16). More than 150 protein spots with increased abundances in response to anaerobic growth with toluene or ethylbenzene were detected by 2D DIGE analysis (see Materials and Methods). For identification of the abundant ones among them, corresponding spots were excised from Coomassie-stained gels. Mass spectrometric analysis yielded average MASCOT scores of at least 100, based on at least three independent analyses. With this procedure, 8 out of the 17 predicted gene products of anaerobic toluene degradation (31) and 7 out of the 11 predicted gene products of anaerobic ethylbenzene degradation (42) were identified. Several other abundant, differential proteins could not be identified. Results from 2D DIGE analysis and mass spectrometric identification are summarized for alkylbenzene-adapted cells in Fig. 4 and Table 3 and for acetophenone-adapted cells in Fig. 5.
In toluene-adapted cells (Fig. 4A) the eight identified toluene-specific proteins were 4- to 155-fold up-regulated; e.g., BbsH was up-regulated by 155-fold. Also, in accordance with the transcriptional analysis, gene products from the ethylbenzene pathway were up-regulated in toluene-adapted cells, although to a lesser extent than the toluene-specific gene products. Further proteins, which are not encoded in the bss and bbs operons but which were nevertheless specifically up-regulated in toluene-adapted cells, were EbA1932 and EbA1936 (exclusively discovered by the global proteomic approach). Their identified coding genes are organized in an operon-like structure and located 7.2 kb upstream of the bbs operon (Fig. 1 and Table 4). Interestingly, the same consensus sequence as that previously described for the promoter regions of the bss and bbs operons (Fig. 1A) (31) is present upstream of the EbA1936 gene, agreeing well with the toluene-specific coinduction.
TABLE 4.
Annotation of newly identified genes located adjacent to the bbs operon of denitrifying strain EbN1
| Open reading frame | Length (amino acids) | INTERPRO/COG Reference(s)a | BLASTP hit used for annotationb
|
Putative function | |||
|---|---|---|---|---|---|---|---|
| Gene | Organismc | E value | Accession no. | ||||
| ebA1926 | 150 | COG0589, IPR006015 | rso4641 | Ralso | 9e-24 | Q8XZN6 | Hypothetical protein Rsc1359 |
| ebA1928 | 799 | Psepu | 0 | Q88100 | Putative transporterd | ||
| ebA1929 | 374 | COG4447, IPR002860 | Psepu | 3e-54 | Q88101 | Hypothetical protein, bnr domain | |
| ebA1932 | 454 | IPR010752 | orf2 | Thaar | 5e-38 | O87937 | Hypothetical protein |
| ebA1936 | 649 | IPR010727 | orf1 | Thaar | 0 | O87938 | Hypothetical protein |
| tnpF | 41 | COG5433 | ispg6 | Porgi | 4e-04 | Q66176 | Transposon (fragment) |
| ebA1938 | 167 | IPR000923 | Hypothetical protein | ||||
References relate to INTERPRO (http://www.ebi.ac.uk/interpro) and COG (http://www.ncbi.nlm.nih.gov/COG) databases.
Hits were obtained from BLASTP comparison of predicted proteins from strain EbN1 with TrEMBL databases (http://srs6.ebi.ac.uk).
Abbreviations of names of organisms are according to the list of organism identification codes (SWISS-PROT). Porgi, Porphyromonas gingivalis W83; Psepu, Pseudomonas putida; Ralso, Ralstonia solanacearum; Thaar, Thauera aromatica.
According to BLASTP against TCDB (http://tcdb.ucsd.edu), EbA1928 shows similarity to members of the resistance-nodulation-cell division (RND) superfamily of H+-driven substrate efflux systems.
When cells were adapted to anaerobic ethylbenzene utilization (Fig. 4B), the seven identified ethylbenzene-specific proteins were strongly up-regulated (4- to 242-fold). In contrast, the electrophoretically detectable proteins of the toluene pathway appeared to be absent. Thus, the proteomic results also support the view that toluene and ethylbenzene sensors of strain EbN1 exhibit different degrees of specificity towards their target molecules. In addition to the previously described gene products of the ethylbenzene pathway (42), the products of orf68 and orf84 were also found to be the most strongly up-regulated in response to ethylbenzene degradation (281- and 28-fold, respectively). In fact, the orf68 product belongs to the abundant proteins detected on the 2D DIGE gels. Specific up-regulation of orf68 and orf84 was also found by DNA microarray analysis (Table 2).
