Abstract
The Snm protein secretion system is a critical determinant of Mycobacterium tuberculosis virulence. However, genes encoding components of this pathway are conserved among all mycobacteria, including the nonpathogenic saprophyte Mycobacterium smegmatis. We show that the Snm system is operational in M. smegmatis and that secretion of its homologous ESAT-6 and CFP-10 substrates is regulated by growth conditions. Importantly, we show that Snm secretion in M. smegmatis requires genes that are homologous to those required for secretion in M. tuberculosis. Using a gene knockout strategy in M. smegmatis, we have also discovered four new gene products that are essential for Snm secretion, including the serine protease mycosin 1. Despite the evolutionary distance between M. smegmatis and M. tuberculosis, the M. smegmatis Snm system can secrete the M. tuberculosis ESAT-6 and CFP-10 proteins, suggesting that substrate recognition is also conserved between the two species. M. smegmatis, therefore, represents a powerful system to study the multicomponent Snm secretory machine and to understand the role of this conserved system in mycobacterial biology.
Throughout evolution, numerous bacterial pathogens have acquired specialized protein secretion pathways to deliver effector proteins to host cells (3, 14, 34). These pathways, which are distinct from the ubiquitous Sec pathway, are critical for mediating interactions during infection to allow pathogen survival in the hostile environment of the host. For example, the type III secretion system is used by many pathogenic gram-negative bacteria and requires the assembly of a complex translocon that transfers bacterial proteins directly from the bacterial cytoplasm into the eukaryotic host cell. Different bacterial pathogens secrete distinct sets of type III effectors, each with specific activities tailored to meet the individual needs of the bacterium (14). The type III secretion system and other Sec-independent secretion pathways in gram-negative bacteria are well-studied, but analogous pathways in gram-positive pathogens are poorly understood (36).
Initial clues of the existence of a Sec-independent protein secretion pathway in Mycobacterium tuberculosis came from studies of the immunodominant protein ESAT-6 (early secretory antigenic target, 6 kDa). This protein, along with its binding partner, CFP-10 (culture filtrate protein, 10 kDa), is efficiently secreted from M. tuberculosis cells but lacks N-terminal signal sequences that would target its secretion through the Sec pathway (28-30). The genes encoding ESAT-6 and CFP-10, esxA and esxB, are cotranscribed in a two-gene operon (1). The predicted function of the genes surrounding this operon led to the hypothesis that neighboring genes may be important for ESAT-6 and CFP-10 secretion (4, 9, 19, 33). Furthermore, this region of the genome, known as the RD1 locus for region of difference 1, is notable, as the esxA/esxB operon and several flanking genes have been deleted from Mycobacterium bovis during the repeated passage that led to the attenuated M. bovis BCG vaccine strain. Attempts to rescue ESAT-6 and CFP-10 export in BCG revealed that only a cosmid containing the entire genomic locus that surrounds the esxA/esxB operon could restore ESAT-6 and CFP-10 secretion (23). These results suggest that ESAT-6 and CFP-10 secretion requires genes both upstream and downstream of the esxA/esxB operon, although the specific genes required for this pathway remained obscure.
Recent genetic studies have demonstrated that individual conserved genes within the RD1 region in M. tuberculosis encode an alternative protein secretion pathway, termed the Snm (secretion in mycobacteria) pathway (10, 13, 31). A screen seeking mutants specifically defective for growth in the mouse model of infection isolated a class of mutants with transposon insertions clustered near the esxA/esxB operon (31). Strikingly, these mutants were capable of synthesizing ESAT-6 and CFP-10 yet were unable to secrete them into the extracellular milieu. The transposons disrupted three genes, snm1 (Rv3870), snm2 (Rv3871), and snm4 (Rv3877), that encode two ATPases and a 12-transmembrane-domain protein, respectively. In similar studies, a proline-rich predicted chromosome-partitioning ATPase (Rv3876 or snm3) was also shown to be required for ESAT-6 and CFP-10 secretion (10).
Evidence that the Snm proteins form a secretory apparatus to assist in ESAT-6 and CFP-10 secretion came from biochemical and yeast two-hybrid studies. Purified ESAT-6 and CFP-10 formed a stable 1:1 dimer, and yeast two-hybrid results have confirmed this strong interaction (17, 25, 31). Furthermore, physical interactions were detected between individual components of the Snm machinery (Snm1 and Snm2) and between the machinery and its secretion substrates (Snm2 and CFP-10) (31). These findings led to our working model that Snm2 recruits the ESAT-6-CFP-10 dimer to the cytoplasmic membrane via its interactions with CFP-10 and Snm1, which has three predicted transmembrane domains. Consistent with the role of ATPases in other protein secretion pathways (12, 38), concerted ATP hydrolysis by Snm1 and Snm2 would promote ESAT-6-CFP-10 translocation through the secretory pore consisting of Snm4 (see Fig. 5).
FIG. 5.
Model of the Snm secretion pathway in M. smegmatis. See Discussion for details of the model. CM, cytoplasmic membrane; PG, peptidoglycan layer; mAG, mycolyl arabinogalactan layer. Transmembrane domains were predicted by TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and topology is inferred based on predicted function and some experimental evidence (6).
