Abstract
Burkholderia cenocepacia is an opportunistic bacterium that infects patients with cystic fibrosis. B. cenocepacia strains J2315, K56-2, C5424, and BC7 belong to the ET12 epidemic clone, which is transmissible among patients. We have previously shown that transposon mutants with insertions within the O antigen cluster of strain K56-2 are attenuated for survival in a rat model of lung infection. From the genomic DNA sequence of the O antigen-deficient strain J2315, we have identified an O antigen lipopolysaccharide (LPS) biosynthesis gene cluster that has an IS402 interrupting a predicted glycosyltransferase gene. A comparison with the other clonal isolates revealed that only strain K56-2, which produced O antigen and displayed serum resistance, lacked the insertion element inserted within the putative glycosyltransferase gene. We cloned the uninterrupted gene and additional flanking sequences from K56-2 and conjugated this plasmid into strains J2315, C5424, and BC7. All the exconjugants recovered the ability to form LPS O antigen. We also determined that the structure of the strain K56-2 O antigen repeat, which was absent from the LPS of strain J2315, consisted of a trisaccharide unit made of rhamnose and two N-acetylgalactosamine residues. The complexity of the gene organization of the K56-2 O antigen cluster was also investigated by reverse transcription-PCR, revealing several transcriptional units, one of which also contains genes involved in lipid A-core oligosaccharide biosynthesis.
The Burkholderia cepacia complex comprises a group of phenotypically similar species which are ubiquitous in nature, as they can be found in the rhizosphere, in fresh and marine water, and in association with plants, animals, and humans (11). Human infections with B. cepacia complex are rare in immunocompetent persons, but they can be devastating in patients with cystic fibrosis (CF) and chronic granulomatous disease (24, 25). Isolates representing all B. cepacia complex species have been recovered from the sputum of CF patients (11), yet some species are more common than others. For instance, B. cenocepacia (formerly genomovar III) isolates comprise about 83% of all B. cepacia complex strains isolated from CF patients in Canada (54) and 50% of the isolates in the United States (41), while the rates of incidence of B. multivorans (formerly genomovar II) in the two countries are 10 and 38%, respectively. Infections with B. cenocepacia in CF patients are often associated with a poor clinical recovery, and some specific strains may be associated with increased rates of morbidity and mortality (10).
Genotyping analyses have demonstrated the existence of clonal lineages of B. cenocepacia transmissible strains that infect multiple CF patients. For instance, the ET12 clone predominates among CF patients in Canada and the United Kingdom (23, 33), and infections with these strains have been associated with high rates of mortality (31, 55). In addition, the PHDC strain has been described as the dominant isolate among infected CF patients in the mid-Atlantic region of the United States (6), while the “Midwest” clone was found in multiple CF patients in Ohio and Michigan (9, 36).
B. cepacia complex isolates are resistant to most clinically useful antimicrobial agents (1, 47). They also overcome the bactericidal effects of components of the innate immune system such as antimicrobial peptides (18), lysozyme, lactoferrin, and phospholipase A2, all of which are secreted by airway epithelial cells (3). In addition, some isolates have the ability to survive intracellularly within macrophages (45, 51), respiratory epithelial cells (4, 34), and amoebae (37, 43). A plethora of potential virulence factors in B. cepacia complex isolates have been described. They include cable pili (52), flagella (57), a type III secretion system (56), surface exopolysaccharide (7), production of melanin (61), catalase (38), up to four types of iron-chelating siderophores (15), proteases and other secreted enzymes (12, 30, 59), quorum sensing systems (27, 40), and the ability to form biofilms (28).
The clinical isolates K56-2, J2315, C5424, and BC7 belong to the ET12 clone, as previously demonstrated by macrorestriction and randomly amplified polymorphic DNA analyses (42). Using a modified signature-tagged mutagenesis technique, we have recently identified over 100 transposon mutants of B. cenocepacia K56-2 that were attenuated for survival in a rat model of chronic lung infection (29). The available genomic sequence of strain J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/) was used to annotate the regions in strain K56-2 flanking the transposon insertions in the survival-defective mutants (29). Among the K56-2 survival-defective mutants, four had insertions in genes of an O antigen synthesis cluster. However, when the DNA sequence of the O antigen lipopolysaccharide (LPS) synthesis cluster in strain J2315 was analyzed, we found an IS402 element inserted within a putative glycosyltransferase gene. We hypothesized that this insertion was responsible for the lack of O antigen production by strain J2315 (19). In this study, we demonstrate that strain K56-2 lacks this insertion sequence (IS) element in the O antigen biosynthesis gene cluster and that the cloned glycosyltransferase gene can restore O antigen synthesis in strains J2315, BC7, and C5424. We also show that strain K56-2 produces an O antigen with a structure identical to that of a minor O antigen found in B. cepacia serotype 4 and report for the first time the transcriptional organization of the O antigen synthesis cluster in ET12 strains. Unlike J2315 and some of the K56-2 transposon mutants defective in O antigen synthesis, we also demonstrate that strain K56-2 is resistant to the bactericidal effect of serum complement.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. cenocepacia strains K56-2, J2315, BC7, and C5424 belong to the ET12 clone. These strains were isolated from patients with CF and in different geographic locations. The construction of the library of transposon mutants in K56-2, the details of the signature-tagged mutagenesis screen, and the identification of the K56-2 transposon mutants 33H3, 32D2, 38C2, and 34D8 with transposon insertions within the O antigen biosynthesis cluster were described elsewhere (29). For cloning experiments, we used the Escherichia coli K-12 strain DH5α [F− φ80lacZ M15 endA recA hsdR(rK−mK−) supE thi gyrA relA Δ(lacZYA-argF)U169]. Bacteria were grown at 37°C in Luria-Bertani (LB) medium supplemented, as required, with 100 μg of trimethoprim (Tp) ml−1 and 50 μg of gentamicin (Gm) ml−1 for B. cenocepacia and 50 μg of Tp ml−1 or 40 μg of kanamycin ml−1 for E. coli.
Serum sensitivity assay.
Bacteria were grown 18 h on LB or Trypticase soy agar with 100 μg of Tp ml−1 as required. Cultures were diluted in phosphate-buffered saline (pH 7.0) supplemented with 1% proteose peptone to a final concentration of 107 CFU/ml. Equal volumes of culture and pooled human serum from healthy volunteers (40% final concentration diluted in phosphate-buffered saline with 1% proteose peptone) were mixed and incubated with shaking at 37°C for 2 h. Cultures with no serum and with 40% serum inactivated by incubation at 56°C for 1 h were used as controls. Aliquots were serially diluted, plated on Trypticase soy agar, and incubated at 37°C for 24 to 48 h.
PCR for detection of the presence of an IS element interrupting the wbxE gene.
The primers used to amplify the region spanning IS402 in the O antigen biosynthesis gene cluster were P1123 (5′-TCGCGACCTCGAGAATGCCG-3′) and P1124 (5′-GTGAAACACCGCAGGCTTGATCG-3′), which annealed to regions immediately flanking the insertion element. PCR amplifications were carried out with Taq polymerase (Roche Diagnostics, Laval, Quebec) and GC resolution buffer (GC-rich PCR system; Roche Diagnostics) in a PTC-0200 DNA engine (MJ Research, Incline Village, Nev.). The conditions for the amplification were 2 min at 95°C, 24 cycles of 95°C for 1 min, 55°C for 35 s, and 72°C for 2 min, and a final extension of 7 min at 72°C. The PCR product was visualized in a 0.7% (wt/vol) agarose gel.
