Abstract
The DsbA-DsbB pathway introduces disulfide bonds into newly translocated proteins. Conversion of the conserved cis proline 151 of DsbA to several hydrophilic residues results in accumulation of mixed disulfides between DsbA and its dedicated oxidant, DsbB. However, only a proline-to-threonine change causes accumulation of mixed disulfides of DsbA with its substrates.
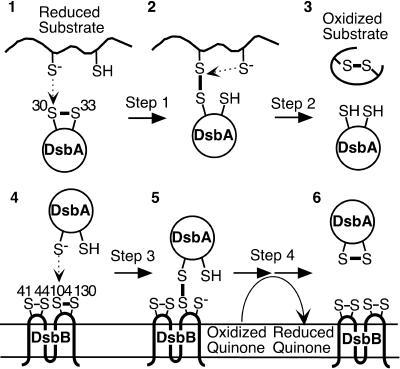
Formation of disulfide bonds is important for the folding, stability, and activity of many proteins exported out of the cytoplasm. In both prokaryotes and eukaryotes, proteins with a thioredoxin fold are responsible for this process (17, 24, 30). In the periplasm of Escherichia coli, DsbA, a thioredoxin superfamily member, introduces disulfide bonds directly into substrate proteins by donating the disulfide bond in its active-site Cys30-Pro31-His32-Cys33 to a pair of cysteines in substrate proteins (Fig. 1, stages 1 to 3) (3, 9, 19). DsbA, in turn, is maintained in its oxidized state (20) by a membrane protein DsbB, which uses its two pairs of essential cysteines (Cys41-Cys44 and Cys104-Cys130) to transfer electrons from DsbA to quinones in the respiratory chain (Fig. 1, stages 4 to 6) (1, 2, 13, 14, 16, 22, 28). Both of these reactions are thought to occur via formation of a mixed disulfide complex between Cys30 of DsbA and one of the cysteines of a substrate (for substrate oxidation) (Fig. 1, stage 2) (6, 7, 18, 33) and between Cys30 of DsbA and Cys104 of DsbB (for reoxidation of DsbA) (Fig. 1, stage 5) (11, 13, 16, 21).
FIG. 1.
Different stages of DsbA function. See main text for description.
We have previously described two mutations which altered residue Pro151 of DsbA and resulted in defects in two different steps in the disulfide bond formation pathway (18). One mutant, P151T, appears to slow down step 2 of the reaction of DsbA with substrate (Fig. 1), thus allowing detection of DsbA in the act of substrate oxidation (Fig. 1, stage 2). The other mutant, P151S, interferes with the transfer of electrons from DsbA to DsbB, accumulating a large amount of a mixed disulfide complex of DsbA and DsbB, a presumed intermediate in the process of reoxidation of DsbA by DsbB (Fig. 1, stage 5) (8, 13, 16). This alteration appears to cause a defect in a step required for resolution of the DsbA-DsbB complex. In DsbA, Pro151 assumes the cis configuration and is positioned very close to the active-site cysteines of DsbA (5, 10, 24, 25).
These mutants are quite useful for identifying intermediates in the enzymatic reactions of DsbA, for identifying the substrates of DsbA, and for analyzing details of the electron transfer pathway. Since a similarly positioned proline is found in nearly all proteins containing thioredoxin-like domains (12, 24, 26), the characterization of similar mutants for these other proteins may be equally useful. In order to begin to determine the utility of this approach, we have examined the effects of altering Pro151 of DsbA to each of the remaining 17 amino acids. We did this in order to see which other amino acid changes, if any, in this residue would give similar phenotypes. Our results may be helpful in the study of other members of the thioredoxin superfamily.
Plasmid constructions.