Bioinformatic analysis of the M. tuberculosis genome has revealed several interesting features of the ESAT-6 and CFP-10 secretion pathway (4, 9, 19, 20, 33). First, the genes that encode ESAT-6 and CFP-10 have duplicated numerous times in the M. tuberculosis genome, resulting in 10 additional operons homologous to esxA/esxB. Interestingly, at 5 of the 11 loci, a conserved set of 10 genes, including snm1-4 homologues, are syntenic with the esx operon. The proximity of the six other genes within these esx loci suggests that they encode additional Snm proteins that function during ESAT-6 and CFP-10 secretion.
Interestingly, the Snm pathway and its esx substrates are not unique to mycobacterial human pathogens, such as M. tuberculosis and Mycobacterium leprae. Transposon mutagenesis in the ectotherm pathogen Mycobacterium marinum uncovered a homologous Snm pathway that was critical for host cell cytolysis and virulence (8). Furthermore, the conserved esx operon and several of the surrounding snm genes have been identified in other gram-positive bacteria, including Bacillus subtilis and Corynebacterium diphtheriae (19). However, only mycobacteria possess the full set of conserved genes from M. tuberculosis that surround the esx operons. Interestingly, Mycobacterium smegmatis, a soil-dwelling, distant relative of M. tuberculosis, contains clear homologues of 5 of the 11 esx loci in M. tuberculosis, including the esxA/esxB locus and all of the surrounding known or putative snm genes (9) (see Fig. 1).
FIG. 1.
M. smegmatis possesses a highly homologous esxA/esxB genomic locus and secretes SmESAT-6 and SmCFP-10 in a medium-dependent manner. (A) The M. smegmatis genomic sequence (http://www.tigr.org/tdb/ufmg/) was obtained from The Institute for Genomic Research, and the M. tuberculosis genomic sequence was obtained from Tuberculist (http://genolist.pasteur.fr/TubercuList/). The numbers below each open reading frame from M. smegmatis denote the encoded protein's percent identity (using the lalign program [http://www.ch.embnet.org/software/LALIGN_form.html]) to its homologue in M. tuberculosis, and the gene names in boldface type represent those included in this study. The asterisks indicate that while the snm3 homologue from M. smegmatis is 51% identical to its counterpart in M. tuberculosis, it lacks the 577-bp N-terminal portion found in the M. tuberculosis gene. (B) Wild-type or ΔesxB cells were grown to late log phase in 7H9 or Sauton's medium, fractionated into the cell-associated pellet (P) fraction and the short-term culture filtrate (S) fraction, and analyzed by Western blotting for the indicated proteins.
As M. smegmatis possesses a highly homologous esxA/esxB genomic locus and has many experimental advantages compared to M. tuberculosis (biochemically and genetically tractable, nonpathogenic, rapid growth), we sought to utilize this organism to understand the mechanism and role of the Snm pathway. In this study, we show that the functionally conserved M. smegmatis Snm pathway secretes its homologous ESAT-6 and CFP-10 substrates (which we designate SmESAT-6 and SmCFP-10) and is regulated by growth conditions. By creating in-frame deletions of the genes surrounding the esxA/esxB locus in M. smegmatis, we have also identified four new Snm proteins that are required for SmESAT-6 and SmCFP-10 secretion.
MATERIALS AND METHODS
Protein preparation and analysis.
M. smegmatis was inoculated from a single colony and grown to mid-log phase (optical density at 600 nm [OD600], 0.6 to 0.8) in 7H9 media supplemented with 0.05% Tween 80 (Fisher Scientific). Cells were washed and inoculated in 7H9 or Sauton's media supplemented with 0.05% Tween 80. Upon reaching mid-log phase (OD600, 0.6 to 0.8), cells were washed and inoculated in 7H9 or Sauton's media without Tween 80 and grown to an OD600 of 0.8. Cells were harvested by centrifugation, and the cultured medium supernatant was filtered through a 0.22-μm-pore-size filter and concentrated with an Amicon Ultra-15 (5,000-molecular-weight cutoff; Millipore). For complementation experiments, medium was supplemented with kanamycin (20 μg/ml; Fisher Scientific). Whole-cell lysates and short-term culture filtrate (corresponding to 5 ml of original culture volume) fractions were precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 4 to 20% Tris-HCl Criterion gels (Bio-Rad). For immunoblotting of M. smegmatis SmESAT-6, polyclonal antibodies were raised against the N-terminal 20 amino acids (aa) plus a C-terminal cysteine (Covance Research Products, Inc.), and the antibody was affinity purified with the Sulfolink kit (Pierce Biotechnology). The antibodies against CFP-10 (K8493) and GroEL (HAT5) were kind gifts from P. Anderson. The antibody against KatG was a kind gift from S. Cole.
Construction of M. smegmatis deletion strains and Southern analysis.
