Abstract
In prokaryotes, disulfides are generated by the DsbA-DsbB system. DsbB functions to generate disulfides by quinone reduction. These disulfides are passed to the DsbA protein and then to folding proteins. To investigate the DsbA-DsbB catalytic system, we performed an in vivo selection for chromosomal dsbA and dsbB mutants. We rediscovered many residues previously shown to be important for the activity of these proteins. In addition, we obtained one novel DsbA mutant (M153R) and four novel DsbB mutants (L43P, H91Y, R133C, and L146R). We also mutated residues that are highly conserved within the DsbB family in an effort to identify residues important for DsbB function. We found classes of mutants that specifically affect the apparent Km of DsbB for either DsbA or quinones, suggesting that quinone and DsbA may interact with different regions of the DsbB protein. Our results are consistent with the interpretation that the residues Q33 and Y46 of DsbB interact with DsbA, Q95 and R48 interact with quinones, and that residue M153 of DsbA interacts with DsbB. All of these interactions could be due to direct amino acid interactions or could be indirect through, for instance, their effect on protein structure. In addition, we find that the DsbB H91Y mutant severely affects the kcat of the reaction between DsbA and DsbB and that the DsbB L43P mutant is inactive, suggesting that both L43 and H91 are important for the activity of DsbB. These experiments help to better define the residues important for the function of these two protein-folding catalysts.
Disulfide bonds are widely found in many exported proteins and contribute to folding and stability of these proteins (10, 11, 34). DsbA, a prokaryotic disulfide catalyst, contains a disulfide at its active site and oxidizes proteins by rapidly donating this disulfide to other proteins. DsbA is perhaps the most oxidizing protein known (23). The active site cysteines in DsbA (Cys30 and Cys33) are separated by two amino acids, Pro31 and His32, constituting a thioredoxin motif. Pro31 enhances the electrostatic interaction of the thiolate ion with the helix dipole (25), while His32 electrostatically stabilizes the thiolate ion (15, 35). These two effects are important for fostering the low pKa of Cys30 and making it so reactive.
DsbB generates disulfides de novo and transfers them to DsbA (4, 9, 28). DsbB has six cysteines, four of which (Cys41, Cys44, Cys104, and Cys130) are essential for its function (16, 20, 24).
DsbB possesses a unique enzymatic activity: it uses the oxidizing power of quinones to generate disulfides (3). DsbB has two substrates: DsbA and quinones. DsbB reoxidizes DsbA and then transfers its electrons to quinones present in the cytoplasmic membrane. Ubiquinone is in turn reoxidized by terminal oxidases, such as cytochrome bd and bo oxidases, which finally transfer electrons to oxygen. Under anaerobic conditions, Escherichia coli switches its immediate electron acceptor from ubiquinone to menaquinone, which is upregulated upon oxygen depletion. Instead of cytochrome oxidases, anaerobic oxidoreductases such as fumarate reductase serve to reoxidize menaquinone (1).
Mutants in a highly conserved residue, Arg48 of DsbB, are only partially active (21). These mutants are defective in β-lactamase oxidation under anaerobic conditions, but not under aerobic conditions, and display a sevenfold increase in Km for ubiquinone while retaining a Km for DsbA similar to that of the wild-type protein. These results suggest that R48 is directly or indirectly important for DsbB's interaction with quinones. A peptide in the second periplasmic domain of DsbB cross-links to a photoreactive quinone analogue, suggesting that the second periplasmic domain of DsbB is also involved in quinone binding (37).
The approach of selecting for spontaneous mutants with disulfide defects, combined with mutating conserved residues in DsbB, allowed us to recover many mutations in DsbA and DsbB. Characterization of these mutations in vivo and in vitro expands our understanding of the interaction of DsbA with DsbB and the binding of DsbB to quinone.
MATERIALS AND METHODS
Ampicillin resistance of dsbA and dsbB mutants.
The plasmid pCLL2100 (29), containing the ccrA gene, was transformed into the wild-type E. coli strain MG1655, resulting in JT272. To test the ampicillin sensitivity of characterized dsbA and dsbB mutants, stocks of P1 lysates were made on pools of bacterial strains PB406 (dsbA CPPC), PB410 (dsbA CSFC), PB416 (dsbA CPSC), PB419 (dsbA CPLC) (8), HK207 (dsbB C41Y), HK209 (dsbB R48C), and HK211 (dsbB R48H) (21) (Table 1) and transduced into JT272 [MG1655(pCLL2100)]. The resulting strains were streaked onto Luria-Bertani (LB) plates containing ampicillin at various concentrations. The plates were incubated at 37°C until colonies were visible and scored for ampicillin resistance.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| HK207 | HK205 dsbB C41Y | 21 |
| HK209 | HK205 dsbB R48C | 21 |
| HK211 | HK205 dsbB R48H | 21 |
| JCB816 | MC1000 dsb+phoR λ malF-lacZ102 zih-12::Tn10 | Lab collection |
