Abstract
Seven new genes designated dsrLJOPNSR were identified immediately downstream of dsrABEFHCMK, completing the dsr gene cluster of the phototrophic sulfur bacterium Allochromatium vinosum D (DSM 180T). Interposon mutagenesis proved an essential role of the encoded proteins for the oxidation of intracellular sulfur, an obligate intermediate during the oxidation of sulfide and thiosulfate. While dsrR and dsrS encode cytoplasmic proteins of unknown function, the other genes encode a predicted NADPH:acceptor oxidoreductase (DsrL), a triheme c-type cytochrome (DsrJ), a periplasmic iron-sulfur protein (DsrO), and an integral membrane protein (DsrP). DsrN resembles cobyrinic acid a,c-diamide synthases and is probably involved in the biosynthesis of siro(heme)amide, the prosthetic group of the dsrAB-encoded sulfite reductase. The presence of most predicted Dsr proteins in A. vinosum was verified by Western blot analysis. With the exception of the constitutively present DsrC, the formation of Dsr gene products was greatly enhanced by sulfide. DsrEFH were purified from the soluble fraction and constitute a soluble α2β2γ2-structured 75-kDa holoprotein. DsrKJO were purified from membranes pointing at the presence of a transmembrane electron-transporting complex consisting of DsrKMJOP. In accordance with the suggestion that related complexes from dissimilatory sulfate reducers transfer electrons to sulfite reductase, the A. vinosum Dsr complex is copurified with sulfite reductase, DsrEFH, and DsrC. We therefore now have an ideal and unique possibility to study the interaction of sulfite reductase with other proteins and to clarify the long-standing problem of electron transport from and to sulfite reductase, not only in phototrophic bacteria but also in sulfate-reducing prokaryotes.
Phototrophic purple and green sulfur-oxidizing bacteria use sulfur compounds as electron donors for reductive carbon dioxide fixation during photolithotrophic growth (7, 10). In these organisms, light energy is used to transfer electrons from sulfur compounds to the level of the more highly reducing electron carriers NAD(P)+ and ferredoxin. In our laboratory we seek to understand oxidative sulfur metabolism in anoxygenic phototrophic bacteria by using the genetically accessible γ-proteobacterium Allochromatium vinosum (formerly Chromatium vinosum [34]) as our model organism. It is a purple sulfur bacterium belonging to the family Chromatiaceae. A. vinosum carries out the complete eight-electron oxidations of sulfide and thiosulfate to sulfate. Intracellularly stored sulfur globules are an obligate intermediate in the process (57). The sulfur in the globules is present in the molecular structure of sulfur chains (58) and is enclosed by a protein envelope, a feature shared by most if not all of the chemotrophic sulfur-oxidizing bacteria that form intracellular sulfur globules (9, 14, 51). Topologically, the sulfur globules of A. vinosum and probably of other members of the Chromatiaceae are located extracytoplasmically, in the periplasm (51).
Of the various steps of oxidative sulfur metabolism, the oxidation of stored or externally added sulfur is arguably the most poorly understood at the molecular level (10, 38). The only gene region known so far to be essential for oxidation of stored sulfur was localized by interposon mutagenesis in A. vinosum (57). Eight open reading frames, designated dsrABEFHCMK, were identified. The dsrAB products form the α2β2-structured sulfite reductase. This protein is closely related to the dissimilatory sulfite reductases from sulfate-reducing bacteria and archaea (31), enzymes which are generally located in the cytoplasm. The adjacent dsrEFHC genes encode small soluble cytoplasmic proteins with hitherto unknown functions. The dsrM-encoded protein is predicted to be a membrane-bound b-type cytochrome, and DsrK exhibits relevant similarity to heterodisulfide reductases from methanogenic archaea. DsrK is predicted to reside in the cytoplasm. Polar insertion mutations immediately upstream of dsrA, in dsrB, dsrH, and dsrM, led to an inability of the cells to oxidize intracellularly stored sulfur whereas the ability of the mutants to oxidize sulfide, thiosulfate, and sulfite under photolithoautotrophic conditions and the ability to undergo photoorganoheterotrophic growth were unaffected.
Although our initial work on the A. vinosum dsr genes allowed the first insights into the oxidation of stored sulfur (57), many points remained enigmatic; these include the way in which the known cytoplasmic or membrane-bound dsr gene products are involved in the degradation of sulfur residing extracytoplasmatically in the periplasm. Taking the described results and remaining questions as our starting point, we have now sequenced the complete dsr gene cluster comprising 15 genes. Biochemical and genetic evidence is presented that confirms a function of the dsr gene products in the oxidation of intracellular sulfur. We purified DsrEFH from A. vinosum and showed that soluble and membrane-bound Dsr proteins form a supercomplex in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. A. vinosum was grown and harvested as described previously (13, 52). Escherichia coli strains were cultured in Luria-Bertani medium (63). Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were included in solid media to identify recombinant plasmids containing inserts in the α portion of lacZ. Antibiotics were used at the following concentrations (in micrograms per milliliter): for E. coli, ampicillin, 100; kanamycin, 50; tetracycline, 5, chloramphenicol, 100; for A. vinosum, kanamycin, 10; streptomycin, 50; ampicillin, 10 (solid media) or 100 (liquid media).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) supE44 λ−thi-1 gyrA relA1 | 26 |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm met (DE3) | Novagen |
| SM10 | KmrsupE44 thi-1 thr-1 recA leuB6 lacY1 tonA21 RP4-2-Tc::Mu-Km::Tn7 in chromosome | 68 |
| HM125 | F− ΔlacX74 galE galK thi rpsL(strA) ΔphoA degP::ΩKanreda51::Tn10(Tetr) rpoH15 | 3 |
| A. vinosum strains | ||
| SM50 | Smr, spontaneous streptomycin-resistant mutant of A. vinosum DSMZ 180T | 13 |
| 21D | Smr, Kmr, dsrB::KmΩ | 57 |
| 39D | Smr, Kmr, dsrL::KmΩ | This work |
| Plasmids | ||
| pBluescript SK II(+) | AprlacZ′, f1 ori | Stratagene |
| pGEM5 Zf(+) | AprlacZ′, f1 ori | Promega |
| pGEM7 Zf(+) | AprlacZ′, f1 ori | Promega |
| pET15b | Apr, His tag (N-terminal) | Novagen |
| pET22b | Apr, His tag (N-terminal) | Novagen |
| pISC-2 | Apr, araC, paraBAD | 3 |
| pISC-2(dsrJ) | Apr, dsrJ in pISC2 | This work |
| pEC86 | Cmr, product of pEC66 and pACYC184 with the ccmABCDEFGH genes | 3 |
| pSUP301 | Apr Kmr; RP4 oriT, p15A ori | 68 |
| pDZ3 | Apr, 3.9-kb EcoRI-XhoI fragment (dsrK to dsrJ) between EcoRI and XhoI of pBluescript SK II(+) | This work |
| pAP40 | Apr, 6.8-kb NcoI fragment (dsrK to dsrN) in NcoI of pGEM5 Zf(+) | This work |
| pAP60 | Apr, 5.85-kb BglII-SphI fragment (dsrJ to ruvB) between BamHI and SphI of pGEM7 Zf(+) | This work |
| pAP38 | Apr, 2.4-kb SalI fragment from pDZ3 in XhoI of pSUP301 | This work |
| pAP39 | Kmr, Kmr cartridge (BamHI) from pHP45Ω in BglII of pAP38 | This work |
| pHP45Ω | Kmr Apr | 22 |
Recombinant-DNA techniques.
Standard methods were used for molecular biological techniques (4, 63). Chromosomal DNA of A. vinosum strains was obtained, and Southern hybridizations were performed overnight at 68°C as described previously (57). PCR amplifications with Taq DNA polymerase were done essentially as described previously (13). Taq polymerase was replaced by Pfu polymerase (used in accordance with the protocol written by Stratagene) when the amplified sequence was designated for the construction of expression plasmids. DNA probes for Southern and Northern hybridization experiments and screening of libraries were digoxigenin labeled by PCR (66). Conjugative plasmid transfer and gene replacement were performed as described previously (13). The genotypes of the A. vinosum recombinants used in this study were confirmed by PCR and Southern hybridization. Double-stranded DNA was sequenced by Sequiserve, Vaterstetten, Germany. Nucleotide sequences were compiled and analyzed with DNASIS (Pharmacia) or Clone Manager (SEC central) software. Similarity searches were conducted using the BLAST algorithm (1). Protein sequences were analyzed using resources available at the ExPASy molecular biology server (http://www.expasy.ch) such as TMpred (32). Signal peptides and protein localization were predicted by the PSORT program package (50). For sequence comparisons, multiple sequence alignments were generated using the ClustalW server (http://www.ebi.ac.uk/clustalw).
Cloning of the complete dsr locus.
The region downstream of dsrK was cloned by using a PCR-amplified fragment from nucleotides 6878 to 7505 of the previously published sequence (57) as a probe to identify several positive clones with a 3.9-kb EcoRI-XhoI insert in a library of 3.5-to 4.5-kb EcoRI-XhoI fragments of chromosomal A. vinosum DNA in the pBluescript vector. Sequencing of the positive clone pDZ3 revealed the presence of dsrK, one complete open reading frame, and one open reading frame that was truncated at the 3′ end. To analyze the missing part of this open reading frame, a clone (pAP40) with a 6.8-kb NcoI fragment was isolated from a library of 6.4- to 7.2-kb NcoI fragments in the pGEM 5 Zf(+)vector. A PCR-amplified fragment covering nucleotides 7546 to 9152 of the complete sequence was used as a probe in this step. Finally, a probe from nucleotides 12065 to 12583 was amplified by PCR to identify plasmid pAP60, which is pGEM7 Zf(+) carrying a 5.85-kb BglII-SphI insert in the vector's BamHI and SphI sites.
Construction of expression plasmids.
