Abstract
To rapidly diagnose infectious organisms causing blood sepsis, bacteria must be rapidly separated from blood, a very difficult process considering that concentrations of bacteria are many orders of magnitude lower than concentrations of blood cells. We have successfully separated bacteria from red and white blood cells using a sedimentation process in which the separation is driven by differences in density and size. Seven mL of whole human blood spiked with bacteria is placed in a 12-cm hollow disk and spun at 3000 rpm for 1 min. The red and white cells sediment more than 30-fold faster than bacteria, leaving much of the bacteria in the plasma. When the disk is slowly decelerated, the plasma flows to a collection site and the red and white cells are trapped in the disk. Analysis of the recovered plasma shows that about 36% of the bacteria is recovered in the plasma. The plasma is not perfectly clear of red blood cells, but about 94% have been removed. This paper describes the effects of various chemical aspects of this process, including the influence of anticoagulant chemistry on the separation efficiency and the use of wetting agents and platelet aggregators that may influence the bacterial recovery. In a clinical scenario, the recovered bacteria can be subsequently analyzed to determine their species and resistance to various antibiotics.
Keywords: Bacterial bloodstream infection, E. coli, Sedimentation, Centrifugation, Human blood, Bacterial separation
1. Introduction
One of the global challenges that face mankind is the evolution of carbapenem-resistant enterobacteriaceae (CRE), as carbapenem antibiotics are considered a “last-resort” antibiotic [1,2]. With CRE blood infections, the mortality rate increases at about 8% for each hour of delay in obtaining a correct diagnosis and appropriate antibiotic treatment [3,4]. Clinical diagnostic protocols usually start with growth of the organisms by 24 h of culture, providing bacteria for species identification and for further growth in antibiotic solutions to determine the antibiotic resistance profile. These time delays result in about a 50% mortality rate for CRE blood infections involving Klebsiella ssp. [5].
A challenge to rapid identification of resistance profile is that symptoms can be manifest at bacterial loadings of only 10–100 colony-forming units per mL (CFU/mL) [6–9].
Thus, microliter volumes of blood (i.e. a finger prick) would not contain sufficient numbers of bacteria if the bacterial loading were only 10 CFU/mL, so several mL of blood are needed.
The need to collect bacteria from blood has led several separation methods. The most practical (for large volumes of whole blood) of these methods generally fall into the broad categories of 1) binding bacteria by selective ligands and subsequent removal, 2) centrifugation, and 3) hydrodynamic separation. These separation techniques have recently been reviewed [10], and only a few will be mentioned herein as background.
Separation by specific binding has been demonstrated on magnetic beads [11–15] and stationary supports (i.e. packed columns, chip-based sensors) [16,17]. Binding ligands include mannose binding lectin, [11,18,19] zinc dipicolylamine [13,20,21], lysozyme [14] and some biopolymers [16]. Many of these methods process microliter volumes of whole blood that often have been heavily diluted. However, some recover bacteria from whole blood. For example, Kang et al. used a fragment of mannose-binding lectin attached to magnetic beads to capture and clear about 90% of the bacteria from whole blood at about 9 mL/min [11]. Similarly, Lee et al. used magnetic beads coated with Zn-bis-dipicolylamine to capture bacteria from undiluted blood with about 80% efficiency, at a processing rate of about 1 mL/min [13].
Separation of bacteria from blood by centrifugation is problematic because the ranges of densities for many bacteria overlap the densities of RBCs. Thus centrifugation will separate bacteria from plasma, but will not separate bacteria from RBCs. However, centrifugation for a short period of time can transiently separate the bacteria from the RBCs because the sedimentation velocity of smaller bacteria is about 30-fold less than the sedimentation velocity of larger RBCs [10]. For horizontal centrifugation of ideal spherical particles that do not interact with each other, the sedimentation velocity, vs, of the particles relative to the fluid is given by
| (1) |
where Dp is the particle diameter, ρp is the particle density, ρf is the fluid density, R is the rotational radius, ω is the rotational angular velocity, g is the gravitational constant, and μ is the fluid viscosity [22]. For non-spherical particles, the Dp can be replaced by an effective diameter, the diameter of a sphere of equal volume. In bacterial-laden blood, this equation is only approximate because the components are not rigid spheres, and their close approach causes their flow fields to interact, particularly in whole blood with normal hematocrits of about 45 [23].
We could find no reports of separating bacteria from blood by sedimentation. But there are several reports of separating bacteria from diluted or whole blood using hydrodynamic separation in microfluidic channels [24–28]. For example, Hou et al. used RBC margination in straight channels to separate bacteria from whole blood with about 80% recovery efficiency [25,28]. Hydrodynamic focusing was used by Mach et al. to recover about 80% of bacteria from diluted blood [26]. Hou et al. used Dean flow microfluidics to recover about 65% of bacteria from slightly diluted blood [25].
While high separation efficiencies can be achieved, these separation processes usually are done on small volumes of diluted blood, and thus microfluidic systems would need to be set up in massively parallel flows to enable processing of mL quantities of whole blood in minutes.
We have developed a fairly simple, rapid and effective method of separating bacteria from large volumes of whole blood using sedimentation principles. A large quantity of human blood (7 mL), spiked with bacteria, is placed in a hollow disk and spun at high rotational velocities for a short time (less than 1 min), during which time the WBCs and RBCs are rapidly sedimented to the periphery of the hollow disk. Then the disk is carefully decelerated and the plasma, which still contains most of the bacteria, is separated from the cells. This paper describes the chemical aspects of this separation system, including the influence of anticoagulants in the blood, the impact of wetting agents, and the use of platelet-coagulating agents. The mechanical aspects will be discussed in a subsequent paper.
