Abstract
To understand the mechanisms of ectoine-induced osmoprotection in Sinorhizobium meliloti, a proteomic examination of S. meliloti cells grown in minimal medium supplemented with ectoine was undertaken. This revealed the induction of 10 proteins. The protein products of eight genes were identified by using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Five of these genes, with four other genes whose products were not detected on two-dimensional gels, belong to the same gene cluster, which is localized on the pSymB megaplasmid. Four of the nine genes encode the characteristic components of an ATP-binding cassette transporter that was named ehu, for ectoine/hydroxyectoine uptake. This transporter was encoded by four genes (ehuA, ehuB, ehuC, and ehuD) that formed an operon with another gene cluster that contains five genes, named eutABCDE for ectoine utilization. On the basis of sequence homologies, eutABCDE encode enzymes with putative and hypothetical functions in ectoine catabolism. Analysis of the properties of ehuA and eutA mutants suggests that S. meliloti possesses at least one additional ectoine catabolic pathway as well as a lower-affinity transport system for ectoine and hydroxyectoine. The expression of ehuB, as determined by measurements of UidA activity, was shown to be induced by ectoine and hydroxyectoine but not by glycine betaine or by high osmolality.
Bacteria have evolved complex stress management strategies to sense and respond to change in their external environment (9, 60). One such environmental parameter is the osmolality of the external growth medium. Bacteria, in principle, require an intracellular osmotic pressure greater than that of the surrounding growth medium to maintain cell turgor, which is generally considered to be the driving force for growth extension and cell division (9, 60). The ability to adapt to changes in the osmolality of the external milieu is therefore of fundamental importance for growth and survival, and thus prokaryotic cells have evolved a number of osmoadaptative mechanisms to cope with elevated osmolality (9, 15). Changes in the external osmolality trigger water fluxes along the osmotic gradient, causing either swelling under hypotonic conditions or plasmolysis and dehydration under hypertonic circumstances (9, 14). To avoid plasmolysis, cells accumulate osmotically active solutes in their cytoplasm, which enables them to reverse the water flux and to restore turgor to a level compatible with growth (9, 15). As a primary response to hyperosmotic stress, bacteria amass large amounts of potassium and its counterion glutamate. This is followed by a dramatic increase in the cytoplasmic concentrations (by synthesis and/or uptake) of a selected group of organic osmolytes, the so-called compatible solutes (9, 37). Intracellular accumulation of compatible solutes as a strategy for adaptation to high environmental osmolality is evolutionarily well conserved in bacteria, archaea, and eukaryotes (9, 10, 13, 16, 37, 56), and important examples of the compatible solutes used are the imino acid proline, the disaccharide trehalose, the trimethylammonium compound glycine betaine (GB), and the tetrahydropyrimidine ectoine (4, 9, 31, 33, 37, 64). With the exceptions of proline and trehalose, the compatible solutes or osmoprotectants used by most bacteria are metabolically inert and their catabolism is unusual. A few phototrophic bacteria can synthesize GB de novo (45, 65), but many bacteria can convert choline to GB in a two-step enzymatic oxidation reaction with glycine betaine aldehyde as the intermediate (6, 12, 40, 54). Additionally, some bacteria can synthesize glycine betaine from carnitine (30, 38, 44). Ectoine is synthesized predominantly by halophilic bacteria, and to our knowledge there is no report on its production by either archaea or eukaryotes (11, 31, 39, 42). This imino acid is produced from l-aspartate-β-semialdehyde in a three step reaction, with l-2,4-diaminobutyrate and Nγ-acetyl-l-2,4-diaminobutyrate as the intermediates (50). However, most nonhalophilic bacteria rely entirely upon uptake of potent osmoprotectants (1, 22, 29, 37), via broadly or narrowly specific osmoporters or a combination of both types (2, 34, 37, 66). In Escherichia coli, two osmoregulated porters mediate uptake of most osmoprotectants: ProP, a single-component carrier, and ProU, an ATP-binding cassette (ABC) transporter, (22, 29). Bacillus subtilis, a gram-positive soil bacterium, possesses five osmoregulated transport systems for osmoprotectants. Three of these (OpuA, OpuB, and OpuC) belong to the ABC transporter superfamily, and two (OpuD and OpuE) are single-component transporters (37). OpuB and OpuE are highly specific; they transport only choline and proline, respectively (35, 64). OpuA and OpuD transport GB and some structurally related osmoprotectants, while OpuC is considered a universal osmoprotectant uptake system, since it mediates the uptake of all tested osmoprotectants (34, 36, 37).
The characterization of osmoprotectant transporters in the soil bacterium Sinorhizobium meliloti is at an early stage in comparison to well-studied model systems such as E. coli and B. subtilis. Nevertheless, two transport systems have been characterized: BetS, a single transmembrane protein, mediates the uptake of GB and proline betaine but not ectoine (8), and, Hut, an ABC histidine transporter, is involved in proline and proline betaine uptake at high affinity and in GB uptake at low affinity (7).
In S. meliloti, osmoprotection by exogenously supplied osmoprotectants differs greatly from that in most studied bacteria. S. meliloti can use ectoine, GB, pipecolate, dimethylsulfoniopropionate (DMSP), sucrose, trehalose, and other disaccharides as osmoprotectants (24, 26, 27, 53, 62, 63); however, with the exception of DMSP, these solutes are all catabolized even under hyperosmotic conditions. Transiently (GB) or durably (DMSP) accumulated osmoprotectants inhibit the de novo synthesis of endogenous osmolytes (glutamate, the dipeptide N-acetylglutaminylglutamineamide, and trehalose), ensuring an energetic economy for the cell.
Ectoine is almost as effective as GB in improving the growth of S. meliloti; in addition, it is not accumulated within the cells but rather is degraded at all medium osmolalities (62). Ectoine transport is inducible, dependent on a periplasmic protein (62), and distinct from GB transport (62). Unlike GB, ectoine does not contribute directly to turgor recovery (26, 27, 62); rather, it stimulates the adaptative capacities of the cell by enhancing the cytosolic levels of two endogenously synthesized osmolytes, glutamate and N-acetylglutaminylglutamineamide (26, 27, 62). The mechanism by which ectoine alleviates osmotic growth inhibition remains unknown (62). These observations raise the question of how the ectoine transporter and the ectoine catabolic pathway contribute to the ectoine-induced osmoprotection mechanism(s).
To gain a deeper insight into this mechanism in S. meliloti, we identified by two-dimensional electrophoresis (2DE) and characterized by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry a number of proteins that were induced when cells were grown in the presence of ectoine. The use of a proteomic approach was facilitated by the completion of the tripartite S. meliloti genome project (20). Among the ectoine-induced proteins, there are a putative periplasmic binding protein, a component of an ABC transport system, and enzymes that seem to be involved in ectoine degradation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are detailed in Table 1. S. meliloti strains were grown aerobically at 30°C in the complex medium MSY (49) to an optical density at 570 nm (OD570) of 1.5 to 1.8; they were then prepared and inoculated in minimal lactate aspartate salts medium (LAS) as previously described (5). E. coli strains were grown aerobically in Luria-Bertani medium (47) at 37°C. For the selection of E. coli strains, ampicillin was added at 50 or 100 μg/ml, tetracycline was added at 10 μg/ml, chloramphenicol was added at 25 μg/ml, and neomycin or kanamycin was added at 50 μg/ml. For the selection of S. meliloti strains, streptomycin was used at 100 μg/ml, tetracycline was used at 5 μg/ml, and neomycin was used at 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | F′ endA1 hsdR17 glnV44 thi-1 recA1 gyrA96 relA1 Δ(lacIZYA-argF)U169 deoR φ80dlac Δ(lacZ)M15 | Bethesda Research Laboratories |
| BL21 | F−gal met r− m−hsdS(λDE3) | Stratagene |
| 102F34 Smr | S. meliloti wild-type derivative; Smr | 53 |
| R3-74 | 102F34, integrated pC5275; Smr Tcr | This work |
| R3-76 | 102F34, integrated pC5271; Smr Tcr | This work |
| R4-39 | 102F34, integrated pC7050; Smr Tcr Nmr | This work |
| Plasmids | ||
| pRK600 | Conjugal transfer helper; Cmr | 18 |
| pSUP102 | ColE1 Mob+; Cmr Tcr | 59 |
| pASK-IBA6 | Overexpression vector | IBA |
| pUIDK3 | Kmr Apr; uidA cassette | 3 |
| pGEMTeasy | PCR cloning vector | Promega |
| pLB22 | pASK-IBA6 ehuB+ | This work |
| pC4870 | 0.71-kb PCR product (with primers) of ehuA in pGEMTeasy; Apr | This work |
| pC4895 | 0.74-kb PCR product (with primers) of eutA in pGEMTeasy; Apr | This work |
| pC6810 | 1.07-kb PCR product (with primers) of ehuA-B in pGEMTeasy; Apr | This work |
| pC5271 | 0.71-kb EcoRI/EcoRI ehuA fragment from pC4870 in pSUP102; Tcr | This work |
| pC5275 | 0.74-kb EcoRI/EcoRI eutA fragment from pC4895 in pSUP102; Tcr | This work |
| pC6910 | 1.067-kb EcoRI/EcoRI ehuA-B fragment from pC6810 in pSUP102; Tcr | This work |
| pC7050 | Cloning of BglII/BglII uidA-Kan cassette from pUIDK3 into pC6910; Kmr Tcr | This work |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Nmr, neomycin resistance; Smr, streptomycin resistance; Tcr, tetracyclin resistance.
