Abstract
Cell extracts of Rhodobacter capsulatus grown on acetate contained an apparent malate synthase activity but lacked isocitrate lyase activity. Therefore, R. capsulatus cannot use the glyoxylate cycle for acetate assimilation, and a different pathway must exist. It is shown that the apparent malate synthase activity is due to the combination of a malyl-coenzyme A (CoA) lyase and a malyl-CoA-hydrolyzing enzyme. Malyl-CoA lyase activity was 20-fold up-regulated in acetate-grown cells versus glucose-grown cells. Malyl-CoA lyase was purified 250-fold with a recovery of 6%. The enzyme catalyzed not only the reversible condensation of glyoxylate and acetyl-CoA to l-malyl-CoA but also the reversible condensation of glyoxylate and propionyl-CoA to β-methylmalyl-CoA. Enzyme activity was stimulated by divalent ions with preference for Mn2+ and was inhibited by EDTA. The N-terminal amino acid sequence was determined, and a corresponding gene coding for a 34.2-kDa protein was identified and designated mcl1. The native molecular mass of the purified protein was 195 ± 20 kDa, indicating a homohexameric composition. A homologous mcl1 gene was found in the genomes of the isocitrate lyase-negative bacteria Rhodobacter sphaeroides and Rhodospirillum rubrum in similar genomic environments. For Streptomyces coelicolor and Methylobacterium extorquens, mcl1 homologs are located within gene clusters implicated in acetate metabolism. We therefore propose that l-malyl-CoA/β-methylmalyl-CoA lyase encoded by mcl1 is involved in acetate assimilation by R. capsulatus and possibly other glyoxylate cycle-negative bacteria.
Acetate is a central intermediate in the overall carbon cycle. The exclusive utilization of acetate or other compounds, which enter the central metabolism at the level of acetyl-coenzyme A (CoA) (such as fatty acids, alcohols, or polyhydroxyalkanoates), poses a unique problem: the conversion of acetyl-CoA to all other cell components. Acetyl-CoA catabolism generally proceeds via the tricarboxylic acid cycle. However, growth is impossible if C4 acids drained from the citric acid cycle for biosynthetic reactions are not replenished. Most facultative anaerobic and aerobic bacteria—including Escherichia coli—use the glyoxylate cycle for acetate assimilation (26, 28). The key enzymes for this pathway are isocitrate lyase (ICL), cleaving isocitrate to succinate and glyoxylate, and malate synthase (MS), condensing glyoxylate and acetyl-CoA to form malate and CoA (Fig. 1A), resulting in the net fixation of two molecules of acetyl-CoA to one molecule of malate. Together with enzymes of the tricarboxylic acid cycle, ICL is responsible for the conversion of acetyl-CoA to glyoxylate. Based on sequence similarities, two groups of malate synthases, MS type A (for acetate assimilation) and MS type G (for glycolate assimilation), can be distinguished. Initially it was thought that only MS type A was involved in acetate assimilation; however, it was recently shown in E. coli, which contains both types of malate synthases, that the gene encoding MS type G is also up-regulated in acetate- but not glucose-grown cells (35). The question of why two isozymes exist is at issue.
FIG. 1.
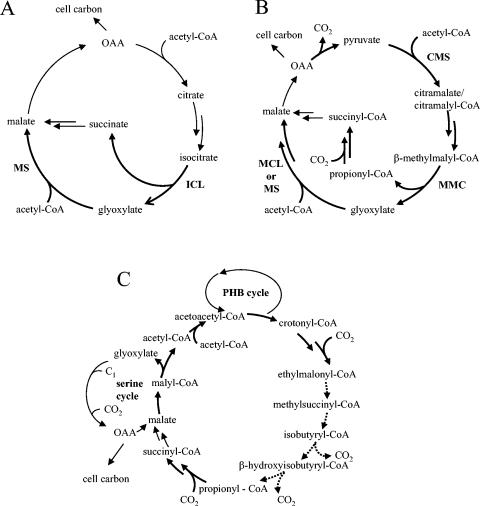
Known and proposed pathways for acetate assimilation. (A) Glyoxylate cycle. The reactions of the key enzymes ICL and MS are marked with bold reaction arrows. (B) Citramalate cycle as proposed by Ivanovsky et al. (25). Reactions catalyzed by citric acid cycle enzymes are marked by nonbold arrows. Proposed key enzymes are citramalate synthase (CMS), β-methylmalyl-CoA lyase (MMC), and MS or MCL. It is not known if the product of the CMS reaction is citramalate or citramalyl-CoA. (C) Assimilation cycle as proposed by Korotkova et al. (29) for M. extorquens. This cycle is interlinked with the polyhydroxybutyrate (PHB) cycle and the serine cycle for the assimilation of C1 compounds. Dashed reaction arrows indicate that the nature of the converting reactions is unknown. OAA, oxaloacetate.
