Abstract
The glyoxylate regeneration cycle (GRC) operates in serine cycle methylotrophs to effect the net conversion of acetyl coenzyme A to glyoxylate. Mutants have been generated in several genes involved in the GRC, and phenotypic analysis has been carried out to clarify their role in this cycle.
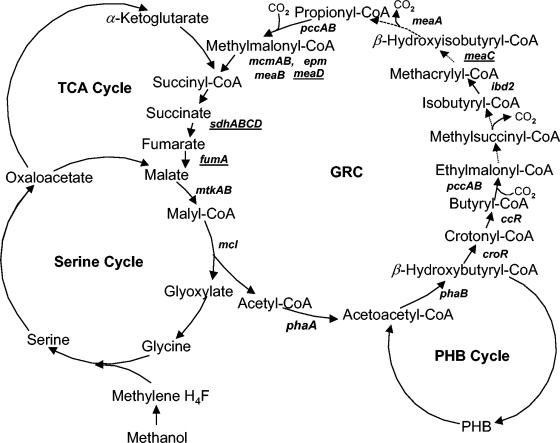
Methylobacterium extorquens AM1 is a facultative methylotroph that utilizes the serine cycle for C1 assimilation (1). In addition to the C1 compounds (methanol and methylamine), M. extorquens AM1 is able to grow on C2 (ethanol and ethylamine), C3 (pyruvate), and C4 (succinate) compounds. The organism does not possess the classical glyoxylate shunt and uses an alternative anapleurotic pathway, the novel glyoxylate regeneration cycle (GRC) (5, 7, 8), to regenerate glyoxylate during growth on C1 and C2 compounds. This pathway involves a series of reactions proceeding via coenzyme A (CoA) derivatives of C3, C4, and C5 carboxylic acids, as shown in Fig. 1. However, a few steps in the cycle have not yet been characterized, and some reactions remain unknown. Specifically, enzymes participating in ethylmalonyl-CoA conversion into isobuturyl-CoA, as well as enzymes involved in the conversion of methacrylyl-CoA into propionyl-CoA, are yet to be revealed, as well as the substrate for MeaA, a putative adenosylcobalamin (AC)-dependent mutase (16). The fate of succinyl-CoA also remains unclear, as no evidence exists for its entrance into the tricarboxylic acid (TCA) cycle. In this work we describe two new genes involved in the GRC and investigate the relationship of the GRC to the TCA cycle.
FIG. 1.
Glyoxylate regeneration cycle in M. extorquens AM1. Gene names are in bold italic. Novel GRC genes described in this work are underlined. Dotted lines indicate reactions catalyzed by enzymes not yet known or detected. For more information, see reference 5.
meaD encodes ACA and is involved in the methylmalonyl-CoA mutase step.
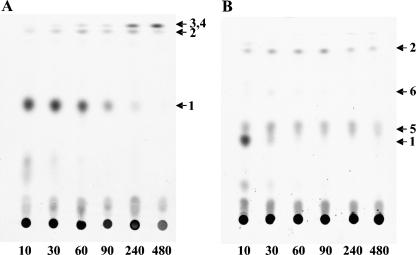
A set of mutants have been isolated previously in a gene designated meaD as part of the random transposon mutagenesis project, showing growth defects on C1 and C2 compounds, characteristic of mutants in the GRC (5, 7, 8). The translated product of meaD shows 38% identity to human ATP:cobalamin adenosyltransferase (ACA) (11). We have previously reported on decreased levels of AC in this mutant (9). AC is a cofactor of methylmalonyl-CoA mutase (MCM), which is a part of the GRC (7). It has also been suggested as a potential cofactor of MeaA, based on its sequence similarity with the sequences of MCM enzymes (16). To directly test for the ACA activity of MeaD, we expressed meaD in Escherichia coli as follows. Primers for meaD PCR amplification were designed so that the coding sequence would be precisely fused to the N terminus of a His6 tag. They were complementary to the first and the last 24 bases of meaD, and they also contained specific restriction sites, NdeI and XhoI, respectively. After PCR amplification and restriction with NdeI and XhoI, the DNA fragment containing meaD was cloned into the pET28a vector (Novagen) using the 5′-NdeI and the 3′-XhoI restriction sites. The resultant plasmid was transformed into the expression strain E. coli BL21 DE3 (Novagen), and the expression was performed as previously described (9). MeaD was purified from cell extracts to homogeneity by nickel affinity chromatography, as described elsewhere (9). The MeaD polypeptide was present at high concentration (data not shown). Size-exclusive chromatography of the purified MeaD on a Superdex 200 HR 10/30 calibrated with suitable marker proteins showed that MeaD had a molecular mass of 50 kDa, indicating that MeaD is present as a dimer. MeaD was tested for ACA activity using the method described in reference 11, and a specific activity of 4.5 mmol/min/mg of protein was determined. To test whether meaD is involved in both MCM and MeaA steps of the GRC or in only one, we followed the fate of 14C-labeled carbon from acetate (4 μCi) in the presence of unlabeled methanol via thin-layer chromatography followed by gas chromatography-mass spectrometry detection, as we described earlier (7). For this experiment, a knockout deletion mutant in meaD was generated, essentially as described earlier (12), to ensure the null function of the gene. The meaD knockout mutant revealed a phenotype similar to the phenotypes of other GRC mutants defective