Pulse-labeling conditions and extraction of protein.
At each labeling time point, 1 ml of culture grown as described above was fed 925 kBq of [35S]methionine-cysteine protein labeling mix (43.475 × 103 GBq · mmol−1; ICN, Orsay, France) for 10 min. The labeling of proteins was stopped by adding 5 μl of a nonradioactive methionine-cysteine solution (2 and 0.4% [wt/vol], respectively). Cells were harvested by centrifugation (12,000 × g, 3 min); resuspended in 100 μl of a buffered solution of 50 mM Tris-HCl [pH 6.8] containing 0.3% (wt/vol) sodium dodecyl sulfate (SDS), 0.5% (vol/vol) β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride; and then heated at 100°C for 5 min. Proteins were precipitated by adding 4 volumes of cold acetone and incubated for 2 h at −20°C. After centrifugation (12,000 × g, 10 min), proteins were solubilized in 50 μl of rehydration buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 0.24% Triton X-100, 20 mM dithiothreitol, 0.48% Biolytes [pH 3 to 10] and traces of bromophenol blue}.
Separation of protein by two-dimensional electrophoresis.
After being harvested by centrifugation, bacterial cells were washed in TE (10 mM Tris, 1 mM EDTA [pH 6.8]). The cell pellet was resuspended in the same buffer with 1 mM phenylmethylsulfonyl fluoride. Cells were disrupted by three passages through a French press, and cell debris was removed by centrifugation at 4°C and 12,000 × g for 30 min. The protein concentration in the supernatant fraction was determined according to the method of Lowry et al. (43). For analytical and preparative 2D gels, 200 and 500 μg of crude protein extract was solubilized in the rehydration solution described above. After electrophoresis of the protein-containing solution for 9 to 16 h at 50 V under low-viscosity paraffin oil, Ready IPG strips (11 cm) (Bio-Rad) covering a pH range of 4 to 7 were subjected to isoelectric focusing as follows: 15 min at 250 V, a linear increase from 250 to 6,000 V over 2.5 h, and a final phase of 6,000 V for 6 h. Strips were kept stored at −80°C or consecutively incubated for 15 min in equilibration buffer I (6 M urea, 2% SDS, 0.375 M Tris-HCl [pH 8.8], 20% glycerol, and 20 mg of dithiothreitol per ml) and for 20 min in equilibration buffer II (6 M urea, 2% SDS, 0.375 M Tris-HCl [pH 8.8], 20% glycerol, and 25 mg of iodoacetamide per ml). The equilibrated strips were then loaded onto the second-dimension polyacrylamide gel, containing 12.5% acrylamide-bisacrylamide (30:0.8, wt/wt). The preparative gels were stained with Coomassie blue R, while the gels containing radiolabeled proteins were dried under vacuum and exposed to a Packard InstantImager electronic autoradiograph for 0.5 to 2 h before exposure to Kodak Hyperfilm-MP for 5 days. The spots were quantified by using the Packard InstantImager software; the relative rate of synthesis of individual spots was calculated by measuring the ratio of radioactivity incorporated in the spot to radioactivity in the whole gel. Results are the means from two to four individual experiments, with standard deviations that did not exceed 20%.
MALDI-TOF MS analysis.
Coomassie blue-stained spots were excised from gels and digested with trypsin. Gel pieces were first destained with 100 μl of a solution of 50% ammonium hydrogenocarbonate (50 mM; pH 8)-50% acetonitrile for 30 to 45 min at 37°C. The destaining solution was removed, and the gels were dried in a vacuum centrifuge (Speed Vac). The gel pieces were rehydrated with 2 μl of trypsin solution (in 50 mM ammonium hydrogenocarbonate; 0.25 mg/ml), and 25 μl of buffer (50 mM ammonium hydrogenocarbonate) was added. After overnight incubation at 37°C, the reaction was stopped by adding 2 μl of 0.5% trifluoroacetic acid. A control extraction (blank) was performed with a piece of the gel from a region of the gel that was free of proteins. Mass spectrometry measurements from liquid solution were conducted with a MALDI-TOF Voyager Elite mass spectrometer (Perspective Biosystems) equipped with a 337-nm nitrogen laser. The analyzer was used in the reflectron mode at an accelerating voltage of 20 kV, a delayed extraction parameter of 130 ns, and a low mass gate of 850 Da. The peptide mass, pI, Mr, and species data were then matched against theoretical values for proteins in the TREMBL-Swiss Prot, National Center for Biotechnology Information, or S. meliloti protein database. The S. meliloti protein database was created after downloading, in a FASTA format, the whole-genome protein sequence of S. meliloti available at the Sinorhizobium BLAST server (http://sequence.toulouse.inra.fr/meliloti.html). The proteins were identified by searching in the National Center for Biotechnology Information or Swiss Trembl database with the MS-Fit software of Protein Prospector (http://prospector.ucsf.edu/ucsfhtml4.0u/msfit.htm) and Profound software from PROWL (http://129.85.19.192/profound_bin/WebProFound.exe). The search parameters allowed for oxidation of methionine and carbamidomethylation of cysteine.
DNA manipulations and plasmid and mutant constructions.
Standard protocols were used for DNA manipulations (57). To mutate ehuA (smb20427) and eutA (smb20431), 710- and 740-bp internal gene fragments, respectively, were PCR amplified by using the chromosomal DNA of S. meliloti as the template and primers Y20427C (5′-TCAAGCGATACGGTCCCC-3′) and Y20427N (5′-TGTGCGCTCCTGTTTGGG-3′) for ehuA and Y20431C (5′-CTGTCGCCCTCTCGCTTGC-3′) and Y20431N (5′-CCGTCCGAGTTGGGGGC-3′) for eutA. The PCR fragments were cloned into pGEMTeasy (Promega), producing plasmids pC4870 and pC4895, respectively. ehuA and eutA internal fragments were transferred as 710- and 740-bp EcoRI/EcoRI fragments, respectively, into the mobilizable plasmid pSUP102 (59), producing pC5271 and pC5275, respectively. To construct mutant strains, the pC5271 and pC5275 plasmids were transferred by conjugation, using E. coli strain DH5α(pRK600) as a helper (18), into S. meliloti 102F34 Smr; these plasmids were recombined into strain 102F34 Smr, creating strains R3-76 and R3-74, respectively.
Construction of a plasmid for overproduction of EhuB.
To construct a plasmid that would allow the overproduction of the S. meliloti EhuB protein in E. coli, we amplified the coding region of ehuB (without the DNA segment encoding the signal sequence) by PCR with chromosomal DNA of S. meliloti as the template and the primers 5′-AAAAAGGGACAAAAAAAAAAGCGCGACGAGAACAAGCTCGAGGAG-3′ and 5′-AAAAAGGGACAAAAAAAAAATATCTTATTTCGCGGCGCAGAGCTTTTC-3′. The resulting 771-bp PCR fragment was cut with BsmFI and inserted into the unique BsmFI site of the expression plasmid pASK-IBA6 (IBA, Göttingen, Germany), generating the ehuB+ plasmid pLB22. The ehuB coding sequence was inserted into the expression vector pASK-IBA6 in frame with an upstream ompA signal sequence and a Strep-TagII affinity peptide. This allowed the secretion of the Strep-TagII-EhuB fusion protein into the periplasm of E. coli, from where it could be recovered by affinity chromatography. In plasmid pLB22 the ehuB coding region is under the control of the anhydrotetracycline-inducible tet promoter present on the vector pASK-IBA6, which allows induction of the transcription of the ehuB gene to high levels. The ehuB coding region of plasmid pLB22 was verified by DNA sequence analysis.
Overproduction and purification of the recombinant S. meliloti EhuB protein in E. coli.