| JCB819 | MC1000 dsbB::kan-5 phoR λ malF-lacZ102 zih-12::Tn10 | Lab collection |
| JT272 | MG1655, pCLL2100 | This work |
| JT357 | JT272, dsbA CPPC zih-12::Tn10 | This work |
| JT358 | JT272, dsbA CSFC zih-12::Tn10 | This work |
| JT359 | JT272, dsbA, CPSC zih-12::Tn10 | This work |
| JT360 | JT272, dsbA CPLC zih-12::Tn10 | This work |
| JT368 | JT272, yihF::insAB_5 | This work |
| JT427 | MG1655 dsbA M153R, pCLL2100 | This work |
| JT491 | MG1655, pCLL2100 non-dsbA or -dsbB mutation | This work |
| JT503 | JT272, yihE::yi22_6 | This work |
| JT656 | BL21 dsbA::kan-5, pJT656 | This work |
| JT720 | JT491, dsbB/pBAD33 | This work |
| JT844 | JT503, yihE/pBAD33 | This work |
| JT855 | JT491, dsbA/pBAD33 | This work |
| JT890 | JT503, λdsbA+ | This work |
| MB31 | D10, pMS421, pBJ41 | Lab collection |
| PB406 | SF100 dsbA CPPC | 8 |
| PB410 | SF100 dsbA CSFC | 8 |
| PB416 | SF100 dsbA CPSC | 8 |
| PB419 | SF100 dsbA CPLC | 8 |
| VL1j | JCB819, pACYC184-lacIq-tet, pWM76 R48A | This work |
| VL2j | JCB819, pACYC184-lacIq-tet, pWM76 Q95A | This work |
| VL3j | JCB819, pACYC184-lacIq-tet, pWM76 Q33A | This work |
| VL4j | JCB819, pACYC184-lacIq-tet, pQE70 | This work |
| VL5j | JCB819, pACYC184-lacIq-tet, pWM76 | This work |
| VL6j | JCB819, pACYC184-lacIq-tet, pWM76 H91A | This work |
| VL14j | JCB819, pACYC184-lacIq-tet, pWM76 Y46F | This work |
| VL15j | JCB819, pACYC184-lacIq-tet, pWM76 R48K | This work |
| pBJ41 | pKK233-2 dsbA | 5 |
| pCLL2100 | pCLL2300 ccrA | 29 |
| pJT651 | pWM76 dsbB L43P | This work |
| pJT653 | pWM76 dsbB R133C | This work |
| pJT655 | pWM76 dsbB L146R | This work |
| pJT656 | pET11a dsbA M153R | This work |
| pVL1 | pWM76 dsbB R48A | This work |
| pVL2 | pWM76 dsbB Q95A | This work |
| pVL3 | pWM76 dsbB Q33A | This work |
| pVL6 | pWM76 dsbB H91A | This work |
| pVL14 | pWM76 dsbB Y46F | This work |
| pVL15 | pWM76 dsbB R48K | This work |
| pWM76 | pQE70 dsbB C8A, C49V | 1 |
| λD69 | λ bam λ 1° Δ(srIλ1-srIλ2) imm-21 nin-5 shn-6O | 5 |
| λdsbA+ | 2.5-kb HindIII from p12-7 cloned into λD69 | 5 |
In vivo selection for chromosomal dsbA and dsbB mutants.
Individual JT272 colonies (1,000) were grown overnight in separate 5-ml LB cultures supplemented with kanamycin (30 μg ml−1) at 37°C. Each culture (10 μl) was spread onto MacConkey (Difco) plates supplemented with ampicillin (15 μg ml−1) and grown until colonies were visible. Colonies were then streaked onto MacConkeyAmp15 plates twice. Ampicillin-resistant colonies were grown in 5-ml LB cultures, and Western blot analysis was carried out on whole-cell extracts with the rabbit polyclonal anti-DsbA antiserum at a 1:4,000 dilution and the ECL Western blotting chemiluminescence detection system (Amersham). Mutants showing wild-type levels of dsbA expression were then subjected to PCR analysis with primers flanking the dsbA and dsbB genes. Mutants with no change in the sizes of dsbA and dsbB were then sequenced to identify the mutated residues.
Cloning DsbA M153R mutant.
The dsbA M153R gene was amplified from the genomic DNA of the strain where methionine 153 had been mutated to arginine in dsbA (JT427). The primers used to amplify the gene were DsbA-Fwd (5′-ATA CAT ATG AAA AAG ATT TGG CTG GCG CTG-3′) and DsbA-Rev (5′-GCC GGA TCC TTA TTT TTT CTC GGA CAG ATA TTT CAC TGT ATC-3′). The PCR product was cloned into pCR2.1 (Invitrogen) and then transformed into INVαF′ cells (Invitrogen). Plasmids were digested with BamHI and NdeI restriction enzymes (Promega) and ligated into predigested pET11a vector (pJT656) cut with the same enzymes. Ligation products were transformed into XL1-Blue cells, and plasmid DNA from ampicillin-resistant colonies was then transformed into BL21 cells (JT656). Plasmid DNA from the transformed BL21 cells was sequenced to confirm its identity.
Purification and reduction of DsbA.
Wild-type DsbA (MB31) and the DsbA M153R mutant (JT656) were purified as described previously (26) over a HiTrap Q HP Sepharose (Amersham Pharmacia Biotech) column and subsequently with a hydroxyapatite column. Periplasmic extracts of DsbA and DsbA M153R were loaded onto 5-ml Q HP columns. A gradient of 10 mM morpholinepropanesulfonic acid (MOPS) (pH 8.0) to 500 mM NaCl-10 mM MOPS (pH 8.0) was used over 500 ml to elute DsbA or DsbA M153R. Fractions containing DsbA or DsbA M153R were then passed over a hydroxyapatite (Bio-Rad) column. Wild-type and mutant DsbA of >99% purity were obtained by running a gradient from 10 mM MOPS (pH 8.0) to 50 mM NaH2PO4-10 mM MOPS (pH 8.0) over 100 ml through a 20-ml hydroxyapatite column. Protein concentration was determined by using an absorption coefficient of 1.028 for a 1-mg/ml solution at an optical density of 280 (12). The proteins were reduced by adding dithiothreitol to a final concentration of 10 mM and incubating the mixture on ice for 1 h. The dithiothreitol was removed by dialyzing against 1 mM EDTA (pH 7.5) overnight at 4°C. The thiol constant was measured by the Ellman assay (31), indicating that DsbA was >98% reduced. Reduced DsbA stored at −70°C retains its redox state for at least a month.