Molecular cloning.
The wbxD and wbxE genes were amplified from B. cenocepacia K56-2 chromosomal DNA by PCR using Pwo polymerase (Roche Diagnostics) and Q solution (Taq DNA polymerase; QIAGEN Inc., Valencia, Calif.). The primers used, P1163 (5′-GCTCTAGATACGCTGTCAGCCGTGCC-3′) and P1164 (5′-GGAATTCCATATGCCGCGATACCAAAAATTTTTGTTCTTTGC-3′), were made on the basis of the published sequence of B. cenocepacia J2315. The amplification conditions were 4 min at 95°C (the enzyme was added at 2.5 min), 28 cycles of 95°C for 40 s, 65.8°C for 40 s, and 72°C for 3 min, and a final extension of 7 min at 72°C. The resulting 3.3-kb product was digested with NdeI and XbaI and ligated to the vector pSCRhaB3. The construction of this expression vector will be reported elsewhere (S. T. Cardona and M. A. Valvano, unpublished data). Transformants carrying recombinant plasmids with the DNA insert were screened by colony PCR using either primers 816 (5′-CGGCATGGGGTCAGGTGGGACCACC-3′) and 824 (5′-GCCCATTTTCCTGTCAGTAACGAGA-3′), which anneal to vector sequences flanking the cloning sites, or primers 824 and 1124. One of these isolated plasmids, designated pXO3, was further examined by digestion with EcoRI to confirm the presence of the DNA insert encoding the wbxE and wbxD genes under the control of the PRHA promoter. The plasmid pXO4, carrying only wbxE, was obtained by digestion of pXO3 with StuI and Asp700 followed by ligation, which resulted in the deletion of the wbxD gene. The plasmid pXO7, carrying only the wbxD gene, was obtained by digestion of pXO3 with NdeI and XhoI followed by treatment with mung bean nuclease and ligation, which resulted in the deletion of the wbxE gene.
Plasmid conjugation into B. cenocepacia.
Plasmids pXO3, pXO4, pXO7, and pSCRhaB3 were conjugated into B. cenocepacia strains J2315, C5424, K56-2, and BC7 by triparental matings (14) using E. coli DH5α(pRK2013) as a helper strain (21). Exconjugants were selected on LB agar plates supplemented with 100 μg of Tp ml−1 and 50 μg of Gm ml−1. In the case of BC7, selection required 500 μg of Tp ml−1 and 50 μg of Gm ml−1.
Construction of additional mutations in the O antigen biosynthesis cluster of B. cenocepacia K56-2.
We constructed polar mutations in wecA and wxz genes by a single-crossover recombination strategy. Briefly, an internal fragment of 300 bp from the coding region of each gene was cloned into the suicide plasmid pGpΩTp to provide homology region for recombination. The construction of this vector will be reported elsewhere (R. S. Flannagan and M. A. Valvano, unpublished data). The resulting plasmid was conjugated into strain K56-2 by triparental mating as described above, and exconjugants containing the integrated mutagenic plasmid were selected on LB agar plates supplemented with 100 μg of Tp ml−1 and 50 μg of Gm ml−1. The presence of the correct insertion was verified by PCR analysis.
Total RNA isolation and RT-PCR analysis.
Total RNA was isolated from the B. cenocepacia strain K56-2 by the method described by Glisin et al. (22). Briefly, B. cenocepacia K56-2 was grown in 25 ml of LB medium to an optical density at 600 nm of 0.6. Bacterial cells were collected and resuspended in 3.5 ml of RNase-free TESS buffer (20 mM Tris [pH 7.6], 10 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate-polyacrylamide gel electrophoresis[SDS]) and lysed by heating at 95°C for 3 min. Cesium chloride powder was added to achieve a final concentration of 1 g ml−1, and the lysate was deposited on top of a CsCl cushion prepared in SW50.1 tubes (Beckman Coulter, Fullerton, Calif.). The cell lysate was centrifuged at 39,000 rpm for 16 h at 20°C in a SW50.1 rotor, and the RNA was recovered as a clear pellet at the bottom of the tube. The RNA pellet was dissolved in 20 mM sodium acetate and 1 mM EDTA, ethanol precipitated, and kept frozen at −20°C. An aliquot of the precipitated RNA was centrifuged, and the pellet was ethanol washed, dried, resuspended in RNase-free water, and treated with DNase I. The DNase I was eliminated by the cleanup protocol of an RNeasy mini kit (QIAGEN), and this RNA was used for the reverse transcription-PCR (RT-PCR) analysis.
Reverse transcription was performed using a Transcriptor reverse transcriptase kit (Roche Diagnostics) with the primers listed in Table 1. The resulting cDNA was subjected to PCR using Taq DNA polymerase (Roche Diagnostics). The conditions for the amplification were 10 cycles of 2 min at 94°C, 10 s at 94°C, 30 s at 54°C, and 2 min at 72°C followed by 30 cycles of 10 s at 94°C, 30 s at 59°C, and 2 min at 72°C and a final extension of 7 min at 72°C.
TABLE 1.
Primers used for RT-PCR analysis
| Primer | Primer sequence | Amplified region(s)a |
|---|---|---|
| P1249 | 5′-TCGATCAGCGGCTGCGCGGC-3′ | 1 |
| P1250 | 5′-GCTGAACCTGCACGACGGGA-3′ | 1 |
| P1251 | 5′-GAGAACGGCAGCACGTTGCG-3′ | 2 |
| P1252 | 5′-CTGGCTCGACGAAGGCGAGA-3′ | 2 |
| P1255 | 5′-GGCACCGGGCAAGGCTACAG-3′ | 3 |
| P1256 | 5′-CAGATCCTCGACGAGACCGG-3′ | 3 |
| P1238 | 5′-GCGCCAGCGTGAGCACTGCA-3′ | 4 |
| P1239 | 5′-GTCGCCGATGACGATGCGCC-3′ | 4, 5 |
| P1241 | 5′-GTCGCGCCGCCGATGACCCG-3′ | 5 |
| P1240 | 5′-CGAGGAGCGCCTTGTCCGGGT-3′ | 6 |
| P1243 | 5′-GCCGGGCAAATGCCGTCGAT-3′ | 6 |
| P1244 | 5′-AATCACCTGCACGACGGGCG-3′ | 7 |
| P1245 | 5′-GGTCCGTTCTCATCCCCGGC-3′ | 7 |
| P1257 | 5′-CACGAACAGCACGACGGCGA-3′ | 8 |
| P1258 | 5′-CGCGGTACGACGACATGGCA-3′ | 8 |
Numbers indicate the regions of the wbc cluster amplified by the primer pairs as indicated in Fig. 1B.
For each PCR, the appropriate controls with water and cDNA synthesized in the absence of reverse transcriptase were included to ensure that the amplifications obtained were a result of cDNA and not of contaminating genomic DNA.
Small-scale LPS preparation and SDS-polyacrylamide gel electrophoresis analysis.