To construct plasmids that express each of the DsbA mutants, substitution mutations were introduced into the dsbA gene of plasmid pHK520 by using a QuikChange site-directed mutagenesis kit (Stratagene) and appropriate mutagenic primers (Table 1). The plasmid pHK520 is a pSC101-derived low-copy-number plasmid carrying dsbA under the lac promoter. This plasmid was constructed by inserting the DsbA-encoding 0.7-kb KpnI-XbaI fragment of pCH3 (11) into pAM238 (pSC101 ori, Specr, lac promoter) (16). Importantly, when the cells were grown on M63 minimal glucose medium (15), pHK520 expressed DsbA at levels comparable to those of DsbA from the chromosome (data not shown). This level of expression is crucial to the success of such studies. We have found that even slight increases in expression of DsbA over wild-type levels can ameliorate or eliminate the phenotypic effects of interesting mutants (V. C. Tam, H. Kadokura, and J. Beckwith, unpublished results).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or features | Reference or source |
|---|---|---|
| Strains | ||
| HK295 | F−Δara-714 galU galK Δ(lac)X74 rpsL thi | 16 |
| HK317 | HK295 ΔdsbA | 18 |
| HK320 | HK295 ΔdsbB | 16 |
| Plasmids | ||
| pAM238 | lac promoter, pSC101-based, Spcr | 16 |
| pDSW206 | Mutations in trc promoter of pTrc99A, pBR322 based, Blar, lacIq | 32 |
| pHK520 | pAM238 DsbA | This study |
| pHK651 | pAM238 DsbA P151T | This study |
| pHK652 | pAM238 DsbA P151S | This study |
| pHK653 | pAM238 DsbA P151A | This study |
| pHK675 | pDSW206 ΔlacIq | This study |
| pHK677 | pHK675 RcsF-c-Myc | This study |
| pLN101 | pAM238 DsbA P151F | This study |
| pLN102 | pAM238 DsbA P151L | This study |
| pLN103 | pAM238 DsbA P151I | This study |
| pLN104 | pAM238 DsbA P151M | This study |
| pLN105 | pAM238 DsbA P151V | This study |
| pLN106 | pAM238 DsbA P151Y | This study |
| pLN107 | pAM238 DsbA P151H | This study |
| pLN108 | pAM238 DsbA P151Q | This study |
| pLN111 | pAM238 DsbA P151N | This study |
| pLN112 | pAM238 DsbA P151K | This study |
| pLN113 | pAM238 DsbA P151D | This study |
| pLN114 | pAM238 DsbA P151E | This study |
| pLN115 | pAM238 DsbA P151C | This study |
| pLN116 | pAM238 DsbA P151W | This study |
| pLN117 | pAM238 DsbA P151R | This study |
| pLN118 | pAM238 DsbA P151G | This study |
Effects of Pro151 mutations on ability of DsbA to oxidize substrate proteins.
To examine the effect of mutations on the ability of DsbA to oxidize substrates, we assessed the oxidative state of two substrates of DsbA (β-lactamase and RcsF) (18) in the mutants. To detect RcsF, we fused it with c-Myc at its C terminus. Plasmid pHK677, which was used to express both β-lactamase and RcsF-c-Myc, was constructed by inserting the RcsF-c-Myc-encoding 460-bp KpnI-XbaI fragment of pHK646 (18) into pHK675 (pBR322 ori, bla, weakened lac promoter). The expression vector pHK675 was generated by deleting the lacIq-containing 872-bp BssHII fragment from pDSW206 (pBR322 ori, bla, weakened trc promoter, lacIq) (32).