2. Materials and methods
2.1. Hollow spinning disk
The 12-cm-diameter hollow disks are built of photopolymerizable acrylates (VeroClear™ Resin) using rapid-prototyping technology (Stratasys Objet30 Prime, Eden Prairie, MN, USA). Disks are designed to spin around a central axis. The surface of the disk radially inward of the annular fluid collection chamber resembles a bowl with a downward slope toward the center of the disk. When the disk begins to spin, the fluid initially placed in the bowl is flung radially outwards into the annular fluid collection chamber. The disk is designed so that when the spinning disk slows to a stop, the separated plasma layer drains towards the bowl around the center of the disk. Disks are designed with a retaining weir to hold back the red cell pack while the plasma drains to the bowl of the disk. The outer-most edge of the weir from the central axis of rotation protrudes into the trough acting as a splitter to separate the plasma layer from the cell pack layer. The other side of the weir (leading edge) is designed with a sharp corner leading into a downward concave slope designed to quickly drain the plasma toward the bowl. The disks are designed to hold 3.5 mL in the trough below the top of the weir, as indicated by the violet-colored area in Fig. 1. Disks are designed with a partial lid at the top of the annular fluid chamber such that 8.5 mL of blood mixture occupies the annular volume from the leading edge of the weir up to the lid. The partial lid is open in the middle so PBS and blood can easily be pipetted in, and plasma withdrawn; and the disk can be easily washed and reused.
Fig. 1.
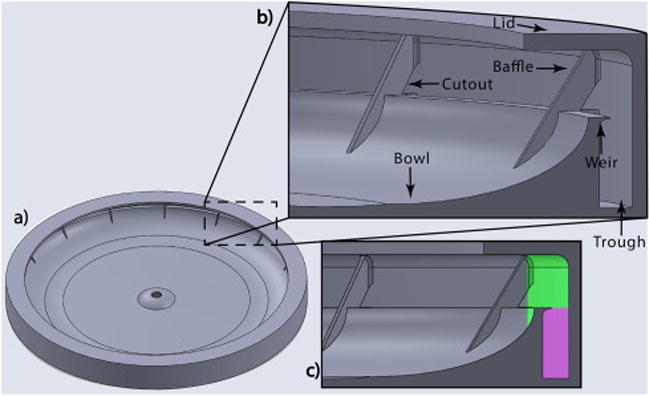
a) Top view of spinning disk; b) Cross-sectional view of inside of spinning disk; c) During spinning annular fluid chamber volume is 8.5 mL. When spinning stops, the upper volume of 5.0 mL (represented by green color) is expected to flow down, while the volume of 3.5 mL (represented by violet color) is retained in the trough by the weir. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For this study, disks were designed with angled baffles extending from the interface between the inside surface of the outer wall and lid of the disk, extending downwards and inwards to the bowl. Baffles were found to increase fluid stability, to decrease shear mixing at the interface between the plasma and the cell pack, and to wick fluid in the plasma layer, pulling it down from the annular fluid chamber into the bowl as the spinning slowed to a stop. Baffles were designed with triangular cutout windows from the inside surface of the outer wall of the disk (see figure) to the tip of the protruding weir, which allowed for flow and circulation behind the weir during the spinning process. The disks in this study had 16 baffles, and all extended in a sloping fashion over the front side of the weir, but they did not extend to the bottom of the trough behind the weir, thus allowing circulation in the trough. The disks were mounted onto a 3D printed platform that was connected by press fit to the spindle of a compact disc motor.
Programmable and repeatable electronic control of the spinning disk mounted on motor is described in detail in the Supplemental Material. The rotational velocity versus time profile for all experiments described herein is shown in Fig. 2.
Fig. 2.
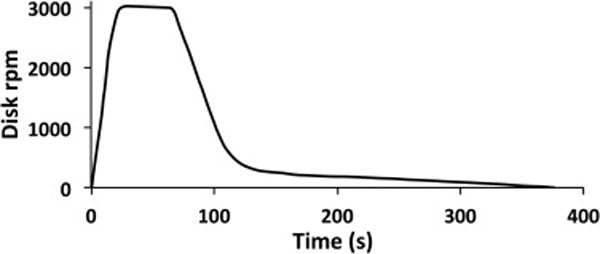
Velocity profile (rpm) of the spinning disk used in these experiments.
Prior to each experiment, the hollow disk was rinsed with water, sprayed copiously with 70% EtOH, agitated for 1 min, and then rinsed again with deionized water. Finally the disk was dried completely using clean compressed air.
2.2. Preparing bacteria
Bacteria was prepared by streaking E. coli (strain BL21 StarDE3) from frozen stock onto a nutrient agar plate and incubating at 37 °C. Twenty-four hrs later, a single colony was selected and transferred via sterile loop into a sterile 250-mL baffled flask containing 50 mL of nutrient broth (Difco, Becton Dickinson, Franklin Lakes, New Jersey), and incubated for 24 h at 37 °C with shaking at about 200 rpm. After 24 h, the bacterial suspension was centrifuged at 8,000xg for 10 min, the pellet was washed with sterile phosphate-buffer saline (PBS), resuspended, pelleted and washed twice more, and then diluted to a target concentration in PBS as estimated by optical density at 660 nm in a spectrometer (Agilent UV/vis spectrophotometer, model Cary 60).
2.3. Preparing blood for spinning
Blood was collected from human volunteers by a qualified phlebotomist directly into vacutainer tubes containing EDTA (BD #366643 10mL, Becton Dickinson, Franklin Lakes, New Jersey), Heparin (BD #367874 10mL) or Citrate (BD # 363083, 4mL), as specified by an IRB-approved protocol at Brigham Young University. Tubes containing blood were stored horizontally in the refrigerator for less than six hours before use. To prepare for spinning, the blood tubes were gently inverted several times by hand and 8 mL were pipetted into a sterile plastic tube (Fisherbrand, Culture Test Tube 17 × 100 mm). 100 μL of diluted E. coli was pipetted into the blood in the sterile tube, producing a concentration of around 106 CFU/mL, which was subsequently quantitated by serial dilution of the blood and plate counting. In some experiments, 115 (μL of various concentrations (0.5–1.1 g/L) of adenosine 5′-diphosphate (ADP) was also added to the blood. The tube was gently inverted by hand several times for about 10 s to ensure good mixing.