The pET series of plasmids was used for the expression of genes in E. coli to (i) confirm the open reading frames suggested by the nucleotide sequence, (ii) produce the proteins encoded by the individual genes of the operon as positive controls for antisera directed against synthetic peptides, and (iii) in the case of DsrC to overproduce the protein for antibody production. Suited restriction sites (NdeI sites) were introduced at the 5′ and 3′ ends of the genes by PCR amplification with modified primers.
Purification of recombinant DsrC for antibody production.
DsrC (in pET15b) was overproduced with an amino-terminal His tag in E. coli BL21 (DE3). A 250-ml flask with 100 ml of Luria-Bertani medium containing 100 μg of ampicillin per ml was inoculated with a 5-ml overnight culture of E. coli BL21(DE3) containing plasmid pET-C (washed twice before inoculation). The cells were cultured at 37°C and 180 rpm. At an optical density at 600 nm of 0.6, formation of recombinant DsrC was induced by addition of 1 mM IPTG. Incubation at 37°C was continued for 2 h. The cells were then harvested by centrifugation at 4°C, and the pellet was resuspended in 5 ml of buffer A (5 mM imidazole, 0.5 M NaCl, 20 mM Tris HCl [pH 7.9]). The cells were disrupted by sonication (1.5 min ml−1) (Cell Disruptor B15; Branson) followed by centrifugation (25,000 × g for 30 min at 4°C). The supernatant was chromatographed on a His-Bind Resin column (Novagen, Madison, Wis.) (3.5 ml; flow rate, 0.5 ml/min) as specified by the manufacturer. The column was washed with buffer A containing 60 mM imidazole followed by a linear gradient of 60 to 400 mM imidazole. DsrC eluted at 230 to 380 mM imidazole. The combined fractions were immediately dialyzed against 20 mM Tris-HCl (pH 8.0) containing 500 mM NaCl followed by concentration to a final volume of not more than 2 ml via Centriprep-10 (Amicon). Final purification was achieved by gel filtration chromatography on Superdex 75 (Amersham Pharmacia Biotech, Uppsala, Sweden) equilibrated with 50 mM Tris-HCl (pH 8.0) containing 500 mM NaCl.
Purification of membrane-associated Dsr proteins from A. vinosum.
Purification of membrane-bound Dsr proteins was monitored by tracking DsrK with the aid of a DsrK-specific antiserum. Thawed cells were resuspended in 20 mM Tris-HCl buffer (pH 7.5) containing 20 mM MgCl2 and some grains of DNase at a ratio of 3 ml of buffer/g (wet weight), disrupted by ultrasonic treatment (1 min ml−1) (Cell Disruptor B15), and centrifuged at 25,000 × g and 4°C for 30 min. The supernatant was subjected to ultracentrifugation (145,000 × g at 4°C for 2 h). The membrane pellet was resuspended in 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0) containing 0.5% thesid (protein concentration, 4.5 mg/ml), stirred for 2 h at 4°C to solubilize membrane proteins, and ultracentrifuged for 45 min. Sodium chloride was slowly added to the supernatant to a final concentration of 280 mM. The solution was then applied to a Q-Sepharose Fast Flow column (Amersham Pharmacia Biotech) (2.6 by 5 cm, flow rate, 4 ml min−1) equilibrated with 50 mM MOPS-KOH (pH 7.0) containing 0.05% thesid. The column was washed with buffer containing 300 mM NaCl, and the protein was eluted with a continuous gradient (260 ml; flow rate, 2 ml min−1) from 300 to 500 mM NaCl. The majority of DsrK eluted at 420 to 470 mM NaCl. These fractions were dialyzed against 5 mM potassium phosphate buffer (pH 6.9) containing 0.05% thesid and applied to a column of ceramic hydroxyapatite (type II, 20 μm [Bio-Rad], 1.6 by 5 cm; flow rate, 2 ml min−1). Protein was eluted in a stepwise gradient of potassium phosphate (pH 6.9) (10, 50, 100, and 400 mM). All solutions contained 0.05% thesid. The majority of DsrK eluted at 100 mM. These fractions were combined, concentrated to 1 ml by ultrafiltration centrifugation (Centricon 50; Amicon) and stored at −20°C. Portions (0.2 ml) of concentrated protein were loaded on a Superose 6 gel filtration column (Amersham Pharmacia Biotech) equilibrated with 50 mM potassium phosphate buffer (pH 6.9) containing 0.05% thesid, and eluted using a flow rate of 0.2 ml min−1. DsrK eluted in a broad peak starting in the void volume of the column, indicating the presence of a large protein complex.
Purification of DsrEFH from A. vinosum.
DsrE, DsrF, and DsrH were detected by specific antisera. Thawed cells were resuspended in 50 mM potassium phosphate buffer (pH 7.5) containing some grains of DNase at a ratio of 3 ml of buffer/g (wet weight), disrupted by ultrasonic treatment, centrifuged, and ultracentrifuged as described above. The supernatant was brought to 40% saturated (NH4)2SO4. After incubation on ice for 10 to 16 h, precipitated protein was separated by centrifugation (30 min at 25,000 × g and 4°C). The supernatant was subjected to hydrophobic interaction chromatography on a low-substitution phenyl-Sepharose matrix (Amersham Pharmacia Biotech) (2.6 by 15 cm) equilibrated with 40% (NH4)2SO4 saturated in 50 mM potassium phosphate buffer (pH 7.5). Bound protein was eluted by decreasing the (NH4)2SO4 concentration in a linear gradient of 350 ml (2 ml min−1). DsrEFH eluted at 20 to 15% (NH4)2SO4. The respective fractions were combined, dialyzed against 10 mM Tris-HCl (pH 7.5), and loaded onto MonoQ HR 5/5 equilibrated with the same buffer. The column was washed with 10 mM Tris-HCl containing 300 mM NaCl, and the proteins were eluted with a linear gradient from 300 to 700 mM NaCl. Fractions with DsrEFH were combined, concentrated to 1.5 ml by ultrafiltration centrifugation (Centriplus YM10: Millipore, Bedford, Mass.), and further purified by gel filtration on Superdex TM200 equilibrated with 10 mM Tris-HCl (pH 7.5) containing 150 mM NaCl. The molecular mass of the protein was estimated by calibration of the column with standard proteins (Sigma Marker low-range; molecular mass, 6,500 to 66,000 Da). The DsrEFH proteins eluted after 76 to 80 ml. The estimated molecular mass of the purified EFH complex was 75 kDa.
Protein techniques.
Immunoblot (Western) analysis (72) was performed by the “semidry” procedure, using the Transblot SD semi-dry transfer apparatus (BioRad, Munich, Germany) and nitrocellulose membranes (Protean BA 85; Schleicher & Schüll, Dessel, Germany) with antibodies raised from rabbits. DsrC antigens were detected with antibodies raised against recombinant DsrC purified from E. coli. DsrE (H2N-VNNSTRLTTPPQDDRH-CONH2), DsrF (H2N-CLTRGQDTKGIGMKNF-CONH2), DsrH (H2N-VNKSPFERNSLESC-CONH2), DsrJ (H2N-DANRNPQPIDQPDQFC-CONH2), DsrO (H2N-SQRLREIPSRQIREDL-CONH2), DsrK (H2N-CDLDDPNEEEETDEAA-CONH2), DsrN (H2N-ADVEMCDRPQGRGYVR-CONH2 and H2N-TVRRGTGIDGSHDGIV-CONH2, simultaneously), and DsrL (H2N-MATSSDEMKMKPTWRC-CONH2) were detected with antisera raised against the given oligopeptides, comprising highly immunogenic epitopes deduced from the nucleotide sequence (Eurogentec, Seraing, Belgium). Antisera against DsrO, DsrJ, DsrK, DsrN, and DsrL were used at a 1:500 dilution, Anti-DsrE was diluted 1:1,000. Antisera against DsrF and DsrH were purified on an EAH Sepharose affinity matrix to which the respective immunogenic peptides were coupled (Eurogentec). The purified antibodies were used at a 1:4,000 dilution. Binding of Anti-DsrF and Anti-DsrH was detected with the ECL Plus Western blotting detection system (Amersham Bioscience, Little Chalfont, United Kingdom); in the other cases, 4-chloro-1-naphthol was used essentially as described previously (63). All of these antibodies were subsequently shown to react specifically against proteins overexpressed from the corresponding genes cloned into E. coli.
The molecular masses of denatured proteins were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using the method of Laemmli (41). Nondenatured or denatured proteins were stained with Coomassie blue or silver stain.
N-terminal amino acid sequences were determined from protein bands in polyacrylamide gels by automated Edman degradation using a Porton 2090e protein sequencer (Beckman Coulter, Fullerton, Calif.) as described previously (9). Briefly, subunits were separated by SDS-PAGE and electrophoretically transferred onto polyvinylidane difluoride membranes, visualized by Comomassie brilliant blue staining, excised, and placed in the sample cartridge of the protein sequencer under a fiberglass disk.
Heme staining.
Heme staining was performed after blotting of proteins separated by SDS-PAGE onto polyvinylidene difluoride membranes (Immobilon-P; Millipore). Heme-dependent peroxidase activity was visualized as described previously (74), using the ECL Plus system (Amersham) as specified by the manufacturer.
UV-visible spectroscopy.
UV-visible spectra of samples in 1-ml quartz cuvettes were recorded using an Agilent Technologies (Böblingen, Germany) 8453 diode array spectrophotometer.
Turnover of reduced sulfur compounds.
Turnover of reduced sulfur compounds was measured in batch culture, and sulfur compounds were quantified as described by Dahl (13).
GenBank accession number.
The nucleotide sequence data reported in this paper have been submitted to GenBank and are available under accession number U84760.
RESULTS
Nucleotide sequence of the complete dsr locus.