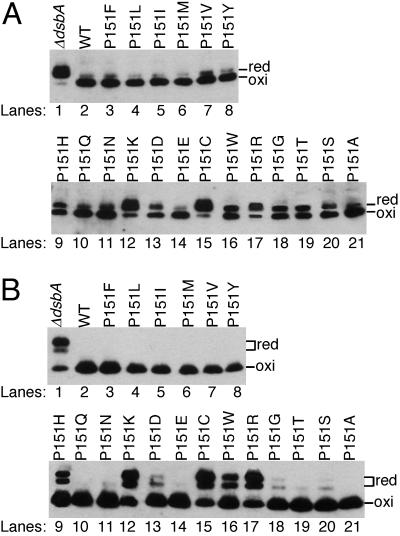
We transformed E. coli strain HK317 (ΔdsbA) with both pHK677 (carrying bla and RcsF-c-Myc) and each of the dsbA mutant plasmids and examined the abilities of the mutants to promote disulfide bond formation in β-lactamase (Fig. 2A) and RcsF-c-Myc (Fig. 2B). To distinguish the oxidized (disulfide-bonded) form from the reduced form of β-lactamase and RcsF-c-Myc by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), cellular proteins were first treated with acid to inhibit thiol-disulfide reactivity and then the free cysteines were alkylated with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (15). This modification retards the mobility of the reduced forms of proteins on gels. In the ΔdsbA strain expressing the wild-type dsbA from the plasmid, both β-lactamase and RcsF-c-Myc were completely oxidized (Fig. 2, lane 2). However, in the absence of the dsbA plasmid, the substrate proteins were mostly reduced (Fig. 2, lane 1), confirming that both proteins are the substrates of DsbA.
FIG. 2.
Capabilities of the DsbA P151 mutants to oxidize the substrate proteins in vivo. Strain HK317 (ΔdsbA) was transformed with both pHK677 (carrying bla and rcsF-c-Myc) and one of the following plasmids: pAM238 (empty vector; lane 1), pHK520 (wild-type DsbA; lane 2), pLN101 (P151F; lane 3), pLN102 (P151L; lane 4), pLN103 (P151I; lane 5), pLN104 (P151M; lane 6), pLN105 (P151V; lane 7), pLN106 (P151Y; lane 8), pLN107 (P151H; lane 9), pLN108 (P151Q; lane 10), pLN111 (P151N; lane 11), pLN112 (P151K; lane 12), pLN113 (P151D; lane 13), pLN114 (P151E; lane 14), pLN115 (P151C; lane 15), pLN116 (P151W; lane 16), pLN117 (P151R; lane 17), pLN118 (P151G; lane 18), pHK651 (P151T; lane 19), pHK652 (P151S; lane 20), and pHK653 (P151A; lane 21). Cells were grown at 30°C in M63 minimal glucose medium, and cellular proteins from the exponential culture were alkylated with AMS and separated with SDS-PAGE. Disulfide bond formation in β-lactamase (Bla; two cysteines) (A) and RcsF-c-Myc (RcsF; four cysteines) (B) was visualized with Western blotting by using anti-Bla (5 Prime 3 Prime, Inc., Boulder, Colo.) and anti-c-Myc (A-14; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) antibodies.
Three mutants (P151K, P151C, and P151R) showed strong defects, leaving a large portion of the substrates in their reduced forms (Fig. 2, lanes 12, 15, and 17) and exhibiting phenotypes that suggest defects in multiple steps of the enzyme reaction (data not shown). One of these, P151C, introduces a third cysteine into the active site region of the protein and may, thus, interfere with the normal redox activity of the protein. The other two, P151R and P151K, introduce basic amino acids into the region, which could alter the ionic state of the cysteines in reduced DsbA (4, 23) and/or disrupt the structure of this region.
However, the rest of the mutants showed weak or no detectable defects in substrate oxidation: they oxidized more than 50% of the two substrates. Notably, the P151F, P151L, P151I, P151M, P151V, P151Y, P151Q, P151E, and P151A mutants oxidized substrates almost as well as the wild-type enzyme (Fig. 2).
In many of the mutants, defects in disulfide bond formation were more pronounced in β-lactamase than in RcsF-c-Myc (e.g., P151S [Fig. 2, lane 20]). This observation may be explained by the fact that β-lactamase can rapidly fold into its active folded structure even in the absence of disulfide bond formation and, once folded, its cysteines are inaccessible to DsbA (3).