Within the group of the rhodospirillaceae there is a large number of species, among them Rhodobacter capsulatus, Rhodospirillum rubrum, and Rhodobacter sphaeroides, which reportedly contain MS activity but lack ICL activity (1, 27). The ability to grow on C2 compounds as the sole carbon source in spite of the absence of ICL has also been observed for other bacteria belonging to the genus Streptomyces (19) and to the group of α-proteobacteria like Paracoccus versutus (10) and Methylobacterium extorquens (3, 31). In all three cases, alternatives to the glyoxylate cycle for acetyl-CoA assimilation are discussed.
Phototrophic purple nonsulfur bacteria can be grouped into three categories with respect to the presence of key enzymes of the glyoxylate cycle in acetate-grown cells: those lacking ICL activity (e.g., R. rubrum), those lacking MS activity (e.g., Rubrivivax gelatinosus), or those which contain both key enzyme activities (e.g., Rhodopseudomonas palustris) (1). Acetate assimilation by R. rubrum has been studied intensively over the last 50 years by labeling studies and on the level of enzyme activities (2, 5, 12, 23, 25). Incorporation of isotopically labeled acetate and bicarbonate into cell components using growing cultures or washed cell suspensions of R. rubrum (12, 23) resulted in a labeling pattern for glutamate, which Hoare argued is not consistent with formation of glutamate from oxaloacetate via the citric acid cycle (12). In addition, bicarbonate is specifically incorporated; however, the overall assimilation of acetate does not result in a net CO2 fixation, pointing to carboxylation and decarboxylation reactions instead (12, 23, 25).
Ivanovsky et al. (25) proposed a new citramalate cycle of acetate assimilation for R. rubrum. The key steps of this cycle are the condensation of acetyl-CoA and pyruvate to citramalate, which is subsequently converted to β-methylmalate/β-methylmalyl-CoA, followed by β-methylmalyl-CoA cleavage to propionyl-CoA and glyoxylate (Fig. 1B). Pyruvate is regenerated from propionyl-CoA by a series of conventional reactions, i.e., carboxylation of propionyl-CoA to methylmalonyl-CoA and conversion of methylmalonyl-CoA via succinyl-CoA to malate or oxaloacetate. (Oxidative) decarboxylation of these C4 compounds yields pyruvate closing the cycle. Glyoxylate condenses with a second molecule of acetyl-CoA to form malate. The proposed citramalate cycle is ICL independent; however, it still demands the presence of MS activity.
The standard assay for MS activity measures the glyoxylate- and acetyl-CoA-dependent formation of free CoA. MS activity can be due to the presence of an MS enzyme of either type A or type G. Furthermore, an apparent MS activity can also be due to the combined action of two other enzymes: malyl-CoA lyase catalyzing the reversible synthesis of malyl-CoA, and a malyl-CoA-hydrolyzing enzyme releasing CoA. In Chloroflexus aurantiacus, such a misleading MS activity may actually be due to the presence of l-malyl-CoA/β-methylmalyl-CoA lyase and citrate synthase, which is known to hydrolyze l-malyl-CoA (15, 17, 21).