The dsr locus of A. vinosum consists of 15 genes dedicated to sulfur metabolism (Fig. 1). We have already reported the cloning and sequencing of the dsrABEFHCMK genes (57). Further sequencing yielded a total of 14,467 bp and revealed the presence of seven further complete open reading frames, termed dsrLJPONRS, downstream of dsrK. These are closely spaced and all are preceded by putative ribosome binding sites. The noncoding regions between the different genes are short (the largest is 105 bp, between dsrK and dsrL), or the genes are located directly adjacent. The dsrS gene is followed by an incomplete open reading frame homologous to ruvB, which is transcribed in the opposite direction. In E. coli the ruvB gene product is part of the RuvABC resolvasome, which catalyzes the resolution of Holliday junctions that arise during genetic recombination and DNA repair (18).
FIG. 1.
(A) Schematic overview of the dsr locus of A. vinosum and two related genes clusters in the green sulfur bacterium C. tepidum. The same names were given to clearly related genes. Predicted functions of gene products are indicated under the genes where appropriate. The positions of insertion sites of the kanamycin Ω interposon in the A. vinosum double-crossover mutants 21D and 39D are indicated. (B) Schematic presentation of the Dsr proteins from A. vinosum. The scheme is based on sequence analysis of the encoding genes and on biochemical information where available.
As already reported (57), two hairpin loop structures with the potential to act as transcription terminators are found between dsrB and dsrE. At 32 nucleotides downstream of the dsrS stop codon, another inverted repeat with a potential for formation of a hairpin loop structure (free energy of formation, −38.9 kJ mol−1) was identified. Poly(T) sequences located directly downstream of such hairpin loops in typical eubacterial rho-independent transcription terminators (60) are lacking in all three cases.
The results of the sequence analyses of the proteins deduced from dsrLJOPNRS are summarized in Table 2.
TABLE 2.
Features of the dsr-encoded proteins
| Feature | DsrL | DsrJ | DsrO | DsrP | DsrN | DsrR | DsrS |
|---|---|---|---|---|---|---|---|
| Calculated mol wt +/− signal peptide | 71,429/− | 16,322/13,631 | 28,901/23,860 | 45,664/− | 50,473/− | 11,436/− | 41,097/− |
| Transport pathway | None | Sec dependent | Tat dependent | None | None | None | None |
| Transmembrane helices | None | 1 in signal peptide | 1 in signal peptide | 10 | None | None | None |
| Predicted cellular localization | Cytoplasm, soluble | Periplasm or membrane | Periplasm | Membrane | Cytoplasm, soluble | Cytoplasm, soluble | Cytoplasm, soluble |
| Predicted cofactor binding sites | Fe-S clusters (N terminus), FAD, 2 × [Fe4S4] at C terminus | 3 × heme c (CXXCH) | Probably 4 × [Fe4S4] | None | None | None | None |
| Conserved domains | Pfam0007: pyridine nucleotide, disulfide ORb; COG0493: NADPH-dependent glutamate synthase β chain | None | COG0437: FeS cluster containing hydrogenase components | Pfam03916, COG557: NrfD, polysulfide reductase | COG1797: CobB, cobyrinic acid a,c-diamide synthase | Pfam 1521: HesB-like domain; COG0316: IscA | None |
| Highest sequence similaritya | Magn029254, TdenA01002524 | Magn029253, TdenA01002523 | TdenA01002522, Magn029252 | TdenA01002521, Magn029251 | Magn028021, TdenA01002520 | TdenA01002519 | TdenA01000965 |
| Further comments | CXXC motif preceding C-terminal ferredoxin domain | 16-Fe ferredoxin | Probable menaquinol:acceptor oxidoreductase | ATP and glutamine binding motifs |
Tden, Thiobacillus denitrificans; Magn, Magnetospirillum magnetotacticum.
OR, oxidoreductase.
DsrL.
DsrL matches the conserved GltD domain (β-subunit of glutamate synthases), which transfers electrons from NAD(P)H to an acceptor protein or protein domain (73). DsrL contains the adenylate binding motif GXGXXG/A/P (77) twice; the first probably interacts with the adenylate portion of flavin adenine dinucleotide (FAD), and the second interacts with the pyridine nucleotide cofactor (54). An alanine or serine, as found in DsrL at the terminal position, is typical for NADP(H) binding proteins (65). The similarity of DsrL to FAD-containing pyridine nucleotide-disulfide oxidoreductases such as glutathione reductase or thioredoxin reductase does not extend to the active sites of the latter enzymes. Only the first cysteine residue of the typical CXXC thioredoxin reductase active-site motif (76) is conserved at the respective position in DsrL. At their amino-terminus the small subunits of glutamate synthases carry two putative [Fe-S] clusters, the first with four and the second with three or four [Fe-S] binding cysteine residues (25, 54). The second cysteine of the first cluster is missing in DsrL. Among the proteins related to DsrL is SudA, the large α-subunit of sulfide dehydrogenase (SudAB) from Pyrococcus furiosus (25). It catalyzes the reduction of polysulfide to H2S and also functions as a ferredoxin:NADP oxidoreductase. Phylogenetic analyses showed that dsrL does not fall into one clade with sudA-homologous sequences (2). DsrL probably has a function distinct from glutamate synthase as well as from sulfide dehydrogenase, since it has a C-terminal ferredoxin extension unrelated to either GltD or SudA and is linked neither to a gltB-like nor to a sudB-like gene (2).
DsrJ, DsrO, and DsrP.
DsrJ appears to be a triheme cytochrome c with a typical sec-dependent leader peptide mediating transport across the cytoplasmic membrane (21). The signal peptide of the related cytochrome AF503 from the archaeal sulfate reducer Archaeoglobus fulgidus (46) is not cleaved off. The probable 16-Fe ferredoxin DsrO is transported to the periplasm via the Tat system (5, 30). The signal peptide of the closely related protein AF499 from A. fulgidus is not present in the mature enzyme (46). Ten transmembrane helices are predicted for DsrP, AF500 from A. fulgidus (46), and HmcC from the eubacterial sulfate reducer Desulfovibrio vulgaris (62), while only eight are found in other related proteins such as E. coli NrfD, the membrane component of formate-dependent nitrite reductase (33). None of the histidine residues of DsrP is located within a probable transmembrane helix, and the protein is therefore not predicted to bind heme b.
DsrN.
DsrN from A. vinosum resembles cobyrinic acid a,c-diamide synthase. This enzyme is part of the vitamin B12 biosynthetic pathway, where it catalyzes the ATP-dependent amidation of cobyrinic acid to cobyrinic acid a,c-diamide (17). An amidated siroheme, siroamide, has been detected in Desulfovibrio species (48), and its has been postulated that the amidation of siroheme is catalyzed by dsrN-encoded proteins (44). It may be noteworthy in this respect that one of the two dsr gene clusters of the green sulfur bacterium Chlorobium tepidum contains not only dsrN but also a gene resembling cysG encoding siroheme synthase (uroporphyrinogen III methyltransferase) followed by a gene representing a coupling between cysG and hemD, the latter encoding uroporphyrinogen III synthase (Fig. 1) (20). The presence of dsrN in the dsr gene clusters of different phototrophic sulfur bacteria raises the possibility that the prosthetic group of sulfite reductase in these organisms is not the classical siroheme but siro(heme)amide.
DsrR and DsrS.
Three possible start codons exist for dsrR (positions 12859, 12907, and 12910). A putative ribosome binding site, AGGGG, is found beginning at nucleotide 12898, suggesting nucleotide 12910 as the most probable translational start. DsrR is related to IscA and to the HesB domain, both of which may be involved in [Fe-S] cluster formation (12). However, two carboxy-terminal cysteine residues conserved in the IscA-HesB domain are lacking in DsrR. The only dsrS homologue is found in the partially sequenced genome of the chemotrophic sulfur oxidizer Thiobacillus denitrificans (NZ_AAFH01000002.1).
Identification and cellular localization of the predicted proteins.
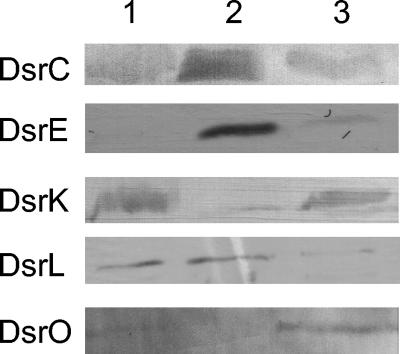
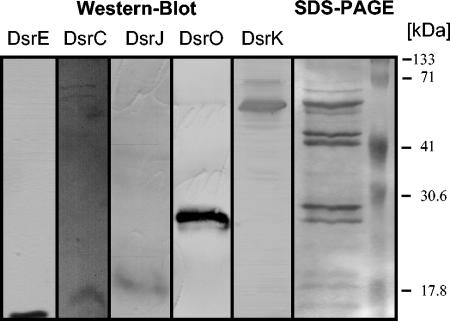
With the exception of the dsrAB-encoded sulfite reductase (64), biochemical information about the proteins encoded by the genes of the A. vinosum dsr operon has so far not been available. Our first aim was therefore to find whether the proteins predicted by the dsr genes are indeed produced in A. vinosum and, if so, to obtain information about their localization within the bacterial cell. We concentrated on the products of the dsr genes that have homologues in the only other known dsr gene clusters from a phototrophic bacterium, i.e., in the green sulfur bacterium C. tepidum. (Fig. 1) (20). Detailed analysis of the dsrR and dsrS gene products were therefore postponed. DsrC, DsrE, DsrK, DsrL, and DsrO were readily and specifically detected after SDS-PAGE of extracts from sulfide/thiosulfate-grown cells by immunoblot analysis (Fig. 2). Anti-C, anti-E, and anti-L antibodies detected antigens of 15, 14 (see Fig. 2 to 4 and 6), and 70 kDa (see Fig. 2 and 3; size not shown), respectively, whose size matched the predicted size of the dsrC, dsrE, and dsrL gene products. As predicted, all three proteins were found almost exclusively in the soluble fraction (Fig. 2). DsrF and DsrH of the predicted sizes were also shown to be soluble and to form a hexameric protein together with DsrE (see below, Fig. 4). The anti-K and anti-O antisera reacted specifically with proteins associated with the membrane fraction (Fig. 2). While anti-K antibodies detected a 57-kDa protein (see Fig. 6) corresponding in size to the complete predicted dsrK gene product, the 24-kDa polypeptide detected with anti-O (see Fig. 6) probably represented a processed form of the complete dsrO-deduced polypeptide (28.9 kDa). In crude soluble or membrane fractions, proteins reactive with anti-N and anti-J antisera could not be observed; this may be due either to a low abundance of the antigens in the cell or to a relatively low specificity of the antisera. However, an anti-N reactive polypeptide of 50 kDa, corresponding in size to the dsrN gene product, was detected in several fractions after hydrophobic interaction chromatography of the soluble A. vinosum proteins (not shown), proving that dsrN is expressed during phototrophic growth on reduced sulfur compounds. An antigen reactive with anti-J antiserum was clearly present in solubilized membrane fractions enriched in DsrK (see Fig. 6), also proving the formation of this dsr gene product in A. vinosum. The apparent molecular mass of the detected protein (14 to 17 kDa, depending on the size markers used) does not allow a clear conclusion about whether the predicted signal peptide is cleaved off (13.6 versus 16.3 kDa).