2.4. Spinning
Preliminary experiments showed that adding 1.5 mL of phosphate-buffer saline (PBS) to the blood increased (by more than 1.5 mL) the amount of plasma recovered. Therefore, just before spinning, 1.5 mL of PBS was pipetted onto the bowl of the disk. Then 7.0 mL of bacteria-spiked blood (sometimes with ADP) was also pipetted into the pool of PBS on the disk, and spinning commenced immediately. The blood was quickly flung to the inside surface of the outer wall of the disk. The thickness of the blood layer was 2.0 mm. Sedimentation of WBCs and RBCs into a cell-pack layer created a clear plasma layer within seconds that grew in thickness with spinning time. Ramp-up in speed to 3000 rpm took 22 s, and the speed was held at 3000 rpm for 45 s. During spinning two distinct vertical layers were observed to form, a transparent yellow cell-free plasma layer and a dark red cell pack layer. Then the disk was decelerated, quickly at first, and then slowly (see Fig. 2). Slow deceleration at low rpm was essential to avoid mixing of the clear plasma with the cell pack. Video clips of the beginning and ending of a spin are shown in the Supplemental Materials.
After the disk stopped spinning, the plasma that flowed down from the weir was collected by pipetting into a pre-weighed sterile tube. Typically there were some small streaks of red cells that flowed down with the plasma; usually the red streaks came from near the baffles (see Fig. 3). The tube containing recovered plasma was weighed and the weight of the plasma was recorded. The spinning disk was then washed and sterilized for subsequent experiments.
Fig. 3.
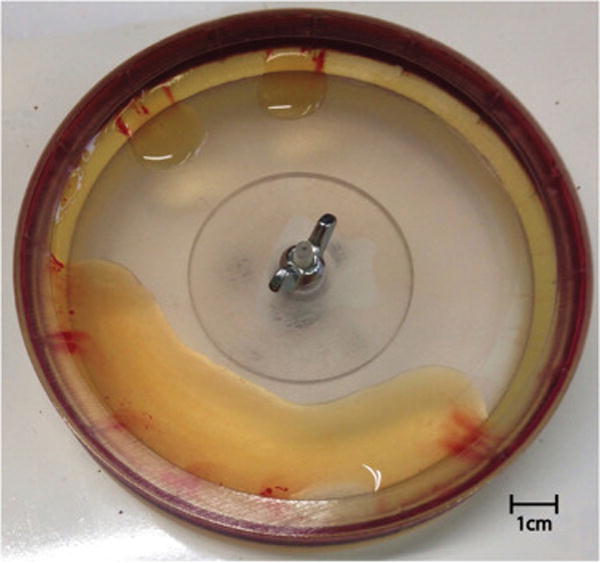
Photograph of the disk and the plasma that flowed down after a typical spin. The double streaks of thin red color originate from each side of a baffle. The red cell pack remains at the periphery of this transparent disk. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5. Quantitation of bacteria, red blood cells and platelets
Bacterial load in the whole blood (before pipetting onto the hollow disk) was determined by serial dilution in PBS and plate counting on nutrient agar plates (37 °C, 24–48 h). The bacterial load in the recovered plasma was also determined by serial dilution and plating. The bacteria loaded onto the hollow disk was calculated as the product of the whole blood CFU/mL, Cb,bl, multiplied by the volume of blood pipetted into the disk (Vbl, 7.0 mL). The bacteria recovered in the plasma was calculated as the product of the plasma CFU/mL, Cb,pl, multiplied by the volume of plasma recovered, Vpl, from each experiment. The efficiency of bacterial recovery, εBR, is the bacteria recovered in plasma divided by the original bacteria in the hollow disk.
| (2) |
Quantitation of RBCs in the whole blood or in recovered plasma was done indirectly by measuring the amount of hemoglobin (Hb) in the sample as follows. In calibration experiments, blood was diluted in PBS and RBCs were measured with a hemocytometer. The optical densities of these samples were measured at 280, 421 and 600 nm wavelengths on a UV/vis spectrometer. A correlation function was generated that related the hemocytometer counts to the optical density values. RBC counts in blood and in recovered plasma were calculated from this correlation.
The red cell reduction (RCR) is defined as the concentration of RBCs in the recovered plasma (CRBC,pl) divided by the concentration of RBCs in the whole blood (CRBC,bl) that was spiked with bacteria.
| (3) |
As will be discussed later, platelets (having a similar size and density as bacteria) did not sediment as quickly as did RBCs and white blood cells (WBCs). Their concentration in recovered plasma was measured by photographing (Zeiss, Axiocam 105) with a microscope (Olympus, BH2-UMA) the appropriate dilutions of the recovered plasma. Platelets were counted from digitized photographs using ImageJ software (NIH) and our own custom MATLAB codes.
3. Results
Whole blood (7 mL) spiked with bacterial suspension and slightly diluted by mixing with 1.5 mL of PBS was successfully separated within one minute into a plasma-rich layer and a cell-dense layer using the hollow disk described above. By carefully decelerating the disk, the plasma-rich layer was separated and collected, and is called hereafter “recovered plasma”. We emphasize that the disk must be carefully decelerated because if the deceleration occurs too rapidly, the recovered plasma mixes back into the cell-dense layer, and the separation is unsuccessful. We believe that the remixing of recovered plasma with the cells is a manifestation of Kelvin-Helmholtz instability [29–31].
Even with careful deceleration to avoid re-mixing, there was usually some “streaking” of red cells into the transparent amber-colored plasma as the disk finally stopped and the separated plasma flowed down for collection (see Fig. 3). The streaks usually originated from the baffles in the disk, and are thought to originate from faster flow of recovered plasma in the region near the baffle, as will be discussed later. However, despite the small amount of streaking, the overall concentration of RBCs was greatly reduced in the recovered plasma. The plasma also contained a large amount of bacteria. The following results present the measured reduction in RBCs, the volume of plasma recovered, the amount of bacteria recovered, and the platelet concentration in the recovered plasma for human blood that was anticoagulated (upon collection) with EDTA, citrate or heparin.