To overproduce the S. meliloti EhuB protein in E. coli, we transformed plasmid pLB22 into E. coli strain BL21. A 10-liter flask containing 5 liters of a defined minimal medium (MMA) (48) supplemented with 100 μg of ampicillin ml−1, 0.5% (wt/vol) glucose as the carbon source, and 0.2% (wt/vol) Casamino Acids was inoculated to an OD578 of 0.1 from an overnight culture of strain BL21(pLB22) prepared in the same growth medium. The cells were grown at 37°C with vigorous stirring until the culture had reached mid-exponential phase (OD578 of 0.5 to 0.8). Increased transcription of the ehuB gene from the plasmid-encoded tet promoter in pLB22 was then induced by the addition of anhydrotetracycline (final concentration, 0.2 μg ml−1). The cells were grown for further 1.5 h to allow EhuB production and were subsequently harvested by centrifugation (10 min, 3,000 × g).
To release periplasmic proteins from the EhuB-overproducing E. coli BL21(pLB22) cells, the cell paste was resuspended in 50 ml of ice-cold buffer P (100 mM Tris-HCl [pH 8], 500 mM sucrose, and 1 mM EDTA) to allow the formation of spheroplasts. After 30 min of incubation on ice, the spheroplasts were separated from the soluble periplasmic protein extract by centrifugation (15 min, 21,000 × g). Insoluble material in the supernatant was subsequently removed by ultracentrifugation (60 min, 120,000 × g). Induction of the expression of the genetically engineered ompA-strep-tag II-ehuB fusion construct in strain BL21(pLB22) led to an appreciable production of the OmpA-Strep-TagII-EhuB fusion protein in addition to a Strep-TagII-EhuB species (see Fig. 6A). However, the former protein species was not released into the soluble periplasmic protein fraction, thereby allowing us to separate the Strep-TagII-EhuB fusion from its unprocessed precursor protein (OmpA-Strep-TagII-EhuB). The cleared, soluble periplasmic protein fraction was then loaded onto a 10-ml Strep-Tactin column (IBA) equilibrated with buffer W (100 mM Tris-HCl, pH 8). After the column was washed with 10 bed volumes of buffer W, the bound Strep-TagII-EhuB proteins were eluted from the affinity resin with buffer E (100 mM Tris-HCl [pH 8], 2.5 mM desthiobiotin). EhuB-containing fractions were collected and dialyzed overnight against 10 liters of 10 mM Tris-HCl (pH 7) at 4°C. The purified EhuB protein was stored at 4°C until further use. In general, approximately 3 mg of pure EhuB protein were obtained per 1 liter of culture used for the overexpression of the ehuB gene in strain BL21(pLB22). Protein concentrations were estimated by using a commercially available kit (bicinchoninic acid protein assay kit; Pierce, Rockford, Ill.).
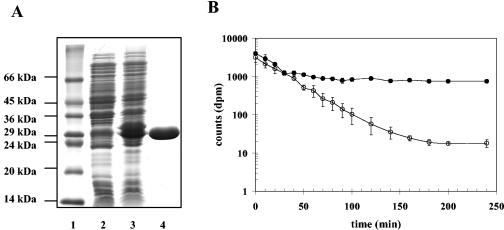
FIG. 6.
Purification of the EhuB ectoine-binding protein and determination of the binding constant of EhuB for ectoine. (A) Expression of the recombinant ehuB gene in E. coli strain BL21(pLB22) was induced by the addition of anhydrotetracycline (0.2 μg ml−1) in exponentially grown cells. Lane 1, molecular mass standard; lane 2, uninduced cell extracts of strain BL21(pLB22); lane 3, induced cell extract of strain BL21(pLB22); lane 4, purified Strep-TagII-EhuB protein (14 μg was loaded onto the gel). The proteins were electrophoretically separated on an SDS-15% polyacrylamide gel and visualized by staining with Coomassie brilliant blue. (B) Release of [14C]ectoine from dialysis tubings containing either no EhuB (○) or purified Strep-TagII-EhuB protein (5 μM) (•), each containing 13 μM [14C]ectoine in a total reaction volume of 250 μl. The graph shows data from two representative experiments. Error bars indicate standard deviations.
Determination of the binding constant of the EhuB protein for ectoine.
To determine quantitatively the binding constant of the recombinant EhuB protein for [14C]ectoine, we used a substrate release assay that measures the retention of [14C]ectoine by the purified EhuB protein from a dialysis bag, according to the procedure described by May et al. (46) for the glycine-betaine-binding protein ProX from E. coli. We calculated the binding constant (KD) of the EhuB protein for its substrate ectoine by determining the velocity of the exit of [14C]ectoine (13 μM) from the dialysis bag in either the presence (v2) or the absence (v1) of 5 μM EhuB protein [P] by using the formula KD = v2 × (1 + [P]/v1), as described by May et al. (46).
Construction of transcriptional fusions to uidA.
The UIDK3 cassette containing a promoterless uidA gene (3) was used to generate a transcriptional fusion in ehuB. The uidA-Km cassette of pUIDK3 (Apr Kmr) was released by BglII and ligated to BglII-digested pC6910, producing the plasmid pC7050, in which the uidA-Km cassette and the ehuAB internal gene fragment were oriented in the same direction. This plasmid was conjugated into strain 102F34 Smr, and recombinants were isolated by selecting for neomycin resistance.
β-Glucuronidase assays.
The β-glucuronidase assays were carried out as described previously (23) with clarified cell lysates obtained by disrupting bacteria with glass beads. Specific β-glucuronidase activities were expressed as micromoles of para-nitrophenol liberated per minute per milligram of protein. The protein concentration was determined by the method of Lowry et al. (43) with bovine serum albumin as a standard. Results are the means from at least three independent experiments, and the standard deviations were less than 10%.
Uptake and intracellular fate of ectoine.
[14C]ectoine (5.5 MBq mmol−1) was prepared biologically as described previously (32). Ectoine uptake was determined as described previously (62) except that 0.45-μm-pore-size nitrocellulose filters (Millipore) were used instead of 0.7-μm-pore-size GF/F fiberglass filters (Whatman). External [14C]ectoine concentrations were varied from 20 to 200 μM. The metabolism of [14C]ectoine was monitored as described previously (62). The results shown are the means from at least three independent experiments, and the standard deviations were less than 10%.
RESULTS
Ectoine-induced proteins.
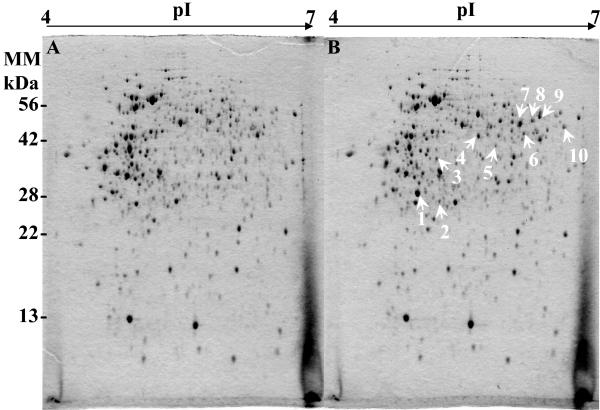
For comparative purposes, we prepared crude protein extracts of soluble proteins from S. meliloti strain 102F34 Smr grown either in LAS or in LAS supplemented with 1 mM ectoine and used them for 2D gel electrophoresis analysis. By comparing the protein profiles of the Coomassie blue-stained gels, we identified 10 protein spots that were not detected in the control (Fig. 1A) and were significantly produced by cells grown in the presence of ectoine (Fig. 1B). All of these protein spots were excised from the gel and examined by MALDI-TOF mass spectrometry to generate peptide mass fingerprints. Eight out of 10 protein spots, numbered in Fig. 1B, could be identified, and putative identities based on peptide mass fingerprint homology are given in Table 2. Among the ectoine-induced polypeptides was the putative ABC transporter amino-acid-binding protein (Smb20428), a periplasmic component of a putative amino acid ABC transport system which also comprises three other components, an ATPase (Smb20427) and two permeases (Smb20429 and Smb20430). The ald (Smc01169) structural gene encodes a probable alanine dehydrogenase protein involved in alanine metabolism. We also found two enzymes involved in amino acid metabolism, a putative aminotransferase protein (Smb20423) and a probable aminotransferase (Sma1855). The group of ectoine-induced proteins with putative functions also included Smb20433, a cyclodeaminase protein, and Smb20434, a hydrolase-peptidase protein. Furthermore, two proteins (Smb20431 and Smb2035) with thus-far-undefined biochemical function were identified as ectoine-induced proteins (Table 2).
FIG. 1.
2DE of soluble proteins synthesized in S. meliloti 102F34 Smr grown in LAS medium without (A) or with (B) 1 mM ectoine. Proteins were visualized by staining with Coomassie brilliant blue. Equivalent amounts (200 μg) of crude proteins were separated by 2DE. The proteins spots indicated by arrows were analyzed by MALDI-TOF mass spectrometry and are listed in Table 2.
TABLE 2.