FIG. 2.
Western blot analysis of proteins fractions of A. vinosum with antibodies against the indicated Dsr proteins. Lanes: 1, crude extracts; 2, soluble fraction; 3, membrane fraction. Proteins of cell extracts (10 μg per lane) were separated by SDS-PAGE.
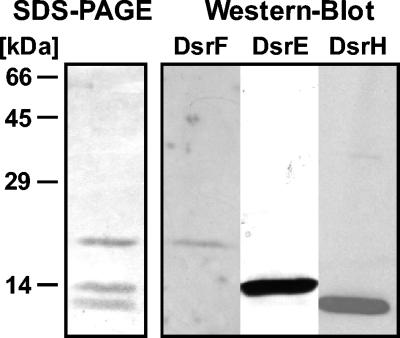
FIG. 4.
SDS-PAGE and Western blot analyses of DsrEFH from A. vinosum. A 1.5-μg portion of purified protein was used for SDS-PAGE and Western blot analyses, respectively.
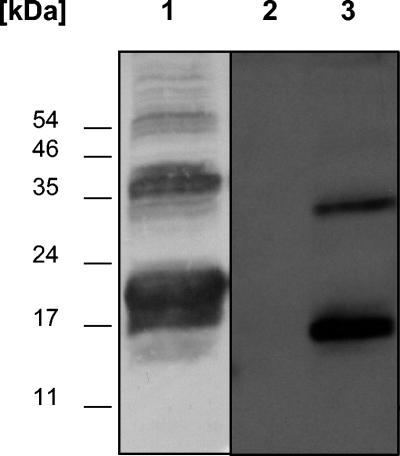
FIG. 6.
Immunochemical analysis of a crude extract of a DsrJ-overproducing E. coli strain (E. coli HM125) containing pEC86 and pISC-2(dsrJ) after SDS-PAGE (lane 1); heme stain of a crude extract of E. coli HM125 containing pEC86 and pISC-2 (lane 2); heme stain of a crude extract of E. coli HM125 containing pEC86 and pISC-2(dsrJ) (lane 3).
FIG. 3.
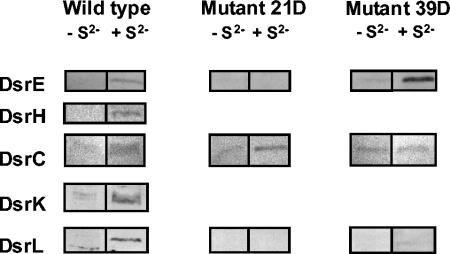
Sulfide-dependent formation of DsrE, DsrH, DsrC, DsrK, and DsrL in A. vinosum wild type and mutants 21D and 39D. A. vinosum was cultivated photoorganoheterotrophically in complex RCV medium in completely filled, closed 100-ml bottles. After the cells reached the late exponential phase, sulfide (4.2 mM) was added. Cells (2 ml) were collected by centrifugation 5 h after sulfide addition, resuspended with 100 μl of 20 mM Tris HCl (pH 7.5) and 50 μl of SDS sample buffer, and incubated at 100°C for 5 min. A 10-μl volume of each sample was subjected to SDS-PAGE and Western blot analysis with antibodies raised against the indicated proteins.
Sulfide induction experiments.
Northern hybridization experiments had previously shown that transcription of dsrAB is induced under sulfur-oxidizing conditions. The dsrC gene was independently and constitutively expressed (57). To these studies we added Western blot analyses and examined the sulfur-dependent regulation of dsr genes with DsrE, DsrH, DsrC, and DsrK as representatives of the old gene products and DsrL as representative of the novel gene products. A. vinosum was grown photoorganoheterotrophically on malate. When the cultures had reached the late exponential phase, 4.2 mM sulfide was added. The sulfide concentration immediately decreased, and uptake appeared to be completed after approximately 5 h, when the concentration of intracellular sulfur reached its maximum. Samples were taken immediately before sulfide addition and immediately after complete sulfide uptake. While DsrE, DsrH, DsrK, and DsrL were hardly detectable in the absence of sulfide, their concentration increased dramatically in sulfur-containing cells (Fig. 3). In contrast, the dsrC gene product was detectable even in the absence of sulfide. In the wild-type, its abundance also appeared to increase under sulfur-oxidizing conditions (Fig. 3).
In summary, our Northern and Western blot analyses and the observation that dsrAB-encoded sulfite reductase is present in easily detectable amounts only in cells grown photolithotrophically on sulfide (64) suggest the conclusion that the dsrABEHKL genes are expressed and the encoded proteins are formed at a low basic level even in the absence of sulfur compounds but that an increased production is specifically induced by sulfide and/or stored sulfur. Concerted regulation of these genes points at cotranscription of all 15 dsr genes, implying that they constitute an operon. The concentration of the 0.9-kb dsrC mRNA and also of the encoded polypeptide is not as strongly influenced by the presence of reduced sulfur compounds as are those of the other genes in the operon.
Construction and characterization of an A. vinosum strain with a mutation in dsrL.
To assess the necessity of the newly sequenced dsr genes for sulfur oxidation, an A. vinosum mutant (39D) carrying a kanamycin Ω interposon (22) in dsrL was constructed (Fig. 1). The interposon usually leads to premature termination of transcription of the affected gene and downstream genes in the same transcription unit. To prove this effect, the presence of gene products encoded upstream and downstream of the interposon insertion site was assessed in mutant 39D and also in mutant 21D (Fig. 3). The latter carries the interposon in dsrB (Fig. 1) (57). Indeed, DsrE and DsrL, which are encoded downstream of dsrB, were not formed in mutant 21D, while DsrE, which is encoded upstream of dsrL, was still present in mutant 39D. Like the wild type, mutant 39D produced DsrE in a sulfide-dependent manner. DsrL was clearly lacking in mutant 39D. DsrC was present in both mutants, again proving its formation independent of the other Dsr proteins. Like all other mutants carrying an interposon in one of the dsr genes, A. vinosum 39D is completely unable to oxidize intracellularly stored sulfur arising from the oxidation of sulfide or thiosulfate. The rates of sulfide and thiosulfate oxidation to sulfur were unaffected in mutant 39D. Data obtained for batch cultures of A. vinosum 39D growing photolithotrophically on sulfide or thiosulfate were almost identical to those measured for the other dsr mutants (57) and are therefore not shown. We can now conservatively state that dsrL and most probably also the downstream dsr genes are indispensable for the oxidation of intracellular sulfur in A. vinosum.
DsrE, DsrF, and DsrH are subunits of a soluble multimeric protein.
Sulfite reductase (DsrAB) is the only dsr-encoded protein from A. vinosum characterized so far (64). We initiated the biochemical characterization of the other dsr gene products by purification of selected soluble and membrane-bound polypeptides. DsrE, DsrF, and DsrH are predicted to be rather small (14.6, 15.6, and 11.1 kDa, respectively), soluble, cytoplasmic proteins that do not contain prosthetic groups (57). Using the specific antiserum for detection, DsrE was purified from the soluble protein of A. vinosum cell extracts. The purified preparation contained three polypeptides with apparent molecular masses of 12, 14, and 18 kDa by SDS-PAGE. These polypeptides were identified as DsrH, DsrE, and DsrF, respectively (Fig. 4). Native PAGE yielded a single protein band for DsrEFH (data not shown). A molecular mass of 75 kDa was determined by analytical gel filtration on Superdex 200, which also yielded no indication that multiple components were present. We thus conclude that DsrEFH form a single tight complex. Analyses of Coomassie brilliant blue-stained gels indicated that the proteins are present in a 1:1:1 ratio. Together, these data suggest an α2β2γ2 structure.
In a supercomplex, DsrK is associated with DsrAB, DsrEFH, DsrC, DsrO, and DsrJ.
Membrane-bound protein complexes containing DsrM- and DsrK-related proteins have been purified from the archaeal sulfate reducers A. fulgidus and A. profundus (46, 47). During purification, the proteins were traced by UV-visible spectroscopy, taking advantage of the presence of b-type heme in the DsrM-related polypeptides. We could not use the same strategy because membranes of the phototrophic A. vinosum contain the cytochrome bc1 complex in abundance; we were unable to detect a new membrane-bound b-type cytochrome in this background. Instead, we traced DsrK by making use of the specific antiserum. The membrane fraction was isolated, and proteins were solubilized with the detergent thesid (0.5%). This treatment solubilized DsrK in sufficient quantity without bringing vast amounts of light-harvesting complexes containing carotenoids and bacteriochlorophyll into solution.