DsbA and DsbA M153R redox potential determination.
The redox potentials of both DsbA and the mutant DsbA M153R proteins were determined by using an adaptation of the method which was used to analyze the redox equilibrium of DsbA with reduced glutathione and oxidized glutathione (36).
Construction of dsbA/pBAD33, dsbB/pBAD33, yihE/pBAD33, and yihF/pBAD33.
The primers used to amplify the dsbA gene were XbaI_DsbA (5′-GCT CTA GAA TGA AAA AGA TTT GGC TGG CGC TG-3′) and DsbA_HindIII (5′-CCC AAG CTT TTA TTT TTT CTC GGA CAG ATA TTT CAC TGT ATC-3′). The primers used to amplify the dsbB gene were XbaI_DsbB-Fwd (5′-GTC TAG AAT GTT GCG ATT TTT GAA CCA ATG TTC AC-3′) and PstI_DsbB-Rev (5′-GCT GCA GTT AGC GAC CGA ACA GAT CAC G-3′). The primers used to amplify the yihE gene were XbaI_YihE (5′-GCT CTA GAA TGA ATA ACA GCG CTT TTA CTT TCC AG-3′) and YihE_PstI (5′-CCC TGC AGT TAA TAC ATA GGT GTT AAT TGC AAA GG-3′). The primers used to amplify the yihF gene were XbaI_YihF (5′-GCT CTA GAG TGG ATA TTC AAA GTT TTG CTG TTT TAT CAG GG-3′) and YihF_HindIII (5′-CCC AAG CTT TTA ATG AAC AAA ACG TCC GGC-3′). The PCR products were cloned into pCR2.1 (Invitrogen) and then transformed into INVαF′ cells (Invitrogen). Plasmids were digested with XbaI and HindIII restriction enzymes (Promega) for dsbA and yihF cloning or with XbaI and PstI for dsbB and yihE cloning and ligated into predigested pBAD33 vector cut with the same enzymes. Ligation products were transformed into XL1-Blue cells, and plasmid DNAs from ampicillin-resistant colonies were sequenced to confirm their identities.
Site-specific mutagenesis.
The dsbB expression plasmid (pWM76) lacking the nonessential cysteines, Cys8 and Cys49, was constructed as previously described and is fully active (1). This plasmid was subsequently mutated with the QuikChange site-directed mutagenesis kit (Stratagene) to obtain the mutants Q33A (pVL3), Y46F (pVL14), R48A (pVL1), R48K (pVL15), H91A (pVL6), Q95A (pVL2), L43P (pJT651), R133C (pJT653), and L146R (pJT655). The primers used for mutagenesis are as follows: DsbB Q33A (5′-GCT GTG GTT CGC GCA TGT GAT GTT ACT GAA A-3′), DsbB Y46F (5′-TGC GTG CTC TGT ATT TTC GAA CGC GTC GCG-3′), DsbB R48A (5′-TAT TTA TGA AGC CGT CGC GTT ATT CGG CGT T-3′), DsbB R48K (5′-TAT TTA TGA AAA AGT CGC GTT ATT CGG CGT T-3′), DsbB H91A (5′-CAG TTA ACT TAC GAG GCC ACC ATG CTT CAG-3′), DsbB Q95A (5′-CAC ACC ATG CTT GCC CTC TAT CCT TCG CCG TTT-3′), DsbB L43P Fwd (5′-CTG AAA CCT TGC GTG CCC TGT ATT TAT GAA CGC-3′), DsbB R133C (5′-GGC GAT TGC GCC GAG TGT CAG TGG G-3′), and DsbB L146R (5′-CTG GAA ATG CCG CAG TGG CGG CTC GG-3′). Mutations were identified by restriction digest screening and confirmed by DNA sequencing.
Purification of DsbB mutants.
Wild-type and DsbB mutants were purified from membranes prepared from the respective overexpression strains as previously described (1). All proteins purified were >95% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
DsbB enzymatic assay.
The apparent Km values for DsbB with DsbA and DsbA M153R were measured under saturating conditions of Q0C10 as previously described (3). The activity of DsbB can be monitored by monitoring the redox state of its in vivo substrate DsbA by fluorescence spectroscopy (2) or by measuring the reduction of quinone photometrically (3).
Determination of in vivo periplasmic redox state in mutant strains.
Mutant plasmids were transformed into JCB819 (dsbB::kan-5) along with a pACYC184-lacIq construct. JCB819 transformed with the empty pQE70 vector and pACYC-lacIq plasmid served as a disulfide-defective control strain. An overnight culture was grown in M63 minimal medium (27) supplemented with 0.1% Casamino Acids and individual amino acids at 40 μg/ml. Disulfide bond exchange during sample preparation was prevented by treatment with 0.1 M thiol-specific agent iodoacetamide (IAM), which rapidly blocks free thiols. Proteins were precipitated with 8% trichloroacetic acid on ice for 60 min and then resuspended in Laemmli loading buffer. The oxidized form of β-lactamase was separated from its reduced form on a 14% Tris-glycine gel (Novex). Western blot assays were performed, and β-lactamase was detected with an anti-β-lactamase antibody (5 Prime-3 Prime, Inc.). The dsbB expression level in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG) was examined by immunoblotting analysis to verify that all of the mutants were producing adequate levels of DsbB.
RESULTS
Reconstruction of in vivo selection.