LPS was extracted as described previously (44). Briefly, cells from overnight plate cultures were suspended in a lysis buffer containing proteinase K and incubated for 16 h at 60°C followed by a hot phenol extraction and a subsequent extraction of the aqueous phase with ether. LPS was resolved by electrophoresis in 14% polyacrylamide gels with a Tricine-SDS system (39, 53) and visualized by silver staining (44). For Western blotting, electrophoresed LPS samples were transferred to nitrocellulose membranes. The membranes were incubated with a 1:1,000 dilution of a rabbit polyclonal anti-B. cepacia O4 serum, kindly provided by H. Monteil (26), followed with an anti-rabbit monoclonal antibody conjugated to alkaline phosphatase (Sigma, St. Louis, Mo.) and detection with 5-bromo,4-chloro,3-indolylphosphate-nitroblue tetrazolium substrate for alkaline phosphatase-based detection systems (Sigma).
LPS purification and structural analysis.
B. cenocepacia strains K56-2 and J2315 were grown in 1 liter of LB medium at 37°C for 24 h, and LPS was purified by the Darveau-Hancock method as described previously (16). The quality of the purified LPS was confirmed by Tricine-SDS-polyacrylamide gel electrophoresis as described above. To elucidate the chemical structure of the B. cenocepacia O antigen polysaccharides, 20 mg of the purified LPS was hydrolyzed in 2% (vol/vol) acetic acid at 100°C for 2 h. The water-soluble products were fractionated on a BioGel P-2 column (Bio-Rad, Hercules, Calif.), yielding a polymeric material with a broad elution profile. This material was refractionated on a long BioGel P-10 column to reduce core constituents from the O antigen.
The compositions of the LPS and isolated O antigen polysaccharide were determined by the preparation and analysis of trimethylsilyl (TMS) methyl glycosides (60). Briefly, the samples were methanolysed by 1 M methanolic-HCl at 80°C for 18 h followed by re-N-acetylation of methyl glycosides by use of pyridine-acetic anhydride in the presence of methanol at 100°C for 1 h. The free hydroxyl groups of re-N-acetylated methylglycosides were trimethyl sialylated using Tri-Sil reagent (Pierce Biotechnology, Rockford, Ill.) at 80°C for 20 min. The volatile TMS methylglycosides were then analyzed by combined gas chromatography-mass spectrometry (GC-MS) using a DB-1 capillary column (J&W Scientific; Agilent Technologies, Palo Alto, Calif.) (30 m long by 0.25 mm inside diameter by 0.25 μm film thickness), and detection was done with a mass-selective detector (Hewlett-Packard HP 5890 GC interfaced to a 5970 MSD). The fatty acids and hydroxy fatty acids were detected as methyl esters and TMS methyl esters, respectively, by GC-MS using the same column. Glycosyl linkages were determined by the preparation and GC-MS analysis of partially methylated alditol acetates. Methylation analysis was performed as described previously (8).
Nuclear magnetic resonance spectroscopy (NMR) spectra were collected using a Varian Inova600 spectrometer. Data were converted to NMRpipe format and viewed and compared using NMRdraw software. The samples were exchanged several times with D2O (Sigma-Aldrich) (99.8%), and final measurements were made in 0.5 ml of D2O solutions (100% D; Cambridge Isotope Laboratories, Andover, Mass.) at 30°C. Proton NMR spectra were measured at 600 MHz with a spectral width of 8 kHz. The gradient correlated spectroscopy (gCOSY) spectra were measured over a spectral width of 2.5 kHz with a data set (t1 × t2) of 256 × 2,048 points with 16 scans. The total correlated spectroscopy and nuclear Overhauser effect spectroscopy spectra were collected using the same data size with 32 scans and mixing times of 80 and 200 ms, respectively. The gradient-sensitive heteronuclear single-quantum coherence (HSQC) experiment was done with spectral widths of 2.5 and 13.9 kHz in the proton and carbon dimensions, respectively, and 64 scans were acquired. The gradient multiple-bond correlation (HMBC) experiment was performed with 128 scans, and the spectral widths were set at 2.5 and 25.0 kHz in the proton and carbon dimensions, respectively.
DNA sequence.
The sequence of the wbxE gene of B. cenocepacia K56-2 was deposited in GenBank under accession number AY633623.
RESULTS AND DISCUSSION
Survival-defective mutants of B. cenocepacia K56-2 display defects in O antigen production and sensitivity to serum complement.
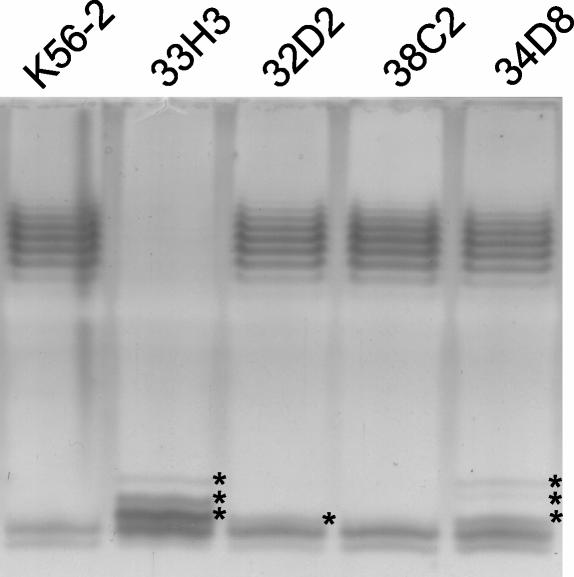
We have recently reported that four mutants in B. cenocepacia K56-2 with a survival defect in the rat model of chronic lung infection had transposon insertions that were mapped to an O antigen LPS biosynthesis cluster (29). The DNA sequence of the K56-2 chromosome flanking the sites of each of these transposon insertions was identical to the sequence on the O antigen biosynthesis cluster found in chromosome 1 of strain J2315, suggesting that both strains carry the same clusters of genes with comparable gene organization characteristics (Fig. 1; also see below). The LPS was extracted from mutants 33H3, 32D2, 38C2, and 34D8 and examined by gel electrophoresis and silver staining (Fig. 2). Mutant 33H3 (carrying a transposon insertion in a wbiI homolog) showed an obvious defect in O antigen production compared to wild-type strain K56-2. Also, the lipid A-core region in this mutant had several additional bands compared to that of K56-2. Mutants 34D8 (insertion in a putative rmlD gene) and 32D2 (insertion in a putative wbiG homolog) produced an apparently normal O antigen but also had several additional bands in the lipid A-core region which were absent from the K56-2 LPS sample (Fig. 2). We interpreted these additional bands in mutants 33H3, 34D8, and 33D2 as lipid A core with substitutions of partial O antigen sugar components, as has been described in other systems (20). Mutant 38C2, in which the transposon insertion is located between divergent genes wbxC and wbxD (Fig. 1), produced an O antigen LPS that appeared identical to that of the wild-type K56-2 (Fig. 2). The LPS phenotype of these mutants is consistent with the location of the transposon insertions in the O antigen cluster. In the case of mutant 33H3, the insertion is in approximately the middle of wbiI, which likely abolishes completely the function of this gene (see below). In contrast, the insertions in mutants 34D8 and 32D2 are located at the very beginning and the very end of the rmlD and wbiG coding regions, respectively (Fig. 1). Since the transposon used, TnMod-OTp (29), does not cause polar effects (T. A. Hunt and M. A. Valvano, unpublished data), it is possible that these insertions not only partially affect the function of these genes but also allow readthrough transcription; thus explaining the formation of a complete O antigen and abnormal lipid-A core at the same time.