M. extorquens is a methylotroph, which uses an ICL-independent serine cycle for the assimilation of C1 compounds (33). The organism contains l-malyl-CoA synthetase (malate thiokinase, l-malate + MgATP + CoA → l-malyl-CoA + MgADP + phosphate) and l-malyl-CoA lyase (MCL) (l-malyl-CoA lyase → acetyl-CoA + glyoxylate), thus being able to regenerate glyoxylate from which the C1 acceptor molecule glycine is made; however, ICL is lacking. The primary carbon fixation product of the serine cycle is acetyl-CoA, from which glyoxylate has to be regenerated in an unknown way. Isolation of a series of mutants, which no longer were able to utilize C1 or C2 compounds but were rescued by the addition of glyoxylate, leads to the proposal of a so-called glyoxylate regeneration pathway from acetyl-CoA by Korotkova et al. (29) (Fig. 1C). Several products of those genes, identified in mutagenesis studies, were shown to be upregulated in methanol-grown M. extorquens (34). Key intermediates of this pathway are β-hydroxyisobutyrate and methylsuccinate, which were detected as labeled intermediates after incubation of cell extracts of M. extorquens with 14C-labeled acetate in the presence of methanol (29).
We show here that acetate-grown R. capsulatus exhibits no ICL activity, but expresses malyl-CoA lyase. The enzyme was purified and shown to catalyze not only the reversible cleavage of l-malyl-CoA but also the reversible cleavage of β-methylmalyl-CoA. We present evidence which points to the involvement of this enzyme in acetate assimilation of R. capsulatus.
MATERIALS AND METHODS
Materials.
Chemicals were obtained from Fluka (Neu-Ulm, Germany), Merck (Darmstadt, Germany), Sigma-Aldrich (Deisenhofen, Germany), or Roth (Karlsruhe, Germany); biochemicals were from Genaxxon (Biberach, Germany), Fermentas (St. Leon-Rot, Germany), or Applichem (Darmstadt, Germany). [2-14C]Propionate was obtained from Hartmann Analytic (Braunschweig, Germany), and [1-14C]acetate was purchased from Amersham Biosciences (Freiburg, Germany).
Bacteria and growth conditions.
R. capsulatus B10, R. palustris, and R. rubrum S1 (laboratory strains) were grown anaerobically in the light (6,000 lx) at pH 6.7 and 30°C on minimal medium containing 22.4 mM NH4Cl, 0.8 mM MgSO4, 0.5 mM CaCl2, 20 mM potassium phosphate buffer (pH 6.7), vitamin solution (36), 1 ml of trace element solution (500 mg of EDTA [EDTA, disodium salt], 300 mg of FeSO4 · 7H2O, 3 mg of MnCl2 · 4H2O, 5 mg of CoCl2 · 6H2O, 1 mg of CuCl2 · 2H2O, 2 mg of NiCl2 · 6H2O, 3 mg of Na2MoO4 · 2H2O, 5 mg of ZnSO4 · 7H2O, and 2 mg of H3BO3 [pH 3])/liter, and the following carbon source: 20 mM sodium acetate, 10 mM NaHCO3 plus 20 mM sodium acetate, 10 mM glucose, or 10 mM fructose. Cells used for regulatory studies were grown in 2-liter bottles (after being transferred at least five times on medium with the same carbon source) and harvested in mid-exponential phase at an optical density of 0.5 to 1.0. Cell material for protein purification was obtained after growth of R. capsulatus in a 12-liter fermenter using 20 mM acetate as the sole carbon source. Cells were harvested by centrifugation at 4°C and 12,000 × g at the end of exponential growth at an optical density of 1.1 to 1.4. R. capsulatus grown in a 12-liter fermenter did not yield a solid pellet but a cell suspension of 100 to 120 ml per 12 liters. Cells were stored in liquid nitrogen until use. E. coli DH5α and BL21(DE3) cells (42) were grown in Luria-Bertani broth.
Syntheses.
l-Malyl-CoA, acetyl-CoA, and propionyl-CoA were synthesized as described elsewhere (14, 17, 21). erythro-β-Methylmalyl-CoA was synthesized enzymatically from propionyl-CoA and glyoxylate by using l-malyl-CoA/β-methylmalyl-CoA lyase from C. aurantiacus as previously described (22).
[1-14C]acetyl-CoA and [2-14C]propionyl-CoA were synthesized enzymatically as reported (21), with the exception that in the case of [2-14C]propionyl-CoA synthesis, 3-hydroxypropionyl-CoA synthetase isolated from Metallosphaera sedula (B. E. Alber and G. Fuchs, unpublished data) was used.