When cells were anaerobically grown with a mixture of toluene and ethylbenzene (Fig. 4C), both sets of proteins were up-regulated. Relative to the cultures adapted to a single alkylbenzene, here the relative protein abundances were lower.
Acetophenone-adapted cells (Fig. 5) are characterized by the absence of EbdC (γ-subunit of ethylbenzene dehydrogenase) and EbdD (predicted chaperone for maturation of ethylbenzene dehydrogenase). This finding agrees well with the absence of ebdA expression (α-subunit of ethylbenzene dehydrogenase) in acetophenone-adapted cells as revealed by the aforementioned real-time RT-PCR experiments (Fig. 3). In contrast, several subunits of acetophenone carboxylase (Apc1 [72-fold], Apc3 [36-fold], Apc4 [104-fold], and Apc5 [6-fold]) and the predicted benzoylacetate CoA-ligase (Bal [11-fold]) were strongly up-regulated in the same cells (Fig. 5). Apparently, the adaptation to acetophenone as the sole organic substrate resulted in the specific formation of only those enzymes, which are involved in its catabolism.
DISCUSSION
Simultaneous toluene and ethylbenzene utilization.
The parallel utilization of toluene and ethylbenzene in cultures was reflected in simultaneous expression of genes from both degradation pathways. Therefore, neither of the two alkylbenzenes is regarded as a preferred substrate, precluding a regulatory process such as catabolite repression at this level. This regulatory behavior of strain EbN1 might reflect an adaptation to low concentrations of the rather insoluble alkylbenzenes, as usually encountered in the natural environment. Repression of these pathways by substrates like succinate might occur, since this is known from aerobic aromatic degradation (11, 16, 18, 50).
Sequential regulation of the ethylbenzene pathway.
The upper and lower parts of the pathway for anaerobic ethylbenzene degradation were previously suggested to be sequentially induced by their respective substrates, ethylbenzene and acetophenone (42). In fact, strain EbN1 can utilize both compounds as growth substrates (39). Such a regulatory scenario is supported by the absence of ebdA expression (Fig. 3) and EbdCD formation (Fig. 5) in acetophenone-adapted cells as well as by the concomitant presence of several subunits of acetophenone carboxylase and of putative benzoylacetate CoA-ligase (Fig. 5). A sequential mode of regulation could be of economic benefit, considering that strain EbN1 probably encounters acetophenone (a plant metabolite) as substrate in the natural soil habitat. Similarly, in Pseudomonas putida, the pathway for aerobic degradation of toluene or xylenes is also divided into two sequentially regulated parts (45).
Substrate specificity of pathway regulation.
Substrate-dependent regulation of aromatic catabolism usually occurs on the transcriptional level and involves a large variety of transcriptional regulators (12, 19). Two different two-component regulatory systems were previously suggested to trigger expression of genes involved in anaerobic toluene (TdiSR system) and ethylbenzene (Tcs2/Tcr2 system) degradation in strain EbN1. Toluene recognition by the TdiSR system was suggested due to the close neighborhood of its coding genes to the bss operon and the high similarity to other two-component regulatory systems for toluene degradation in related strains (31). Even though the Tcs2/Tcr2 and TdiSR systems are also highly similar to each other, their sensor components differ markedly in one of the PAS domains implicated in substrate sensing. This finding, together with the close proximity of the tcs2/tcr2 genes to the catabolic operons of the ethylbenzene pathway, suggested that Tcs2 recognizes ethylbenzene (42). In addition, a third two-component system (Tcs1/Tcr1 system) was suggested to be implicated in recognition of acetophenone due to its similarity to previously described p-hydroxyacetophenone-sensing systems of plant pathogenic bacteria (42). The present study suggests a strict specificity of the predicted toluene sensor, which is contrasted by relaxed specificities of the predicted ethylbenzene (Tcs2) and acetophenone (Tcs1) sensors. One may speculate that the sensory pocket receiving toluene in the TdiS protein cannot accommodate the larger ethylbenzene molecule. In contrast, toluene may access the sensory pockets of Tcs1 and Tcs2. Alternatively, a cross-regulation between TdiSR and the other two systems may occur. The relaxed specificity of the ethylbenzene and acetophenone sensors could explain the low-level expression of all “ethylbenzene genes” in cells grown with toluene, suggesting that toluene acts as a gratuitous inducer for the ethylbenzene-specific operons. The in vivo experiments with toluene-adapted cells (Fig. 2B) indicated that the capacity to degrade ethylbenzene had to be induced under these conditions. Thus, the low-level expression of ethylbenzene genes may facilitate metabolic adaptation when ethylbenzene becomes available. Indeed, the maximal consumption rate of ethylbenzene was reached earlier in toluene-adapted cells (around 20 h) (Fig. 2B) than that of toluene in ethylbenzene-adapted cells (>30 h) (Fig. 2C). Future investigations might aim to define the sensory domain and molecular mechanism which are responsible for alkylbenzene recognition. In this respect, it should be noted that benzylsuccinate rather than toluene was only recently suggested as the true inducer of tut (corresponds to bss) expression in T. aromatica strain T1, since cells grown with pyruvate expressed tut genes upon addition of benzylsuccinate (9). Such a regulatory scenario appears unlikely in strain EbN1, since an up-regulation of the bssA expression (real-time RT-PCR) or formation of Bss/Bbs proteins (proteomics) was not observed in cells grown with a mixture of pyruvate (10 mM) and benzylsuccinate (1 mM) (data not shown).