S. meliloti ectoine-induced proteins analyzed by MALDI-TOF mass spectrometry
| Spot no. | Calculated Mr/pIa | Theoretical Mr/pIb | Mouse scorec | Peptides matchedd | Sequence coverage (%)c | Proteinc | Genomic accession designatione,f |
|---|---|---|---|---|---|---|---|
| 1 | 27.7/4.7 | 29.58/5.03 | 1.34e+09 | 18/136 | 81 | Putative ABC transporter amino-acid-binding protein | smb20428 |
| 2 | 28.5/5.0 | 28.20/5.44 | 4.85e+03 | 9/69 | 25 | Hypothetical arylmalonate decarboxylase protein | smb20431 |
| 3 | 37.9/4.8 | 35.34/4.95 | 2.12e+10 | 21/112 | 56 | Hypothetical protein | smb20435 |
| 4 | 41.7/5.2 | Not identified | |||||
| 5 | 40.9/5.3 | 34.71/5.72 | 3.05c+08 | 17/145 | 57 | Putative cyclodeaminase protein | smb20433 |
| 6 | 44.1/5.7 | 43.53/5.79 | 5.5e+07 | 18/96 | 45 | Putative hydrolasepeptidase protein | smb20434 |
| 7 | 48.6/5.6 | Not identified | |||||
| 8 | 50.0/5.7 | 49.89/5.86 | 1.12e+09 | 15/178 | 39 | Putative aminotransferase protein | smb20423 |
| 9 | 50.0/5.8 | 51.00/6.19 | 2.51e+12 | 18/108 | 43 | Probable aminotransferase | sma1855 |
| 10 | 46.3/5.9 | 39.16/6.06 | 8.25e+10 | 18/84 | 53 | Probable alanine dehydrogenase oxidoreductase protein | ald or smc01169 |
Observed molecular weight and isoelectric point obtained from 2DE calibrated with known standards (Bio-Rad). Mr is in thousands.
Theoretical Mr and pI calculated for the predicted amino acid sequence of the protein.
Mouse score and sequence coverage are indicated in order to facilitate judgement of the reliability of the data.
Number of peptides observed in mass spectra contributing to mouse score and probability of occurrence as calculated by MS-FIT and Profound, respectively, using peptide mass fingerprint data.
Based on the S. meliloti 1021 genomic database (http://sequence.toulouse.inra.fr/meliloti).
Smc, Sma, and Smb refer to products of open reading frames located on the chromosome and on symbiotic plasmids A and B, respectively.
Genetic organization of genes encoding ectoine-induced proteins.
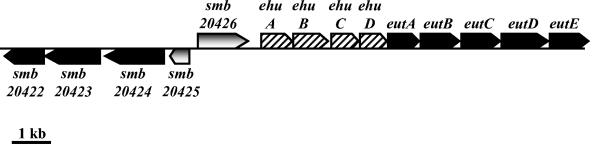
S. meliloti has three replicons: the chromosome (3.7 Mb) and two megaplasmids, pSymA (1.4 Mb) and pSymB (1.7 Mb) (61). Most of the genes encoding ectoine-induced proteins are located in the same region of the megaplasmid pSymB: smb20428, smb20431, smb20433, smb20434, and smb20435 genes are all oriented in the same direction. Four other genes whose products were not detected on our 2D gels belong to this gene cluster; they are smb20427, smb20429, smb20430, and smb20432. Because most of these genes were induced by ectoine, we named the ABC transporter genes ehuA, ehuB, ehuC, and ehuD (formerly smb20427, smb20428, smb20429, and smb20430, respectively) (Fig. 2). They are components of the ehu system (for ectoine-hydoxyectoine uptake). ehuA encodes an ATPase, ehuB encodes a periplasmic binding protein, and ehuC and ehuD encode permeases. Downstream of the ehu transporter genes are five genes, which were named eutABCDE for ectoine utilization (Fig. 2). On the basis of sequence homologies, eutA, -B, -C, -D, and -E encode a hypothetical arylmalonate decarboxylase, a putative threonine dehydratase, a putative cyclodeaminase, a putative hydrolase-peptidase, and a hypothetical protein that might be involved in ectoine catabolism, respectively (Fig. 2). Upstream of the ehu transporter genes, there is a hypothetical transcriptional regulator gene (smb20426) of the GntR family, which is oriented in the same direction as the ehuABCD genes (Fig. 2). Adjacent to the smb20426 gene, a gene (smb20425) that encodes a putative Lrp/AsnC transcriptional regulator is found. Downstream of the smb20425 gene is the smb20423 gene, whose product, a putative aminotransferase, was also detected on our 2D gels as an ectoine-induced protein. The smb20422 and smb20424 genes, encoding a putative oxidoreductase and a putative succinate semialdehyde dehydrogenase, respectively, flank the smb20423 gene. These genes are oriented in the same direction as smb20425 (Fig. 2). As determined from BLAST searches of available databases, the S. meliloti EhuA, -B, -C, and -D components of the Ehu transporter exhibit high homologies to components of putative amino acid uptake systems, and EutABCDE exhibit high homologies to proteins with unknown functions and to putative amino acid catabolic enzymes. All of them were identified in the sequenced genomes of some strains belonging to the rhizobium group, such as Mesorhizobium loti (http://www.kazusa.or.jp/rhizobase/) and Agrobacterium tumefaciens (http://www.tigr.org/) and in a sequenced genome of Pseudomonas putida strain KT2440 (http://www.tigr.org/).
FIG. 2.
Genetic organization of the S. meliloti ehuABCD-eutABCDE locus. The map was redrawn from a contig at http://sequence.toulouse.inra.fr/meliloti.html. Arrows above and below the line represent open reading frames directed from left to right and vice versa, respectively.
Phenotypes of ehuA and eutA mutants.
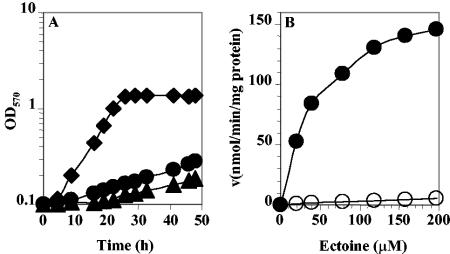
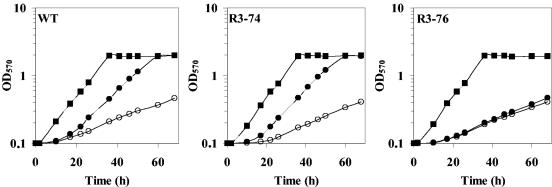
To investigate the role of the ehu-eut gene cluster in ectoine uptake and catabolism in S. meliloti, plasmid integration mutants of ehuA (R3-76) and eutA (R3-74) were constructed (see Materials and Methods). The resulting mutants were tested for their ability to utilize ectoine as a sole carbon and nitrogen sources. As shown in Fig. 3A, the two mutants were highly affected in utilization of ectoine as the sole energy substrate. Growth rate values showed a clear difference from that of the wild type (WT) (Fig. 3A). The growth rate decreased from 0.23 generation · h−1 for the WT to 0.029 and 0.033 generation · h−1 for the R3-76 and R3-74 mutants, respectively. The growth rates of the mutants showed no difference from that of the WT when cells were grown in minimal medium with lactate and aspartate as carbon and nitrogen sources, respectively (see Fig. 5). The R3-76 (ehuA) and R3-74 (eutA) mutants, which are disrupted into the putative ATP-binding protein of the Ehu transport system and the hypothetical arylmalonate decarboxylase, respectively, grew more slowly than the WT in minimal medium supplemented with 10 mM ectoine. The ability of the R3-76 and R3-74 mutants to still utilize ectoine, albeit inefficiently, as a carbon and a nitrogen source strongly suggests that S. meliloti possesses at least one additional transport system for uptake of ectoine into the cell and one ectoine-degrading pathway in addition to EutABCDE.
FIG. 3.
Growth curves and kinetics of transport. (A) Growth curves of S. meliloti wild-type strain 102F34 Smr (⧫) and derivatives R3-76 (ehuA) (▴) and R3-74 (eutA) (•) on minimal medium with ectoine as the sole carbon and nitrogen source. (B) Transport of [14C]ectoine by ectoine-induced cells of S. meliloti strains 102F34 Smr (•) and R3-76 (ehuA) (○). Cells were grown in LAS medium in the presence of 100 μM ectoine. The uptake of [14C]ectoine was measured in LAS medium. The rate of [14C]ectoine uptake at different external [14C]ectoine concentrations is shown. From these data a Km of 48 μM and a Vmax of 182 nmol/min/mg of protein for the wild type and a Km of 200 μM and a Vmax of 10 nmol/min/mg of protein for the ehuA mutant were calculated.
FIG. 5.