3.1. Red cell reduction
Although not strictly essential, elimination of RBCs is very desirable because RBCs may interfere with the subsequent processing and identification of the recovered bacteria. The recovered plasma still contained a reduced, but significant number of RBCs, as shown in Fig. 4. On average, gender of the blood donor had a significant effect on the number of the red blood cells in the recovered plasma. Blood from male donors generally had a greater RBC concentration than the blood from females, and this gender difference was manifest in higher number of red blood cells in the recovered plasma from male donors compared to that found in the recovered plasma from female donors. The average concentrations of RBCs in the blood of male and female donors were 5.46 × 109/mL and 5.14 × 109/mL. As the mean RBC concentrations (averaged over all donors of same gender) in recovered plasma were 4.21 × 108/mL and 2.06 × 108/mL, respectively, this represents an average RCR (red cell reduction) of 0.077 and 0.040 for male and female blood, and an average elimination of about 94% of the RBCs.
Fig. 4.
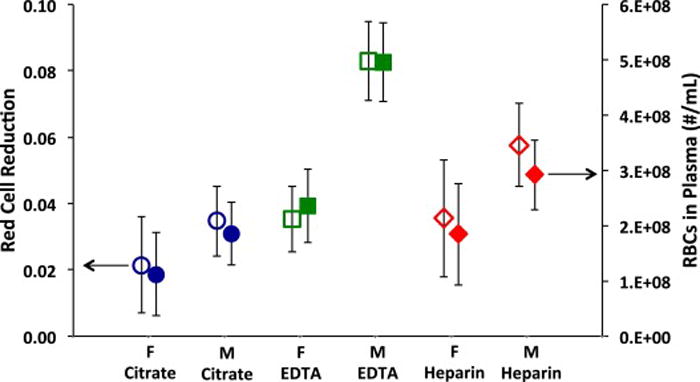
RBCs in recovered plasma versus the gender of the donor and the type of anticoagulant used. The left axis represents the red cell reduction (RCR) in the recovered plasma. The right axis represents the concentration of RBCs in the recovered plasma. The bacterial concentration ranged from 1.0 × 103 to 1.7 × 108 CFU/mL. The initial red cell concentrations in the slightly diluted blood had an average of 4.1 × 109 RBCs/mL. M = male; F = female. The mean values and 95% confidence intervals are represented (n> 15).
The type of anticoagulant made a difference in RCR value. Sodium citrate anticoagulant produced the greatest reduction in RBCs, and EDTA produced the least reduction. Male blood anticoagulated with EDTA had particularly high RBC counts (4.96 × 108/mL). When RBC counts were averaged across genders, the mean average (and 95% CI) for blood treated with citrate was 1.50 × 108/mL ± 0.49 × 108/mL, slightly and significantly less (p = 0.039) than the average for heparin-treated blood of 2.35 × 108 ± 0.62 × 108. The average for blood collected in EDTA tubes was 3.82 × 108 ± 0.55 × 108, which was significantly greater (p = 0.002) than blood collected in citrate tubes.
3.2. Plasma recovery
As shown in Fig. 5, neither the type of anticoagulant nor the gender of the donor produced a significant effect on the volume of the recovered plasma. The average volume of the recovered plasma in the experiments are 3.86 ± 0.19 mL and 3.81 ± 0.22 mL for female and male citrated blood, respectively; 3.70 ± 0.14 mL and 3.57 ± 0.17 for EDTA-treated females and male blood, respectively; and 3.81 ± 0.20 mL and 3.74 ± 0.19 mL for heparinized female and male blood, respectively. An important observation is that about 3.7mLof plasma were recovered from a volume of8.5mL of blood and PBS. The geometrically calculated volume behind the weir is 3.5 mL. Thus about 1.3 mL of fluid did not drain down for collection. Non-recovered liquid was observed on top of the weir, and in corners formed by baffles and the lid.
Fig. 5.
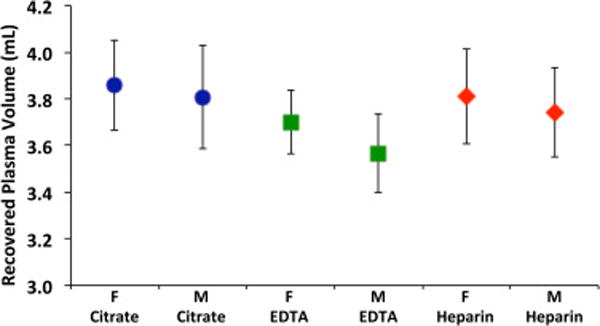
Volume of recovered plasma versus the gender of the donor and the type of anticoagulant used. M = male; F= female. The mean values and 95% confidence intervals are represented (n> 18).
Because the plasma did not readily wet the photopolymerized disk, we did several experiments with wetting agents in an attempt to cause more plasma to flow down from the top of the weir and corners of the hollow disk. Wetting agents that we applied included γ-aminobutyric acid, sebacic acid and Pluronic P105. These were administered by spraying or soaking the disk, and then wiping or blowing dry, immediately before the blood was pipetted into the hollow disk. While these agents changed the contact angle of water on a freshly photopolymerized and washed surface, when blood was spun and the disk eventually slowed to a stop, the plasma at the edge of the weir had a contact angle of greater than 90° and did not wet or spread on the weir. None of these treatments produced any significant change in the amount of plasma recovered or in the rate of drainage of the plasma.
3.3. Bacterial recovery
The average εBR (efficiency of bacterial recovery) was approximately 0.36 in the experiments that used EDTA as anticoagulant. There was no statistically significant difference between bacteria recovery using male blood 0.37 ± 0.03 (n = 60) and female blood 0.33 ± 0.04 (n = 32) (mean ± 95% CI, p = 0.09,). When citrate or heparin was used as an anticoagulant, we could neither accurately nor repeatedly measure the CFUs in the recovered plasma; we attribute this to inhibition of bacterial growth due to hemoglobin [32,33], as will be discussed later. Thus we cannot report a εBR from blood anticoagulated with citrate nor heparin in these experiments using bacterial loadings on the order of 106 CFU/mL blood or less.