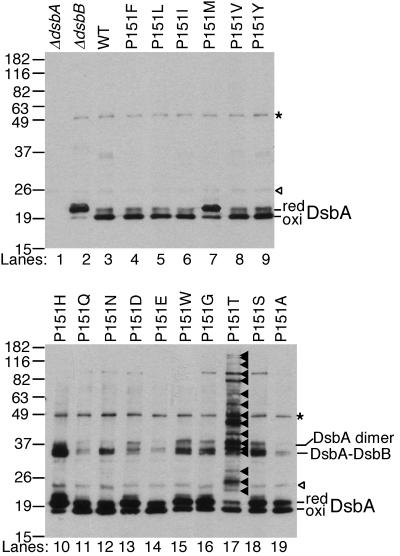
To characterize the 16 mutants that showed substantial in vivo activity, we examined the redox state of DsbA by probing the AMS-alkylated lysate with antibody to DsbA (Fig. 3). We made the following observations on the implications of the combined data for this collection of mutants.
FIG. 3.
In vivo redox states of the DsbA mutants. AMS-alkylated lysates of the following strains were separated by SDS-PAGE and analyzed with Western blotting using antibody to DsbA (15). Lanes: 1, HK317/pAM238; 2, HK320 (ΔdsbB)/pAM238; 3, HK317/pHK520; 4, HK317/pLN101; 5, HK317/pLN102; 6, HK317/pLN103; 7, HK317/pLN104; 8, HK317/pLN105; 9, HK317/pLN106; 10, HK317/pLN107; 11, HK317/pLN108; 12, HK317/pLN111; 13, HK317/pLN113; 14, HK317/pLN114; 15, HK317/pLN116; 16, HK317/pLN118; 17, HK317/pHK651; 18, HK317/pHK652; 19, HK317/pHK653. Open arrowheads, nonspecific bands; asterisks, a mixed-disulfide complex formed between DsbA and another protein (H. Kadokura and J. Beckwith, unpublished results); closed arrowheads in lane 17, DsbA-substrate complexes.
P151T.
This amino acid alteration, characterized previously (18), is the only one of all those tested here to cause a significant defect in resolution of DsbA-substrate complexes (Fig. 1, step 2). The mutant strain, thus, accumulates numerous DsbA-substrate complexes (Fig. 3, lane 17) (18). The P151T and P151S proteins, which exhibit very different phenotypes, showed the same standard redox potential of −152 and −151 mV, respectively, indicating that the redox potentials alone cannot explain the phenotypes of these two mutations (18). Thus, some structural feature resulting from this specific amino acid change appears to prevent the second cysteine of the substrate from resolving this complex (12, 18).
P151H, P151S, P151N, P151W, and P151G.
These five mutants (P151S was characterized previously [18]) accumulate a major band with an apparent molecular mass of 36 kDa, in addition to the reduced and oxidized forms of DsbA (Fig. 3, lanes 10, 12, 15, 16, and 18). Since this band was recognized by anti-DsbB antibody and disappeared when samples were treated with reductant (data not shown), it represents a mixed disulfide complex between DsbB and DsbA. Further coexpression of wild-type DsbA from the chromosome to compete for oxidation of substrate proteins resulted in disappearance of most of the complex (data not shown), indicating that the DsbA-DsbB complex accumulates when mutant DsbA is actively oxidizing substrate proteins. Thus, the DsbA-DsbB complex forms in the process of oxidation of DsbA by DsbB.
The P151H and P151S changes result in the greatest accumulation of DsbA-DsbB complex. The side chains of serine, threonine, or histidine residues each have the potential to form a hydrogen bond with the sulfhydryl group of a cysteine residue. Such bonding, in the context of a DsbA-DsbB or DsbA-substrate complex, may alter resolvability of mixed disulfides.
P151F, P151L, P151I, P151V, P151Y, and P151M.