To generate deletion strains, the 5′ and 3′ 800-bp flanks surrounding each gene were amplified by PCR, sequence analyzed, and introduced into pjsc232, creating an AflII site at the junction between the flanks. This strategy created an in-frame gene deletion which encodes a truncated gene product (see Table 2 for precise truncated sizes of each deleted gene). These plasmids were introduced as described previously (11), and integrants were selected by growth on 7H10 agar containing kanamycin. Single colonies were picked and grown to late log phase in liquid 7H9 media without kanamycin, and recombinants were selected by growth on 7H10 agar with 5% sucrose. Deletion generation was confirmed by PCR and Southern analysis. Strains are listed in Table 1, and more details of Southern analysis are given in Table 2.
TABLE 2.
Details of Southern blot analysis of M. smegmatis mutant strains
| Strain | Enzyme(s) | Probe | Expected size of wild-type gene (bp) | Expected size of Δ gene (bp) | Wild-type gene product size (aa) | Δ gene product size (aa) |
|---|---|---|---|---|---|---|
| ΔSm3866 (snm5) | Aflll, Sphl | 847-bp 3′ flank | 4,076 | 2,374 | 294 | 31 |
| ΔSm3868 | Aflll, Sphl | 831-bp 5′ flank | 4,076 | 2,311 | 574 | 38 |
| ΔSm3869 (snm6) | Aflll, BstBl | 886-bp 5′ flank | 17,751 | 5,618 | 479 | 29 |
| Δsnm1 (Sm3870) | Aflll, Ncol | 871-bp 3′ flank | 3,822 | 1,546 | 742 | 32 |
| Δsnm2 (Sm3871) | Aflll, Sphl | 841-bp 3′ flank | 3,879 | 1,316 | 593 | 20 |
| ΔesxB (cfp-10) | Aflll, Sphl | 820-bp 3′ flank | 4,619 | 3,741 | 100 | 13 |
| ΔesxA (esat-6) | Aflll, Sphl | 842-bp 3′ flank | 4,619 | 3,419 | 95 | 9 |
| Δsnm4 (Sm3877) | Aflll, Sphl | 855-bp 5′ flank | 4,619 | 2,851 | 498 | 22 |
| ΔSm3882c (snm7) | Aflll, Ncol | 885-bp 3′ flank | 7,552 | 1,758 | 464 | 16 |
| ΔmycP1 (Sm3883c) | Pvul | 865-bp 5′ flank | 2,294 | 1,695 | 449 | 15 |
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) |
|---|---|
| Strains | |
| mc2 155 | Wild-type |
| SCM17 | ΔSm3866 (snm5) |
| SCM25 | ΔSm3868 |
| SCM29 | ΔSm3869 (snm6) |
| SCM30 | Δsnm1 (Sm3870) |
| SCM26 | Δsnm2 (Sm3871) |
| SCM27 | ΔesxB (cfp-10 Sm3874) |
| SCM31 | ΔesxA (esat-6 Sm3875) |
| SCM32 | Δsnm4 (Sm3877) |
| SCM28 | ΔSm3882c (snm7) |
| SCM21 | ΔmycP1 (snm8 Sm3883c) |
| SCM67 | Wild-type + pMJ31 |
| SCM72 | ΔSm3866 + pMJ31 |
| SCM41 | ΔSm3866 + pSEC43 (Sm3866-myc) |
| SCM73 | ΔSm3869 + pMJ31 |
| SCM45 | ΔSm3869 + pSEC47 (Sm3869-myc) |
| SCM79 | ΔSm3882c + pMJ31 |
| SCM120 | ΔSm3882c + pSEC80 (Sm3882c-2×HA) |
| SCM80 | ΔmycP1 + pMJ31 |
| SCM122 | ΔmycP1 + pSEC82 (mycP1-2×HA) |
| SCM76 | ΔesxB + pMJ31 |
| SCM141 | ΔesxB + pSEC108 (esxB) |
| SCM77 | ΔesxA + pMJ31 |
| SCM142 | ΔesxA + pSEC109 (esxA) |
| SCM143 | ΔesxA + pMH406 (M. tuberculosis esxA/B) |
| Plasmids | |
| pMJ13 | groEL2 promoter, C-terminal in-frame 2× HA tag, Kanr |
| pMJ31 | groEL2 promoter, C-terminal in-frame myc tag, Kanr |
| pSEC43 | groEL2 promoter, Sm3866-myc, Kanr |
| pSEC47 | groEL2 promoter, Sm3869-myc, Kanr |
| pSEC108 | groEL2 promoter, untagged esxB, Kanr |
| pSEC109 | groEL2 promoter, untagged esxA, Kanr |
| pSEC72 | mycP1 promoter, C-terminal in-frame 2× HA tag, Kanr |
| pSEC80 | mycP1 promoter, Sm3882c-2×HA, Kanr |
| pSEC82 | mycP1 promoter, mycP1-2×HA, Kanr |
| pMH406 | mop promoter, M. tuberculosis esxA/B, Kanr (see reference 10) |
Construction of snm and esx complementation plasmids.