Effect of exogenously provided ectoine on the growth of S. meliloti strains 102F34 Smr (WT), R3-74 (eutA), and R3-76 (ehuA) at elevated salinity. Cells were grown aerobically at 30°C in LAS medium containing 0 M NaCl (▪), 0.5 M NaCl (○), or 0.5 M NaCl and 1 mM ectoine (•).
Identification of Ehu, an ectoine uptake system.
To characterize the contribution of the Ehu transport system to ectoine uptake in S. meliloti, the WT and the R3-76 mutant strains were grown in LAS medium supplemented with 100 μM ectoine as an inducer of ectoine transport activity. Cells were grown to mid-exponential phase and harvested by centrifugation; after washing, the uptake of radiolabeled ectoine was measured with ectoine concentrations ranging from 20 to 200 μM. For the WT strain, we found that the transport rate was half maximal at 48 μM (Km) and that the maximal rate of transport (Vmax) was 182 nmol/min/mg of protein, indicating the presence of a high-affinity ectoine transport system (Fig. 3B). In the mutant R3-76, the apparent Km of ectoine uptake by the cells was found to be 200 μM, with a Vmax of 10.3 nmol/min/mg of protein (Fig. 3B). These data suggest that the Ehu transporter plays a major role in the overall ectoine uptake activities in S. meliloti but that, in addition to the Ehu system, at least one other ectoine uptake system is operating in this microorganism.
Also, we measured the rate of uptake of ectoine, used at final concentration of 158 μM, in the WT and in the R3-76 and R3-74 mutants. Cells were grown overnight in LAS medium in either the presence or absence of 100 μM ectoine. Under noninduced conditions, all of the strains exhibited the same ectoine uptake activity, with rates of 10, 10, and 9 nmol/min/mg of protein for the WT and the R3-74 (eutA) and R3-76 (ehuA) mutant strains, respectively. When the cells were cultivated in the presence of ectoine, the ectoine uptake activity was strongly stimulated in the WT and in the R3-74 mutant (141 and 157 nmol/min/mg if protein, respectively), whereas in the R3-76 mutant, the ectoine transport was barely increased (10 nmol/min/mg of protein). These data suggest that only Ehu is induced by ectoine and that the minor ectoine uptake activity is not under ectoine control.
Induction of ehu by ectoine and hydroxyectoine.
To study the expression of the ehu gene cluster, the ehuB gene was fused to the uidA reporter gene in the mutant strain R4-39 and the induction of ehuB was monitored by measuring the level of β-glucuronidase activity in cells grown in LAS medium in either the presence or absence of ectoine or other osmoprotectants. The UidA activity was measured at time zero, and then the culture was divided into four portions. At 10 min, 1 mM ectoine was added to the first culture, 1 mM hydroxyectoine was added to the second, 1 mM GB was added to the third, and nothing was added to the fourth. The expression of ehuB was monitored for 200 min, and every 15 to 30 min, samples were taken (Fig. 4A). In the absence of an osmoprotectant or in the presence of GB, the expression of ehuB did not vary significantly; it remained at a low level, between 1.2 and 2 μmol/min/mg of protein. When ectoine or hydroxyectoine was added to the cultures, the expression of ehuB increased linearly with time; it increased from 1 μmol/min/mg of protein for the control to 9.2 and 8.5 μmol/min/mg of protein at 200 min after the addition of ectoine and hydroxyectoine, respectively (Fig. 4A). These results are in agreement with those obtained by proteomic analysis and also by measurement of ectoine transport activity under inducing and noninducing conditions. In all, they confirm that ehu is strongly induced by ectoine and its structural derivative hydroxyectoine.
FIG. 4.
Expression of ehuB in S. meliloti. (A) Induction of ehuB by ectoines. Cells of strain R4-39 were grown in LAS medium to mid-exponential phase and then divided into four portions. Samples were taken at various time intervals and assayed for UidA (or β-glucuronidase [GUS]) activity. One culture was untreated (◊), 1 mM ectoine was added to the second culture (⧫), 1 mM hydroxyectoine was added to the third culture (▵), and 1 mM glycine betaine was added to the fourth culture (▴). (B) Influence of ectoine concentration and medium salinity on the induction of ehuB by ectoine. Cells were grown to mid-exponential phase in LAS medium without (◊) or with (⧫) 0.5 M NaCl, with different external ectoine concentrations, and then were assayed for UidA activity.
The effects of the ectoine concentration and the salinity of the medium on the expression of ehuB were also examined. The cells were cultivated in LAS medium in the presence or absence of 0.5 M NaCl and in the presence of increasing concentrations of ectoine, ranging from 0 to 250 μM. The cells were harvested by centrifugation at mid-exponential growth phase, and after washing, the β-glucuronidase activity in cellular extracts was measured. In the absence of ectoine, the β-glucuronidase activity was 1.2 μmol/min/mg of protein; the expression of ehuB increased hyperbolically with increased ectoine concentration in the medium. The expression of ehuB was enhanced 4-, 14-, and 17-fold when the culture medium was supplemented with 10, 100, and 250 μM ectoine, respectively (Fig. 4B). Interestingly, the addition of 0.5 M NaCl did not influence the induction of ehuB by ectoine, since the curves for induction of ehuB, at low and high osmolalities according to the concentration of the ectoine in the medium, were similar (Fig. 4B). These data indicate that ehu expression is regulated not by osmotic stress but by ectoine and hydroxyectoine, the substrates of this transport system.
Osmoprotection of the WT and the ehuA and eutA mutants by ectoine.
To assess the contribution of the Ehu transporter to cell osmoprotection by ectoine, the WT strain and the ehuA and eutA mutant strains were cultivated in LAS medium and in LAS medium with 0.5 M NaCl in either the presence or the absence of 1 mM ectoine (Fig. 5). At low osmolality, the three strains exhibited the same growth rate, i.e., 0.125 generation · h−1. The growth of the three strains was strongly impaired at high osmolality in the absence of ectoine; the growth rate of salt-stressed cells was 0.042 generation · h−1 for both the WT strain and the ehuA and eutA mutant strains (Fig. 5). The addition of 1 mM ectoine strongly alleviated growth inhibition in the WT and R3-74 (eutA) but not in R3-76 (ehuA) (Fig. 5); the growth rate was 0.042 generation · h−1 for R3-76, but it increased from 0.042 to 0.1 generation · h−1 for the WT and R3-74 (Fig. 5). Glycine betaine, a potent osmoprotectant in S. meliloti (63), was more efficient than ectoine in protecting the three strains against osmotic stress (data not shown). Hence, the Ehu transporter of S. meliloti is indispensable for ectoine-induced osmoprotection in S. meliloti.
EhuB is a high-affinity ligand-binding protein for ectoine.
Database searches showed that the EhuB protein is most likely a soluble, periplasmic ligand-binding protein, since it is initially produced with an N-terminal signal sequence and it exhibits amino acid sequence homology to various putative amino-acid-binding proteins (data not shown). These features suggest that EhuB might serve as a periplasmic ligand-binding protein for the uptake of ectoine via the EhuABCD ABC transport system. To test binding of ectoine by EhuB experimentally, we expressed the ehuB gene in E. coli with a Strep-TagII affinity tag and purified the heterologously overproduced fusion protein by affinity chromatography to apparent homogeneity (Fig. 6A). We then used the purified EhuB protein (5 μM) for initial binding assays, according to the procedure of Richarme and Kepes (55), with radiolabeled [14C]ectoine at a final substrate concentration of 18 μM. Under these conditions, the EhuB protein bound ectoine avidly. Unlabeled ectoine effectively competed with [14C]ectoine binding to EhuB, whereas glycine betaine (at a 1,000-fold excess) was not able to reduce [14C]ectoine binding to EhuB (data not shown). To determine quantitatively the binding constant of EhuB for ectoine, we used a substrate release assay that measures the retention of [14C]ectoine by the purified EhuB protein by use of a dialysis bag (46). A 5 μM concentration of the EhuB protein was combined with 13 μM [14C]ectoine in a total volume of 250 μl in a dialysis bag that was immersed in 1 liter of 10 mM Tris-HCl (pH 7) buffer and stirred. Radiolabeled ectoine was rapidly released from the dialysis bag when no EhuB protein was present but was only slowly released in the presence of EhuB (Fig. 6B), indicating that this protein was able to bind and retain the ligand ectoine. From repeated release assays we calculated a binding constant of the EhuB protein for ectoine of 0.5 ± 0.2 μM. Hence, the EhuB protein from S. meliloti functions as a high-affinity ligand-binding protein for ectoine.
Effect of ehuA and eutA gene inactivation on expression of the other ectoine-induced genes.