for growth on C1 and C2 compounds, and the phenotype was reversed by the addition of glyoxylate (5, 7, 8). The following labeled products were detected in the meaD mutant: β-hydroxybutyrate, β-hydroxyisobutyrate, methylsuccinate, and methylmalonate (Fig. 2). No succinate was detected, indicating that the cycle must be interrupted at the MCM step. The data on the presence of labeled methylmalonate in this mutant imply that MeaA must not require MeaD for its activity. This conclusion was further supported by testing the distribution of 14C from [1-14C]butyrate (final concentration, 0.04%; 0.2 μCi per assay mixture) between CO2 and biomass, as previously described (7). We have shown previously that mutants in the known GRC genes fall into two groups with regard to [1-14C]butyrate labeling pattern: the first group includes mutations in mcmA and meaB as well as the wild-type strain and these strains accumulate the majority of 14C (65 to 70%) as 14CO2, while the second group includes pccA, pccB, ibd2, and meaA mutants, which accumulate only 18 to 27% of radioactivity as 14CO2 (7). Mutants in the first group were blocked in reactions following the decarboxylation step (catalyzed by an unknown decarboxylase) (Fig. 1), and mutants in the second group were blocked in reactions preceding the decarboxylation step (7). In similar experiments, the meaD mutant accumulated levels of radioactivity in 14CO2 similar to the levels reported for the wild-type strain (about 70%), confirming that MeaD is involved in the MCM step of the cycle and is not essential for the MeaA step. It is possible that MeaA binds a modified form of AC. Another explanation for the labeling pattern in the meaD mutant would be that MeaD is specific to MCM and another gene participates in biosynthesis of the cofactor for MeaA.
FIG. 2.
Autoradiograms of labeled intermediates accumulated after exposure of whole cells of meaC (A) and meaD (B) mutants to [14C]acetate and methanol, separated by thin layer chromatography (Kieselgel 60; Merck), using the mobile phase of isopropyl ether-formic acid-water (90:7:3). Numbers at the bottom are time (in seconds). Products detected: 1, β-hydroxybutyrate; 2, methylsuccinate; 3, ethylmalonate; 4, butyrate; 5, β-hydroxyisobutyrate; 6, methylmalonate. Ethylmalonate and butyrate were cut out as a single spot and identified by mass spectrometry (8).
meaC is involved in a step between methylsuccinyl-CoA and propionyl-CoA.
Another transposon insertion mutant showing growth defects on C1 and C2 compounds has been described in a gene designated as meaC (13). The polypeptide translated from meaC shows similarity with acyl dehydratases, based on COG analysis (National Center for Biotechnology Information). One of the steps in the GRC, the conversion of methacrylyl-CoA into 3-hydroxyisobuturyl-CoA, would require an enzyme possessing hydratase activity, and the gene for this step has not been identified (Fig. 1). We tested for a possible role of meaC in this step by following the fate of 14C-labeled carbon from acetate and also by following 14C distribution from 14C-labeled butyrate, as described above for the meaD mutant. For these experiments, a knockout deletion mutant in meaC was generated essentially as described in reference 12 to ensure the null function of MeaC. The phenotype of the resultant mutant was similar to the phenotype of the MeaD mutant and other GRC mutants (5, 7, 8). When incubated with 14C-labeled acetate in the presence of unlabeled methanol, the mutant accumulated β-hydroxybutyrate, butyrate, ethylmalonate, and methylsuccinate but not β-hydroxyisobutyrate, methylmalonate, or succinate (Fig. 2). These data place the reaction catalyzed by MeaC between methylsuccinyl-CoA and β-hydroxyisobutyryl-CoA. These results agree with the data from the label distribution experiments in which the meaC mutant incorporated only about 14% of the radioactivity into CO2, indicating that the reaction catalyzed by MeaC precedes the decarboxylation step. There are two steps between methylsuccinyl-CoA and β-hydroxyisobutyryl-CoA catalyzed by unknown enzymes; however, only one of them would require a hydratase or dehydratase. Thus, it is most likely that MeaC catalyzes the hydration of methacrylyl-CoA. However, we were unable to directly test the hydratase activity of MeaC due to our inability to obtain MeaC in the soluble fraction in expression experiments similar to those described above for MeaD (data not shown).