Complementation vectors were derived from pMV261.kan, a high-copy episomal plasmid in which transcription is driven by the constitutive groEL2 promoter (32). C-terminal 2× hemagglutinin (HA) (pMJ13) or Myc (pMJ31) tags were created by PCR and ligated into pMV261.kan. Each of the snm and esx genes were amplified by PCR from wild-type M. smegmatis genomic DNA, sequenced, and introduced into pMJ13 or pMJ31, creating C-terminally tagged constructs.
To complement the Sm3882c and mycP1 mutant secretion defect, the groEL2 promoter in pMJ13 was replaced with the PCR-amplified 505-bp predicted promoter immediately upstream of the mycP1 gene. To generate esxA and esxB complementation constructs, a stop codon was introduced immediately after the open reading frame by using the QuikChange site-directed mutagenesis kit (Stratagene). The pMH406 M. tuberculosis esxA/B complementation plasmid was a kind gift from D. Sherman. Strains and plasmids are listed in Table 1.
RESULTS
Similarity of esxA/esxB loci in M. smegmatis and M. tuberculosis.
The M. smegmatis genomic sequence was obtained from The Institute for Genomic Research (http://www.tigr.org/tdb/ufmg/), and our preliminary annotation revealed that the esxA/esxB genomic locus is highly conserved between M. smegmatis and M. tuberculosis (Fig. 1A). The loci are organized in a similar fashion, and M. smegmatis possesses strong homologues for all of the known and putative snm genes (9). There are, however, some interesting differences between the two loci. For example, BLAST searches of a 4,459-bp region 5′ of the Sm3881 gene revealed three genes that encode putative transposases that are absent in M. tuberculosis. It is possible that these transposases and their associated transposons may have mediated the extensive duplication and/or horizontal gene transfer of the esx locus. Furthermore, the M. smegmatis snm3 homologue encodes a product lacking the proline-rich N terminus of its counterpart in M. tuberculosis. Despite these differences, however, the overall conservation of the esxA/esxB locus suggests that SmESAT-6 and SmCFP-10 may be secreted by M. smegmatis (9).
Analysis of SmESAT-6 and SmCFP-10 secretion under various growth conditions in M. smegmatis.
To test our hypothesis, we examined SmESAT-6 and SmCFP-10 secretion into the culture media during growth in standard 7H9 liquid media. Wild-type M. smegmatis was grown to log phase, cells were removed by centrifugation, and the supernatant was filtered and concentrated into the short-term culture filtrate (STCF) fraction. Proteins were extracted from the cell-associated and STCF fractions, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western blotting with polyclonal antibodies raised against M. smegmatis SmESAT-6 and M. tuberculosis CFP-10. As shown in Fig. 1B, when cultured in 7H9 media, SmESAT-6 and SmCFP-10 are present in the cell-associated whole-cell lysate fraction but absent from the STCF fraction (lanes 1 and 2). Both proteins have apparent molecular masses similar to their homologues in M. tuberculosis (molecular mass of ESAT-6, 8.4 kDa; molecular mass of CFP-10, 11.5 kDa) (15, 27). Thus, although SmESAT-6 and SmCFP-10 were clearly expressed by M. smegmatis cells grown in 7H9 media, they did not appear to be secreted.
For technical reasons, STCF fractions are typically prepared from M. tuberculosis cultures grown in Sauton's medium, and we reasoned that SmESAT-6 and SmCFP-10 secretion in M. smegmatis may be inhibited by the growth conditions present in 7H9. Therefore, to mimic more closely the in vitro culturing conditions used for M. tuberculosis (10, 30, 31), we also prepared STCF fractions from M. smegmatis cultured in Sauton's medium. Strikingly, both SmESAT-6 and SmCFP-10 are secreted into the STCF fraction when grown in this media (Fig. 1B, lanes 3 and 4). Cell autolysis did not contribute significantly to the protein profiles in the STCF fractions, as two intracellular proteins, KatG and GroEL, which accumulate extracellularly under prolonged growth conditions in 7H9 and Sauton's media (24, 37; data not shown), were exclusively cell associated at the time that all STCF fractions were harvested (Fig. 1B, lower panels). To show that our antibodies specifically recognized SmESAT-6 and SmCFP-10, we analyzed extracts prepared from a ΔesxB mutant (see below), as deletion of esxB in M. tuberculosis abolishes both SmESAT-6 and SmCFP-10 expression (31). As expected, the bands corresponding to SmESAT-6 and SmCFP-10 were missing in extracts from the M. smegmatis ΔesxB mutant (Fig. 1B, lanes 5 to 8). STCF protein profiles were qualitatively similar between 7H9 and Sauton's media (data not shown), suggesting that this medium dependence is specific for SmESAT-6 and SmCFP-10 export. Based on these findings, we conclude that SmESAT-6 and SmCFP-10 secretion into the STCF fraction, but not expression, is dependent on the growth media; therefore, we cultured M. smegmatis in Sauton's media for the remainder of these studies.
Construction of snm and esx in-frame deletion strains.