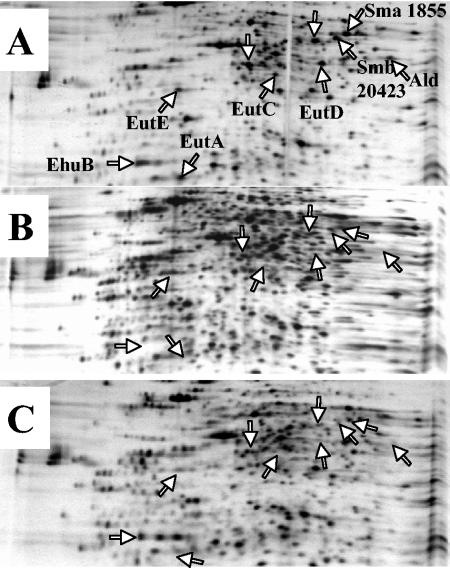
To examine the effect of the inactivation of the ehuA and eutA genes on the expression of the other genes induced in the presence of ectoine, an analysis of protein patterns by 2DE was undertaken. WT, R3-74 (eutA), and R3-76 (ehuA) cells were grown in LAS medium; after a doubling time, 1 mM ectoine was added to each culture and proteins were radiolabeled with [35S]methionine-cysteine for 1 h after ectoine addition (see Materials and Methods). As shown previously, the induction of EhuB, EutA, EutC, EutD, EutE, Ald, Sma1855, Smb20423 and two unidentified proteins was observed in the WT (Fig. 7A). In R3-76 (ehuA), all of the proteins encoded by genes localized downstream from the inactivated gene ehuA were absent from the 2D gel, as were the proteins encoded by other ectoine-induced genes localized on pSymA, on pSymB, or on the chromosome (Fig. 7B). In the R3-74 strain, only EhuB was produced, its encoding gene being upstream of the inactivated gene. All of the other proteins induced in the presence of ectoine were not detected on the 2D gel (Fig. 7C). It should be noted that the protein profiles of the three strains were identical in the absence of ectoine (data not shown).
FIG. 7.
Two-dimensional analysis of proteins expressed in S. meliloti strains 102F34 Smr (A), R3-76 (B), and R3-74 (C) in LAS medium supplemented with 1 mM ectoine. Cells were pulse-labeled 1 h after ectoine addition. Whole-cell protein extracts were analyzed by 2DE followed by autoradiography. Arrows indicate ectoine-induced polypeptides displaying differences in their relative rate of synthesis.
Taken together, these data strongly suggest that the ectoine-induced genes which are localized in the same gene cluster (ehu-eut) comprise an operon. This suggestion is corroborated by the fact that the inactivation, by plasmid insertion, of the ehuA or eutA gene has a polar effect on the expression of genes that are downstream of the insertion point.
Fate of ectoine in the WT and the ehuA and eutA mutants.
To investigate the fate of ectoine in S. meliloti, cells of the WT and the R3-74 and R3-76 mutant strains were grown to mid-exponential growth phase in LAS medium (containing 10 mM lactate and 10 mM aspartate) with either no or 0.5 M NaCl added. Cells were harvested, washed, and then concentrated to an OD570 of 1 in an isotonic LAS medium; [14C]ectoine (158 μM) was added to the cell suspension. The cultures were incubated for 2 h at 30°C with shaking, and the radioactivity of ethanol extracts and insoluble materials was further analyzed.
At low osmolality, almost all of the radiolabeled ectoine was taken up by WT cells; about 70% of the total radioactivity was recovered in the insoluble materials, whereas the ethanol-soluble fraction and the supernatant represented only 1.8 and 1.6%, respectively. The radioactivity recovered in the supernatant did not correspond to ectoine but rather corresponded to a nonpolar catabolic product (data not shown). In R3-76 (ehuA), only 34% of the [14C]ectoine was transported by the cells; 66, 10, and 15% were recovered in the supernatant (as ectoine), in the ethanol-soluble fraction, and in the ethanol-insoluble fraction, respectively. In R3-74 (eutA), which was the mutant that was not affected in ectoine transport activity, only 9% of the total radioactivity was recovered in the supernatant, corresponding well to ectoine (data not shown), and 91% of the [14C]ectoine was imported by the cells. Of this, the major part (48%) was found in the insoluble materials, whereas 26% was in the ethanol-soluble fraction.
At high osmolality, the fate of ectoine in the WT, ehuA, and eutA strains was quite similar to that observed in the strains at low osmolalilty. Indeed, 71, 4, and 3% of the total radioactivity were found in ethanol-insoluble materials, the ethanol-soluble fraction, and the supernatant, respectively, for the WT. Thin-layer chromatography analysis of the supernatant revealed that all of the radiocarbon detected was associated with a nonpolar compound (data not shown). In R3-76, the major ectoine transport activity, which depends on the Ehu transporter, was absent, and the residual transport activity in S. meliloti seemed to be inhibited by high osmolality; indeed only 11% of supplied [14C]ectoine was internalized after 2 h, in comparison to 34% at low osmolality. Whereas 89% (as ectoine) was in the supernatant, only 4 and 3% were recovered in ethanol-soluble fraction and in insoluble materials, respectively. In R3-74, 48, 30, and 10% were found in insoluble materials, the ethanol-soluble fraction, and the supernatant (as ectoine), respectively.
Thin-layer chromatography analysis of the different ethanol-soluble fractions showed that in the WT, the small amounts of radioactivity found in the ethanol-soluble fraction were attributable not to ectoine but rather to breakdown products, under either low- or high-osmolality conditions (data not shown); in contrast, for the mutants we observed that more than 80% of the radioactivity of the ethanol-soluble fractions was in ectoine, whatever the medium salinity (data not shown). These data confirm the main role of Ehu in the uptake of ectoine at low or high osmolality in S. meliloti, and the impairment of the ehuA mutant in ectoine catabolism likely resulted from a polar effect of the mutation. In addition, the inactivation of eutA, which exerts a polar effect on the expression of eutBCDE genes (Fig. 7C), showed that the ability of R3-74 cells to degrade ectoine was reduced but not completely abolished; this suggests that S. meliloti possesses at least one additional ectoine catabolic pathway besides the EutA, -B, -C, -D, and -E enzymes composing the first pathway.
DISCUSSION
In this study, we report the identification, in S. meliloti, of the first ectoine-induced pathway involved in the uptake and catabolism of ectoine. The involvement of this pathway in the assimilation of ectoine was verified by the construction of gene disruption mutants. Most of the ectoine-induced genes are located on the megaplasmid pSymB in the same gene cluster, which is composed of the ehuABCD-eutABCDE genes (Fig. 2). These genes are oriented in the same direction, whereas the gene smb20423, which is also induced by ectoine, is divergently oriented and flanked by the smb20422 and smb20424 genes. Other ectoine-induced genes, such as smb1855 and ald, are localized on the megaplasmid pSymA and on the chromosome, respectively. ehuABCD-eutABCDE probably function as an operon, since the inactivation of either ehuA or eutA by plasmid insertion had a polar effect on the expression of genes located downstream. Moreover, sequence analysis of intergenic regions showed that there is no copy of RIME1 (51), a rhizobium-specific intergenic mosaic element that contains two large inverted repeats; indeed, only an enterobacterial repetitive intergenic consensus amplified sequence was found downstream of the eutE gene (21). These observations further support the assumption that ehuABCD-eutABCDE constitute an operon. The inactivation of ehu showed that this system is necessary to trigger osmoprotection of S. meliloti in the presence of ectoine and that it is responsible for 95% of the ectoine uptake activity in S. meliloti (Fig. 3B). The components of the Ehu transporter share homology with components of ABC transporters of amino acids in bacteria but not with transporters of osmoprotectants. In various bacteria, several transport systems are known to specifically transport organic molecules for osmoprotection; some of them have a strict specificity and others have a broad specificity but have a high affinity for GB, the most potent osmoprotectant in all studied bacteria. This is the case, for example, for ProP and ProU, two transport systems of E. coli that mediate uptake of all known osmoprotectants (9, 14). In B. subtilis, five transport systems (OpuA, OpuB, OpuC, OpuD, and OpuE) are involved in the uptake of osmoprotectants, but only OpuC mediates uptake of all know osmoprotectants (37). In most studied bacteria, ectoine is transported by universal osmoprotectant transporter and usually with low affinity (32, 33). A few high-affinity ectoine transport systems, such as the Tea transporter in Halomonas elongata (28) and the EctP transporter in Corynebacterium glutamicum (52), are involved in ectoine uptake, either alone or together with other osmoprotectants, respectively.