GRC overlaps with the TCA cycle.
In our original description of the GRC in M. extorquens AM1, we suggested that a sequence of the reactions of the TCA cycle between succinyl-CoA and malate would be required for the cycle to operate (7). The enzymes involved would be succinyl-CoA synthase (SCS) or succinyl-CoA hydrolase (SCH), succinate dehydrogenase (SDH), and fumarase (Fum), as seen in Fig. 1. Analysis of the almost-complete sequence of the genome of M. extorquens AM1 has shown that a single gene cluster is present and predicted to encode SCS (scsAB) and a single gene cluster is present that is predicted to encode SDH (sdhABCD), while two genes predicted to encode Fum (fumA and fumB) are present (4). The gene for SCH could not be identified, as no such gene is known for any organism. Previous attempts to mutate sdhA or fumA by selecting recombinants on methanol or succinate did not result in knockout mutants, suggesting that both sdhA and fumA are essential for growth on either substrate (17, 18). We reasoned that if sdhA and fumA were indeed involved in glyoxylate regeneration, selecting on methanol in the presence of glyoxylate should result in knockout mutants. Indeed, by selecting on methanol in the presence of 1 mM glyoxylate, double-crossover knockout mutants were obtained by using the donors employed in the previous studies (17, 18). These new mutants were tested for growth on methanol or succinate. Both mutants grew poorly on methanol, but also grew even more poorly on succinate, compared to wild-type growth rates (data not shown). Growth on methanol was stimulated by the addition of glyoxylate, as is typical of mutants in the GRC. Activity measurements confirmed the nature of the mutants. While SDH (assayed as described in reference 14) was detected in the wild type grown on succinate, methanol, or methanol supplemented with glyoxylate (12 ± 2 mU on either substrate [mean ± standard deviation]), it was not detectable in the sdhA mutant grown on methanol supplemented with glyoxylate (<1 mU). Fumarase activity (measured as the increase in absorbance at 340 nm in the following reaction mixture: 100 mM Tris-HCl [pH 8.0], 10 mM fumarate, 1 mM NAD, 2 U of malate dehydrogenase [all from Sigma] and cell extract) was not detectable in the fumA mutant grown on methanol supplemented with glyoxylate (<0.5 mU), while it was found in extracts of wild-type cells grown on succinate, methanol, or methanol supplemented with glyoxylate (90 ± 10, 40 ± 5, and 30 ± 5 mU, respectively).
To test the function of the second fumarase, FumB, a donor plasmid was constructed to create a mutation in fumB as described in reference 12. Transconjugants were selected on succinate, methanol, or methanol supplemented with glyoxylate. Double-crossover recombinants were selected on all the substrates based on their tetracycline sensitivity (3), and these were confirmed by diagnostic PCR analysis (data not shown). The mutants grew on both succinate and methanol, without glyoxylate addition, indicating that fumB is not essential in either the TCA cycle or the GRC. Levels of fumarase activity measured in this mutant (70 ± 10 mU on succinate and 30 ± 5 mU on methanol) were similar to the levels found in wild-type cells. In proteomic analysis of M. extorquens AM1, both FumA and FumB proteins have been detected (10). However, mutagenesis analysis implies that only one gene, fumA, is an essential gene. To test the function of SCS in both the TCA cycle and the GRC, a knockout mutant in scsA was generated essentially as described in reference 3. Double-crossover mutants were selected on both methanol and succinate and the mutants had a wild-type growth phenotype, indicating that SCS is not an essential gene for either the TCA cycle or the GRC. We were not able to demonstrate SCS activity in M. extorquens AM1. Two different tests were employed: the hydroxylamine assay as described in reference 2 and the disappearance of CoA from a reaction mixture containing Tris-HCl (pH 8.0; 100 mM), succinate (10 mM), MgCl2 (10 mM), ATP (5 mM), and CoA (0.1 mM) (all from Sigma), and cell extract. CoA was detected with Ellman's reagent (15). These data point to SCS not being expressed or expressed at low, undetectable levels. No SCS polypeptides have been detected via the proteomics approach (10). SCH activity was tested in the following reaction mixture: Tris-HCl (pH 8.0), 100 mM; succinyl-CoA, 1 mM; CoA, 0.5 mM; DTNB, 1 mM (all from Sigma); and cell extract. High levels of SCH activity (330 ± 30 and 360 ± 35 mU, respectively) were measured in both the wild-type strain and the SCS mutant cultures grown on succinate, and lower levels (130 ± 15 and 140 ± 15 mU, respectively) were measured in cultures grown on methanol. These data suggest that SCH may be the major enzyme converting succinyl-CoA into succinate in both the TCA cycle and the GRC, or that SCS (if expressed) and SCH have redundant functions.