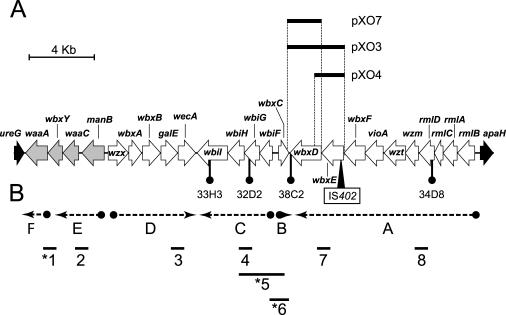
FIG. 1.
(A) Genetic organization of a ca. 29-kb region of the ET12 genome in B. cenocepacia strains K56-2 and J2315 containing genes for core-lipid A and O antigen biosynthesis. The flanking genes ureG and apaH are indicated in black. The four genes represented with gray shading encode proteins putatively involved in lipid A-core biosynthesis. waaA, 3-deoxy-d-manno-octulosonic acid transferase; wbxY, conserved hypothetical protein; waaC, heptosyltransferase I; manB, phosphomannomutase; wzx, O antigen exporter; wbxA, glycosyltransferase; wbxB, glycosyltransferase; galE, UDP-glucose epimerase; wecA, UDP-N-acetylglucosamine 1-P transferase; wbiI, nucleotide sugar epimerase-dehydratase; wbiH, UDP-UDP-N-acetylglucosamine 1-P transferase; wbiG, nucleotide sugar epimerase-dehydratase; wbiF, glycosyltransferase; wbxC, acetyltransferase; wbxD, glycosyltransferase; wbxE, glycosyltransferase; vioA, nucleotide sugar aminotransferase; wzt, ATPase; wzm, permease; rmlDCAB, dTDP-rhamnose biosynthesis. The locations of the transposon insertions in the K56-2 mutants 33H3, 32D2, 38C2, and 34D8, as well as the location of the IS402 element in strain J2315, are indicated. The boundaries of the DNA inserts from plasmids pXO3, pXO4, and pXO7 are depicted. (B) Transcriptional organization of the LPS cluster of B. cenocepacia K56-2. Dotted lines with arrowheads indicate the transcriptional regions (regions A through F) deduced from the expected PCR products for the RT-PCR analysis (indicated by numbers 1 to 8). Asterisks indicate that no PCR product was obtained.
FIG. 2.
Electrophoretic profiles of LPS extracted from K56-2 mutants 33H3, 32D2, 38C2, and 34D8 in comparison to the profile of the parental K56-2 strain. The loading of the LPS was standardized on the basis of the number of cells used to prepare the samples. Samples were run in 14% polyacrylamide gels in a Tricine-SDS system and developed by silver staining as described in Materials and Methods. Asterisks indicate novel bands probably corresponding to core-lipid A replaced with partial O antigen subunits.
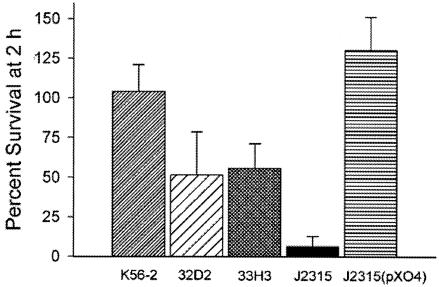
To determine whether an association existed between LPS O antigen profiles in these mutants and attenuation in vivo, we also examined their ability to survive in the presence of serum complement. These experiments were conducted with mutants 33H3 (no O antigen production) and 32D2 (O antigen production and abnormal core-lipid A), as they represented the two phenotypes we observed. A comparison of serum killing assays conducted using 40% fresh and heat-inactivated pooled human serum revealed that the survival of mutants 32D2 and 33H3 after 2 h in fresh human serum was reduced to approximately 50% of the survival of the parental strain K56-2 (Fig. 3). In contrast, strain J2315 was highly sensitive to serum (5% survival), while the same strain containing the complementing plasmid pXO4, which reconstitutes O antigen expression (see below), was serum resistant. From these results, we conclude that an intact O antigen is required for serum resistance. The difference in the serum sensitivities of strains 33H3 and 32D2 in comparison with that of strain J2315 may be due to the presence in the mutants of partial O antigen components attached to core-lipid A. In addition, differences in other surface components may account for the high serum sensitivity of strain J2315. For instance, strain J2315 cannot produce capsular exopolysaccharide (46). In contrast, we have obtained survival-defective mutants in strain K56-2 with an insertion in the capsular exopolysaccharide gene cluster (29), suggesting that strain K56-2 as well as the mutants 33H3 and 32D2 may possess a capsule. Further studies are required to fully understand the basis of serum resistance in strain K56-2.
FIG. 3.
The serum sensitivity of wild-type K56-2 and mutants 33H3 and 32D2 and mutants J2315 and J2315(pXO4) was determined by incubating bacterial cultures in 40% fresh and heat-inactivated pooled human serum. The percentages of survival were calculated by comparing the growth (measured in CFU per milliliter) of each strain in heat-inactivated serum (100% survival). Bars represent the means of at least three experiments; standard errors are also indicated.
Absence of O-specific polysaccharide from B. cenocepacia strain J2315 is associated with the presence of the IS402 in the O antigen biosynthesis gene cluster.
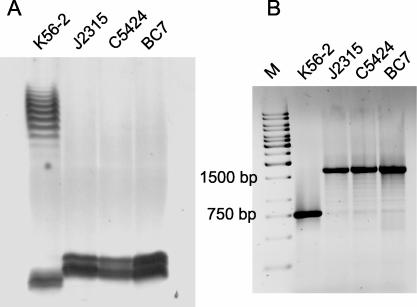
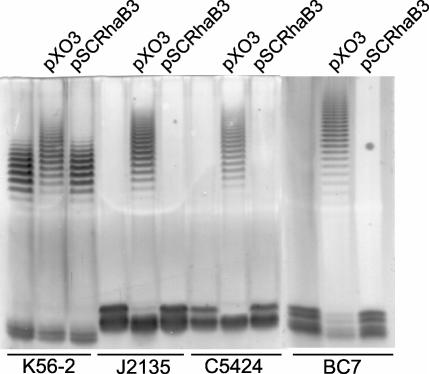
It was previously reported that B. cenocepacia J2315 produces an LPS that lacks O antigen (19). Analysis of the LPS profiles by silver staining revealed that K56-2 was the only isolate in our collection of ET12 strains that had a LPS with a characteristic ladder-like O-polysaccharide banding pattern (Fig. 4A), while the other strains (J2315, C5424, and BC7) displayed only a core-lipid A band. The core-lipid A band in strain J2315 was higher than that in strain K56-2, which could have been due to the presence of a truncated O antigen subunit (see below). An inspection of the sequence of the O antigen gene cluster from strain J2315 revealed an IS402 element that was inserted within the wbxE gene, which encodes a putative glycosyltransferase (Fig. 1). To investigate whether failure to produce O antigen in B. cenocepacia J2315, C5424, and BC7 correlated with the presence of the insertion element within the O antigen biosynthesis cluster, we designed primers that were complementary to regions flanking IS402. Figure 4B shows that a 1.7-kb product was amplified from DNA preparations of strains J2315, C5424, and BC7, while a 750-bp product was obtained from K56-2 DNA. The difference in the masses of these fragments was consistent with the presence of the 900-bp IS402 element in the genome of strains J2315, C5424, and BC7, strongly suggesting that the same gene is interrupted in all these strains. The PCR results mirror the O antigen LPS expression profiles, as K56-2 was the only ET12 strain that can form O antigen and does not have the IS402 element inserted in the O antigen biosynthesis region.