3.4. Platelet recovery
It is less desirable to have platelets in the recovered plasma since these may interfere with subsequent analysis of the bacteria by adhering to or clogging channels or filters. The number of the platelets in the recovered plasma was calculated from multiple microphotographs as explained in the materials and methods section. As seen in Fig. 6, the fewest platelets were observed when the blood was anticoagulated with citrate, and the most were observed using EDTA-treated blood. However, due to the large scatter in platelet numbers between individuals, the differences are not statistically significant. Donors were not screened for use of aspirin or other platelet-affecting drugs. We observed no difference associated with the gender of the donor.
Fig. 6.
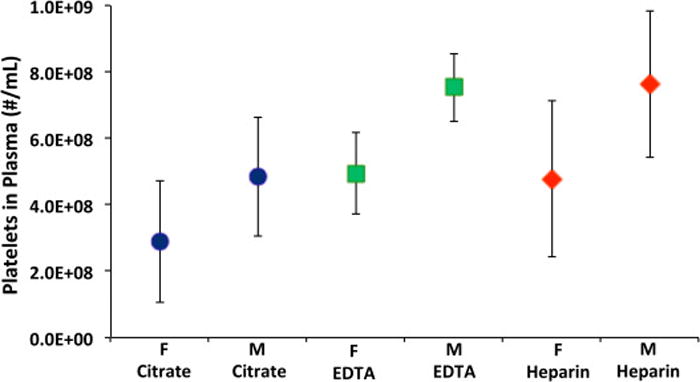
Concentration of platelets in the recovered plasma as a function of anticoagulant and gender. The ADP concentration in these experiments ranged from 0 to 1.1 g/l. M = male; F = female. The mean values and 95% confidence intervals are represented (n>6).
Adenosine diphosphate (ADP) is known to cause platelet aggregation in anticoagulated blood [34]. Several experiments were done in which ADP was added to the blood samples at concentrations of 0.5–1.1 g/L before spinning to aggregate the platelets, as large aggregates would sediment out faster than bacteria and individual platelets. This concentration is used in other procedures to aggregate platelets in blood. However, the resulting platelet counts in recovered plasma were not statistically different than without ADP.
4. Discussion
We were successful in using a sedimentation process on a hollow spinning disk to separate the plasma from fresh human blood containing E. coli bacteria. The separation of all RBCs is not complete, but it is sufficient to remove the majority of RBCs and nearly all of the WBCs, while leaving much of the bacteria and platelets in the recovered plasma. In these experiments using slightly diluted blood, we recovered on the order of 36% of the bacteria that was inoculated at fairly high concentrations into the blood, in the range of 102 to 108 CFU/mL. While higher than expected to be found in patients with clinical manifestation of sepsis, these concentrations were used in these initial experiments so the bacteria recovery could be quantified using plate-counting techniques.
The elimination of WBCs was essential so that subsequent molecular analysis of the bacteria DNA would not be contaminated with human DNA. Although the vast majority of RBCs do not contain DNA, their removal was desired to allow easier subsequent processing of the recovered plasma sample through filters that might be blocked by high RBC concentrations.
The reduction or elimination of platelets was also desired as they could block or clog filters or microfluidic channels. However, we found that in these experiments the platelet function was sufficiently blocked by the anti-coagulants in that they did not form large aggregates and did not appear to adhere to surfaces. We tried to remove platelets by reversing their anti-aggregation with ADP. However, addition of ADP at the concentrations employed for platelet aggregometry made no significant difference in platelet concentration in recovered plasma, perhaps because the rate of forming large aggregates is on the time scale of minutes [34], and in these experiments the spinning started in less that a minute following addition of ADP.
While a complete separation of bacteria and RBCs would be desirable, the physics of separation by sedimentation shows that not all bacteria can be recovered in separated plasma, and that increasing bacterial recovery also produces more unwanted RBCs in the recovered plasma. Thus for clinical application, this process needs to be optimized to recover sufficient numbers of bacteria for subsequent analysis, and yet have adequately low RBC and platelet counts that the plasma can be processed to isolate the bacteria for molecular analysis of its DNA to identify the species and resistance profile. We found that there are several chemical and physical parameters that govern the reduction of RBCs and the amount of bacterial recovery, and thus influence their balance for a clinical process. The final optimization of spinning speed, time, disk size and more will depend on the tolerance of the clinical detection process to interference from blood cells and plasma proteins and on the detection sensitivity of the process that identifies the bacteria and its resistance profile.
4.1. General disk design
The design of the hollow disk, its spinning, and its carefully controlled deceleration are keys to efficient RBC reduction while collecting as much bacteria as possible. In preliminary experiments we made many iterations of disk design and finally arrived at the design used in these experiments (Fig. 1). This design has a weir and several baffles that are designed to retain the cell pack from flowing down and yet maximize the volume of recovered plasma that flows down and is collected.
The weir is a barrier designed to retain the red cell pack so it does not flow to the center of the disc when spinning ceases. In these experiments, the weir height is designed to retain 3.5 mL of volume, which is roughly the volume of RBCs in 7.0 mL of male blood. In preliminary experiments we examined many shapes of the upper weir surface and arrived at the design used in this experiment. The steep front edge promoted quick flow of plasma over the weir, and the pointed profile of the back edge of the weir seemed to reduce the amount of streaking of red cell pack into the recovered plasma.
During the spinning, a separation of clear plasma and cell pack was visually observed through the transparent plastic forming the roof of the hollow disk. The thickness of the cleared layer of plasma increased during the spinning. After 1 min, if the disk was decelerated too rapidly, the red layer of cells mixed back into the clarified plasma, and a fluid of red blood color flowed down when the disk stopped spinning. This is referred to as “mixing” and is easily differentiated from “streaking”, in which the plasma layer and cell pack remained separated, but with thin streaks of red as explained in the following paragraph. We think that mixing is caused by a Kelvin-Helmholtz instability [29–31] at the interface between the plasma and cell pack, which have differing densities and viscosities. Mixing during deceleration has been observed by others in smaller-scale blood separation systems [35].