To test whether the Snm secretion pathway functions similarly in both M. smegmatis and M. tuberculosis, we sought to make M. smegmatis strains that lacked each of the homologues of the snm1, snm2, snm4, esxA, and esxB genes. In-frame deletion constructs were created by PCR and inserted into a vector that contained the sacB counterselectable marker. In the first of a two-step homologous recombination procedure, wild-type M. smegmatis was transformed with these constructs to yield merodiploid strains that contained the sacB gene flanked by both the wild-type and mutated copy of the targeted gene in the genome (Fig. 2A; Table 1) (21). These strains were then grown in liquid culture and subsequently plated on solid media containing sucrose to select for cells in which the sacB gene had been deleted. Individual colonies were isolated and tested for proper deletion of the targeted gene by PCR. In all cases, the distribution of recombination events that led either to retention of the wild-type or the deletion allele was nearly 50:50, demonstrating that none of the genes are essential for viability (data not shown). Furthermore, these mutations had no obvious effect on bacterial growth, as the growth rate of each mutant strain was identical to that of the wild-type strain (data not shown). Accurate deletion of each gene was confirmed by Southern blot analysis, which revealed the expected patterns for the deletion strains (Fig. 2B; Table 2), and sequence analysis of the integrated DNA confirmed that the deletions were in frame and that no additional mutations arose during the construction.
FIG. 2.
In-frame deletion strategy and Southern blot analysis of mutants. (A) The in-frame deletion strategy involved PCR amplification of 800-bp 5′ and 3′ flanks to the wild-type (wt) gene of interest. The oligonucleotides were appropriately engineered to introduce a unique AflII restriction digest site to ensure that the ligated 5′ and 3′ flanks would express an in-frame truncated gene product. Recombination between the genome and either 800-bp flank produced a kanamycin-resistant merodiploid strain, and counterselection on sucrose for recombinants selected for either the desired in-frame deletion or the restored wild-type gene product. (B) Southern blot analysis of each deletion (Δ) strain revealed the expected bands. Genomic DNA was digested with AflII and one additional enzyme to confirm that the suicide plasmid had recombined in the proper location and that the AflII site and, thus, the correct reading frame were intact. For the mycP1 Southern blot analysis, genomic DNA was digested with PvuI alone. More detail is given in Table 2.
Conservation of Snm machinery between M. tuberculosis and M. smegmatis.
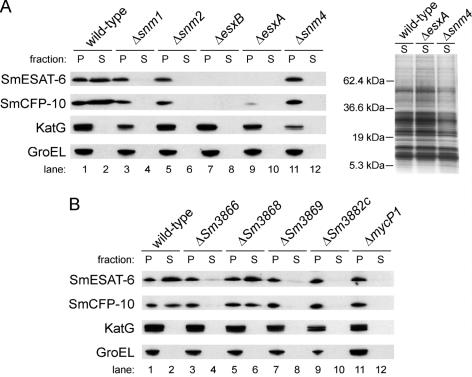
As predicted, Western blot analysis of proteins extracted from the snm1, snm2, and snm4 deletion strains showed that SmESAT-6 and SmCFP-10 secretion, but not synthesis, is blocked in these mutants (Fig. 3A, lanes 1 to 6, 11, and 12). Furthermore, SmESAT-6 and SmCFP-10 are absent in their respective deletion strains (Fig. 3A, lanes 7 to 10). In M. tuberculosis, the loss of either esxA or esxB alone abolishes the expression of both ESAT-6 and CFP-10, though polar effects cannot be excluded (31). In-frame deletions of the esxA and esxB homologues in M. smegmatis show a similar relationship: esxB is required for the expression of SmESAT-6 and esxA is required for full expression of SmCFP-10. While low levels of SmCFP-10 are apparent in the lysate fraction of the esxA mutant (Fig. 3A, lane 9), the expression of both homologues is required for substantial expression of both SmESAT-6 and SmCFP-10. Silver staining of the STCF fractions from the mutant strains revealed indistinguishable secretion profiles compared to the wild-type strain, which suggests that these mutations do not cause gross secretion defects (Fig. 3A, right panel). In sum, these results clearly demonstrate that a functional Snm secretion apparatus exists in M. smegmatis and requires the components homologous to those required in M. tuberculosis.
FIG. 3.
M. smegmatis secretes SmESAT-6 and SmCFP-10 in an Snm-dependent manner, and four additional uncharacterized genes are also required for SmESAT-6 and SmCFP-10 secretion. (A and B) Wild-type and various mutant cells were grown to late log phase in Sauton's medium, fractionated into the cell-associated pellet (P) fraction and the short-term culture filtrate (S) fraction, and analyzed by Western blotting for the indicated proteins or by silver staining (right panel of panel A).
Identification of novel snm mutants.