Conclusions.
In this report we have defined new genes and confirmed previously identified genes that contribute to the novel pathway for regeneration of glyoxylate that is a part of C1 and C2 metabolism in isocitrate lyase-negative serine cycle methylotrophs. Evidence suggests the distribution of this pathway might be much broader than initially thought, because genes specific to the pathway are identifiable in a variety of nonmethylotrophic alpha-proteobacteria by BLAST analyses with available genomic sequences (data not shown), and because, in streptomycetes, some of these genes have been demonstrated to play a role in C2 metabolism (6). The new genes described here, meaD encoding ACA, an enzyme essential for biosynthesis of AC, which is a cofactor for MCM, and meaC, which may encode the methacrylyl-CoA dehydratase, provide more members of this metabolic pathway. In addition, we have shown that SDH and one of the two Fum enzymes (FumA) are essential enzymes and function in both the TCA cycle and the GRC, while the second Fum (FumB) is not essential but may have a role in the organism's fitness. FumB may be responsible for the slow growth on methanol and succinate of the FumA mutant, while the biochemical basis of the residual growth of the SdhA mutant on methanol or succinate is less clear. Nonspecific oxidation of succinate may take place, or malate synthesis may be bypassed via other intermediates. We demonstrate that SCS, an enzyme traditionally involved in the TCA cycle, is not essential in M. extorquens AM1, while SCH, the gene for which remains unidentified, is suggested to have a major function in both the TCA cycle and the GRC.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM58933).
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Aoshima, M., M. Ishii, and Y. Igarashi. 2004. A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:751-761. [DOI] [PubMed] [Google Scholar]
- 3.Chistoserdov, A. Y., L. V. Chistoserdova, W. S. McIntire, and M. E. Lidstrom. 1994. The genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chistoserdova, L., S. W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdova, L. V., and M. E. Lidstrom. 1996. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiology 142:1459-1468. [DOI] [PubMed] [Google Scholar]
- 6.Han, L., and K. A. Reynolds. 1997. A novel alternative anaplerotic pathway to the glyoxylate cycle in streptomycetes. J. Bacteriol. 179:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korotkova, N., L. Chistoserdova, V. Kuksa, and M. E. Lidstrom. 2002. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korotkova, N., and M. E. Lidstrom. 2001. A connection between poly-β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 183:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korotkova, N., and M. E. Lidstrom. 2004. MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation. J. Biol. Chem. 279:13652-13658. [DOI] [PubMed] [Google Scholar]
- 10.Laukel, M., M. Rossignol, G. Borderies, U. Völker, and J. A. Vorholt. 2004. Comparison of the proteome of Methylobacterium extorquens AM1 grown under methylotrophic and non-methylotrophic growth conditions. Proteomics 4:1247-1264. [DOI] [PubMed] [Google Scholar]
- 11.Leal, N. A., S. D. Park, P. E. Kima, and T. Bobik. 2003. Identification of the human and bovine ATP:Cob(I)alamin adenosyltransferase cDNAs based on complementation of a bacterial mutant. J. Biol. Chem. 278:9227-9243. [DOI] [PubMed] [Google Scholar]
- 12.Marx, C. J., and M. E. Lidstrom. 2002. A broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 13.Marx, C. J., B. N. O'Brien, J. Breezee, and M. E. Lidstrom. 2003. Novel methylotrophy genes of Methylobacterium extorquens AM1 identified by using transposon mutagenesis including a putative dihydromethanopterin reductase. J. Bacteriol. 185:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy, P., and M. M. Weber. 1986. Solubilization, purification, and characterization of succinate dehydrogenase from membranes of Mycobacterium phlei. J. Bacteriol. 167:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riddles, P. W., R. L. Blakeley, and B. Zerner. 1979. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid)—a reexamination. Anal. Biochem. 94:75-81. [DOI] [PubMed] [Google Scholar]
- 16.Smith, L. M., W. G. Meijer, L. Dijkhuizen, and P. Goodwin. 1996. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology 142:657-684. [DOI] [PubMed] [Google Scholar]
- 17.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C3 and C4 metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 18.Van Dien, S. J., Y. Okubo, M. T. Hough, N. Korotkova, T. Taitano, and M. E. Lidstrom. 2003. Reconstruction of C3 and C4 metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology 146:601-609. [DOI] [PubMed] [Google Scholar]