While the plasma was flowing down, sometimes there were thin streaks of red color that flowed into the transparent amber-colored plasma, which we call “streaking”. While there was no mixing in this set of experiments using the disk design of Fig. 1 and using the deceleration profile shown in Fig. 2, streaking was observed to some extent in most (but not all) experiments. Typically the first volume of plasma to flow down was transparent amber in color, as if devoid of RBCs. Then streaks of red started to flow with the plasma, originating from the edges of the baffles that extended beyond the weir. We believe that streaking is caused by laminar flow of the plasma that drags some of the adjacent red cell pack with it. We cannot tell if the baffles somehow wick the red cell pack into the plasma, or if there is higher local velocity near the baffles that drags a very small amount of the red cell pack into the plasma. During iterations on the disk design, we found that the amount of streaking is reduced by putting triangular windows in the back of the baffles above the weir height (see Fig. 1) and by not extending the baffles to the bottom of the trough. We posit that these openings in the region of the red cell pack allowed some flow of the red cell pack that relaxed the red cell pack while the plasma was coming down, and reduced any localized flow of the red cell pack, leading to less streaking.
Sixteen baffles were selected for this disk design, which is a rough compromise between fewer baffles that produced more streaking and more baffles that made washing the disk onerous.
The weir and baffles have not yet been optimized in any systematic experimental design. We expect that with more design development and experimentation we can further increase the amount of plasma that flows down, and minimize the amount of streaking and mixing of the cell pack into the recovered plasma.
4.2. General plasma recovery
It is curious that the amount of blood and PBS was 8.5 mL, and the volume behind the weir was 3.5 mL. Yet we consistently obtained only about 3.7 mL of plasma. Visual observation showed that there was liquid on the top of the weir, in the upper corners of the back wall and roof, and the corners between baffles and walls. In the version of the disk used in these experiments, the corners were rounded with fillets, which reduced plasma retention, but did not eliminate it. Since εBR is proportional to plasma recovery, we know that more design changes should be done to promote more plasma recovery. Adjusting the edge and slope of the weir, and increasing the roof distance appear to be the most influential parameters for future iterations.
We tried to increase plasma recovery by putting wetting agents on the surface, as the acrylic disk does not wet readily. Initial experiments of wiping, dipping or spraying the disk with γ-aminobutyric acid, sebacic acid, or Pluronic P105 did not statistically increase the plasma recovery; nor did they appear to significantly change wettability.
4.3. Total bacterial recovery
The efficiency of bacterial recovery (εBR) is influenced by many parameters. We have already mentioned parameters that govern plasma recovery. Of course the disk design, the spinning speed, and the time for sedimentation are perhaps the most important, as increasing the latter two parameters will spin more of the bacteria into the red cell pack and preclude recovery. This report focuses on the chemical aspects of this separation system that affect recovery. We already mentioned that wetting agents and platelet aggregation agents had little influence, at least in the ranges investigated.
The most significant chemical parameter appears to be the type of anticoagulant used during blood collection. EDTA, sodium citrate and heparin were examined. EDTA and citrate inhibit coagulation by chelating calcium and inhibiting calcium dependent processes such as platelet aggregation. Heparin binds to antithrombin III and interferes with fibrinogen polymerization [36]. Heparin also binds to bacteria [16,37], so its use in the separation process may be problematic if heparin causes aggregation of bacteria, such that they sediment faster as a group than as individual bacteria. On the other hand, clinical blood samples have such a low bacterial loading that aggregation by heparin might be extremely rare.
The data suggest that the use of EDTA leaves more RBCs and platelets in the recovered plasma, which is less desirable. Unfortunately the use of citrate and heparin appeared to inhibit bacterial growth during serial dilution and plating, and therefore we do not have quantitative comparisons of bacterial recovery using these 3 anticoagulants, and it may be possible that citrate may be more useful in bacterial recovery and RBC reduction in clinical applications, as long as the bacteria do not need to be cultured.
We attributed the inhibition of bacterial growth to the lack of EDTA in the presence of RBCs. In exploratory experiments, there was no inhibition of bacterial growth on nutrient plates in clean plasma from blood anticoagulated with citrate, heparin or EDTA in which the RBCs were entirely removed from plasma by centrifugation in test tubes for 10 min. Yet adding back the centrifuged RBCs into the centrifuged plasma inhibited bacterial growth on plates when the blood was originally anticoagulated with citrate or heparin. But no inhibition was observed when adding back RBCs to blood anticoagulated with EDTA. The hemoglobin within RBCs is known to inhibit bacterial growth [32,33]. When recovered plasma with a low concentration of RBCs is diluted and plated to grow the bacteria into colonies, the bacteria are deposited on the plate along with high numbers of RBCs. We speculate that these RBCs eventually lyse on the agar plate, and the nutrient feeding the bacteria contains Hb, which inhibits growth. Apparently EDTA somehow interferes with the action of Hb on bacteria, but we have found nothing in the literature to support this proposition.
We consider it remarkable that bacteria can be recovered at such a high concentration, given that the bacteria start randomly distributed throughout the disk, surrounded by billions of much larger discoidal and deformable red blood cells that are packed closely, leaving very little room for the escape of a bacterium from the surrounding cage of RBCs. When the spinning starts, the RBCs start sedimenting as a group toward the back wall of the hollow disk. This concerted movement of nearly half the volume displaces the plasma fluid away from the back wall. The slightly smaller bacteria are caught between the fluid flow pushing them away from the back wall and the gauntlet of RBCs that close in upon them and impede their migration with the flow of the displaced plasma. The closer the bacteria are to the back wall of the disk, the higher will be the probability that they will be trapped by the condensing field of RBCs before they are pushed by plasma to the clear plasma phase. This scenario leads to the proposal that if the RBCs were not packed so closely together, more bacteria would escape to the plasma phase. Thus we hypothesize that a higher εBR could be attained if the blood were diluted even more before spinning. However, more dilution and the corresponding larger blood volume would create a longer sedimentation distance, a longer sedimentation time (spin time), and larger volume of waste. Optimization of the process depends on constraints or needs, such as the process time, process volume and waste volume.