Since M. smegmatis secretes SmESAT-6 and SmCFP-10 in an Snm-dependent fashion, we sought to utilize this powerful model system to discover new components of the Snm pathway. To this end, we generated in-frame deletions of the five putative snm genes from the esxA/esxB locus by using the same two-step strategy described above (Fig. 2). Strikingly, Western blot analysis of protein extracts harvested from these deletion strains revealed that four of the five genes, Sm3866, Sm3869, Sm3882c, and mycP1 (Sm3883c) are required for SmESAT-6 and SmCFP-10 secretion (Fig. 3B). Interestingly, Sm3883c encodes a previously identified membrane-anchored serine protease termed mycosin 1 (2, 6). Sm3866 shares limited homology with ABC ATPases involved in type I secretion (35), and Sm3869 encodes a membrane protein with predicted ATP/GTP binding sites. Finally, Sm3882c is likely in an operon with mycP1 and encodes an uncharacterized membrane protein. We propose that the previously uncharacterized snm genes be named snm5 (Sm3866), snm6 (Sm3869), and snm7 (Sm3882c). Interestingly, a predicted AAA ATPase, Sm3868, is not required for SmESAT-6 and SmCFP-10 secretion despite its high sequence conservation (76% identity) between M. smegmatis and M. tuberculosis. In summary, these results identify four additional genes that are required for the Snm pathway, and they suggest that a large, multicomponent secretion apparatus functions during SmESAT-6 and SmCFP-10 secretion in M. smegmatis.
Complementation of novel snm and esx mutants.
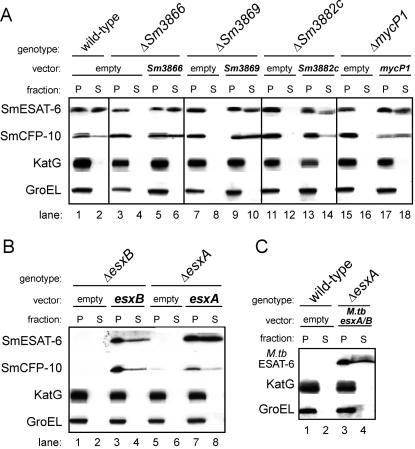
To confirm that our newly discovered snm genes are indeed functioning during SmESAT-6 and SmCFP-10 secretion, we sought to complement their secretion defects. Constitutive expression with the M. tuberculosis groEL2 promoter of tagged Sm3866 and Sm3869 in the appropriate deletion strain fully restored SmESAT-6 and SmCFP-10 secretion compared to the deletion strains containing the empty vector (Fig. 4A, lanes 1 to 10). Expression of Sm3882c and mycosin 1 from this promoter caused cell toxicity (data not shown). Therefore, as both genes are likely in an operon (Fig. 1), we engineered a new complementation plasmid in which the constitutive groEL2 promoter was replaced with the 505-bp predicted mycP1 promoter region. As shown in Fig. 4B, lanes 11 to 18, these complemented Sm3882c and mycP1 mutants secrete SmESAT-6 and SmCFP-10 to approximately wild-type levels. Furthermore, cell autolysis induced by exogenous gene expression is not responsible for the complemented phenotype, as KatG and GroEL are both exclusively cell associated in all strains (Fig. 4, lower panels). These results prove that Sm3866, Sm3869, Sm3882c, and mycosin 1 function during SmESAT-6 and SmCFP-10 secretion.
FIG. 4.
Complementation of mutant strains with individual genes confirms that the Snm secretion pathway functions in M. smegmatis. (A and B) Various deletion strains were transformed with either an empty vector (pMJ13) or a multicopy episomal complementation plasmid constitutively expressing each individual gene C-terminally tagged with myc. In the case of Sm3882c and mycP1 complementation, the complementation plasmid expressed the individual genes tagged with 2× HA from the presumed endogenous promoter (505-bp fragment upstream of mycP1). (C) To test for M. tuberculosis (M.tb) ESAT-6 and CFP-10 secretion from M smegmatis, cells were transformed with pMH406, which complements the M. tuberculosis esxA/B deletion (10; data not shown). All cells were cultured in Sauton's medium with kanamycin, fractionated, and analyzed by Western blotting.
To rule out the possibility of polar effects in the ΔesxA and ΔesxB mutant strains, we used an episomal plasmid containing the appropriate untagged gene driven by the groEL2 promoter for complementation analysis. Expression of SmCFP-10 and SmESAT-6 in their respective deletion strains rescued the synthesis and secretion of both SmCFP-10 and SmESAT-6 compared to vector-transformed knockout strains (Fig. 4B, lanes 1 to 8). Interestingly, C-terminally tagged versions of SmESAT-6 and SmCFP-10 were inefficiently secreted, although the reason for this is unclear (data not shown).
We next investigated whether the M. tuberculosis ESAT-6 and CFP-10 homologues could be secreted by the M. smegmatis system (Fig. 4C). Importantly, the M. tuberculosis ESAT-6 monoclonal antibody, which readily detects secreted M. tuberculosis ESAT-6 (31), does not recognize endogenous M. smegmatis SmESAT-6 (lanes 1 and 2). Introduction of an episomal plasmid, in which expression of M. tuberculosis esxA and esxB is driven by the synthetic mop promoter (10), into ΔesxA cells resulted in synthesis and secretion of the M. tuberculosis homologues (lanes 3 and 4). This result demonstrates that substrate recognition is similar between the two species. Moreover, our results demonstrate a high degree of functional conservation of the Snm secretory apparatus between M. smegmatis and M. tuberculosis.