After anion-exchange chromatography on Q-Sepharose and chromatography on ceramic hydroxyapatite, the DsrK-containing sample was subjected to gel filtration on Superose 6. DsrK eluted in a broad peak starting in the void volume of the column, indicating the presence of a large protein complex. SDS-PAGE revealed that gel filtration chromatography did not result in the removal of any of the polypeptide bands present in the samples after purification on hydroxyapatite. This pointed at a specific association of the proteins present in the samples.
The protein solution then purified was green and had a UV-visible spectrum reminiscent of sulfite reductase, with absorption maxima at 413 and 595 nm, typical for siroheme-containing proteins (64). The spectrum did not yield evidence for the presence of heme b or c in the preparation. On SDS-PAGE, the fractions after hydroxyapatite chromatography yielded five major polypeptides with apparent molecular masses of 57, 45, 42, 29, and 24 kDa (Fig. 5). In addition, several minor bands in the size range from 12 to 18 kDa were present.
FIG. 5.
SDS-PAGE and Western blot analyses of DsrK-containing fractions (5 μg of protein) after anion-exchange and hydroxyapatite chromatography.
The amino-terminal sequence of the 57-kDa polypeptide was determined to be AKATFEVP, which conforms exactly to the sequence deduced from dsrK without the initial methionine. The 45- and 42-kDa polypeptides yielded the sequences AIDKHA and N-methyl-AEMREXIESG, respectively. The residue indicated as X corresponds to Cys, which is unstable unless derivatized to form a more stable thioether (8). The sequences match exactly the N termini of the polypeptides deduced from dsrA and dsrB, respectively. The initiator methionine appears to be cleaved off in both cases. The unusual posttranslational modification of the N-terminal alanine residue of DsrB was previously observed during sequencing of DsrB from another strain of A. vinosum (34a). Other examples of proteins with N-terminal N-methylated amino acids are mainly structural proteins and include components of bacterial flagella and ribosomal proteins, for which it has been suggested that N-terminal methylation plays a role in aggregation, with the methylated N terminus buried in the interface between interacting subunits (71). The presence of both subunits of sulfite reductase in the DsrK-containing sample explains the observed UV-visible spectrum. The N-terminal amino acid sequence of the 29-kDa band was determined to be PADLIAPSILSANPA. It did not conform to any of the polypeptides encoded in the dsr locus but exhibited significant similarity to the N termini of ribulose-3-phosphate epimerases, e.g., from Haemophilus influenzae (23).
Since we were unable to obtain N-terminal sequence information for the other polypeptide bands in the preparation, we checked the presence of further dsr-encoded proteins via Western blot analyses. Indeed, we obtained signals with anti-DsrE, anti-DsrC, anti-DsrJ, and anti-DsrO (Fig. 5). The association of all of these proteins with DsrK appeared to be specific since they were not detected in fractions eluting from the anion-exchange and hydroxyapatite columns that did not contain DsrK (data not shown). Since DsrJ is predicted to bind three heme c moieties, heme staining of the bands present in the DsrK-enriched fractions was attempted. However, this was unsuccessful, showing that DsrJ was present only in substoichiometric amounts. None of the bands could be assigned to the predicted membrane proteins DsrM or DsrP.
DsrJ contains covalently bound heme c.
To prove the presence of heme c in DsrJ, the respective gene was overexpressed in E. coli HM125, a strain especially designed for the overexpression of heme c containing proteins under aerobic conditions (3). The complete dsrJ gene including the potential signal peptide-coding sequence was cloned into the pISC-2 vector, which allows expression of genes under control of the arabinose-inducible pBAD promotor (3). Cells were cultivated aerobically, induced with arabinose, harvested, and disrupted as described previously (3). Immunochemical detection clearly showed the presence of two broad bands around 18 and 37 kDa in the overexpressing strain (Fig. 6). These bands were absent in cells harvested before induction with arabinose (data not shown). The size of these bands fits with that calculated for monomeric DsrJ (16.3 kDa) and dimeric unprocessed DsrJ (32.6 kDa), indicating first that the signal peptide of a large fraction of DsrJ is not cleaved off in E. coli and second that recombinant DsrJ has a tendency to form multimers even in the presence of SDS. Heme staining of proteins separated by SDS-PAGE unambiguously revealed the presence of heme in the E. coli strain containing plasmid pISC-2 with dsrJ, while a strain containing the unaltered pISC-2 vector did not contain any proteins with covalently attached heme (Fig. 6). This observation indicates that heme is covalently attached to the overproduced protein DsrJ. The polypeptides reacting positively on heme staining appeared to be smaller (15 and 32 kDa) than the majority of the protein that reacted with the anti-J serum. Obviously, only part of the recombinant DsrJ underwent correct maturation, including transport to the periplasm, cleavage of the signal peptide, and heme insertion. Nevertheless, our experiments allow the conclusion that DsrJ is a c-type cytochrome with covalently bound heme.
DISCUSSION
In the present study, the sequence of the dsr gene cluster of A. vinosum was completed and seven new genes, dsrLJOPNRS, were characterized. Interposon mutagenesis proved an essential role for the encoded proteins in the oxidation of sulfur formed as an intermediate during the oxidation of sulfide and thiosulfate (57; also see above). By immunochemical techniques, we were able to prove the formation of all tested dsr-encoded proteins in A. vinosum. With the exception of DsrC, which appeared to be present constitutively, the formation of the tested Dsr gene products was greatly enhanced by sulfide. Hybridizations with gene probes directed against dsrA, dsrEFHC, dsrM, and dsrK had previously shown for several different species of purple sulfur bacteria that the dsr genes have a very similar if not identical arrangement to that in A. vinosum (16). In addition, sequences of genes similar to those in the A. vinosum dsr locus have become available from the whole-genome sequence of the green sulfur bacterium C. tepidum (20). They are organized in two clusters, dsrCABLKMPOJ and dsrCABLEFHN (Fig. 1). The ubiquitous presence of the dsr genes in anoxygenic phototrophic sulfur bacteria stresses the importance of their products for oxidative sulfur metabolism. In addition, we can conclude that the mechanism of sulfur oxidation appears to be similar for anoxygenic phototrophs forming intracellular (A. vinosum) and extracellular (C. tepidum) sulfur globules. The duplication of dsrCABL in C. tepidum may hint to a central role of the respective proteins.
Recently, genome sequence data for the chemotrophic sulfur oxidizer Thiobacillus denitrificans ATCC 25259 has become available through GenBank (NZ_AAFH01000005, NZ_AAFH010000021). This organism contains two dsr gene clusters in one of which (TdenA01002530 to Tden A01002518) the genes dsrEFHCMKLJOPNR are organized exactly as in A. vinosum. Since the T. denitrificans genome sequence is not yet complete, it remains unclear whether these genes are preceded by dsrAB. As is evident from Table 2, the A. vinosum and T. denitrificans dsr genes are very closely related.
In dissimilatory sulfate-reducing bacteria and archaea, i.e., in organisms that obtain energy from performing the opposite overall reaction as compared to phototrophic sulfur oxidizers, dsr-related genes appear to be located in at least two independent operons. The sulfite reductase-encoding genes dsrAB can be linked with dsrN and in one case also with dsrC (42, 44). dsrC constitutes an independent transcriptional unit in D. vulgaris (36) and is also located apart from dsrAB in the hyperthermophilic archaeal sulfate reducer Ag. fulgidus (40). In all sulfate reducers studied so far (15, 37, 42-44), dsrAB are linked with the dsrD gene encoding a protein of around 9 kDa with a B- and Z-DNA binding motif (49). In Bilophila wadsworthia, an organism able to reduce sulfite but not sulfate, dsrD is even fused to dsrB (45). In phototrophic sulfur oxidizers, a related gene does not appear to be present (20), indicating a specific function for the dsrD gene product in the reduction of sulfite. In addition to the dsrAB clusters, genes related to dsrMKJOP are present in the hme gene cluster of A. fulgidus (40, 46) and in the hmc gene cluster of D. vulgaris (62). So far, it appears that the dsrE, dsrF, and dsrH genes have no equivalents in sulfate reducers. The encoded polypeptides might therefore be important for the system to operate in the oxidative direction. A dsrL-homologous gene has so far also not been observed in any of the dsr-related gene clusters in dissimilatory sulfate reducers. Homologues of many of the genes in the A. vinosum dsr gene cluster are also present in two groups in the genome sequence of the magnetotactic bacterium Magnetospirillum magnetotacticum (GenBank accession no. NZ_AAAP00000000), indicating that this organism is able to perform some type of energy-yielding sulfur metabolism. Summarizing, we can state that prokaryotes containing sulfite reductase, irrespective of whether they are sulfur oxidizers or sulfate reducers, conserve a core set of dsr-related genes, namely, dsrAB encoding sulfite reductase itself, dsrC encoding a cytoplasmic protein with strictly conserved cysteine residues, dsrMKOPJ encoding a membrane-spanning electron-transporting complex, and dsrN.
DsrN is most probably involved in the biosynthesis of siro(heme)amide from siroheme (44). Glutamine serves as the amino-group donor in the process. It is noteworthy in this respect that dsrL encodes a protein resembling the small subunit of bacterial glutamate synthases. If DsrL retains glutamate synthase activity, as has been described for a related protein from Pyrococcus kodakaraensis (35), it could theoretically provide the glutamine for siroheme amidation. However, experimental evidence for such a function of DsrL is lacking. On the basis of its amino acid sequence, it is more likely that DsrL is able to transfer two reducing equivalents between NADPH + H+/NADP+ and an as yet unidentified acceptor/donor via the protein-bound FAD cofactor. Several related proteins function as NADPH:feredoxin oxidoreductases (6, 25). With its additional C-terminal ferredoxin domain, DsrL would carry this electron-transporting unit within the same polypeptide chain. DsrL also contains a CXXC motif immediately preceding the ferredoxin domain. This motif may be involved in a redox reaction through thiol-disulfide exchange. In summary, DsrL could well be involved in electron transfer from or to sulfite reductase or the potential (hetero)disulfide reductase DsrK and associated proteins.