FIG. 4.
Association of O antigen production with the absence of an insertion element in the O antigen biosynthesis gene cluster of strain K56-2. (A) LPS electrophoretic profiles for B. cenocepacia ET12 strains. Samples were run in 14% polyacrylamide gels in a Tricine-SDS system and developed by silver staining. (B) PCR amplification experiment to investigate the presence of IS402 interrupting the wbxE gene on B. cenocepacia in ET12 strains. Amplified products were run in a 0.7% agarose gel. M, 1-kb DNA ladder.
Reconstitution of O antigen synthesis in strain J2315.
The DNA sequence of the 750-bp amplicon obtained from strain K56-2 revealed an open reading frame corresponding to the WbxE polypeptide and did not have the IS402 element. With this information we could determine that wbxE encodes a polypeptide of 456 amino acids that was homologous to a glycosyltransferase of the RfaG family (see Table S1 in the supplemental material). By comparison with the J2315 genomic sequence in the same region, we concluded that the insertion element was inserted in a manner that would have caused a truncation of the WbxE polypeptide after Val84 (data not shown).To demonstrate that the interruption of the wbxE gene is responsible for the absence of O antigen from strains J2315, C5424, and BC7, we cloned the wbxE and wbxD genes of strain K56-2 into the vector pSCRhaB3. This experiment generated the plasmid pXO3, in which both genes were expressed under the control of the rhamnose-inducible PRHA promoter (Fig. 1). The wbxD gene was included in the cloning experiment to avoid the possibility of a polar effect of the IS402 insertion on this gene.
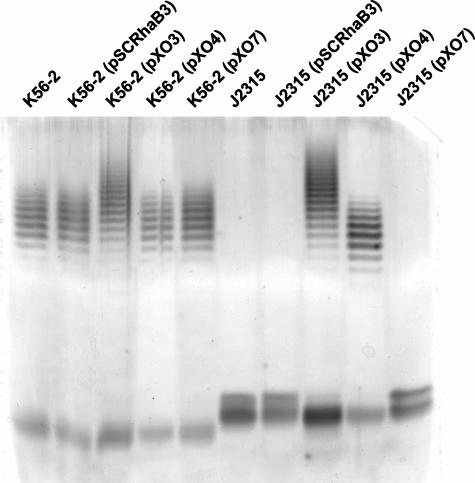
The plasmid pXO3 was mobilized by triparental mating into strains J2315, BC7, and C5424. Figure 5 shows that these strains formed O antigen in the presence of pXO3 but not in the presence of the pSCRhaB3 vector control. Thus, we conclude from these results that the lack of O antigen production in the ET12 strains J2315, BC7, and C5424 is due to a mutation in the wbxE gene caused by the insertion of the IS402 element. However, the O antigen in the complemented strains displayed a larger amount of high-molecular-weight polymers than the O antigen of the parental strain K56-2. This upshift in the O antigen banding profile was also observed with strain K56-2 conjugated with pXO3, but it was absent from strain K56-2 containing the vector alone. These differences in the O antigen migration patterns could be due to an increased gene dosage of the recombinant wbxD and wbxE genes in the strains that carry pXO3. To determine which gene was involved in this phenomenon we constructed pXO4 and pXO7, both derivatives of pXO3 lacking wbxD and wbxE, respectively. Figure 6 shows that pXO4 also complemented O antigen expression in strain J2315, but in this case the migration profile of the O antigen bands was identical to the profile of the parental K56-2 strain. Similar results were obtained with pXO4 in strain C5424 (data not shown). pXO7 did not complement the O antigen expression in J2315 or affect the migration profile of the O antigen bands in strain K56-2 (Fig. 6). Taken together, these results demonstrate that the IS402 in wbxE is nonpolar with respect to the expression of wbxD and also suggest that the bands of higher polymerization found in the strains carrying the pXO3 plasmid are due to an unbalanced stoichiometry of both wbxD and wbxE.
FIG. 5.
LPS electrophoretic profiles for B. cenocepacia ET12 strains in the presence of the complementing plasmid pXO3 or vector control pSCRhaB3.
FIG. 6.
LPS electrophoretic profiles for B. cenocepacia ET12 strains K56-2 and J2315 in the presence of the vector control pSCRhaB3 and complementing plasmids pXO3, pXO4, and pXO7.
Organization of the O-specific polysaccharide biosynthesis gene cluster in B. cenocepacia strains J2315 and K56-2.
A DNA sequence of approximately 29 kb, containing a cluster of genes involved in lipid A-core oligosaccharide and O antigen biosynthesis, was retrieved from the B. cenocepacia J2315 genome database. This region is located on chromosome 1, between nucleotides 3,398,496 and 3,427,242, and it is flanked by ureG (encoding a urease subunit component) and apaH (encoding a putative diadenosine tetraphosphatase). Computer-assisted analysis using the programs Artemis (50) and BLASTP (2) revealed 24 open reading frames (see Table S1 in the supplemental material) organized in five predicted transcriptional units. Most of the open reading frames were preceded by ribosome binding sites (see Table S2 in the supplemental material), and many of them were separated by small intergenic regions or had overlaps with the adjacent gene. With the exception of the IS402 element, discrete PCR amplifications from strain K56-2 genomic DNA at various points of this cluster indicated that the same gene organization is present in both strains, including the presence of the same flanking genes (data not shown), as expected from their shared clonal lineage.
The transcriptional organization of the O antigen biosynthesis gene cluster in strain K56-2 was experimentally determined by nested RT-PCR amplifications. We prepared cDNA from RNA isolated from B. cenocepacia K56-2 and amplified it by PCR using primer pairs that would allow detection of cotranscription (Fig. 1B). We particularly investigated the regions where the open reading frames have more than 20 bp of noncoding intergenic regions. Lack of amplification of the expected fragment in regions 1, 5, and 6 (Fig. 1B) indicated the absence of cotranscription and suggested that the contiguous genes in these regions are part of distinct transcriptional units. By contrast, cotranscription was detected in regions 2, 3, 4, 7, and 8 (Fig. 1B). Region 8 spans a 67-bp intergenic region between rmlD and wzm. This intergenic sequence has the potential to form a stem-loop structure (data not shown), which may play a regulatory role in gene expression. Alternatively, this region may contain a weak promoter that the RT-PCR cannot detect since there may be readthrough from the upstream transcript. Additional bands were observed in some of the PCRs, which may have been due to nonspecific annealing due to the high G+C content of some of the primers. These experiments suggest that there is a large transcriptional unit that spans rmlBACD-wzm-wzt-vioA-wbxFED (Fig. 1B, region A). The gene wbxC is singly transcribed in the opposite direction (Fig. 1B, region B), while the genes wbiFGHI form another transcriptional unit (Fig. 1B, region C). The remaining genes of the cluster are organized in three additional transcriptional units. They include the genes wzx-wbxAB-galE-wecA, manB-waaC-wbxY, and waaA, corresponding to regions D, E, and F, respectively, in Fig. 1B. The analysis of the predicted homologies of the gene products permitted us to establish functional assignments for the majority of the polypeptides (Fig. 1 legend; also see Table S1 in the supplemental material).