To further dissect the mechanism of interaction between DsbA, DsbB, and their substrate molecules, we set out to develop an in vivo selection for chromosomal dsbA and dsbB mutants to determine the role and importance of non-active-site residues for their functions. The selection depends on the unique properties of the zinc-dependent β-lactamase CcrA (18, 32). This β-lactamase is active only in the absence of disulfide bonds, thus allowing a powerful and specific selection for mutants that are defective in disulfide bond formation simply by demanding growth on ampicillin-containing plates (29). Wild-type E. coli cells carrying a plasmid containing the ccrA gene are sensitive to ampicillin, since DsbA catalyzes the formation of aberrant disulfide bond linkages in CcrA, causing it to be unstable. Mutations in DsbA or DsbB will allow for functional CcrA and, hence, ampicillin resistance.
To determine whether mutations varying in severity confer different levels of ampicillin resistance, we tested the ampicillin resistance of wild-type E. coli containing pCLL2100 (CcrA+) and various chromosomal dsbA and dsbB point mutants that were known to have partial defects. The results shown in Table 2 suggest that we should be able to obtain weak chromosomal dsbA and dsbB mutants by simply plating CcrA-containing strains on ampicillin-containing plates. This should allow us to select for mutations at sites in addition to those in the active site cysteines. This combined with in vitro characterization should allow us obtain a more complete picture of residues important for the activity of DsbA and DsbB.
TABLE 2.
Active site mutants show various ampicillin sensitivitiesa
| Strains | Result of growth on LB + ampicillin concn (μg/ml) of:
|
|||||
|---|---|---|---|---|---|---|
| 0b | 10 | 20 | 40 | 100 | 200 | |
| JT272 CPHC | +++ | ± | − | − | − | − |
| JT357 CPPC | +++ | +++ | +++ | + | ± | ± |
| JT358 CSFC | +++ | +++ | + | ± | − | − |
| JT359 CPSC | +++ | +++ | + | − | − | − |
| JT360 CPLC | +++ | +++ | ++ | + | − | − |
The active site CPHC of wild-type DsbA was mutated to CPPC, CSFC, CPSC, or CPLC. Wild-type E. coli (MG1655) strains carrying a CcrA plasmid and possessing the various chromosomal mutations in the active site of DsbA were streaked onto LB plates containing various concentrations of ampicillin. Sizes of colonies after an overnight incubation at 37°C are denoted as follows: +++, large colonies; ++, medium-sized colonies; +, small colonies; ±, tiny colonies; −, lack of growth.
Growth on LB alone.
In vivo selection for chromosomal dsbA and dsbB mutants.
The vast majority of ampicillin-resistant mutants recovered from our in vivo selection were null (278 clones), truncated (36 clones), insertion (3 clones), and mutants with reduced DsbA production (81 clones), with only 61 showing wild-type levels of DsbA production as well as the correct size of DsbA. Of these 61 mutants, 16 showed insertions in dsbB (data not shown), and the remaining 45 mutants that displayed wild-type sizes of dsbA and dsbB were sequenced to determine which residues were mutated.
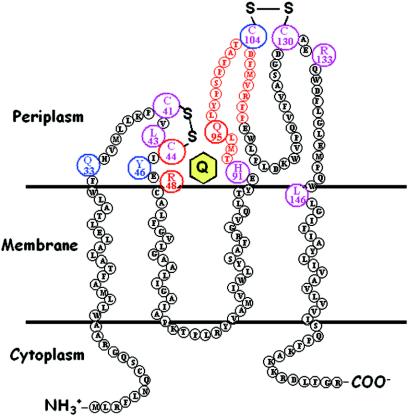
Sequencing results indicate that many of the mutations are in the important active site C-X-X-C motif of DsbA (12 independent isolates). These active site residues have been previously characterized (8, 13, 38, 39). The multiple Cys30, Pro31, His32, and Cys33 mutants recovered in our selection emphasize the importance of these residues in the function of DsbA. The recovery of these active site mutants reiterates the importance of the active site residues in the function of DsbA in vivo. However, because we are interested in the study of residues other than the previously characterized ones, we went on to focus on the novel DsbA M153R mutant we recovered. The majority of dsbB point mutants recovered from the selection are in its active site cysteines (Cys41, Cys44, Cys104, and Cys130) (10 independent isolates). A previous study revealed four cysteines essential for DsbB activity, Cys41, Cys44, Cys104, and Cys130 (20). Among the DsbB mutants we recovered in our selection are three with a mutaion in Arg48. A genetic screen previously revealed Arg48 as a potential residue for quinone interactions (21). The recovery of these cysteine and arginine mutants in our selection ties in very nicely with previous results demonstrating their importance in the function of DsbB. In addition to the cysteine and arginine mutants, we recovered 4 novel DsbB mutants, L43P, H91Y, R133C, and L146R, in a total of 14 independent isolates (Fig. 1).
FIG. 1.
Topological model of the transmembrane structure of DsbB. Important DsbB amino acid residues are shown as enlarged circles. These include the catalytic cysteines, residues recovered from our in vivo selection for disulfide defective mutants, and those we subjected to site-directed mutagenesis. C44, R48, and Q95 are implicated in quinone binding and are shown as red circles. The quinone is shown as a yellow hexagon. Q33, Y46, and C104 are implicated in DsbA binding and are shown as blue circles. Residues important for the catalytic mechanism or proper folding of DsbB are shown in purple. The cysteines C41, C44, C104, C130, and H91 appear to be vital for catalysis. Of these, two appear to have multiple roles. C44 is implicated in both catalysis and quinone binding, so it is shown as a red circle surrounding purple letters. C104 is implicated in both catalysis and DsbA binding, so it is shown as a blue circle surrounding purple letters. R133 and L146 are probably important for proper folding, as substitutions in these residues result in no protein production.