Enzyme assays.
All enzyme assays were performed at 30°C. One unit corresponds to one micromole of substrate converted per minute.
Malate synthase.
MS activity was measured by the method of Srere et al. (40) by following the glyoxylate- and acetyl-CoA-dependent release of CoA. The standard reaction mixture (0.5 ml) contained 200 mM potassium morpholinopropanesulfonic acid (MOPS/KOH [pH 7.2]), 5 mM MgCl2, 0.25 mM 5-5′-dithiobis(2-nitrobenzoic acid) (DTNB, from 10 mM stock solution in 0.5 M potassium phosphate [pH 7.2], 1 mM EDTA), 1 mM acetyl-CoA, and protein. The reaction was started by the addition of 10 mM sodium glyoxylate and followed at 412 nm (ɛ412 = 13,600 M−1 cm−1). In R. palustris, MS activity is inhibited by DTNB, and the assay was modified accordingly: DTNB was omitted from the reaction mixture (1 ml), and at various time points samples (200 μl) were withdrawn and heated to 60°C for 1 min. After centrifugation for 5 min at 14.000 × g and 4°C in order to remove precipitated protein, 5 μl of DTNB stock solution and 295 μl of H2O was added, and the absorption was measured at 412 nm to determine the amount of free CoA formed.
Isocitrate lyase.
A spectrophotometric assay was used to measure isocitrate-dependent formation of glyoxylate. The standard reaction mixture (0.5 ml) contained 100 mM MOPS/KOH (pH 7.2), 5 mM MgCl2, 2 mM dithioerythritol, 3.5 mM phenylhydrazine, and protein. The reaction was started by the addition of 2 mM isocitrate and followed at 324 nm (ɛ324 for glyoxylate phenylhydrazone was determined as 15,000 M−1 cm−1).
l-Malyl-CoA cleavage.
The cleavage of malyl-CoA into acetyl-CoA and glyoxylate was monitored spectrophotometrically by following the formation of the glyoxylate phenylhydrazone derivative at 324 nm. The standard assay mixture (0.5 ml) contained 200 mM MOPS/KOH (pH 7.5), 5 mM MgCl2, 3.5 mM phenylhydrazinium chloride, 0.5 mM l-malyl-CoA, and protein. In the case of purified enzyme, 0.25 mM l-malyl-CoA was used.
β-Methylmalyl-CoA cleavage.
The activity was measured according to the malyl-CoA lyase assay with 0.25 mM β-methylmalyl-CoA as the substrate in an assay volume of 0.25 ml.
Formation of l-malyl-CoA.
l-Malyl-CoA formation from acetyl-CoA and glyoxylate was coupled to citrate synthase-catalyzed hydrolysis of l-malyl-CoA (17). The standard assay mixture (0.5 ml) contained 200 mM MOPS/KOH (pH 7.5), 5 mM MgCl2, 0.25 mM DTNB, 1 mM acetyl-CoA, 10 U of citrate synthase, and protein. Ten units of citrate synthase was sufficient to determine activities of up to 12.5 nmol min−1 of l-malyl-CoA formation. The reaction was started by the addition of 10 mM glyoxylate, and the formation of the CoA-DTNB disulfide was monitored at 412 nm.
Protein concentrations were determined by the method of Bradford (6) using bovine serum albumin as a standard. Protein concentrations of the purified MCL from R. capsulatus were also estimated by using the A280 and the extinction coefficient (ɛ280 = 1,225 liters g−1 cm−1) calculated from the deduced amino acid sequence of the mcl1 gene (16). Both methods agreed well. For determination of Km values, the concentration of one substrate was varied while keeping the concentration of the other substrate constant; the concentrations given were saturating. Data were analyzed by nonlinear regression using the Michaelis-Menten equation and the Prism 4.0 software package (GraphPad Software, Inc., San Diego, Calif.).
HPLC analysis.