The presence of wzm and wzt genes, which encode a predicted two-component ABC transporter, suggests that the O antigen produced by ET12 strains is exported across the plasma membrane by the ABC transport pathway. In this pathway, the O antigen subunit is polymerized in the cytosol and exported across the membrane by a two-component ABC transporter (for a review, see reference 48). This conclusion is consistent with the absence of a wzy gene encoding a polymerase and is also consistent with the presence of wbxF encoding a large protein with two glycosyltransferase domains. These large glycosyltransferases are usually present in strains producing O antigens that are exported by the ABC pathway and are generally involved in the extension of the O antigen (48). In addition, the wbiH gene encodes a predicted transmembrane protein with the features of members of the WecA family. These proteins are involved in the transfer of N-acetylhexosamines to an undecaprenol-phosphate intermediate to initiate the assembly of the O antigen subunits (58). The WbiH polypeptide has strong similarities with gene products from the B. mallei and B. pseudomallei exopolysaccharide clusters (5, 17) and also with WbpL from Pseudomonas aeruginosa (49). Remarkably, there is also a WecA homolog in the transcriptional region D that is closely related to UDP-N-acetylglucosamine 1-phosphate transferases. In addition, this region contains a gene whose predicted product is similar to proteins of the Wzx family, which are involved in the translocation of the O antigen subunit across the plasma membrane prior to their polymerization by the Wzy polymerase. We investigated the role of wecA and wzx genes in the biosynthesis and assembly of the O polysaccharide in strain K56-2 by constructing derivative strains with polar mutations in both genes. O antigen synthesis was not affected in the mutants (data not shown), suggesting that the O antigen in K56-2 is produced by an ABC transport pathway.
The genes present in the transcriptional units E and F (Fig. 1) are likely involved in the formation of part of the inner-core oligosaccharide. This is particularly the case for waaA and waaC; the gene products of both have strong amino acid sequence identity with 3-deoxy-d-manno-octulosonic acid and heptose transferases, respectively. The role of manB in relation with core synthesis cannot be assessed until the structure of the core is completely determined, but it is likely that this gene encodes a function also required for core oligosaccharide assembly. The gene wbxY is the only gene of this region that encodes a protein conserved in other bacteria but without a known function. It is likely that the product of wbxY is involved in lipid A-core biosynthesis or modification, as the gene order manB-waaC-wbxY-waaA is conserved in other Burkholderia species whose genomes are presently being sequenced (http://genome.jgi-psf.org/microbial/).
The O antigen in B. cenocepacia ET12 strains is made of a trisaccharide repeating unit.
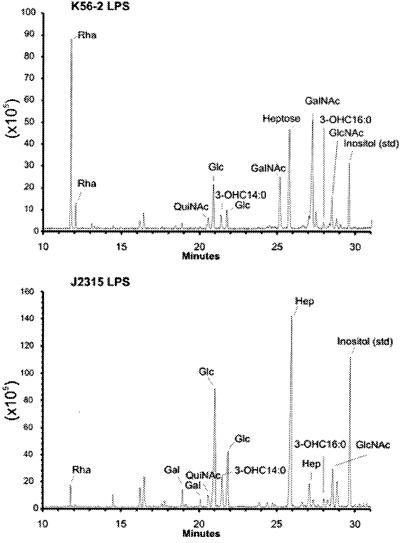
To understand the functions of the gene products encoded by the O antigen biosynthesis gene cluster in ET12 strains we elucidated the chemical structure of the O antigenic polysaccharide repeat produced by strain K56-2. The compositions of the LPS of strains K56-2 and J2315 were determined, and the GC-MS profiles are shown in Fig. 7. The LPS from strain K56-2 contains rhamnose (Rha) and N-acetylgalactosamine (GalNAc) that are not observed in the LPS from strain J2315, indicating that these glycosyl residues are part of the O antigen polysaccharide that is present in strain K56-2 and is not present in the O antigen-defective strain J2315 (Fig. 7). The LPS from J2315 does contain a small amount of Rha and galactose (Gal), which are likely part of the core oligosaccharide region, as has been previously reported for B. cepacia (32). Alternatively, since IS402 is inserted in a predicted glycosyltransferase gene it is also possible that the Rha and Gal in the J2315 core correspond to an incomplete O unit added to core-lipid A. This is consistent with the higher molecular mass of the lipid A-core band in J2315 (Fig. 4A).
FIG. 7.
The GC-MS profiles of the glycosyl residues present in strain K56-2 (top panel) and strain J2315 (bottom panel). The various TMS methyl glycoside peaks are as labeled. Inositol was added to each sample as an internal control. The y axis denotes ion intensity.
The strain K56-2 LPS was hydrolyzed under mildly acidic conditions (2% acetic acid, 100°C for 1 h), and the water-soluble products were fractionated on a Bio-Gel P-2 column, yielding a polymeric material that gave a broad elution profile (data not shown). This material was refractionated on a Bio-Gel P-10 column, obtaining one peak with no heterogeneity. Glycosyl composition analysis of the purified polysaccharide was performed, and the results are shown in Table 2. As with the analysis of the intact LPS, Rha and GalNAc were identified as the major constituents; lower, but significant, amounts of Hep and Glc were also present due, presumably, to the core oligosaccharide component expected to be present at the reducing end of this polysaccharide.
TABLE 2.
The glycosyl linkages of the O-chain polysaccharide isolated from B. cenocepacia strain K56-2
| Permethylated sugara | Linkageb | Relative mole (%)c |
|---|---|---|
| 2,3,4-Me3Rha | T-Rhap | 2 |
| 2,3-Me2Rha | →4)-Rhap | 30 |
| 2,3,4,6-Me4Glc | T-Glcp | 4 |
| 2,3,4-Me3Glc | →6)-Glcp | 3 |
| 2,3,6-Me3Glc | →4)-Glcp | 2 |
| 2,6,7-Me3Hep | →3,4)-Hepp | 4 |
| 2,3,4,6,7-Me5Hep | T-Hepp | 8 |
| 4,6-Me2GalNAc | →3)-GalpNAc | 47 |
Trace amounts of 3,7-linked Hep, 7-linked Hep, and T-Gal were also present.
T,terminal residue.
The observed ratio of 4-linked Rha: 3-linked GalNAc is less than expected, mainly due to the stability of the (1→3)-linkage between the GalNAc residues.
Methylation analysis of the polysaccharide showed the presence of 4-substituted rhamnopyranose and 3-substituted 2-acetamido-2-deoxy-galactopyranoside as major permethylated sugar constituents in a molar ratio of 1:2, together with minor constituents probably arising from the core oligosaccharide, namely, terminal Glc, 2-substituted Glc, 3,4-substituted Hep, and terminal Hep (Table 2). These results indicated that this polysaccharide consisted of a trisaccharide repeating unit composed of one 4-substituted Rha and two 3-substituted GalNAc residues.