DISCUSSION
In this study, we have presented the first functional characterization of the Snm alternative protein secretion pathway in a nonpathogenic bacterium. M. smegmatis possesses an esx/snm locus homologous to that of M. tuberculosis and secretes SmESAT-6 and SmCFP-10 into the extracellular milieu. Our genetic analysis of this locus revealed that SmESAT-6 and SmCFP-10 secretion strictly requires homologues of snm1, snm2, and snm4, which are required for secretion in M. tuberculosis. Furthermore, the M. smegmatis Snm machinery secretes the M. tuberculosis ESAT-6 and CFP-10 homologues. Therefore, the Snm system appears to function analogously in both organisms, providing an opportunity to study this pathway in a more tractable organism.
Indeed, by studying the Snm pathway in M. smegmatis, we have identified four additional snm genes and contributed to our working model of the Snm alternative protein secretion pathway (Fig. 5). Of particular interest is our finding that the serine protease mycosin 1 is required for Snm secretion. Given its predicted periplasmic location, it may act like the Sec pathway signal peptidase which removes N-terminal signal sequences upon secretion (18). However, as SmESAT-6 and SmCFP-10 show no obvious size change upon export, we hypothesize that mycosin 1 may act as a regulator of Snm secretion. For example, its proteolytic activity may either degrade an Snm inhibitor or activate an Snm protein, perhaps in the cell wall (2, 6). Given the functional conservation between the Snm pathway in M. smegmatis and M. tuberculosis, it is likely that mycP1 and snm5-7 will also be required for ESAT-6 and CFP-10 secretion in M. tuberculosis. Of course, verification will require mutation of these individual genes in M. tuberculosis. Importantly, we were able to complement all of the newly found snm mutants with individual genes, confirming that the proteins function during SmESAT-6 and SmCFP-10 secretion in M. smegmatis.
An unexpected finding of these experiments was that SmESAT-6 and SmCFP-10 are secreted during growth in Sauton's medium but not in 7H9. Because there are numerous differences between Sauton's and 7H9 medium, it seems likely that the Snm system is responsive to one or more components of the growth medium. Our efforts to determine the precise component responsible for this regulation have been unsuccessful. For example, while Sauton's medium, compared to 7H9, has no dextrose, calcium, or zinc, supplementing Sauton's medium with these components did not suppress SmESAT-6 and SmCFP-10 secretion (data not shown). Our efforts to understand this potential regulation are ongoing; however, this is the first suggestion that the Snm pathway may be regulated and reveals a potential similarity with Sec-independent secretion pathways from gram-negative bacteria, which also require strict medium conditions for active protein export (5, 26).
The unique mycobacterial cell wall represents an enormous barrier to protein secretion, and how ESAT-6 and CFP-10 are translocated across all of the layers of the thick mycobacterial cell wall remains unknown. In addition to the peptidoglycan layer, mycobacteria possess hydrophobic long-chain mycolic acids esterified to a polymeric arabinogalactan layer (Fig. 5). There are at least two models for how ESAT-6 and CFP-10 are exported across the cell wall. First, the Snm system may translocate these proteins only across the cytoplasmic membrane, leaving them to diffuse across the cell wall, perhaps through porins or passive processes (16, 13, 22). Alternatively the Snm system may function analogously to type III secretion and provide a continuous conduit through which ESAT-6 and CFP-10 are translocated from the bacterial cytosol directly across all cell wall layers.
Finally, why may the Snm system be conserved in M. smegmatis yet absolutely critical for M. tuberculosis virulence? We envision several possibilities. First, the pathway may play an identical and fundamental role in both organisms, perhaps in cell-to-cell communication. In support of this, Flint et al. (7) recently reported that M. smegmatis snm mutants display increased conjugation efficiency and present evidence that Snm secretion suppresses conjugation in trans. Although conjugal transfer has not been detected in M. tuberculosis, it is plausible that both organisms may utilize ESAT-6 and CFP-10 as density-dependent signals to coordinate cell activities. Finally, in accordance with their different environmental niches, each species may secrete additional substrates besides ESAT-6 and CFP-10. Thus, the Snm pathway, like other Sec-independent pathways, may be modular, allowing substrates to evolve for the particular needs of each organism. Thus, studying the Snm pathway of M. smegmatis will not only help delineate the mechanism of Snm secretion but perhaps uncover the evolutionarily conserved role of the Snm pathway.
Acknowledgments
We thank M. Jain for providing complementation plasmids and all members of the Cox lab for helpful discussions and critical reading of the manuscript.
S.E.C. is supported by a National Science Foundation Graduate Research Fellowship and the Achievement Rewards for College Scientists Scholarship Program. J.S.C. gratefully acknowledges the support of the Pew Scholars Program in the Biomedical Sciences and the Sandler Family Supporting Foundation. This work was supported by National Institutes of Health grant AI68540.