High-performance liquid chromatography (HPLC) analysis of malyl-CoA and methylmalyl-CoA formation. The reaction mixture (1.25 ml) contained 200 mM MOPS/KOH (pH 7.5), 5 mM MgCl2, 0.5 mM acetyl-CoA plus 20-kBq [1-14C]acetyl-CoA (or 0.5 mM propionyl-CoA plus 20-kBq [2-14C]propionyl-CoA) and 3 μg of purified MCL. The addition of 10 mM glyoxylate started the reaction. Samples of 200 μl were taken after 0, 2, 5, 15, and 30 min, and the reaction was stopped by addition of 10 μl of 25% HCl. Precipitated protein was removed by centrifugation, and samples were applied to a Lichrospher 100 C-18 reversed-phase column (125 by 4 mm; Merck) equilibrated with 1% acetonitrile in 50 mM potassium phosphate buffer (pH 6.8). The column was developed using a 30-ml linear increasing gradient of 1 to 8% acetonitrile and a flow rate of 1 ml min−1. The formation of l-malyl-CoA was confirmed by adding 10 U of citrate synthase after 15 min, which specifically hydrolyzes l-malyl-CoA but not d-malyl-CoA to malate (15). The retention times of standards were as follows: acetyl-CoA, 18 min; propionyl-CoA, 24 min; malyl-CoA, 10 min; β-methylmalyl-CoA, 8 min; CoA, 9 min.
For rate determination of product formation in cell extracts, the formed CoA-esters were hydrolyzed. Samples were prepared as described above, with the exception that the samples were hydrolyzed by adding 40 μl of 4 M KOH and incubated for 20 min at 60°C. Samples were acidified by addition of 10 μl of 25% HCl. Precipitated protein was removed by centrifugation, and samples were applied to an Aminex HPX-87H column (300 by 7.8 mm; Bio-Rad, München, Germany) equilibrated with 2.5 mM H2SO4 at 0.4 ml min−1. The same conditions were used to develop the column. The retention times of standards were as follows: mesaconate, 35 min; oxaloacetate, 13 and 15 min; citrate, 14.5 min; acetate, 24 min; malate, 17.5 and 30 min; glyoxylate, 16 min; pyruvate, 19 min; propionate, 29 min; fumarate, 32 min; β-methylmalate, 18 and 39 min.
Heterologous expression of the icl gene from R. capsulatus and production of the protein in E. coli.
Chromosomal DNA from R. capsulatus was isolated using standard techniques. Two synthetic oligonucleotides were designed to amplify the complete icl gene using chromosomal R. capsulatus DNA as a template: the forward primer, 5′-GGAATTCCATATGACGAAAAGACAGACTTT-3′ introduces an NdeI site (underlined) at the initiation codon; the reverse primer, 5′-GGAACTGCAGCCTCCCTTGCTTCACTCTG-3′ introduces a PstI site (underlined) after the stop codon. PCR was performed for 30 cycles, including denaturation at 94°C for 1 min, annealing at 60°C for 2 min, and extension at 72°C for 2 min. The PCR product was isolated and cloned into pUC19. The nucleotide sequence of the PCR product was confirmed to ensure that no errors had been introduced. The plasmid was digested with NdeI and PstI, and the fragment containing the icl gene was ligated into pT7/7 (43), resulting in plasmid pICL004. Competent E. coli BL21(DE3) cells (42) were transformed with pICL004, grown at 37°C in Luria-Bertani broth containing 100 μg of ampicillin ml−1, and induced at an optical density of 0.8 with 0.4 mM isopropyl thiogalactopyranoside (IPTG). After additional growth for 3 h, the cells were harvested and stored in liquid nitrogen until use. Thawed cell paste from 10 ml of cell culture was resuspended in 0.6 ml of 100 mM Tris/HCl (pH 7.8), 5 mM MgCl2, and 20 μg of DNase I ml−1. After addition of 1 g of glass beads (diameter, 0.1 to 0.25 mm), the cell solution was treated in a mixer mill (type MM2; Retsch, Haare, Germany) for 8 min at 30 Hz. The supernatant obtained after centrifugation (10 min, 12,000 × g, 4°C) was used in mixing experiments with cell extract from acetate-grown R. capsulatus.
Purification of malyl-CoA lyase from R. capsulatus.