Five of these six hydrophobic substitutions exhibited the least effect on DsbA activity, showing efficient oxidation of substrates (Fig. 2) and no alteration of the redox state of DsbA (Fig. 3). Nevertheless, they, as well as the other substitution mutants, did show varying degrees of hypersensitivity to Cd2+, a phenotype typical of dsbA and dsbB mutants (data not included) (29, 31). In the structures that have been reported for mixed disulfide complexes between human thioredoxin and two substrate peptides, the ring of the analogous proline forms van der Waals contacts with the sulfur of the cysteine of the substrate involved in the mixed disulfide bond (27). Thus, the hydrophobic amino acids substituted for DsbA's Pro151 may suffice for formation of similar contacts. These contacts may be important for the proper DsbA-DsbB interactions (see below; P151M) and resolution of mixed disulfides between either DsbA and substrate or DsbA and DsbB.
The exception to the lack of effects of the hydrophobic substitutions is the methionine substitution, which accumulates a larger amount of the reduced form of DsbA without accumulation of DsbA-DsbB complex (P151M; Fig. 3, lane 7), suggesting a defect in a step required for DsbA-DsbB complex formation. This difference may be related to the large size of the methionine residue.
Concluding remarks.
Our previous work suggested that mutating the conserved cis proline residue of thioredoxin superfamily members might be useful for studies on the mechanisms involved in electron transfer pathways, particularly in the detection of mixed disulfide intermediates in these pathways. Here we have shown that alteration of proline 151 to five other amino acid residues causes the accumulation of mixed disulfide complexes between DsbA and its dedicated oxidant DsbB. Thus, the specificity of the defect is not limited to a single specific mutational alteration, and such mutants may have similarly useful properties in other systems. The same is not true for alterations that cause accumulation of DsbA-substrate complexes, where only the P151T change shows such a phenotype. Further studies with other thioredoxin family members will be necessary to determine whether the change from proline to threonine has similar effects, whether other changes might work, or whether the phenomenon is only seen in a subset of these proteins.
Acknowledgments
We are grateful to Peter Metcalf and George Georgiou for comments on the manuscript. We also thank the lab members for suggestions and discussion.
This work was supported by NSF grant DBI-0243489 to L.N., NIH grant GM41883 to J.B., and in part by the Leadership Alliance to L.N. J.B. is an American Cancer Society Research Professor.
REFERENCES
- 1.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 2.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 4.Blank, J., T. Kupke, E. Lowe, P. Barth, R. B. Freedman, and L. W. Ruddock. 2003. The influence of His94 and Pro149 in modulating the activity of V. cholerae DsbA. Antioxid. Redox Signal. 5:359-366. [DOI] [PubMed] [Google Scholar]
- 5.Charbonnier, J. B., P. Belin, M. Moutiez, E. A. Stura, and E. Quéméneur. 1999. On the role of the cis-proline residue in the active site of DsbA. Protein Sci. 8:96-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darby, N. J., and T. E. Creighton. 1995. Catalytic mechanism of DsbA and its comparison with that of protein disulfide isomerase. Biochemistry 34:3576-3587. [DOI] [PubMed] [Google Scholar]
- 7.Frech, C., M. Wunderlich, R. Glockshuber, and F. X. Schmid. 1996. Preferential binding of an unfolded protein to DsbA. EMBO J. 15:392-398. [PMC free article] [PubMed] [Google Scholar]
- 8.Grauschopf, U., A. Fritz, and R. Glockshuber. 2003. Mechanism of the electron transfer catalyst DsbB from Escherichia coli. EMBO J. 22:3503-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grauschopf, U., J. R. Winther, P. Korber, T. Zander, P. Dallinger, and J. C. Bardwell. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947-955. [DOI] [PubMed] [Google Scholar]