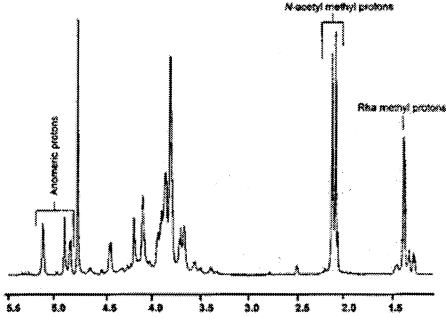
The 1H NMR spectrum (Fig. 8) of the K56-2 O-specific polysaccharide revealed the presence of three anomeric signals at 5.09, 4.86, and 4.80 ppm, together with N-acetyl methyl singlets at 2.02 and 2.07 ppm and a C6 Rha methyl doublet at 1.35 ppm. These results are consistent with the presence in the O unit of two residues of GalNAc and one residue of Rha, respectively, and with data obtained from methylation analysis indicating that the polysaccharide consists of a Rha1GalNAc2 trisaccharide repeating unit. This conclusion was also supported by the 1H-13C correlations obtained from the HSQC spectrum (data not shown), which contained three signals in the anomeric regions at 103.0/4.86, 94.24/5.09, and 102.4/4.81 ppm together with signals from N-acetyl methyl groups at 22.7/2.07 and 22.7/2.02 ppm and a C6 Rha methyl signal at 17.4/1.35 ppm. The proton and carbon NMR assignments of the three glycosyl residues present in the repeating unit were made through COSY and total correlated spectroscopy analyses (data not shown), together with the HSQC spectral spectra described above. The results are given in Table 3. The chemical shifts of H-4 and C-4 of the Rha residue and H-3 and C-3 of the GalNAc residues were downfield, in agreement with the methylation data which showed that Rha was substituted at O-4 and GalNAc was substituted at O-3. With these assignments the sequence of the glycosyl residues was determined using HMBC and nuclear Overhauser effect spectroscopy analyses (data not shown). The HMBC spectrum shows a correlation between H-1 (4.86 ppm) of α-L-Rhap and C-3 (71.1 ppm) of α-D-GalpNAc, H-1 (5.09 ppm) and C-3 of β-D-GalpNAc (75.2 ppm), and H-1 of β-D-GalpNAc (4.81 ppm) and C-4 of α-L-Rhap (80.8). The NOESY spectrum confirmed these same interglycosyl correlations between the residues (data not shown). Therefore, we concluded that the sequence of the O antigen repeat is as follows: →4)-α-L-Rhap-(1→3)-α-D-GalpNAc-(1→3)-β-D-GalpNAc-(1→). This structure has been previously reported as a minor O polysaccharide in B. cepacia serotype O4 (13). A Western blot experiment with O4-specific polyclonal rabbit antiserum confirmed a positive reaction with purified K56-2 LPS (data not shown).
FIG. 8.
The proton spectrum of the O-chain polysaccharide isolated from strain K56-2. The anomeric, N-acetyl methyl, and Rha methyl protons are indicated. The remaining glycosyl residue ring protons resonate between 3.5 and 4.7 ppm. A complete proton assignment was determined as described in the text and as given in Table 3.
TABLE 3.
The proton and carbon NMR assignments of the O-chain polysaccharide isolated from B. cenocepacia strain K56-2
| 1H/13C | Protein and carbon NMR assignment for:
|
||
|---|---|---|---|
| →4)-α-Rha | →3)-α-GalNAc | →3)-β-GalNAc | |
| H1/C1 | 4.86/103.0 | 5.09/94.2 | 4.81/102.4 |
| H2/C2 | 3.77/77.1 | 4.40/48.9 | 4.05/51.8 |
| H3/C3 | 3.90/70.8 | 3.77/71.1 | 3.82/75.2 |
| H4/C4 | 3.66/80.8 | 4.05/68.9 | 4.14/63.9 |
| H5/C5 | 3.83/68.3 | ND/NDa | 3.55/75.8b |
| H6/C6 | 1.35/17.4 | 3.78-3.83/61.5c | 3.78-3.83/61.4c |
ND, not determined.
These assignments were deduced from the HMBC data.
These assignments were obtained from the HSQC data.
Concluding remarks.
We have demonstrated in this work that the O antigen expression of O antigen-deficient B. cenocepacia strains of the ET12 lineage can be reconstituted by the complementation of a mutated gene in the O antigen cluster. We also determined the genetic organization of these genes and the chemical structure of the O antigen subunit in strain K56-2. Our results indicate the O antigen synthesis cluster is very complex and also contains genes encoding lipid A-core components. Two remarkable aspects of the gene organization of the O antigen biosynthesis cluster in ET12 strains cluster are the presence of two genes encoding putative initiating enzymes, WecA and WbiH, and the presence of genes encoding proteins involved with two different export pathways for the O antigen subunits, the ABC transport pathway (represented by the putative proteins Wzm and Wzt) and the Wzy-dependent pathway (represented by the putative Wzx protein). Reasoning on the basis of the chemical structure of the O repeat we can conclude that the genes rmlBACD encode the enzymes for the synthesis of dTDP-rhamnose and that wbiI is probably involved in the synthesis of UDP-GalNAc. The wbxE is also a glycosyltransferase, but its specificity remains uncertain at this time. Also, the presence of vioA and wbxC, encoding a transaminase and acetylase, respectively, suggests the presence of an acetamido-dideoxy sugar that is probably necessary either for the assembly of the O antigen subunit onto the lipid A-core oligosaccharide or for the termination of the elongation step, an important characteristic of O antigens assembled by the ABC transporter pathway (48). Further analyses, including the elucidation of the lipid A-core structure in strains J2315, are under way in our laboratories and will allow us to elucidate the complete pathway of O antigen biosynthesis and assembly in ET12 B. cenocepacia strains.
To our knowledge this is the first report on the organization of the O antigen LPS cluster in B. cenocepacia. The O antigen-defective strain J2315 is considered to be one of the first isolates obtained that belongs to this clone. However, due to the chronic nature of B. cenocepacia infections in CF patients, it is not possible to ascertain whether the strain lost its O antigen prior to or after colonization in the airways. An analogous situation has been documented for infections of CF patients with P. aeruginosa, where the pathogen exhibits large chromosomal inversions and adapts to a complex phenotype that among other features includes O antigen deficiency (35). Therefore, as it is possible that O antigen expression in B. cenocepacia isolates, as in the case of P. aeruginosa, may only be needed for the initial stages after the establishment of the infection. The role of O antigen in the initial steps of colonization is presently being studied in our laboratories.
Supplementary Material
Acknowledgments
We thank H. Monteil for the gift of B. cepacia O typing antisera, S. Cardona and R. Flannagan for supplying pSCrhaB3 and pGpΩTp, respectively, C. Bhatt for technical assistance, and E. Vinés and R. Flannagan for useful discussions and technical advice.