The buffer concentration of thawed cell suspension from a 12-liter fermenter (estimated 15 g [wet weight]) was adjusted to 50 mM MOPS/KOH (pH 7.5) and 5 mM MgCl2 and passed through a chilled French pressure cell at 1,100 lb/in2. The cell lysate was centrifuged at 16,000 × g for 20 min at 4°C, and the supernatant was ultracentrifuged for 1 h at 100,000 × g. All purification steps were done aerobically at 4°C.
(i) Ammonium sulfate precipitation.
Saturated ammonium sulfate solution was added to the supernatant to a final concentration of 35% (NH4)2SO4, stirred for 20 min, and centrifuged at 34,000 × g for 20 min. Saturated ammonium sulfate solution was added to the supernatant to a final concentration of 55% (NH4)2SO4, stirred for 20 min, and centrifuged at 34,000 × g for 20 min. The pellet was resuspended in 15 ml of 100 mM MOPS/NaOH (pH 7.5) and 5 mM MgCl2 and dialyzed overnight against 1 liter of 10 mM MOPS/NaOH (pH 7.5) and 5 mM MgCl2.
(ii) DEAE chromatography.
The dialyzed protein solution from step i was applied to a 50-ml DEAE-Sepharose column (Amersham Biosciences) equilibrated with buffer A (20 mM MOPS/NaOH [pH 7.5]). After being washed with 150 ml of buffer A containing 100 mM KCl, the column was developed with a 400-ml increasing linear gradient of 100 to 500 mM KCl at 2 ml min−1. The peak of activity eluted between 180 and 230 mM salt and the active fractions were pooled. Solid ammonium sulfate was added to the pooled DEAE-fractions to a final concentration of 1 M and equilibrated overnight.
(iii) Butyl-Sepharose chromatography.
The mixture from step ii was loaded onto a 20-ml butyl-Sepharose column (Amersham Biosciences) equilibrated with buffer B (10 mM MOPS/NaOH [pH 7.5]) containing 1 M (NH4)2SO4. After being washed with 60 ml of wash buffer, the column was developed with a 400-ml deceasing linear gradient of 1 to 0 M (NH4)2SO4 at 1 ml min−1. Active fractions (between 780 and 720 mM salt) were pooled, and the (NH4)2SO4 concentration was adjusted to 1 M with buffer B containing 2 M (NH4)2SO4.
(iv) Resource phenyl chromatography.
The enzyme solution from step iii was applied to a Resource phenyl column (1 ml of Resource-PHE; Amersham Biosciences) equilibrated with buffer B containing 1 M (NH4)2SO4. After being washed with 5 ml of wash buffer, the column was developed with a 50-ml deceasing linear gradient of 1 to 0 M (NH4)2SO4. MCL activity eluted between 690 and 510 mM (NH4)2SO4. Active fractions were pooled.
(v) Gel filtration chromatography.
The volume of the active pool from step iv was reduced to 1 to 2 ml by ultrafiltration (Centricon 30) and applied to a 120-ml column (Superdex 200; Amersham Biosciences) equilibrated with 20 mM MOPS/KOH (pH 6.8) and 100 mM KCl at a flow rate of 0.5 ml min−1. l-Malyl-CoA lyase was eluted with a retention time of 66 ml.
Step iv was repeated, and the purified enzyme was stored with 10% (wt/wt) glycerol at −20°C.
Divalent metal cation dependency.
Purified malyl-CoA lyase was dialyzed against 10 mM MOPS/KOH (pH 6.8), 0.2 mM EDTA for 6 h (total volume, 3,000× sample volume) followed by dialysis against 10 mM MOPS/KOH (pH 6.8) to remove EDTA (total volume, 2,000× sample volume). Enzyme activity was determined by using the malyl-CoA cleavage assay, omitting MgCl2, and adding 5 mM MnCl2, 5 mM MgCl2, 5 mM CoCl2, 1 mM CaCl2, or 1 mM ZnSO4 to the reaction mixture.
Molecular mass determination.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (32), using 12% gels. Proteins were stained with Coomassie blue according to the method of Zehr et al. (47). The native molecular mass was determined by fast protein liquid chromatography, using a 25-ml Superdex 200 gel filtration column (Amersham Biosciences) calibrated with catalase (230 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), and ovalbumin (45 kDa). Protein samples (0.2 ml) were applied to the column, equilibrated with 20 mM MOPS/KOH (pH 6.8) and 100 mM NaCl, and eluted with the same buffer with a flow rate of 0.2 ml min−1.