The recovery of all of these mutants in DsbA and DsbB verifies that the selection is able to give rise to mutations in residues important for the functions of DsbA and DsbB.
Properties of DsbB mutants.
First, the four novel DsbB (L43P, H91Y, R133C, and L146R) mutants we recovered in our selection were analyzed. One of the novel mutants contains a mutation that alters His91. A similar mutant, H91A, was independently generated by site-directed mutagenesis and will be described below. Due to a lack of sensitive DsbB antibody to determine the expression level of dsbB in the other three point mutants we recovered, we analyzed their overproduction by SDS-PAGE and found that only JT658 (L43P) overproduces DsbB (data not shown). The remaining mutants were not overproduced, presumably because they affect the folding or stability of DsbB and were not further characterized.
Site-directed mutants of dsbB.
To further study the structure and function of DsbB, we chose to mutate four conserved residues based on a sequence alignment: Q33, Y46, H91, and Q95. We also mutated arginine 48 to lysine (pVL15) to explore the consequences of a protein that contains a more conservative substitution than the previously reported R48H variant. Q33 (pVL3), H91 (pVL6), and Q95 (pVL2) were each mutated to the chemically neutral residue alanine, while Y46 was mutated to phenylalanine (pVL14) (Fig. 1).
The DsbB mutants, R48A, Q95A, Q33A, H91A, Y46F, and R48K, were purified as previously described (1). All of these mutants overexpressed to the same extent as the wild type and are found in the membrane fraction.
The purified DsbB mutants were analyzed for their enzymatic parameters by using purified reduced DsbA and dodecyl quinone as substrates (Table 3). The mutants can be separated into three groups based on different kinetic behaviors compared to wild-type DsbB. The first group includes mutants Q33A and Y46F, which show a greater than fivefold change in apparent Km for DsbA. This suggests that these residues are involved directly or indirectly in DsbA binding. It should be noted here that the apparent Km for quinone displayed by this group of proteins is likely to represent a substantial overestimate. Since these mutants have a substantially increased apparent Km for DsbA, it is not practical to use a saturating amount of DsbA in the reactions. Thus, the higher apparent Km (Q) values of Q33A, Y46F, R48K, and H91A compared to that of wild-type protein cannot yet be judged to be significant.
TABLE 3.
Kinetic analysis of DsbB mutantsa
| Enzyme | Km (DsbA) | Km (Q) | kcat |
|---|---|---|---|
| WT | 3.5 | 1 | 3.7 |
| Q33A | 40 | 5 | 2.7 |
| Y46F | 18 | 1.8 | 4.2 |
| R48K | 6.2 | 1.1 | 0.9 |
| R48H | 3.3 | 7.1 | 1.6 |
| H91A | 8.8 | 1.7 | 0.68 |
| Q95A | 6.5 | 16 | 9 |
Kinetic analysis was performed as described in Materials and Methods with reduced purified DsbA and dodecyl quinone (Q) as two substrates. Values of apparent Km are in micromolar; kcat is in nanomoles of DsbA/nanomole of DsbB/second. Wild type (WT) refers to WM76, the wild-type DsbB with C8A and C49V substitutions. Results shown in bold are significantly different from that of the wild type. No significant activity was detected for R48A and L43P. R48H values are quoted from reference 21.
The second group of mutants, R48K and H91A, both exhibit a >10-fold decrease in catalytic activity compared to wild-type DsbB and no significant effect on the apparent Km for DsbA or the apparent Km for quinone. The L43P mutant is inactive. This implies that these three residues, L43, R48, and H91, are important for the activity of DsbB. L43 could also be important for the structure of DsbB. R48 was previously implicated in quinone binding because of the strongly increased apparent Km for quinone observed for an R48H mutant protein. That the more conservative lysine substitution at this position has very little if any effect on apparent Km for quinone suggests that conservation of charge is important for quinone interaction. The strongly decreased kcat observed for both the R48K and R48H mutations provides further evidence for the importance of this residue in DsbB's catalytic activity.
The third group is made up of a single member, Q95A. This mutant shows a higher apparent Km for quinone and no major changes in the other two kinetic parameters. This implies that this residue is involved either directly or indirectly in quinone binding.
Redox status of periplasm is altered in DsbB mutant strains.
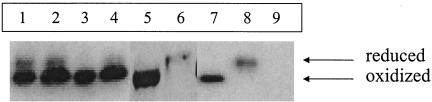
To directly test how efficiently disulfide bond formation proceeds in the periplasm of the mutant strains, we examined the redox status of a periplasmic protein β-lactamase. This protein does not require its disulfide for stability, and its oxidation status is maintained once the protein is folded. The ratio of oxidized to reduced β-lactamase present in the periplasm at steady state provides a good measure of the efficiency of the disulfide catalytic machinery. We found that the redox state of β-lactamase varies in different mutant strains, as indicated in Fig. 2. Compared to the wild-type control, it appears that Y46F, R48A, and Q95A exhibit a significantly lower DsbB enzymatic efficiency. H91A is severely impaired, as shown by the majority of β-lactamase present in the reduced form.
FIG. 2.