In S. meliloti, the osmoprotectant transport seems differ from that observed in other bacterial models. For example, an amino acid transporter such as the Hut system, an OpuC-like transporter with a high affinity for histidine, is able to mediate the uptake of GB, proline betaine, and proline (7). S. meliloti also shows ectoine transport activity that is induced only by the substrate and depends on a periplasmic protein (62). Ehu, characterized in this study, is a binding-protein-dependent transport system. Our data conclusively show that the Ehu ABC transporter is responsible for the main ectoine transport activity observed in S. meliloti (Fig. 3B). It is induced by its substrates (ectoine and hydroxyectoine) but not by elevated osmolality, which also does not affect its activity (Fig. 4A and B). We have purified the periplasmic substrate-binding protein (EhuB) of the Ehu transporter and demonstrated that it binds ectoine with high affinity (KD = 0.5 ± 0.2 μM) (Fig. 6), but this protein does not recognize GB as one of its substrates. Preliminary binding assays with the purified EhuB protein also indicate that EhuB serves as a hydroxyectoine-binding protein (unpublished results). This is consistent with the observation that transcription of the structural genes for the Ehu transporter is induced by both ectoine and hydroxyectoine (Fig. 4A), compounds that are very similar in chemical structure.
The difference between S. meliloti and other bacterial models is not limited to the nature and the specificity of the osmoprotectant uptake systems, but also includes the behavior and the fate of the osmoprotectants. In most studied bacteria, osmoprotectants are classically considered to be metabolically inert compounds; they are transported through specialized transport systems and accumulated within the cytoplasm to reach molar concentrations. Interestingly, when the cells are exposed to a hypo-osmotic environment, they expel these compounds through mechanosensitive channels to prevent a strong water influx, a buildup of turgor, and a rupture of the cell wall (9). In S. meliloti, osmoprotectants can be classified into two families: accumulated solutes, such as those observed in most bacteria, and nonaccumulated solutes, which have been described only for this bacterium (25). S. meliloti has adopted a different strategy for the second group: the osmoprotection by a solute is concomitant with its use as an energy and carbon substrate. The osmoprotective effect seems to occur through the stimulation of the stress metabolic pathways, which involve not only compatible solute synthesis but also restoration of homeostasis under hyperosmotic constraints. The fate of ectoine in S. meliloti resembles that of amino acids and sugars in other bacteria, where the ABC transporter genes are often expressed together with genes involved in the metabolism of the transported compound. The inactivation of eutA (Fig. 7) and of other eut genes (unpublished data) by plasmid insertion showed that the eut gene products are involved in the catabolism of ectoine. The mutants are affected in their ability to use ectoine as a sole carbon and nitrogen source, but the metabolism of ectoine in these mutants is not completely abolished. This suggests that the tripartite genome of S. meliloti contains other genes encoding ectoine degradation enzymes.
Ectoine is a compatible solute produced by several halophilic bacteria (11, 31, 39). Some of these are soil bacteria which could be considered potential ectoine suppliers for S. meliloti. Since S. meliloti is a nonproducer of ectoine, its conservation of a metabolic pathway dedicated to ectoine transport and catabolism implies that ectoine is found in its close environment. Ectoine is not known to be produced by animals or plants. Curiously, two nodule-specific M. loti compounds were found in nodules of the legume Lotus tenuis induced by strain NZP2037. One, called rhizolotine, has been characterized as the riboside of a novel α-hydroxyimino acid containing a 1,4,5,6-tetrahydropyrimidine ring; this compound can be considered a derivative of ectoine (58). It was degraded by M. loti NZP2037 but not by strains of S. meliloti SU47, Rhizobium trifolii NZP561, or A. tumefaciens C58 (58). This suggests that compounds structurally related to ectoine might be synthesized in nodules of symbiotic association between rhizobia and legume host plants, such as S. meliloti and its host plants Medicago sativa, Medicago truncatula, and Melilotus alba. Recently, a proteomic study of bacterial nodules of M. truncatula and M. alba showed that EhuB (Smb20428) as well as 12 other ABC transporter proteins were found in nodule bacteria (17), whereas in cultured cells a total of 84 different ABC-type transporter proteins were present (17). S. meliloti is known to possess 430 ABC transporter-type genes (19, 20). This result strongly suggests that Ehu is of particular importance to nodule bacteria, but the nature of the substrates exchanged between the plant and the bacteroid through this system remains to be determined.
The analysis of sequenced genomes showed that ehuA-eutABCDE cluster was entirely conserved, not always with the same genetic organization, in the symbiotic bacterium M. loti and also in other, free-living bacteria such as A. tumefaciens and P. putida (data not shown). This suggests that these rhizobacteria are able to use ectoine and other structurally related compounds as sole carbon and nitrogen sources. However, more studies are needed to determine the role and the nature of the substrates for ehuABCD-eutABCDE gene products in S. meliloti when this bacterium is in a free-living state and/or in a symbiotic association.
Among the ectoine-induced genes, the ald gene encodes an alanine dehydrogenase; this enzyme catalyses the conversion of alanine into pyruvate and ammonium and vice versa, utilizing β-NAD+ as a cofactor. The induction of this gene in the presence of ectoine is intriguing, but one can suppose that alanine or ammonium is a catabolic product of ectoine. Ammonium can be produced by deamination of ectoine or of one of its catabolism products, whereas alanine could be produced by transfer of an amino group from an ectoine catabolic product onto pyruvate by a transaminase enzyme. A study with Rhizobium leguminosarum showed that alanine dehydrogenase (AldA) production is stimulated by carboxylic acids such as succinate, malate, and pyruvate, although the strongest stimulant is alanine. Those authors originally proposed that AldA may have a subtle role in balancing the intracellular levels of organic acids and alanine, but they concluded that this enzyme instead contributes to net alanine synthesis in laboratory cultures (41). Ald of S. meliloti displays 85% amino acid identity with its R. leguminosarum homologue and might show a similar regulation. Obviously, biochemical and genetic studies are necessary to determine the precise role of alanine dehydrogenase in the catabolism of ectoine.
In conclusion, the Ehu ABC-type transporter system but not the ectoine catabolic pathway, is essential to osmoprotection of S. meliloti cells. However, ectoine accumulated in eut mutant strains or degraded in the WT strain is still osmoprotective. Indeed, the biochemical characterization of enzymes involved in ectoine degradation will help us to understand the beneficial effect of ectoine catabolism on stressed S. meliloti cells.
Acknowledgments
We thank Céline Henry and Alain Guillot from INRA, Jouy en Josas, France, for performing the mass spectrometry analysis. S. Georgeault, C. Monnier, M. C. Savary, and M. Uguet are acknowledged for their technical assistance. We are indebted to V. Koogle for helpful language improvement.
This work was supported by the Centre National de la Recherche Scientifique and the Ministère de la Recherche et de l'Education Nationale, the Deutsche Forschungsgemeinschaft through SFB-395 and the Graduiertenkolleg “Proteinfunktion auf atomarer Ebene,” The Max-Planck Institute for Terrestrial Microbiology (Marburg), and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Angelidis, A. S., and G. M. Smith. 2003. Three transporters mediate uptake of glycine betaine and carnitine by Listeria monocytogenes in response to hyperosmotic stress. Appl. Environ. Microbiol. 69:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baliarda, A., H. Robert, M. Jebbar, C. Blanco, and C. Le Marrec. 2003. Isolation and characterization of ButA, a secondary glycine betaine transport system operating in Tetragenococcus halophila. Curr. Microbiol. 47:347-351. [DOI] [PubMed] [Google Scholar]
- 3.Bardonnet, N., and C. Blanco. 1992. ′uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol. Lett. 93:243-248. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, T., M. Jebbar, Y. Rassouli, S. Himdi-Kabbab, J. Hamelin, and C. Blanco. 1993. Ectoine accumulation and osmotic regulation in Brevibacterium linens. J. Gen. Microbiol. 139:129-138. [Google Scholar]
- 5.Bernard, T., J.-A. Pocard, B. Perroud, and D. Le Rudulier. 1986. Variations in the response of salt-stressed Rhizobium strains to betaines. Arch. Microbiol. 143:359-364. [Google Scholar]
- 6.Boch, J., B. Kempf, R. Schmid, and E. Bremer. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boncompagni, E., L. Dupont, T. Mignot, M. Østerås, A. Lambert, M. C. Poggi, and D. Le Rudulier. 2000. Characterization of a Sinorhizobium meliloti ATP-binding cassette histidine transporter also involved in betaine and proline uptake. J. Bacteriol. 182:3717-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boscari, A., K. Mandon, L. Dupont, M. C. Poggi, and D. Le Rudulier. 2002. BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti. J. Bacteriol. 184:2654-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bremer, E., and R. Krämer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatibles solutes in bacteria, p. 79-97. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 10.Burg, M. B. 1997. Renal osmoregulatory transport of compatible organic osmolytes. Curr. Opin. Nephrol. Hypertens. 6:430-433. [DOI] [PubMed] [Google Scholar]
- 11.Cánovas, D., C. Vargas, M. I. Calderón, A. Ventosa, and J. J. Nieto. 1998. Characterization of the genes for the biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halomonas elongata DSM 3043. Syst. Appl. Microbiol. 21:487-497. [DOI] [PubMed] [Google Scholar]
- 12.Cánovas, D., C. Vargas, S. Kneip, M. J. Moron, A. Ventosa, E. Bremer, and J. J. Nieto. 2000. Genes for the synthesis of the osmoprotectant glycine betaine from choline in the moderately halophilic bacterium Halomonas elongata DSM 3043, USA. Microbiology 146:455-463. [DOI] [PubMed] [Google Scholar]
- 13.Chen, T. H., and N. Murata. 2002. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 5:250-257. [DOI] [PubMed] [Google Scholar]
- 14.Csonka, L. N., and W. Epstein. 1996. Osmoregulation. p1210-1213. In F C. Neidhardt et al. (ed.). Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington D.C.