Further gene products involved in anaerobic alkylbenzene degradation.
Another valuable outcome of the DNA microarray or proteomics approach was that due to their differential expression, more genes could be related to either of the two investigated pathways. The genes orf68 and orf57 are located directly upstream and downstream of the ebdA and ped genes, respectively (Fig. 1B). The rather large intergenic regions between orf68 and ebdA (267 bp) and between ped and orf57 (150 bp) indicate that orf68 and orf57 may not be part of the ebd operon, which was previously suggested to consist of the ebdABCD and ped genes (42). Nevertheless, they are likely to also be involved in anaerobic ethylbenzene degradation, since their expression was most highly up-regulated in cells adapted to ethylbenzene (by 22.5- and >200-fold according to DNA-microarray and 2D DIGE analysis, respectively, in the case of orf68). The predicted apc operon of the lower part of the ethylbenzene pathway probably extends beyond bal, since adjacent genes orf92, orf90, orf87, and orf84 were most highly up-regulated in response to growth with ethylbenzene. However, their function remains elusive so far. Since selected genes of a β-oxidation complex (fadABE) and a thioesterase (orf206) encoded in the 15-kb distance to the ebd operon were not up-regulated during growth with ethylbenzene (Table 2), the missing benzoylacetyl-CoA thiolase remains unaccounted for. In toluene-adapted cells, five additional genes (c2B001, c2A200, c2A203, c2A204, and c2A308) were specifically up-regulated according to DNA microarray analysis. These genes are located directly downstream of bssH, and the short intergenic distances (0 to 22 bp) of c2B001, c2A200, c2A203, and c2A204 already suggested that they were part of the bss operon (31). Considering the specific expression of bbsJ, ebA1936 and ebA1932 genes anaerobic toluene degradation might also involve more gene products than previously thought. EbA1932 and EbA1936 are highly similar to the products of orf1 and orf2 from T. aromatica K172, where they are located adjacent to the tdiSR genes. Due to the presence of transposase genes (istAB) and a transposon gene fragment (tnpF4) downstream of the ebA1932 gene and another transposon gene fragment (tnpF) upstream of the ebA1936 gene (Fig. 1A and Table 4), one may speculate that these genes have been translocated in the genome of strain EbN1 when compared to the organization in T. aromatica K172. Since the products of both genes belong to the most abundant proteins on the 2D DIGE gel, they are probably involved in anaerobic toluene degradation; however, an erroneous formation cannot be excluded. The close proximity of a resistance-nodulation-cell division-type transporter-encoding gene (ebA1928) may point to an involvement in coping with toxic toluene concentrations, as recently suggested for BssH (31). These results also indicate an involvement of more than two transcriptional units in the case of both pathways.
Acknowledgments
We thank Daniela Lange (Bremen) and Özlem Ogras (Berlin) for technical assistance and Alfred Beck (Berlin) for bioinformatical support. We are grateful to Friedrich Widdel for supporting proteomic work at the MPI in Bremen.