In vivo β-lactamase redox state in DsbB mutants. Overnight cultures of mutant strains and appropriate control strains in M63 (without Cys) medium (26) supplemented with 0.1% Casamino Acids are trapped with IAM and analyzed by SDS-PAGE and immunoblotting with antibody against β-lactamase. Oxidized and reduced forms of β-lactamase migrate differently due to the attachment of IAM to the free thiols of reduced proteins. The gene coding for β-lactamase is carried on the pQE70 vector. Lane 1: JCB819 containing R48A (VL1j). Lane 2: JCB819 containing Q95A (VL2j). Lane 3: JCB819 containing Q33A (VL3j). Lane 4: JCB819 containing Y46F (VL14j). Lane 5: JCB819 containing R48K (VL15j). Lane 6: JCB819 containing H91A (VL6j). Lane 7: JCB819 containing wild-type dsbB (VL5j). Lane 8: JCB819 containing pQE70 (VL4j). Lane 9: JCB816 alone.
Untranslated leader sequence mutants of dsbB.
Two mutants obtained from the genetic selection show altered sequences in the untranslated leader sequence of dsbB. These two mutants both have a single-base substitution in the untranslated leader sequence region, leading to increased CcrA activity and cadmium sensitivity, likely because of a decreased expression of dsbB. Wild-type DsbB provided in trans complements these two phenotypes.
DsbA M153R has altered kinetic properties.
In addition to many of the residues previously defined as important in DsbA function (8, 14, 35), one additional residue, M153R, was identified by mutation. To characterize the DsbA M153R mutation in vitro, we cloned the gene, overproduced the protein, and purified it. We found that DsbB exhibits an apparent Km for the purified M153R mutant, which is 6- to 10-fold higher than the reported apparent Km values for wild-type DsbA. However, DsbB has an increased kcat of 12.2 s−1 for the DsbA M153R protein compared to a kcat of 3.7 s−1 for the wild-type DsbA protein. The catalytic efficiency (kcat/Km) for reoxidizing the mutant DsbA M153R (3.5 × 105 M−1 s−1) is therefore only slightly less than that for wild-type DsbA (6.5 × 105 M−1 s−1) despite major changes in both Km and kcat. That there is a greatly increased apparent Km for DsbB suggests that methionine 153 of DsbA could be involved either directly or indirectly in DsbB interaction.
The mutation from methionine to arginine also does not drastically affect the redox properties of the M153R mutant form of DsbA. The mutant has a slightly more reducing redox potential for DsbA M153R (−157 mV) than for wild-type DsbA (−138 mV), which was measured in parallel experiments. The M153R substitution thus does not seem to affect the folding or oxidizing potential of DsbA to a great extent, so we examined the effect of DsbA M153R on physiological substrates.
Phenotypic characterization of DsbA M153R.
It has been previously observed that dsbA-null mutants are nonmotile, are resistant to M13 phage infection, have low levels of alkaline phosphatase activity, and are sensitive to cadmium (5, 9, 30, 33). To test whether the DsbA M153R mutant strain (JT427) is deficient in vivo in its role of catalyzing the formation of disulfide bonds, its phenotypes for the above properties were examined. DsbA M153R is motile and M13 sensitive and shows only slightly reduced levels of alkaline phosphatase activity (data not shown), suggesting that M153R has, at most, a very slight defect in disulfide bond formation. We found however, that the novel mutant DsbA M153R is sensitive to 0.1 mM cadmium and that the sensitivity can be rescued by providing wild-type DsbA in trans. Wild-type bacteria are resistant to levels of cadmium up to 0.5 mM. DsbA- and DsbB-null mutants are sensitive to 0.01 mM cadmium. Thus, the DsbA M153R mutant is slightly sensitive to cadmium. Mutations in dsbA and dsbB enhance the sensitivity of a gram-negative bacterium to cadmium because periplasmic proteins in these mutants acquire disulfide bonds at a slower rate (30, 33). The cadmium sensitivity is likely due to a competition between cadmium binding to thiols, which inactivates the protein, and disulfide bond formation, which simultaneously promotes the folding of the protein and eliminates the thiols from cadmium attack. The above results seem to suggest that lack of motility, resistance to M13 phage infection, and a dramatic reduction in alkaline phosphatase activity require a severe defect in DsbA. A weak mutant like DsbA M153R is thus able to display a phenotype close to that of the wild type in these assays. Sensitivity to cadmium however, is a less stringent phenotype, and even a weak DsbA mutant appears to exhibit such a phenotype.
Insertions in upstream and downstream genes affect dsbA expression.
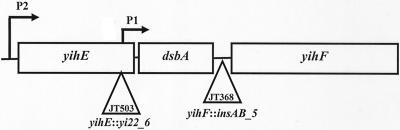
We recovered two mutants that contained insertion sequence (IS) elements upstream and downstream of dsbA in our ccrA selection (Fig. 3). JT503 has an IS element, yi22_6, in the gene upstream of dsbA known as yihE. JT368 has an IS element, insAB_5, in the promoter of the downstream gene yihF. In both cases, the protein coding sequence of DsbA is unaffected.
FIG. 3.
Genomic organization of dsbA region. There are two promoters, P1 and P2, for dsbA (6). P1 is located in the distal portion of the upstream gene yihE, while P2 is located upstream of yihE. The promoter for yihF is located between the dsbA and yihF open reading frames. JT503 (yihE::yi22_6) contains an insertion in the distal portion of the upstream gene yihE, while JT368 (yihF::insAB_5) contains an insertion in the promoter for yihF.
JT503 shows heightened sensitivity to cadmium.
Overexpressing yihE in JT503 (JT844) does not complement the cadmium sensitivity or ampicillin resistance phenotype of the mutant, but a single copy of dsbA on a λD69 clone (λdsbA+) (JT890) does, suggesting that the insertion in yihE negatively affects the expression of dsbA, as has previously been observed (7). The IS element in yihE is thus likely to lead to a defect in dsbA expression that went undetected in our initial screen for DsbA levels by Western blot analysis. Western blot analysis of the JT368 mutant which contains an insertion sequence in the promoter of the yihF gene downstream of dsbA showed that dsbA expression is reduced slightly (data not shown), indicating that it too is negatively affecting the expression of dsbA possibly by affecting transcriptional termination.