Purification of DsrE from the soluble fraction of A. vinosum showed that this protein is a subunit of an α2β2γ2-structured holoprotein consisting of DsrE, DsrF, and DsrH. DsrE belongs to the Cluster of Orthologous Group COG1553 (National Center for Biotechnology Information database, National Library of Medicine, National Institutes of Health), for which structural information is available. The crystal structure of YchN from E. coli, a protein of unknown function, has been described (67). It shows a dimer of homotrimers arranged in a cylindrical ring structure. This homohexameric structure is reminiscent of the heterohexameric structure of DsrEFH when we keep in mind that DsrE, DsrF, and DsrH have sequence similarity (57). In several members of the protein family, including YchN, there are two conserved cysteine residues (Cys78 and Cys81 in YchN). Cys78 is absolutely invariant, and the structural environment around Cys78 implies that it is a putative active-site residue. For members of the YchN family sharing the CXXC motif, a function as a thiol:disulfide oxidoreductase would theoretically be possible. However, in DsrE only the first of these two cysteine residues, Cys78, is conserved. MTH1491 from the archaeon Methanobacterium thermoautotrophicum is another protein related to DsrE that has been crystallized (11). This protein forms a homotrimer. Interestingly, the corresponding gene is located adjacent to a putative soxB gene in M. thermoautotrophicum. The SoxB protein is part of the thiosulfate-oxidizing multienzyme complex of Paracoccus pantotrophus (24). Moreover, for MTH1491, an interaction with sulfate has been suggested (11).
Most of the Dsr proteins were found to be localized in cell fractions predicted from the gene sequences: sulfite reductase has already been purified from the soluble fraction by others (64), and DsrC, DsrEFH, DsrL, and DsrN also appeared to be soluble. However, although DsrK, DsrJ, and DsrO are predicted not to form transmembrane helices but to be soluble proteins, they were detected exclusively in the membrane fraction, indicating a tight attachment to integral membrane proteins, probably DsrM and/or DsrP. Indeed, the copurification of DsrK, DsrJ, and DsrO indicates a specific association of the proteins and suggests that DsrMKJOP assemble a membrane-spanning electron-transporting complex as depicted in Fig. 1. This idea is supported by the purification of the related Hme complex from A. fulgidus (46). We have so far not been able to detect DsrM and DsrP in our preparations. In the Hme complex from A. fulgidus, the DsrP-related polypeptide was also missing and the 34-kDa DsrM-homologous polypeptide was hardly detectable in boiled samples (46). This was attributed to protein aggregation, which is typical for integral membrane proteins. These authors also reported that the 16-kDa DsrJ-homologous protein was present only in substoichiometric amounts. In some preparations, the protein was completely absent. The situation with the preparation from A. vinosum appears to be similar. While DsrJ was clearly detected with a specific antiserum, a corresponding band could not be clearly identified in Coomassie blue-stained gels, nor could the predicted covalently bound heme be found in the samples. The presence of covalently attached heme in this protein was, however, verified for recombinant DsrJ produced in E. coli.
On the basis of predicted amino acid sequences and gene arrangement, the A. fulgidus Hme and the D. vulgaris Hmc complexes are the most similar to the proposed DsrMKJOP complex from A. vinosum. All three complexes contain a periplasmic cytochrome c subunit (DsrJ and Af503 with 3 hemes, HmcA with 16 hemes), a periplasmic iron-sulfur-cluster-containing subunit (DsrO, AF499, and HmcB), an integral membrane subunit (DsrP, AF500, and HmcC), a second membrane-bound protein possibly containing hemes b (DsrM, AF501, and HmcE), and a cytoplasmic subunit carrying iron-sulfur clusters with similarities to HdrD, the active-site subunit of heterodisulfide reductase (DsrK, AF502, and HmcF). The Hmc, Hme, and Dsr complexes contain subunits and redox cofactors on the periplasmic side and also on the cytoplasmic side of the membrane. It has been proposed that the Hmc complex has subunits that act as menaquinone reductase (HmcABC) and subunits acting as menaquinol oxidase (HmcEF) (61), whereas the A. fulgidus Hme complex was proposed to function as a bidirectional menaquinol oxidoreductase which can transfer electrons to both a cytoplasmic acceptor (via the HdrD-like subunit) or a periplasmic acceptor (via the c-type cytochrome) (46).
The exact physiological function of the Hme and Hmc complexes in dissimilatory sulfate reducers has not been elucidated. It has been proposed that these and related transmembrane complexes from sulfate-reducing bacteria are involved in the transport of electrons originating from oxidative metabolic pathways to the cytoplasmically located enzymes of sulfate reduction, namely, APS reductase and sulfite reductase (29, 46, 47, 56, 61, 62). Indeed, several studies support the role of the D. vulgaris Hmc complex in transmembrane electron transport by linking periplasmic hydrogen oxidation to cytoplasmic sulfate reduction (19, 39, 55, 69). Likewise, evidence has been provided that Hme functions as menaquinol:acceptor oxidoreductase in A. fulgidus, mediating the electron transfer from the quinone pool to an as yet unidentified electron carrier in the cytoplasm, which in turn is thought to function in its reduced form as an electron donor of the enzymes of sulfate reduction (46). In the phototrophic sulfur oxidizers A. vinosum and C. tepidum, the genes for sulfite reductase are located adjacent to those encoding Hmc- and Hme-related transmembrane complexes (Fig. 1). Our Northern and Western analyses of A. vinosum wild-type and mutant strains clearly showed that the respective genes are coordinately expressed (57) (Fig. 3), supporting the notion that the encoded gene products may interact physiologically. Furthermore, we obtained experimental evidence for a close interaction of sulfite reductase (DsrAB) from A. vinosum with the Hmc/Hme-related membrane-bound Dsr complex (Fig. 5). Sulfite reductase itself is a soluble enzyme and as such has been purified from the soluble cell fraction (64). However, part of the enzyme obviously resides in the membrane fraction. After chromatographic separation of solubilized membrane proteins, sulfite reductase is detected exclusively in samples containing DsrK and other components of the Dsr complex (DsrO, DsrJ). The same holds true for DsrE and DsrC. That part of these soluble proteins that is attached to the membrane appears to be specifically associated with DsrK. These observations point at a close in vivo interaction and formation of a supercomplex between sulfite reductase, DsrC, DsrEFH, and the DsrKMPOJ complex in A. vinosum. The proposed interaction could very well be such that electrons are transferred from sulfite reductase through DsrEFH and/or DsrC via thiol/disulfide interchanges to the Dsr complex, with DsrK probably representing a (hetero)disulfide reductase. The observed close association of sulfite reductase and the Dsr complex in A. vinosum strongly supports the proposed electron transfer from the Hmc-Hme complexes to sulfite reductase, the major enzyme of the sulfate reduction pathway, in dissimilatory sulfate reducers. It may be interesting in this respect that sulfite reductase from Desulfovibrio desulfuricans is present in both soluble and membrane-bound forms (70). On the basis of our results, it would appear likely that the association of sulfite reductase with the membrane in this sulfate reducer is also mediated by a Dsr/Hmc/Hme-related complex. Although not visible after SDS-PAGE analysis of sulfite reductase purified from the membrane fraction (70), the respective transmembrane complex might have been present in substoichiometric amounts. This would explain the different behavior of D. desulfuricans sulfite reductase purified from the soluble and membrane fractions, i.e., different stimulation by CO and, most importantly, different reactivity with cytochrome c3. Only sulfite reductase isolated from the membrane fraction was able to accept electrons from cytochrome c3. The latter is located in the bacterial periplasm, and a direct interaction with the cytoplasmically located sulfite reductase is not possible in vivo or in vitro unless mediated by a membrane-spanning electron-transporting unit.
We previously suggested two different models for the function of Dsr proteins in oxidative sulfur metabolism (57). In the first, it was assumed that extracytoplasmically stored sulfur is made available for oxidation by the cytoplasmic sulfite reductase by reduction to the level of sulfide and diffusion as H2S or transport in an unknown form and by an unknown mechanism over the cytoplasmic membrane. DsrK and DsrM were suggested to accept electrons from sulfite reductase. With knowledge of the complete dsr gene cluster and by analogy to the proposed function of Dsr-homologous proteins in sulfate reducers (29, 46, 62), it indeed appears likely that sulfite reductase and the DsrMKJOP complex react such that electrons released by sulfite reductase are fed into photosynthetic electron flow via the membrane complex. The system in the phototrophic sulfur oxidizers would thus operate opposite to that in sulfate reducers. DsrM could operate as a quinone reductase, DsrP could operate as a quinol oxidase, and finally, the c-type cytochrome DsrJ would be reduced. From here, electrons could be transferred to HiPIP, the primary electron donor to the photosynthetic reaction center (75). Alternatively, the system could work as suggested for A. fulgidus (46). Here, the membrane complex is thought to represent two rather independent modules transferring electrons from the quinone pool to the two different sides of the membrane. When we apply this suggestion to the situation in A. vinosum, it could be envisioned that electrons are transferred from sulfite reductase to quinone via DsrMK and that quinol would then be reoxidized at the cytochrome bc1 complex and thereby directly fed into the primary donor of the reaction center. Alternatively, electrons from quinol could be used to reduce NAD via NADH:quinone oxidoreductase in a reaction driven by the transmembrane proton gradient (Δp). The second unit, consisting of the DsrP, DsrO, and DsrJ proteins, would transfer electrons originating from periplasmic oxidation processes into the quinone pool. Theoretically, these could stem from the sulfur compounds that are oxidized in the periplasm (sulfide [59] and at least part of sulfite [13]). However, sulfide and sulfite oxidation have been shown to proceed independently of the presence of Dsr proteins (57). Regardless of whether electrons are transferred only into the quinone pool via DsrMK or further onto periplasmic electron acceptors via the complete complex DsrMKJOP, such a scenario would not leave a role for the potential NADPH:acceptor oxidoreductase DsrL. One option would be that DsrL is involved in the reductive release of sulfide, the substrate of sulfite reductase, from a carrier molecule, possibly an organic perthiol. The resulting thiol could then be shuttled to the extracytoplasmic space, where it picks up sulfur from the stored sulfur globules and thereby regenerates the perthiol, which is transported back into the cytoplasm.