N-terminal amino acid analysis.
An aliquot (2 μg) of the protein solution from the last purification step was precipitated with 8% trichloroacetic acid, separated by SDS-12% PAGE, and transferred to an Immobilon-Psq transfer membrane (Millipore, Eschborn, Germany). N-terminal sequencing was performed by TopLab (Martinsried, Germany) using a Procise 492 sequencer (Applied Biosystems). The phenylthiohydantoin derivatives were identified with an online Applied Biosystems 140C phenylthiohydantoin analyzer.
Computer analysis.
The BLAST program was used to search databases at the National Center for Biotechnology Information (NCBI; Bethesda, Md.) and the Department of Energy (DOE) Joint Genome Institute (Walnut Creek, Calif.).
RESULTS
Isocitrate lyase activity in cell extracts of R. capsulatus grown on acetate.
Phototrophic purple nonsulfur bacteria can be grouped into three categories with respect to the presence of key enzyme activities of the glyoxylate cycle in acetate-grown cells: those lacking ICL activity (e.g., R. sphaeroides), those lacking MS activity (Rubrivivax gelatinosus), and those which contain both key enzyme activities (e.g., R. palustris) (1). In cell extracts of acetate-grown R. palustris, both ICL and apparent MS activities were detected, whereas R. rubrum showed no ICL activity (Table 1). These results confirm those of previous studies, which showed that R. palustris uses the glyoxylate cycle and R. rubrum uses an unknown pathway for acetate assimilation (1, 5, 12, 23, 25). Also, no ICL activity was detected in cell extracts of acetate- and glucose-grown R. capsulatus.
TABLE 1.
Regulation of key enzymes of the glyoxylate cycle and related enzymes in some phototrophic proteobacteriaa
| Organism | Growth substrate | Activity (nmol min−1 mg−1) of:
|
||
|---|---|---|---|---|
| Isocitrate lyase | Apparent malate synthaseb | Malyl-CoA lyasec | ||
| R. capsulatus | Acetate/bicarbonate | <3 | 200 ± 8 | 52 ± 4 |
| Glucose | <1 | 40 ± 3 | <2 | |
| R. rubrum | Acetate/bicarbonate | <1 | 40 ± 5 | 35 ± 1 |
| Fructose | <1 | 8 ± 2 | 10 ± 1 | |
| R. palustris | Acetate/bicarbonate | 155 ± 15 | 240 ± 20 | <1 |
Specific activities were obtained by linear regression of protein dependency. The error given is the standard deviation.
Malate synthase activity was measured as glyoxylate-dependent release of CoA from acetyl-CoA and is indistinguishable from a combination of malyl-CoA lyase with a malyl-CoA hydrolyzing enzyme.
Malyl-CoA lyase was measured in the direction of malyl-CoA cleavage.
A search of the almost-complete R. capsulatus genome for an icl-like gene, however, resulted in one hit on contig 2G06-2D11. The deduced amino acid sequence of the identified gene has 58% identity to ICL from R. palustris and is downstream of a gene with a deduced amino acid sequence 47% identical to MS type A from E. coli, indicating that R. capsulatus has the genetic potential for a functioning glyoxylate cycle. ICL from R. capsulatus was produced in E. coli using a T7 promoter and polymerase system. The specific activity of ICL in cell extracts of E. coli harboring plasmid ICL004 after induction with IPTG was 1.0 U mg−1. Cell extracts of the recombinant E. coli and acetate-grown R. capsulatus were mixed to an expected final ICL activity of 0.05 U mg−1 and incubated at 30°C. After addition of cell extract of R. capsulatus or after 30 min of incubation, no loss of ICL activity was observed, indicating that ICL is not inhibited by any factors present in cell extracts of R. capsulatus. This control indicates that ICL is lacking and that its gene is probably not expressed in acetate-grown cells of R. capsulatus.
Apparent malate synthase activity in cell extracts of R. capsulatus.