Non-DsbA- or -DsbB-linked mutation.
Upon sequencing the upstream and downstream regions of the dsbA and dsbB open reading frames, we found one mutant that not only had wild-type dsbA and dsbB open reading frames but also did not possess any nearby upstream or downstream mutations. This mutant, JT491, is cadmium resistant like wild-type MG1655. The introduction of dsbA/pBAD33 (JT855) or dsbB/pBAD33 (JT720) clones failed to complement the ampicillin resistance phenotype of the mutant (data not shown), suggesting that there could be another gene in E. coli affecting the formation of aberrant disulfides in CcrA. The recovery of a non-DsbA- or -DsbB-linked mutant raises the possibility that there may be either another player in the Dsb system or another way to affect the overall redox status of the periplasm. However, the only phenotype of this mutation was ampicillin resistance. No other indications of a generalized defect in disulfide bond formation were seen. In our view, this makes it likely that this mutation affects the rate of disulfide bond formation in CcrA indirectly. For example, zinc insertion may be enhanced so that insertion effectively competes with Dsb-catalyzed disulfide inactivation of the zinc binding cysteine. It is also possible that this mutation affects the folding or expression of CcrA, ampicillin permeability, or transport.
DISCUSSION
Formation of intramolecular disulfide bonds is an assisted process mediated by the strongly oxidizing periplasmic protein DsbA (5, 22). Reoxidation of DsbA is carried out by the inner membrane protein DsbB (4, 28). The Cys104-Cys130 bond in DsbB's second periplasmic domain is thought to be the direct donor of disulfides to DsbA (23), and the first periplasmic domain has been implicated in quinone binding (21).
The results presented here, however, implicate residues from the first periplasmic domain in the interaction with DsbA and residues from the second periplasmic domain in quinone interaction. Thus, the surface involved in DsbA and quinone interaction may involve residues from both the first and second periplasmic domains. For instance, Q33 and Y46, two highly conserved amino acid residues in the first domain, appear important for interaction with DsbA. However, C104 present in the second periplasmic domain is found in a disulfide cross-link with DsbA, and this mixed disulfide is thought to be an intermediate in the transfer of disulfide bonds from DsbA to DsbB (21). Thus, both the first and second periplasmic domains may be involved with DsbA interaction.
Mutation of the Q95 residue, which lies at the beginning of the second periplasmic domain, results in a significant defect in quinone binding without major effects on DsbA binding. This residue is found within a peptide previously implicated in quinone binding because it cross-links with a photoreactive quinone analogue (37). These results, in conjunction with previously published evidence that C44 and R48 are involved in quinone binding, suggest that the quinone binding surface has elements from both the first periplasmic domain (C44 and R48) and the second (Q95) (19, 21). The regions of DsbB implicated in quinone binding are shown in Fig. 1, as are those implicated in DsbA interaction.
Our studies show that a conserved His91 residue, found within the second periplasmic domain, is important for DsbB catalytic activity. The H91A mutant shows severe defects in disulfide bond formation in vivo, and in our kinetic study, H91A shows a specific defect in its catalytic activity (10-fold decrease in kcat) without changing the apparent Km for DsbA or quinone substantially. This suggests that His91 is likely involved in the catalytic mechanism of dithiol-disulfide interchange rather than in DsbA or quinone binding. The imidazole group may, for instance, serve as a proton donor in the active site of DsbB, participating in the electron transfer between Cys41-Cys44 and quinone. All residues implicated as being important for the catalytic activity or proper folding of DsbB are shown in Fig. 1.
In summary, the mutants we recovered in our genetic selection add to the knowledge of the DsbA-DsbB system. Previously, it was thought that the first periplasmic domain of DsbB was involved in quinone interaction and that the second periplasmic domain was involved in DsbA interaction. Our results now indicate that residues from both the first and second periplasmic domains of DsbB are involved in both DsbA and quinone interaction. The M153R mutant of DsbA appears to affect DsbA-DsbB interaction. Leu43, a highly conserved residue adjacent to the catalytic Cys44 residue in DsbB, could have a structural role or be involved in the catalytic mechanism of DsbB, while His91, a conserved residue in the second periplasmic domain, is involved in the catalytic mechanism of DsbB.
Acknowledgments
This work was supported by an NIH grant to J.C.A.B.