In a second model (57), it was proposed that DsrK is involved in reduction of the perthiol and obtains the required electrons from quinol via DsrM. While this is still a possibility, such a scenario would not explain the presence of DsrJOP. The model would furthermore not be able to explain how electrons resulting from the sulfite reductase reaction are fed into the photosynthetic electron chain. A transfer of electrons directly to NADP+ via DsrL would be a shortcut not involving photosynthetic electron transport.
With both DsrM and DsrP being integral membrane proteins probably reactive with quinone and quinol, it can be envisaged that a concerted action of DsrMJKOP generates a proton gradient through a redox loop mechanism, as has been suggested for related complexes from sulfate reducers (56). If we assume that it comes to a net extrusion of protons to the periplasm, this would contribute to Δp. As discussed previously such a system would be subject to thermodynamic back pressure mediated by light-dependent Δp (57). This effect would be especially strong under conditions where a high Δp is generated by cyclic electron flow, i.e., when the electron transport chain is overreduced due to limiting CO2 (electron acceptor) or due to high concentrations of highly reduced electron donors (e.g. external hydrogen sulfide). Indeed, the oxidation of sulfide in batch cultures of purple sulfur bacteria proceeds in a biphasic manner (7) the oxidation of stored sulfur to sulfite (E0′ for the HSO3−/S0 couple is −40 mV) and further to sulfate starts only when sulfide is almost completely oxidized to stored sulfur (E0′ for the S0/HS− couple is −270 mV).
Our Dsr complex preparations contain a prominent band of 29 kDa (Fig. 5) that, upon amino-terminal sequencing, exhibited striking similarity to ribulose-3-phosphate epimerases. We do not know whether this protein is a contamination or specifically associated with Dsr proteins. Interestingly, a gene encoding a closely related protein is present in C. tepidum. In this organism, deletion of a gene encoding a ribulose bisphosphate carboxylase-like protein causes, among other effects, a decreased rate of stored sulfur oxidation; i.e., the mutant forms significantly more sulfur globules than the wild type does (27, 28). All these observations may gain importance when we keep in mind that the sulfur chains in the sulfur globules of phototrophic bacteria most probably carry organic residues at one or both ends (58).
In conclusion, we completed the sequence of the dsr operon from the purple sulfur bacterium A. vinosum. Besides siro(heme)amide sulfite reductase, the dsr genes encode several proteins carrying redox cofactors and several proteins carrying conserved cysteine residues that could undergo thiol/disulfide interchanges or act as sulfur substrate binding molecules. In addition, DsrN is probably involved in biosynthesis of the sulfite redcutase prosthetic group. A sulfide/sulfur-induced formation in A. vinosum could be verified for almost all Dsr proteins by Western blotting. DsrKJO were purified together from the membrane fraction, proving the predicted formation of a transmembrane complex by DsrMKJOP. This complex was intimately associated with sulfite reductase, DsrC, and DsrEFH. Although the exact nature of the interaction between these proteins is still unclear, we now have an ideal and unique possibility to study it and to clarify the long-standing problem of electron transport from and to sulfite reductase, not only in phototrophic bacteria but also in sulfate reducing prokaryotes (7, 10, 53).
Acknowledgments
This work was supported by grant Da 351/3-1 from the Deutsche Forschungsgemeinschaft. A. S. Pott-Sperling was supported by a scholarship of the Studienstiftung des Deutschen Volkes.
Excellent technical assistance by Monika Kräling and Birgitt Hüttig is gratefully acknowledged. We thank Hans-Georg Sahl (Bonn, Germany) for production of antiserum against DsrC. We are indebted to Hans G. Trüper for stimulating discussions.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, J. O., and A. J. Roger. 2002. Evolutionary analyses of the small subunit of glutamate synthase: gene order conservation, gene fusions, and prokaryote-to-eukaryote lateral gene transfers. Eukaryot. Cell 1:304-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arslan, E., H. Schulz, R. Zufferey, P. Kunzler, and L. Thöny-Meyer. 1998. Overproduction of Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251:744-747. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors. Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 6.Bossi, R. T., A. Aliverti, D. Raimondi, F. Fischer, G. Zanetti, D. Ferrari, N. Tahallah, C. S. Maier, A. J. R. Heck, M. Rizzi, and A. Mattevi. 2002. A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase. Biochemistry 41:8807-8818. [DOI] [PubMed] [Google Scholar]
- 7.Brune, D. C. 1989. Sulfur oxidation by phototrophic bacteria. Biochim. Biophys. Acta 975:189-221. [DOI] [PubMed] [Google Scholar]
- 8.Brune, D. C. 1992. Alkylation of cysteine with acrylamide for protein sequence analysis. Anal. Biochem. 207:285-290. [DOI] [PubMed] [Google Scholar]
- 9.Brune, D. C. 1995. Isolation and characterization of sulfur globule proteins from Chromatium vinosum and Thiocapsa roseopersicina. Arch. Microbiol. 163:391-399. [DOI] [PubMed] [Google Scholar]
- 10.Brune, D. C. 1995. Sulfur compounds as photosynthetic electron donors, p. 847-870. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Christendat, D., V. Saridakis, Y. Kim, P. A. Kumar, X. Xu, A. Semesi, A. Joachimiak, C. H. Arrowsmith, and A. M. Edwards. 2002. The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum. Protein Sci. 11:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cupp-Vickery, J. R., J. J. Silberg, D. T. Ta, and L. E. Vickery. 2004. Crystal structure of IscA, an iron-sulfur cluster assembly protein from Escherichia coli. J. Mol. Biol. 338:127-137. [DOI] [PubMed] [Google Scholar]
- 13.Dahl, C. 1996. Insertional gene inactivation in a phototrophic sulphur bacterium: APS- reductase-deficient mutants of Chromatium vinosum. Microbiology 142:3363-3372. [DOI] [PubMed] [Google Scholar]
- 14.Dahl, C. 1999. Deposition and oxidation of polymeric sulfur in prokaryotes, p. 27-34. In A. Steinbüchel (ed.), Biochemical principles and mechanisms of biosynthesis and biodegradation of polymers. Wiley-VCH, Weinheim, Germany.
- 15.Dahl, C., N. M. Kredich, R. Deutzmann, and H. G. Trüper. 1993. Dissimilatory sulphite reductase from Archaeoglobus fulgidus: physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. J. Gen. Microbiol. 139:1817-1828. [DOI] [PubMed] [Google Scholar]
- 16.Dahl, C., G. Rákhely, A. S. Pott-Sperling, B. Fodor, M. Takács, A. Toth, M. Kräling, K. Györfi, Á. Kovács, J. Tusz, and K. Kovács. 1999. Genes involved in hydrogen and sulfur metabolism in phototrophic sulfur bacteria. FEMS Microbiol. Lett. 180:317-324. [DOI] [PubMed] [Google Scholar]
- 17.Debussche, L., D. Thibaut, B. Cameron, J. Crouzet, and F. Blanche. 1990. Purification and characterization of cobyrinic acid a,c-diamide synthase from Pseudomonas denitrificans. J. Bacteriol. 172:6239-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickman, M. J., S. M. Ingleston, S. E. Sedelnikova, J. B. Rafferty, R. G. Lloyd, J. A. Grasby, and D. P. Hornby. 2002. The RuvABC resolvasome. Eur. J. Biochem. 269:5492-5501. [DOI] [PubMed] [Google Scholar]
- 19.Dolla, A., J. K. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174:143-151. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, J. A., K. E. Nelson, L. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fekkes, P., and A. J. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fellay, R., J. Frey, and H. M. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vivo insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 23.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. Fitzhugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrman, N. S. M. Geohagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 24.Friedrich, C. G., A. Quentmeier, F. Bardischewsky, D. Rother, R. Kraft, S. Kostka, and H. Prinz. 2000. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182:4677-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagen, W. R., P. J. Silva, M. A. Amorim, P.-L. Hagedoorn, H. Wassink, H. Haaker, and F. T. Robb. 2000. Novel structure and redox chemistry of the prosthetic groups of the iron-sulfur flavoprotein sulfide dehydrogenase from Pyrococcus furiosus; evidence for a [2Fe-2S] cluster with Asp(Cys) ligands. J. Biol. Inorg. Chem. 5:527-534. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 27.Hanson, T. E., and F. R. Tabita. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. USA 98:4397-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson, T. E., and F. R. Tabita. 2003. Insights into the stress response and sulfur metabolism revealed by proteome analysis of a Chlorobium tepidum mutant lacking the Rubisco-like protein. Photosynth. Res. 78:231-248. [DOI] [PubMed] [Google Scholar]
- 29.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw, and C. M. Fraser. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554-559. [DOI] [PubMed] [Google Scholar]
- 30.Hensel, M., A. P. Hinsley, T. Nikolaus, G. Sawers, and B. C. Berks. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32:275-287. [DOI] [PubMed] [Google Scholar]
- 31.Hipp, W. M., A. S. Pott, N. Thum-Schmitz, I. Faath, C. Dahl, and H. G. Trüper. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate- reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891-2902. [DOI] [PubMed] [Google Scholar]
- 32.Hofman, K., and W. Stoffel. 1993. TMbase — a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166-171. [Google Scholar]
- 33.Hussain, H., J. Grove, L. Griffiths, S. Busby, and J. Cole. 1994. A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Mol. Microbiol. 12:153-163. [DOI] [PubMed] [Google Scholar]
- 34a.Johnson, T. 1998. M.S. thesis. Arizona State University, Tempe.