In conclusion we have shown a process that can recover about 36% of the bacteria in human blood by allowing an imperfect separation of RBCs and platelets from plasma. The amount of RBCs could probably be decreased further by sedimenting longer, but this would also decrease bacterial recovery. The use of EDTA, citrate and heparin as anticoagulants were examined, and EDTA corresponds to the poorest red blood cell reduction. In these experiments we were unable to determine the effect of citrate and heparin on bacteria recovery, as they interfered with bacterial counting by plate growth. Use of EDTA also correlated with higher platelet counts in the recovered plasma, but this increase may not be statistically significant. There was no significant effect on bacterial recovery or RBC reduction when using ADP as a platelet aggregator, even at concentrations used in platelet aggregometry. The amount of plasma recovered was determined by the disk geometry, and was not influenced by the anticoagulant or any wetting agents on the surface of the disk. While the εBR is low compared to some processes that use microfluidic channels or magnetic beads, the strong advantages of this simple sedimentation technique include the rapid processing, the large blood volume, the small dilution and waste, and the inexpensive hardware and consumables.
We envision the eventual application of this technology in clinical diagnosis of blood infections and the rapid identification of the infectious organism and its antibiotic resistance profile. We anticipate that rapid separation of bacteria from blood (in min) will be followed by filtration and washing (seconds) to remove the plasma. Then the bacteria could be lysed (along with residual platelets and RBCs) in minutes, and the DNA recovered for rapid molecular detection of unique identifying genes by PCR or other techniques. Our device is small and inexpensive, and the “used” disk (containing packed blood cells) can be disposed of. The simple and inexpensive technology could be the front end of “point-of-care” processes in clinics and hospitals, requiring only 7mL of blood, and avoiding sending samples to centralized laboratories.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grant number R01AI116989.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.colsurfb.2017.03.027.
References
- 1.CDC. Morbidity and Mortality Weekly Report. CDC; Atlanta, GA: 2013. Vital Signs: Carbapenem-Resistant Enterobacteriaceae; pp. 165–170. [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman KE, Lessler J, Cosgrove SE, Harris AD, Lautenbach E, Han JH, Milstone AM, Massey CJ, Tamma PD. Antibacterial resistance leadership, a clinical decision tree to predict whether a bacteremic patient is infected with an extended-Spectrum beta-lactamase-producing organism. Clin Infect Dis. 2016;63(7):896–903. doi: 10.1093/cid/ciw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweizer ML, Furuno JP, Harris AD, Johnson JK, Shardell MD, McGregor JC, Thom KA, Cosgrove SE, Sakoulas G, Perencevich EN. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis. 2011;11:279. doi: 10.1186/1471-2334-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 5.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, Stuber F. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol (Berl) 2008;197(3):313–324. doi: 10.1007/s00430-007-0063-0. [DOI] [PubMed] [Google Scholar]
- 7.Kreger BE, Craven DE, Carling PC, McCabe WR. Gram-negative bacteremia. III. Reassessment of etiology, epidemiology and ecology in 612 patients. Am J Med. 1980;68(3):332–343. doi: 10.1016/0002-9343(80)90101-1. [DOI] [PubMed] [Google Scholar]
- 8.Leggieri N, Rida A, Francois P, Schrenzel J. Molecular diagnosis of bloodstream infections: planning to (physically) reach the bedside. Curr Opin Infect Dis. 2010;23(4):311–319. doi: 10.1097/QCO.0b013e32833bfc44. [DOI] [PubMed] [Google Scholar]
- 9.Yagupsky P, Nolte FS. Quantitative aspects of septicemia. Clin Microbiol Rev. 1990;3(3):269–279. doi: 10.1128/cmr.3.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitt WG, Alizadeh M, Husseini GA, McClellan DS, Buchanan CM, Bledsoe CG, Robison RA, Blanco R, Roeder BL, Melville M, Hunter AK. Rapid separation of bacteria from blood-review and outlook. Biotechnol Prog. 2016;32(4):823–839. doi: 10.1002/btpr.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, Mammoto T, Berthet JB, Tobin H, Cartwright MJ, Watters AL, Rottman M, Waterhouse A, Mammoto A, Gamini N, Rodas MJ, Kole A, Jiang A, Valentin TM, Diaz A, Takahashi K, Ingber DE. An extracorporeal blood-cleansing device for sepsis therapy. Nat Med. 2014;20(10):1211–1216. doi: 10.1038/nm.3640. [DOI] [PubMed] [Google Scholar]
- 12.Kang JH, Um E, Diaz A, Driscoll H, Rodas MJ, Domansky K, Watters AL, Super M, Stone HA, Ingber DE. Optimization of pathogen capture in flowing fluids with magnetic nanoparticles. Small. 2015;11(42):5657–5666. doi: 10.1002/smll.201501820. [DOI] [PubMed] [Google Scholar]
- 13.Lee JJ, Jeong KJ, Hashimoto M, Kwon AH, Rwei A, Shankarappa SA, Tsui JH, Kohane DS. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. 2014;14(1):1–5. doi: 10.1021/nl3047305. [DOI] [PubMed] [Google Scholar]