REFERENCES
- 1.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 2.Bader, M., W. Muse, T. Zander, and J. Bardwell. 1998. Reconstitution of a protein disulfide catalytic system. J. Biol. Chem. 273:10302-10307. [DOI] [PubMed] [Google Scholar]
- 3.Bader, M. W., T. Xie, C. A. Yu, and J. C. Bardwell. 2000. Disulfide bonds are generated by quinone reduction. J. Biol. Chem. 275:26082-26088. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 6.Belin, P., and P. L. Boquet. 1994. The Escherichia coli dsbA gene is partly transcribed from the promoter of a weakly expressed upstream gene. Microbiology 140:3337-3348. [DOI] [PubMed] [Google Scholar]
- 7.Belin, P., E. Quemeneur, and P. L. Boquet. 1994. A pleiotropic acid phosphatase-deficient mutant of Escherichia coli shows premature termination in the dsbA gene. Use of dsbA::phoA fusions to localize a structurally important domain in DsbA. Mol. Gen. Genet. 242:23-32. [DOI] [PubMed] [Google Scholar]
- 8.Bessette, P. H., J. Qiu, J. C. Bardwell, J. R. Swartz, and G. Georgiou. 2001. Effect of sequences of the active-site dipeptides of DsbA and DsbC on in vivo folding of multidisulfide proteins in Escherichia coli. J. Bacteriol. 183:980-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doig, A. J., and D. H. Williams. 1991. Is the hydrophobic effect stabilizing or destabilizing in proteins? The contribution of disulphide bonds to protein stability. J. Mol. Biol. 217:389-398. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, H. F. 1997.. Protein disulfide isomerase and assisted protein folding. J. Biol. Chem. 272:29399-29402. [DOI] [PubMed] [Google Scholar]
- 12.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 13.Grauschopf, U., J. R. Winther, P. Korber, T. Zander, P. Dallinger, and J. C. Bardwell. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947-955. [DOI] [PubMed] [Google Scholar]
- 14.Guddat, L. W., J. C. Bardwell, R. Glockshuber, M. Huber-Wunderlich, T. Zander, and J. L. Martin. 1997. Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci. 6:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guddat, L. W., J. C. Bardwell, T. Zander, and J. L. Martin. 1997. The uncharged surface features surrounding the active site of Escherichia coli DsbA are conserved and are implicated in peptide binding. Protein Sci. 6:1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilhot, C., G. Jander, N. L. Martin, and J. Beckwith. 1995. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc. Natl. Acad. Sci. USA 92:9895-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber-Wunderlich, M., and R. Glockshuber. 1998. A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold. Des. 3:161-171. [DOI] [PubMed] [Google Scholar]
- 18.Hussain, M., A. Carlino, M. J. Madonna, and J. O. Lampen. 1985. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J. Bacteriol. 164:223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inaba, K., Y. H. Takahashi, and K. Ito. 2004. DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J. Biol. Chem. 279:6761-6768. [DOI] [PubMed] [Google Scholar]
- 20.Jander, G., N. L. Martin, and J. Beckwith. 1994. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 13:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadokura, H., M. Bader, H. Tian, J. C. Bardwell, and J. Beckwith. 2000. Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:10884-10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamitani, S., Y. Akiyama, and K. Ito. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishigami, S., and K. Ito. 1996. Roles of cysteine residues of DsbB in its activity to reoxidize DsbA, the protein disulphide bond catalyst of Escherichia coli. Genes Cells 1:201-208. [DOI] [PubMed] [Google Scholar]
- 24.Kishigami, S., E. Kanaya, M. Kikuchi, and K. Ito. 1995. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J. Biol. Chem. 270:17072-17074. [DOI] [PubMed] [Google Scholar]
- 25.Kortemme, T., and T. E. Creighton. 1995. Ionisation of cysteine residues at the termini of model alpha-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 253:799-812. [DOI] [PubMed] [Google Scholar]
- 26.Martin, J. L., G. Waksman, J. C. Bardwell, J. Beckwith, and J. Kuriyan. 1993. Crystallization of DsbA, an Escherichia coli protein required for disulphide bond formation in vivo. J. Mol. Biol. 230:1097-1100. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Missiakas, D., C. Georgopoulos, and S. Raina. 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc. Natl. Acad. Sci. USA 90:7084-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen, B. A., Y. Gluzman, and F. P. Tally. 1991. Escherichia coli chromosomal mutations that permit direct cloning of the Bacteroides fragilis metallo-beta-lactamase gene, ccrA. Mol. Microbiol. 5:1211-1219. [DOI] [PubMed] [Google Scholar]
- 30.Rensing, C., B. Mitra, and B. P. Rosen. 1997. Insertional inactivation of dsbA produces sensitivity to cadmium and zinc in Escherichia coli. J. Bacteriol. 179:2769-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riddles, P. W., R. L. Blakeley, and B. Zerner. 1983. Reassessment of Ellman's reagent. Methods Enzymol. 91:49-60. [DOI] [PubMed] [Google Scholar]
- 32.Sabath, L. D., and E. P. Abraham. 1966. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem. J. 98:11C-13C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stafford, S. J., D. P. Humphreys, and P. A. Lund. 1999. Mutations in dsbA and dsbB, but not dsbC, lead to an enhanced sensitivity of Escherichia coli to Hg2+ and Cd2+. FEMS Microbiol. Lett. 174:179-184. [DOI] [PubMed] [Google Scholar]
- 34.Taniyama, Y., R. Kuroki, F. Omura, C. Seko, and M. Kikuchi. 1991. Evidence for intramolecular disulfide bond shuffling in the folding of mutant human lysozyme. J. Biol. Chem. 266:6456-6461. [PubMed] [Google Scholar]
- 35.Warwicker, J., and P. J. Gane. 1996. Calculation of Cys 30 delta pKa's and oxidising power for DsbA mutants. FEBS Lett. 385:105-108. [DOI] [PubMed] [Google Scholar]
- 36.Wunderlich, M., and R. Glockshuber. 1993. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 2:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie, T., L. Yu, M. W. Bader, J. C. Bardwell, and C. A. Yu. 2002. Identification of the ubiquinone-binding domain in the disulfide catalyst disulfide bond protein B. J. Biol. Chem. 277:1649-1652. [DOI] [PubMed] [Google Scholar]
- 38.Yu, J., S. McLaughlin, R. B. Freedman, and T. R. Hirst. 1993. Cloning and active site mutagenesis of Vibrio cholerae DsbA, a periplasmic enzyme that catalyzes disulfide bond formation. J. Biol. Chem. 268:4326-4330. [PubMed] [Google Scholar]
- 39.Zapun, A., L. Cooper, and T. E. Creighton. 1994. Replacement of the active-site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 33:1907-1914. [DOI] [PubMed] [Google Scholar]