- 34.Imhoff, J. F., J. Süling, and R. Petri. 1998. Phylogenetic relationships among the Chromatiaceae, their taxonomic reclassification and description of the new genera Allochromatium, Halochromatium, Isochromatium, Marichromatium, Thiococcus, Thiohalocapsa, and Thermochromatium. Int. J. Syst. Bacteriol. 48:1129-1143. [DOI] [PubMed] [Google Scholar]
- 35.Jongsreejit, B., R. N. Z. A. Rahman, S. Fujiwara, and T. Imanaka. 1997. Gene cloning, sequencing and enzymtic properties of glutamate synthase from the hyperthermophilic archaeon Pyrococcus sp. KOD1. Mol. Gen. Genet. 254:635-642. [DOI] [PubMed] [Google Scholar]
- 36.Karkhoff-Schweizer, R. R., M. Bruschi, and G. Voordouw. 1993. Expression of the γ- subunit gene of desulfoviridin-type dissimilatory sulfite reductase and of the α- and β- subunit genes is not coordinately regulated. Eur. J. Biochem. 211:501-507. [DOI] [PubMed] [Google Scholar]
- 37.Karkhoff-Schweizer, R. R., D. P. W. Huber, and G. Voordouw. 1995. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl. Environ. Microbiol. 61:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly, D. P., J. K. Shergill, W. P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 71:95-107. [DOI] [PubMed] [Google Scholar]
- 39.Keon, R. G., R. Fu, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 40.Klenk, H.-P., R. A. Clayton, J.-F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 42.Larsen, O., T. Lien, and N. K. Birkeland. 2001. A novel organization of the dissimilatory sulfite reductase operon of Thermodesulforhabdus norvegica verified by RT-PCR. FEMS Microbiol. Lett. 203:81-85. [DOI] [PubMed] [Google Scholar]
- 43.Larsen, O., T. Lien, and N.-K. Birkeland. 1999. Dissimilatory sulfite reductase from Archaeoglobus profundus and Desulfotomaculum thermocisternum: phylogenetic and structural implications from gene sequences. Extremophiles 3:63-70. [DOI] [PubMed] [Google Scholar]
- 44.Larsen, O., T. Lien, and N.-K. Birkeland. 2000. Characterization of the desulforubidin operons from Desulfobacter vibrioformis and Desulfobulbus rhabdoformis. FEMS Microbiol. Lett. 186:41-46. [DOI] [PubMed] [Google Scholar]
- 45.Laue, H., M. Friedrich, J. Ruff, and A. M. Cook. 2001. Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183:1727-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mander, G. J., E. C. Duin, D. Linder, K. O. Stetter, and R. Hedderich. 2002. Purification and characterization of a membrane-bound enzyme complex from the sulfate-reducing archaeon Archaeoglobus fulgidus related to heterodisulfide reductase from methanogenic archaea. Eur. J. Biochem. 269:1895-1904. [DOI] [PubMed] [Google Scholar]
- 47.Mander, G. J., A. J. Pierik, H. Huber, and R. Hedderich. 2004. Two distinct heterodisulfide reductase-like enzymes in the sulfate-reducing archaeon Archaeoglobus profundus. Eur. J. Biochem. 271:1106-1116. [DOI] [PubMed] [Google Scholar]
- 48.Matthews, J. C., R. Timkovich, M. Y. Liu, and J. LeGall. 1995. Siroamide: a prosthetic group isolated from sulfite reductases in the genus Desulfovibrio. Biochemistry 34:5248-5251. [DOI] [PubMed] [Google Scholar]
- 49.Mizuno, N., G. Voordouw, K. Miki, A. Sarai, and Y. Higuchi. 2003. Crystal structure of dissimilatory sulfite reductase D (DsrD) protein — possible interaction with B- and Z- DNA by its winged helix motif. Structure (Cambridge) 11:1133-1140. [DOI] [PubMed] [Google Scholar]
- 50.Nakai, K., and M. Kaneshisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-111. [DOI] [PubMed] [Google Scholar]
- 51.Pattaragulwanit, K., D. C. Brune, H. G. Trüper, and C. Dahl. 1998. Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch. Microbiol. 169:434-444. [DOI] [PubMed] [Google Scholar]
- 52.Pattaragulwanit, K., and C. Dahl. 1995. Development of a genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch. Microbiol. 164:217-222. [Google Scholar]
- 53.Peck, H. D., Jr., and J. LeGall. 1982. Biochemistry of dissimilatory sulphate reduction. Philos. Trans. R. Soc. Lond. Ser. B 298:443-466. [DOI] [PubMed] [Google Scholar]
- 54.Pelanda, R., M. A. Vanoni, M. Perego, L. Piubelli, A. Galizzi, B. Curti, and G. Zanetti. 1993. Glutamate synthase genes of the diazotroph Azospirillum brasilense. Cloning, sequencing, and analysis of functional domains. J. Biol. Chem. 268:3099-3106. [PubMed] [Google Scholar]
- 55.Pereira, I. A. C., C. V. Romao, A. V. Xavier, J. LeGall, and M. Teixeira. 1998. Electron transfer between hydrogenases and mono- and multiheme cytochromes in Desulfovibrio ssp. J. Biol. Inorg. Chem. 3:494-498. [Google Scholar]
- 56.Pires, R. H., A. I. Lourenco, F. Morais, M. Teixeira, A. V. Xavier, L. M. Saraiva, and I. A. C. Pereira. 2003. A novel membrane-bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim. Biophys. Acta 1605:67-82. [DOI] [PubMed] [Google Scholar]
- 57.Pott, A. S., and C. Dahl. 1998. Sirohaem-sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144:1881-1894. [DOI] [PubMed] [Google Scholar]
- 58.Prange, A., R. Chauvistre, H. Modrow, J. Hormes, H. G. Trüper, and C. Dahl. 2002. Quantitative speciation of sulfur in bacterial sulfur globules: X-ray absorption spectroscopy reveals at least three different speciations of sulfur. Microbiology 148:267-276. [DOI] [PubMed] [Google Scholar]
- 59.Reinartz, M., J. Tschäpe, T. Brüser, H. G. Trüper, and C. Dahl. 1998. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170:59-68. [DOI] [PubMed] [Google Scholar]
- 60.Reynolds, R., R. M. Bermudez-Cruz, and M. J. Chamberlin. 1992. Parameters affecting transcription termination by Escherichia coli RNA. I. Analysis of 13 rho-independent terminators. J. Mol. Biol. 224:31-51. [DOI] [PubMed] [Google Scholar]
- 61.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146:551-571. [DOI] [PubMed] [Google Scholar]
- 62.Rossi, M., B. R. Pollock, M. W. Reiji, R. G. Keon, R. Fu, and G. Voordouw. 1993. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J. Bacteriol. 175:4699-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 64.Schedel, M., M. Vanselow, and H. G. Trüper. 1979. Siroheme sulfite reductase from Chromatium vinosum. Purification and investigation of some of its molecular and catalytic properties. Arch. Microbiol. 121:29-36. [Google Scholar]
- 65.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38-43. [DOI] [PubMed] [Google Scholar]
- 66.Seibl, R., H.-J. Höltke, R. Rüger, A. Meindl, H. G. Zauchau, R. Raßhofer, H. Wolf, N. Arnold, J. Wienberg, and C. Kessler. 1990. Nonradioactive labeling and detection of nucleic acids. Biol. Chem. Hoppe-Seyler 371:939-951. [DOI] [PubMed] [Google Scholar]
- 67.Shin, D. H., H. Yokota, R. Kim, and S.-H. Kim. 2002. Crystal structure of a conserved hypothetical protein from Escherichia coli. J. Struct. Funct. Genom. 14:53-66. [DOI] [PubMed] [Google Scholar]
- 68.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 69.Steger, J. L., C. Vincent, J. D. Ballard, and L. R. Krumholz. 2002. Desulfovibrio sp. genes involved in the respiration of sulfate during metabolism of hydrogen and lactate. Appl. Environ. Microbiol. 68:1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steuber, J., H. Cypionka, and P. M. H. Kroneck. 1994. Mechanism of dissimilatory sulfite reduction by Desulfovibrio desulfuricans. Purification of a membrane-bound sulfite reductase and coupling with cytochrome c3 and hydrogenase. Arch. Microbiol. 162:255-260. [Google Scholar]
- 71.Stock, A., S. Clarke, C. Clarke, and J. Stock. 1987. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 220:8-14. [DOI] [PubMed] [Google Scholar]
- 72.Towbin, H., T. L. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vanoni, M. A. and B. Curti. 1999. Glutamate synthase: a complex iron-sulfur flavoprotein. Cell. Mol. Life Sci. 55:617-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vargas, C., A. G. McEwan, and J. A. Downie. 1993. Detection of c-type cytochromes using enhanced chemiluminescence. Anal. Biochem. 209:323-326. [DOI] [PubMed] [Google Scholar]
- 75.Vermeglio, A., J. Li, B. Schoepp-Cothenet, N. Pratt, and D. B. Knaff. 2002. The role of high-potential iron protein and cytochrome c(8) as alternative electron donors to the reaction center of Chromatium vinosum. Biochemistry 41:8868-8875. [DOI] [PubMed] [Google Scholar]
- 76.Waksman, G., T. S. Krishna, C. H. J. Williams, and J. Kuriyan. 1994. Crystal structure of Escherichia coli thioredoxin reductase refined at 2 Å resolution. Implications for a large conformational change during catalysis. J. Mol. Biol. 236:800-816. [PubMed] [Google Scholar]
- 77.Wierenga, R. K., P. Terpstra, and W. G. J. Hol. 1988. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]