REFERENCES
- 1.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144:3195-3203. [DOI] [PubMed] [Google Scholar]
- 2.Brown, G. D., J. A. Dave, N. C. Gey van Pittius, L. Stevens, M. R. Ehlers, and A. D. Beyers. 2000. The mycosins of Mycobacterium tuberculosis H37Rv: a family of subtilisin-like serine proteases. Gene 254:147-155. [DOI] [PubMed] [Google Scholar]
- 3.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta, N., G. L. Lykken, M. C. Wolfgang, and T. L. Yahr. 2004. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 53:297-308. [DOI] [PubMed] [Google Scholar]
- 6.Dave, J. A., N. C. Gey van Pittius, A. D. Beyers, M. R. Ehlers, and G. D. Brown. 2002. Mycosin-1, a subtilisin-like serine protease of Mycobacterium tuberculosis, is cell wall-associated and expressed during infection of macrophages. BMC Microbiol. 2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint, J. L., J. C. Kowalski, P. K. Karnati, and K. M. Derbyshire. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 101:12598-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, L. Y., S. Guo, B. McLaughlin, H. Morisaki, J. N. Engel, and E. J. Brown. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677-1693. [DOI] [PubMed] [Google Scholar]
- 9.Gey Van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C gram-positive bacteria. Genome Biol. 2:0044.1-0044.18. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guinn, K. M., M. J. Hickey, S. K. Mathur, K. L. Zakel, J. E. Grotzke, D. M. Lewinsohn, S. Smith, and D. R. Sherman. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinds, J., E. Mahenthiralingam, K. E. Kempsell, K. Duncan, R. W. Stokes, T. Parish, and N. G. Stoker. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145:519-527. [DOI] [PubMed] [Google Scholar]
- 12.Holroyd, C., and R. Erdmann. 2001. Protein translocation machineries of peroxisomes. FEBS Lett. 501:6-10. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 100:12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15:1725-1752. [DOI] [PubMed] [Google Scholar]
- 15.Mattow, J., P. R. Jungblut, E. C. Muller, and S. H. Kaufmann. 2001. Identification of acidic, low molecular mass proteins of Mycobacterium tuberculosis strain H37Rv by matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. Proteomics 1:494-507. [DOI] [PubMed] [Google Scholar]
- 16.Niederweis, M. 2003. Mycobacterial porins-new channel proteins in unique outer membranes. Mol. Microbiol. 49:1167-1177. [DOI] [PubMed] [Google Scholar]
- 17.Okkels, L. M., and P. Andersen. 2004. Protein-protein interactions of proteins from the ESAT-6 family of Mycobacterium tuberculosis. J. Bacteriol. 186:2487-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 19.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily and a new gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 20.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 21.Pavelka, M. S., Jr., and W. R. Jacobs, Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 23.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9:533-539. [DOI] [PubMed] [Google Scholar]
- 24.Recht, J., A. Martinez, S. Torello, and R. Kolter. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182:4348-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renshaw, P. S., P. Panagiotidou, A. Whelan, S. V. Gordon, G. R. Hewinson, R. A. Williamson, and M. D. Carr. 2002. Conclusive evidence that the major T-cell antigens of the M. tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterisation of the structural properties of ESAT-6, CFP-10 and the ESAT-6-CFP-10 complex: implications for pathogenesis and virulence. J. Biol. Chem. 277:21598-21603. [DOI] [PubMed] [Google Scholar]
- 26.Rietsch, A., M. C. Wolfgang, and J. J. Mekalanos. 2004. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect. Immun. 72:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenkrands, I., K. Weldingh, S. Jacobsen, C. V. Hansen, W. Florio, I. Gianetri, and P. Andersen. 2000. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis 21:935-948. [DOI] [PubMed] [Google Scholar]
- 28.Skjot, R. L., T. Oettinger, I. Rosenkrands, P. Ravn, I. Brock, S. Jacobsen, and P. Andersen. 2000. Comparative evaluation of low-molecular-mass proteins from Mycobacterium tuberculosis identifies members of the ESAT-6 family as immunodominant T-cell antigens. Infect. Immun. 68:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnenberg, M. G., and J. T. Belisle. 1997. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect. Immun. 65:4515-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis requires a novel specialized secretion system. Proc. Natl. Acad. Sci. USA 100:13001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 33.Tekaia, F., S. V. Gordon, T. Garnier, R. Brosch, B. G. Barrell, and S. T. Cole. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329-342. [DOI] [PubMed] [Google Scholar]
- 34.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, S. A., O. L. Shedd, K. C. Ray, M. H. Beins, J. P. Jorgensen, and M. J. Blaser. 1998. Campylobacter fetus surface layer proteins are transported by a type I secretion system. J. Bacteriol. 180:6450-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tullius, M. V., G. Harth, and M. A. Horwitz. 2001. High extracellular levels of Mycobacterium tuberculosis glutamine synthetase and superoxide dismutase in actively growing cultures are due to high expression and extracellular stability rather than to a protein-specific export mechanism. Infect. Immun. 69:6348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye, Y., H. H. Meyer, and T. A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652-656. [DOI] [PubMed] [Google Scholar]