This study was supported by Amersham Biosciences, Bruker Daltonics, the Max-Planck Society, and the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Achong, G. R., A. M. Rodriguez, and A. M. Spormann. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, H. A., H. A. Johnson, M. Reinhard, and A. M. Spormann. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J. Bacteriol. 178:5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beller, H. R., and A. M. Spormann. 1998. Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J. Bacteriol. 180:5454-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boll, M., G. Fuchs, and J. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Champion, K. M., K. Zengler, and R. Rabus. 1999. Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J. Mol. Microbiol. Biotechnol. 1:157-164. [PubMed] [Google Scholar]
- 7.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 8.Coschigano, P. W. 2000. Transcriptional analysis of the tutE tutFDGH gene cluster from Thauera aromatica strain T1. Appl. Environ. Microbiol. 66:1147-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coschigano, P. W., and B. J. Bishop. 2004. Role of benzylsuccinate in the induction of the tutE tutFDGH gene complex of T. aromatica strain T1. FEMS Microbiol. Lett. 231:261-266. [DOI] [PubMed] [Google Scholar]
- 10.Coschigano, P. W., T. S. Wehrman, and L. Y. Young. 1998. Identification and analysis of genes involved in anaerobic toluene metabolism by strain T1: putative role of a glycine free radical. Appl. Environ. Microbiol. 64:1650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl, S., I. Steiner, and U. Gerischer. 2002. Multiple operons connected with the catabolism of aromatic compounds in Acinetobacter sp. strain ADP1 are under carbon catabolite repression. J. Mol. Microbiol. Biotechnol. 4:389-404. [PubMed] [Google Scholar]
- 12.Diaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11:467-475. [DOI] [PubMed] [Google Scholar]
- 13.Diehl, F., S. Grahlmann, M. Beier, and J. D. Hoheisel. 2001. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 29:E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty, N. S., B. H. Littman, K. Reilly, A. C. Swindell, J. M. Buss, and N. L. Anderson. 1998. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis 19:355-363. [DOI] [PubMed] [Google Scholar]
- 15.Duboc-Toia, C., A. K. Hassan, E. Mulliez, S. Ollagnier-de Choudens, M. Fontecave, C. Leutwein, and J. Heider. 2003. Very high-field EPR study of glycyl radical enzymes. J. Am. Chem. Soc. 125:38-39. [DOI] [PubMed] [Google Scholar]
- 16.Duetz, W. A., S. Marqués, B. Wind, J. L. Ramos, and J. G. Van Andel. 1996. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitations in chemostat culture. Appl. Environ. Microbiol. 62:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gade, D., J. Thiermann, D. Markowsky, and R. Rabus. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. Soluble proteins of the marine bacterium Pirellula sp. strain 1. J. Mol. Microbiol. Biotechnol. 5:240-251. [DOI] [PubMed] [Google Scholar]
- 18.Galan, B., A. Kolb, J. L. Garcia, and M. A. Prieto. 2001. Superimposed levels of regulation of the 4-hydroxyphenylacetate catabolic pathway in Escherichia coli. J. Biol. Chem. 276:37060-37068. [DOI] [PubMed] [Google Scholar]
- 19.Gerischer, U. 2002. Specific and global regulation of genes associated with the degradation of aromatic compounds in bacteria. J. Mol. Microbiol. Biotechnol. 4:111-121. [PubMed] [Google Scholar]
- 20.Haas, S., M. Vingron, A. Poustka, and S. Wiemann. 1998. Primer design for large scale sequencing. Nucleic Acids Res. 26:3006-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood, C. S., G. Burchardt, H. Hermann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 22.Heider, J., A. M. Spormann, H. R. Beller, and F. Widdel. 1999. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22:459-473. [Google Scholar]
- 23.Hermuth, K., B. Leuthner, and J. Heider. 2002. Operon structure and expression of the genes for benzylsuccinate synthase in Thauera aromatica strain K172. Arch. Microbiol. 177:312-318. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, H. A., and A. M. Spormann. 1999. In vitro studies on the initial reactions of anaerobic ethylbenzene mineralization. J. Bacteriol. 181:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, H. A., D. A. Pelletier, and A. M. Spormann. 2001. Isolation and characterization of anaerobic ethylbenzene dehydrogenase, a novel Mo-Fe-S enzyme. J. Bacteriol. 183:4536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane, S. R., H. R. Beller, T. C. Legler, and R. T. Anderson. 2002. Biochemical and genetic evidence of benzylsuccinate synthase in toluene-degrading, ferric iron-reducing Geobacter metallireducens. Biodegradation 13:149-154. [DOI] [PubMed] [Google Scholar]