- 15.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-606. [DOI] [PubMed] [Google Scholar]
- 16.da Costa, M. S., H. Santos, and E. A. Galinski. 1998. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 61:117-153. [DOI] [PubMed] [Google Scholar]
- 17.Djordjevic, M. A. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859-1872. [DOI] [PubMed] [Google Scholar]
- 18.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 21.Gilson, E., W. Saurin, D. Perrin, S. Bachellier, and M. Hofnung. 1991. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 19:1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouesbet, G., M. Jebbar, R. Talibart, T. Bernard, and C. Blanco. 1994. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology 140:2415-2422. [DOI] [PubMed] [Google Scholar]
- 23.Gouesbet, G., A. Trautwetter, S. Bonnassie, L. F. Wu, and C. Blanco. 1996. Characterization of the Erwinia chrysanthemi osmoprotectant transporter gene ousA. J. Bacteriol. 178:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouffi, K., T. Bernard, and C. Blanco. 2000. Osmoprotection by pipecolic acid in Sinorhizobium meliloti: specific effects of D and L isomers. Appl. Environ. Microbiol. 66:2358-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouffi, K., and C. Blanco. 2000. Is the accumulation of osmoprotectant the unique mechanism involved in bacterial osmoprotection? Int. J. Food Microbiol. 55:171-174. [DOI] [PubMed] [Google Scholar]
- 26.Gouffi, K., N. Pica, V. Pichereau, and C. Blanco. 1999. Disaccharides as a new class of nonaccumulated osmoprotectants for Sinorhizobium meliloti. Appl. Environ. Microbiol. 65:1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gouffi, K., V. Pichereau, J. P. Rolland, D. Thomas, T. Bernard, and C. Blanco. 1998. Sucrose is a nonaccumulated osmoprotectant in Sinorhizobium meliloti. J. Bacteriol. 180:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grammann, K., A. Volke, and H. J. Kunte. 2002. New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J. Bacteriol. 184:3078-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haardt, M., B. Kempf, E. Faatz, and E. Bremer. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. 246:783-786. [DOI] [PubMed] [Google Scholar]
- 30.Jebbar, M., C. Champion, C. Blanco, and S. Bonnassie. 1998. Carnitine acts as a compatible solute in Brevibacterium linens. Res. Microbiol. 149:211-219. [DOI] [PubMed] [Google Scholar]
- 31.Jebbar, M., G. Gouesbet, S. Himdi-Kabbab, C. Blanco, and T. Bernard. 1995. Osmotic adaptation in Brevibacterium linens: differential effects of proline and glycine betaine on cytoplasmic osmolyte pool. Arch. Microbiol. 163:380-386. [Google Scholar]
- 32.Jebbar, M., R. Talibart, K. Gloux, T. Bernard, and C. Blanco. 1992. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J. Bacteriol. 174:5027-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jebbar, M., C. von Blohn, and E. Bremer. 1997. Ectoine functions as an osmoprotectant in Bacillus subtilis and is accumulated via the ABC-transport system OpuC. FEMS Microbiol. Lett. 154:325-330. [Google Scholar]
- 34.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 36.Kempf, B., and E. Bremer. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701-16713. [DOI] [PubMed] [Google Scholar]
- 37.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 38.Kleber, H. P. 1997. Bacterial carnitine metabolism. FEMS Microbiol. Lett. 147:1-9. [DOI] [PubMed] [Google Scholar]
- 39.Kuhlmann, A. U., and E. Bremer. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landfald, B., and A. R. Strøm. 1986. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodwig, E., S. Kumar, D. Allaway, A. Bourdes, J. Prell, U. Priefer, and P. Poole. 2004. Regulation of l-alanine dehydrogenase in Rhizobium leguminosarum bv. Viciae and its role in pea nodules. J. Bacteriol. 186:842-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louis, P., and E. A. Galinski. 1997. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143:1141-1149. [DOI] [PubMed] [Google Scholar]
- 43.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 44.Lucchesi, G. I., T. A. Lisa, C. H. Casale, and C. E. Domenech. 1995. Carnitine resembles choline in the induction of cholinesterase, acid phosphatase, and phospholipase C and in its action as an osmoprotectant in Pseudomonas aeruginosa. Curr. Microbiol. 30:55-60. [DOI] [PubMed] [Google Scholar]
- 45.Mackay, M. A., R. S. Norton, and L. J. Borowitzka. 1984. Organic osmoregulatory solutes in cyanobacteria. J. Gen. Microbiol. 130:2177-2191. [Google Scholar]
- 46.May, G., E. Faatz, M. Villarejo, and E. Bremer. 1986. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol. Gen. Genet. 205:225-233. [DOI] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.O'Gara, F., and K. T. Shanmugam. 1976. Control of symbiotic nitrogen fixation in rhizobia. Regulation of NH4+ assimilation. Biochim. Biophys. Acta 451:342-352. [DOI] [PubMed] [Google Scholar]
- 50.Ono, H., K. Sawada, N. Khunajakr, T. Tao, M. Yamamoto, M. Hiramoto, A. Shinmyo, M. Takano, and Y. Murooka. 1999. Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic eubacterium, Halomonas elongata. J. Bacteriol. 181:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Østerås, M., J. Stanley, and T. M. Finan. 1995. Identification of Rhizobium-specific intergenic mosaic elements within an essential two-component regulatory system of Rhizobium species. J. Bacteriol. 177:5485-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peter, H., B. Weil, A. Burkovski, R. Krämer, and S. Morbach. 1998. Corynebacterium glutamicum is equipped with four secondary carriers for compatible solutes: identification, sequencing, and characterization of the proline/ectoine uptake system, ProP, and the ectoine/proline/glycine betaine carrier, EctP. J. Bacteriol. 180:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pichereau, V., J.-A. Pocard, J. Hamelin, C. Blanco, and T. Bernard. 1998. Differential effects of dimethylsulfoniopropionate, dimethylsulfonioacetate, and other S-methylated compounds on the growth of Sinorhizobium meliloti at low and high osmolarities. Appl. Environ. Microbiol. 64:1420-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pocard, J. A., N. Vincent, E. Boncompagni, L. T. Smith, M. C. Poggi, and D. Le Rudulier. 1997. Molecular characterization of the bet genes encoding glycine betaine synthesis in Sinorhizobium meliloti 102F34. Microbiology 143:1369-1379. [DOI] [PubMed] [Google Scholar]
- 55.Richarme, G., and A. Kepes. 1983. Study of binding protein-ligand interaction by ammonium sulfate-assisted adsorption on cellulose esters filters. Biochim. Biophys. Acta 742:16-24. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, M. F. 2000. Osmoadaptation and osmoregulation in archaea. Front. Biosci. 5:796-812. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Scott, D. B., R. Wilson, G. J. Shaw, A. Petit, and J. Tempé. 1987. Biosynthesis and degradation of nodule-specific Rhizobium loti compounds in Lotus nodules. J. Bacteriol. 169:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other Gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 60.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 61.Sobral, B. W., R. J. Honeycutt, A. G. Atherly, and M. McClelland. 1991. Electrophoretic separation of the three Rhizobium meliloti replicons. J. Bacteriol. 173:5173-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talibart, R., M. Jebbar, G. Gouesbet, S. Himdi-Kabbab, H. Wroblewski, C. Blanco, and T. Bernard. 1994. Osmoadaptation in rhizobia: ectoine-induced salt tolerance. J. Bacteriol. 176:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talibart, R., M. Jebbar, K. Gouffi, V. Pichereau, G. Gouesbet, C. Blanco, T. Bernard, and J. A. Pocard. 1997. Transient accumulation of glycine betaine and dynamics of endogenous osmolytes in salt-stressed cultures of Sinorhizobium meliloti. Appl. Environ. Microbiol. 63:4657-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 65.Waditee, R., Y. Tanaka, K. Aoki, T. Hibino, H. Jikuya, J. Takano, and T. Takabe. 2003. Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. J. Biol. Chem. 278:4932-4942. [DOI] [PubMed] [Google Scholar]
- 66.Wood, J. M., E. Bremer, L. N. Csonka, R. Krämer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A 130:437-460. [DOI] [PubMed] [Google Scholar]