- 14.Lopes AL, Cardoso J, Santos FR Dos, Silva AC, Stets MI, Zanchin NI, Soares MJ, Krieger MA. Development of a magnetic separation method to capture sepsis associated bacteria in blood. J Microbiol Methods. 2016;128:96–101. doi: 10.1016/j.mimet.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Chu YW, Engebretson DA, Carey JR. Bioconjugated magnetic nanoparticles for the detection of bacteria. J Biomed Nanotechnol. 2013;9(12):1951–1961. doi: 10.1166/jbn.2013.1701. [DOI] [PubMed] [Google Scholar]
- 16.Mattsby-Baltzer I, Bergstrom T, McCrea K, Ward R, Adolfsson L, Larm O. Affinity apheresis fortreatment of bacteremia caused by Staphylococcus aureus and/or methicillin-resistant S. aureus (MRSA) J Microbiol Biotechnol. 2011;21(6):659–664. [PubMed] [Google Scholar]
- 17.Wang SQ, Inci F, Chaunzwa TL, Ramanujam A, Vasudevan A, Subramanian S, Ip ACF, Sridharan B, Gurkan UA, Demirci U. Portable microfluidic chip for detection of Escherichia coli in produce and blood. Int J Nanomed. 2012;7:2591–2600. doi: 10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Didar TF, Cartwright MJ, Rottman M, Graveline AR, Gamini N, Watters AL, Leslie DC, Mammoto T, Rodas MJ, Kang JH, Waterhouse A, Seiler BT, Lombardo P, Qendro EI, Super M, Ingber DE. Improved treatment of systemic blood infections using antibiotics with extracorporeal opsonin hemoadsorption. Biomaterials. 2015;67:382–392. doi: 10.1016/j.biomaterials.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 19.Neth O, Jack DL, Dodds AW, Holzel H, Klein NJ, Turner MW. Mannose-binding lectin binds to a range of clinically relevant microorganisms and promotes complement deposition. Infect Immun. 2000;68(2):688–693. doi: 10.1128/iai.68.2.688-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matosziuk AS, Harney KW, MacRenaris TJ, Meade WM. Synthesis, characterization, and in vitro testing of a bacteria-targeted MR contrast agent. Eur J Inorg Chem. 2012;12:2099–2107. doi: 10.1002/ejic.201101362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leevy WM, Johnson JR, Lakshmi C, Morris J, Marquez M, Smith BD. Selective recognition of bacterial membranes by zinc(II)-coordination complexes. Chem Commun. 2006;15:1595–1597. doi: 10.1039/b517519d. [DOI] [PubMed] [Google Scholar]
- 22.Hiemenz R, Rajagopalan RG. Principles of Colloid and Surface Chemistry. 3rd. Marcel Dekker; New York: 1997. [Google Scholar]
- 23.Harrison RG, Todd P, Rudge SR, Petrides DP. Bioseparations Science and Engineering. Oxford University Press; New York: 2003. [Google Scholar]
- 24.Hou HW, Wu L, Amador-Munoz DP, Vera MP, Coronata A, Englert JA, Levy BD, Baron RM, Han J. Broad spectrum immunomodulation using biomimetic blood cell margination for sepsis therapy. Lab Chip. 2016;16(4):688–699. doi: 10.1039/c5lc01110h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou HW, Bhattacharyya RP, Hung DT, Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip. 2015;15(10):2297–2307. doi: 10.1039/c5lc00311c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mach AJ, Di Carlo D. Continuous scalable blood filtration device using inertial microfluidics. Biotechnol Bioeng. 2010;107(2):302–311. doi: 10.1002/bit.22833. [DOI] [PubMed] [Google Scholar]
- 27.Wu ZG, Willing B, Bjerketorp J, Jansson JK, Hjort K. Soft inertial microfluidics for high throughput separation of bacteria from human blood cells. Lab Chip. 2009;9(9):1193–1199. doi: 10.1039/b817611f. [DOI] [PubMed] [Google Scholar]
- 28.Hou H Wei, Gan HY, Bhagat AA, Li LD, Lim CT, Han J. A microfluidics approach towards high-throughput pathogen removal from blood using margination. Biomicrofluidics. 2012;6(2):24115–2411513. doi: 10.1063/1.4710992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper AP, Boyd WGC. Shear-flow instability at the interface between two viscous fluids. J Fluid Mech. 1983;128:507–528. [Google Scholar]
- 30.Yih CS. Instability due to viscosity stratification. J Fluid Mech. 1967;27(2):337–352. [Google Scholar]
- 31.Goldstein S. On the stability of superposed streams of fluids of different densities. Proc R Soc Lond Ser A. 1931;132(820):524–548. [Google Scholar]
- 32.Parish CA, Jiang H, Tokiwa Y, Berova N, Nakanishi K, McCabe D, Zuckerman W, Xia MM, Gabay JE. Broad-spectrum antimicrobial activity of hemoglobin. Bioorg Med Chem. 2001;9(2):377–382. doi: 10.1016/s0968-0896(00)00263-7. [DOI] [PubMed] [Google Scholar]
- 33.Liepke C, Baxmann S, Heine C, Breithaupt N, Standker L, Forssmann WG. Human hemoglobin-derived peptides exhibit antimicrobial activity: a class of host defense peptides. J Chromatogr B: Analyt Technol Biomed Life Sci. 2003;791(1–2):345–356. doi: 10.1016/s1570-0232(03)00245-9. [DOI] [PubMed] [Google Scholar]
- 34.Hall MW, Goodman PD, Solen KA, Mohammad SF. Formation of occlusive platelet aggregates in whole blood caused by low concentrations of ADP. ASAIO J. 2000;46(6):693–695. doi: 10.1097/00002480-200011000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Amasia M, Madou M. Large-volume centrifugal microfluidic device for blood plasma separation. Bioanalysis. 2010;2(10):1701–1710. doi: 10.4155/bio.10.140. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg RD. Heparin, antithrombin, and abnormal clotting. Annu Rev Med. 1978;29:367–378. doi: 10.1146/annurev.me.29.020178.002055. [DOI] [PubMed] [Google Scholar]
- 37.McCrea K, Ward R, LaRosa SP. Removal of Carbapenem-Resistant Enterobacteriaceae (CRE) from blood by heparin-functional hemoperfusion media. PLoS One. 2014;9(12):e114242. doi: 10.1371/journal.pone.0114242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