- 10.Guddat, L. W., J. C. Bardwell, T. Zander, and J. L. Martin. 1997. The uncharged surface features surrounding the active site of Escherichia coli DsbA are conserved and are implicated in peptide binding. Protein Sci. 6:1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilhot, C., G. Jander, N. L. Martin, and J. Beckwith. 1995. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc. Natl. Acad. Sci. USA 92:9895-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heras, B., M. A. Edeling, H. J. Schirra, S. Raina, and J. L. Martin. 2004. Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide. Proc. Natl. Acad. Sci. USA 101:8876-8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inaba, K., Y. H. Takahashi, and K. Ito. 2004. DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J. Biol. Chem. 279:6761-6768. [DOI] [PubMed] [Google Scholar]
- 14.Jander, G., N. L. Martin, and J. Beckwith. 1994. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 13:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadokura, H., M. Bader, H. Tian, J. C. Bardwell, and J. Beckwith. 2000. Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:10884-10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadokura, H., and J. Beckwith. 2002. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 21:2354-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111-135. [DOI] [PubMed] [Google Scholar]
- 18.Kadokura, H., H. Tian, T. Zander, J. C. Bardwell, and J. Beckwith. 2004. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303:534-537. [DOI] [PubMed] [Google Scholar]
- 19.Kamitani, S., Y. Akiyama, and K. Ito. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishigami, S., Y. Akiyama, and K. Ito. 1995. Redox states of DsbA in the periplasm of Escherichia coli. FEBS Lett. 364:55-58. [DOI] [PubMed] [Google Scholar]
- 21.Kishigami, S., E. Kanaya, M. Kikuchi, and K. Ito. 1995. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J. Biol. Chem. 270:17072-17074. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, T., S. Kishigami, M. Sone, H. Inokuchi, T. Mogi, and K. Ito. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 94:11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lappi, A. K., M. F. Lensink, H. I. Alanen, K. E. Salo, M. Lobell, A. H. Juffer, and L. W. Ruddock. 2004. A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J. Mol. Biol. 335:283-295. [DOI] [PubMed] [Google Scholar]
- 24.Martin, J. L. 1995. Thioredoxin—a fold for all reasons. Structure 3:245-250. [DOI] [PubMed] [Google Scholar]
- 25.Martin, J. L., J. C. Bardwell, and J. Kuriyan. 1993. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature 365:464-468. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy, A. A., P. W. Haebel, A. Torronen, V. Rybin, E. N. Baker, and P. Metcalf. 2000. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat. Struct. Biol. 7:196-199. [DOI] [PubMed] [Google Scholar]
- 27.Qin, J., G. M. Clore, W. P. Kennedy, J. Kuszewski, and A. M. Gronenborn. 1996. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure 4:613-620. [DOI] [PubMed] [Google Scholar]
- 28.Regeimbal, J., S. Gleiter, B. L. Trumpower, C. A. Yu, M. Diwakar, D. P. Ballou, and J. C. Bardwell. 2003. Disulfide bond formation involves a quinhydrone-type charge-transfer complex. Proc. Natl. Acad. Sci. USA 100:13779-13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rensing, C., B. Mitra, and B. P. Rosen. 1997. Insertional inactivation of dsbA produces sensitivity to cadmium and zinc in Escherichia coli. J. Bacteriol. 179:2769-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sevier, C. S., and C. A. Kaiser. 2002. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 3:836-847. [DOI] [PubMed] [Google Scholar]
- 31.Stafford, S. J., D. P. Humphreys, and P. A. Lund. 1999. Mutations in dsbA and dsbB, but not dsbC, lead to an enhanced sensitivity of Escherichia coli to Hg2+ and Cd2+. FEMS Microbiol. Lett. 174:179-184. [DOI] [PubMed] [Google Scholar]
- 32.Weiss, D. S., J. C. Chen, J.-M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zapun, A., J. C. Bardwell, and T. E. Creighton. 1993. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32:5083-5092. [DOI] [PubMed] [Google Scholar]