This work was supported by the Special Program Grant Initiative “In Memory of Michael O'Reilly” funded by the Canadian Cystic Fibrosis Foundation and the Cardiovascular and Respiratory Health Institute of the Canadian Institutes of Health Research (M.A.V.), grant MOP64356 from the Canadian Institutes of Health Research (M.A.V.), grant AI050230 from the National Institutes of Health (J.B.G.), grant GOLDBE03P0 from the Cystic Fibrosis Foundation (J.B.G.), and a grant from the U.S. Department of Energy (DE-FG02-93ER20097) to the Complex Carbohydrate Research Center. T.A.H. and S.L. were recipients of a Graduate Studentship Award from the Canadian Cystic Fibrosis Foundation and a Postgraduate Award (PGSA) from the Natural Sciences and Engineering Research Council of Canada, respectively. A.D.V-D. was supported by the National Institutes of Health through a University of Virginia Infectious Diseases Training grant (AI07046). M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aaron, S. D., W. Ferris, D. A. Henry, D. P. Speert, and N. E. Macdonald. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 161:1206-1212. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baird, R. M., H. Brown, A. W. Smith, and M. L. Watson. 1999. Burkholderia cepacia is resistant to the antimicrobial activity of airway epithelial cells. Immunopharmacology 44:267-272. [DOI] [PubMed] [Google Scholar]
- 4.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burtnick, M. N., P. J. Brett, and D. E. Woods. 2002. Molecular and physical characterization of Burkholderia mallei O antigens. J. Bacteriol. 184:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. S., K. A. Witzmann, T. Spilker, R. J. Fink, and J. J. LiPuma. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643-649. [DOI] [PubMed] [Google Scholar]
- 7.Chung, J. W., E. Altman, T. J. Beveridge, and D. P. Speert. 2003. Colonial morphology of Burkholderia cepacia complex genomovar III: Implications in exopolysaccharide production, pilus expression, and persistence in the mouse. Infect. Immun. 71:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 9.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing: a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185:1454-1462. [DOI] [PubMed] [Google Scholar]
- 10.Coenye, T., T. Spilker, A. Martin, and J. J. LiPuma. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 12.Corbett, C. R., M. N. Burtnick, C. Kooi, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149:2263-2271. [DOI] [PubMed] [Google Scholar]
- 13.Cox, A. D., C. J. Taylor, A. J. Anderson, M. B. Perry, and S. G. Wilkinson. 1995. Structures of the two polymers present in the lipopolysaccharide of Burkholderia (Pseudomonas) cepacia serogroup O4. Eur. J. Biochem. 231:784-789. [DOI] [PubMed] [Google Scholar]
- 14.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885-2890. [DOI] [PubMed] [Google Scholar]
- 15.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darveau, R. P., and R. E. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeShazer, D., P. J. Brett, and D. E. Woods. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081-1100. [DOI] [PubMed] [Google Scholar]
- 18.Devine, D. A. 2003. Antimicrobial peptides in defence of the oral and respiratory tracts. Mol. Immunol. 40:431-443. [DOI] [PubMed] [Google Scholar]
- 19.Evans, E., I. R. Poxton, and J. R. W. Govan. 1999. Lipopolysaccharide chemotypes of Burkholderia cepacia. J. Med. Microbiol. 48:1-8. [DOI] [PubMed] [Google Scholar]
- 20.Feldman, M. F., C. L. Marolda, M. A. Monteiro, M. B. Perry, A. J. Parodi, and M. A. Valvano. 1999. The activity of a putative polyisoprenol-linked sugar translocase(Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129-35138. [DOI] [PubMed] [Google Scholar]
- 21.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glisin, V., R. Crkvenjakov, and C. Byus. 1974. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry 13:2633-2637. [DOI] [PubMed] [Google Scholar]
- 23.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 24.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: Medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 25.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidt, A., H. Monteil, and C. Richard. 1983. O and H serotyping of Pseudomonas cepacia. J. Clin. Microbiol. 18:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 28.Huber, B., K. Riedel, M. Köthe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 29.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia (formerly Burkholderia cepacia genomovar III) genes required for bacterial survival in vivo. Infect. Immun. 72:4010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson, M. L., I. R. Poxton, and J. R. W. Govan. 1998. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect. Immun. 66:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 32.Isshiki, Y., U. Zahringer, and K. Kawahara. 2003. Structure of the core-oligosaccharide with a characteristic D-glycero-α-D-talo-oct-2-ulosylonate-(2→4)-3-deoxy-D-manno-oct-2-ulo sonate [α-Ko-(2→4)-Kdo] disaccharide in the lipopolysaccharide from Burkholderia cepacia. Carbohydr. Res. 338:2659-2666. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keig, P. M., E. Ingham, and K. G. Kerr. 2001. Invasion of human type II pneumocytes by Burkholderia cepacia. Microb. Pathog. 30:167-170. [DOI] [PubMed] [Google Scholar]
- 35.Kresse, A. U., S. D. Dinesh, K. Larbig, and U. Römling. 2003. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 47:145-158. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, A., S. Dietrich, W. Schneider, R. Jacobson, F. P. Downes, B. E. Robinson-Dunn, R. Honicky, J. Smith, and R. Martin. 1997. Genetic relatedness of Burkholderia (Pseudomonas) cepacia isolates from five cystic fibrosis centers in Michigan. Respir. Med. 91:485-492. [DOI] [PubMed] [Google Scholar]
- 37.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 6:1127-1138. [DOI] [PubMed] [Google Scholar]
- 38.Lefebre, M. D., and M. A. Valvano. 2001. Catalases and superoxide dismutases in strains of the Burkholderia cepacia complex and their roles in resistance to reactive oxygen species. Microbiology 147:97-109. [DOI] [PubMed] [Google Scholar]
- 39.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 40.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 42.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marolda, C. L., B. Hauröder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 44.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 172:3590-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreira, L. M., P. A. Videira, S. A. Sousa, J. H. Leitao, M. V. Cunha, and I. Sa-Correia. 2003. Identification and physical organization of the gene cluster involved in the biosynthesis of Burkholderia cepacia complex exopolysaccharide. Biochem. Biophys. Res. Commun. 312:323-333. [DOI] [PubMed] [Google Scholar]
- 47.Nzula, S., P. Vandamme, and J. R. W. Govan. 2002. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 50:265-269. [DOI] [PubMed] [Google Scholar]
- 48.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocchetta, H. L., L. L. Burrows, J. C. Pacan, and J. S. Lam. 1998. Three rhamnosyltransferases responsible for assembly of the A-band D- rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28:1103-1119. [DOI] [PubMed] [Google Scholar]
- 50.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 51.Saini, L., S. Galsworthy, M. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 52.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 54.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tablan, O. C., T. L. Chroba, D. V. Schidlow, J. W. White, K. A. Hardy, P. H. Giligan, W. M. Morgan, L. A. Chow, W. J. Martone, and W. R. Jarvis. 1985. Pseudomonas cepacia colonization in patients with cystic fibrosis: risk factors and clinical outcome. J. Pediatr. 107:382-387. [DOI] [PubMed] [Google Scholar]
- 56.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 71:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomich, M., C. A. Herfst, J. W. Golden, and C. D. Mohr. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect. Immun. 70:1799-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valvano, M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8:S452-S471. [DOI] [PubMed] [Google Scholar]
- 59.Vasil, M. L., D. P. Krieg, J. S. Kuhns, J. W. Ogle, V. D. Shortridge, R. M. Ostroff, and A. I. Vasil. 1990. Molecular analysis of hemolytic and phospholipase C activities of Pseudomonas cepacia. Infect. Immun. 58:4020-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.York, W. S., A. G. Darvill, M. McNeil, T. T. Stevenson, and P. Albersheim. 1985. Isolation and characterization of plant cell walls and cell-wall components. Methods Enzymol. 118:3-40. [Google Scholar]
- 61.Zughaier, S. M., H. C. Ryley, and S. K. Jackson. 1999. A melanin pigment purified from an epidemic strain of Burkholderia cepacia attenuates monocytic respiratory burst activity by scavenging superoxide anion. Infect. Immun. 67:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.