- 27.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kniemeyer, O., and J. Heider. 2001. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J. Biol. Chem. 276:21381-21386. [DOI] [PubMed] [Google Scholar]
- 29.Kniemeyer, O., and J. Heider. 2001. (S)-1-phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 176:129-135. [DOI] [PubMed] [Google Scholar]
- 30.Krieger, C. J., W. Roseboom, S. P. J. Albracht, and A. M. Spormann. 2001. A stable organic free radical in anaerobic benzylsuccinate synthase from Azoarcus sp. strain T. J. Biol. Chem. 276:12924-12927. [DOI] [PubMed] [Google Scholar]
- 31.Kube, M., J. Heider, J. Amann, P. Hufnagel, S. Kühner, A. Beck, R. Reinhardt, and R. Rabus. 2004. Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 181:182-194. [DOI] [PubMed] [Google Scholar]
- 32.Leuthner, B., C. Leutwein, H. Schulz, P. Hörth, W. Haehnel, E. Schiltz, H. Schägger, and J. Heider. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol. Microbiol. 28:615-628. [DOI] [PubMed] [Google Scholar]
- 33.Leuthner, B., and J. Heider. 2000. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of β-oxidation of the intermediate benzylsuccinate. J. Bacteriol. 182:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leutwein, C., and J. Heider. 2001. Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J. Bacteriol. 183:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leutwein, C., and J. Heider. 2002. (R)-benzylsuccinyl-CoA dehydrogenase of Thauera aromatica, an enzyme of the anaerobic toluene catabolic pathway. Arch. Microbiol. 178:517-524. [DOI] [PubMed] [Google Scholar]
- 36.Oelmüller, U., N. Krüger, A. Steinbüchel, and G. C. Friedrich. 1990. Isolation of procaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73-81. [Google Scholar]
- 37.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabus, R., and J. Heider. 1998. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch. Microbiol. 170:377-384. [Google Scholar]
- 39.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96-103. [DOI] [PubMed] [Google Scholar]
- 40.Rabus, R., and F. Widdel. 1996. Utilization of alkylbenzenes during anaerobic growth of pure cultures of denitrifying bacteria from crude oil. Appl. Environ. Microbiol. 62:1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabus, R., D. Gade, R. Helbig, M. Bauer, F. O. Glöckner, M. Kube, H. Schlesner, R. Reinhardt, and R. Amann. 2002. Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp. strain 1 by a proteomic approach. Proteomics 2:649-655. [DOI] [PubMed] [Google Scholar]
- 42.Rabus, R., M. Kube, A. Beck, F. Widdel, and R. Reinhardt. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178:506-516. [DOI] [PubMed] [Google Scholar]
- 42a.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 43.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 44.Ramos, J. L., S. Marques, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas Tol plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:241-372. [DOI] [PubMed] [Google Scholar]
- 45.Sachs, L. 1993. Statistische Methoden, p. 23-27 and 74-81. Springer Verlag, Berlin, Germany.
- 46.Spormann, A. M., and F. Widdel. 2000. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 11:85-105. [DOI] [PubMed] [Google Scholar]
- 47.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 48.Steinfath, M., W. Wruck, H. Seidel, H. Lehrach, U. Radelof, and J. O'Brien. 2001. Automated image analysis for array hybridization experiments. Bioinformatics 17:634-641. [DOI] [PubMed] [Google Scholar]
- 49.Torres, B., G. Porras, J. L. Garcia, and E. Diaz. 2003. Regulation of the mhp cluster responsible for 3-(3-hydroxyphenyl)propionic acid degradation in Escherichia coli. J. Biol. Chem. 278:27575-27585. [DOI] [PubMed] [Google Scholar]
- 50.Tschech, A., and G. Fuchs. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 148:213-217. [DOI] [PubMed] [Google Scholar]
- 51.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
- 52.Widdel, F., A. Boetius, and R. Rabus. 2003. Anaerobic biodegradation of hydrocarbons including methane. .In A. Balows, H. G. Trüper, W. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. [Online.] Springer Science Online, Heidelberg, Germany. http://springerlink.metapress.com.