Malate synthase activity was tested in cell extracts of R. capsulatus as the glyoxylate-dependent release of CoA from acetyl-CoA. Such activity was easily detected in acetate-grown cells (0.20 U mg−1), where it was five times higher than in glucose-grown cells (Table 1). However, the release of free CoA set in only after a short lag phase of 0.5 min and after up to 1.5 min for ammonium sulfate-precipitated protein. Incubation of 14C-labeled acetyl-CoA and glyoxylate with protein after the ammonium sulfate precipitation step followed by HPLC analysis indicated that malyl-CoA was formed as a transient free intermediate and was further hydrolyzed to malate and free CoA (data not shown). l-Malyl-CoA lyase (MCL) activity, which catalyzes the reversible cleavage of malyl-CoA into acetyl-CoA and glyoxylate, was detected in cell extracts of acetate- but not glucose-grown cells. In cell extracts of the ICL-negative R. rubrum, apparent MS and MCL activities were also present, and both activities were up-regulated in acetate- but not fructose-grown cells (Table 1). In cell extracts of acetate-grown R. palustris, which uses the glyoxylate cycle for acetate assimilation and was used as a control, MS activity but no MCL activity could be detected (Table 1). These results suggest that the apparent MS activity in R. capsulatus and R. rubrum is mainly or exclusively due to the combined action of l-malyl-CoA lyase and a malyl-CoA-hydrolyzing enzyme.
Purification of l-malyl-CoA lyase from R. capsulatus.
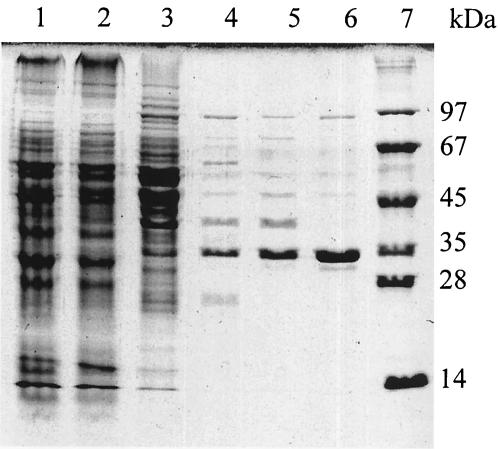
MCL activity was at least 20-fold higher in acetate-grown than glucose-grown R. capsulatus (Table 1), suggesting a role in acetate assimilation. About 90% MCL activity was recovered in the soluble fraction after ultracentrifugation of cell extract for 1 h at 100,000 × g. The enzyme was purified 250-fold with a recovery of 6% (Table 2). After the last purification step, contaminant proteins amounted to less than 10% as judged by denaturing gel electrophoresis (Fig. 2). The specific activity (15 U mg−1) was 10-fold higher than that of the purified malyl-CoA/methylmalyl-CoA lyase from C. aurantiacus (22) and about half of the specific activity of MCL from M. extorquens (18). The enzyme was stable for several weeks at −20°C in the presence of 10% (wt/wt) glycerol. An apparent malate synthase activity was no longer measurable after the DEAE purification step.
TABLE 2.
Purification of l-malyl-CoA lyase from photoheterotrophically acetate-grown R. capsulatus
| Purification step | Protein (mg) | Total activity (μmol min−1)a | Sp act (μmol min−1 mg−1) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 500 | 29 | 0.06 | 1 | 100 |
| 35-55% (NH4)2SO4 | 250 | 20 | 0.08 | 1.3 | 69 |
| DEAE-Sepharose | 28.8 | 16 | 0.6 | 10 | 55 |
| Butyl-Sepharose | 3.5 | 15.2 | 4.3 | 72 | 52 |
| Resource-PHE 1 | 2.5 | 12 | 4.8 | 80 | 41 |
| Gel filtration | 0.5 | 2.8 | 5.6 | 93 | 10 |
| Resource-PHE 2 | 0.12 | 1.8 | 15 | 250 | 6 |
Activities for malyl-CoA lyase were determined by measuring glyoxylate formation from malyl-CoA.
FIG. 2.
SDS-12% PAGE of l-malyl-CoA lyase from R. capsulatus at various steps during purification. Lanes: 1, 22 μg of cell extract (100,000 × g); lane 2, 20 μg of 35 to 55% (NH4)2SO4 precipitate; lane 3, 12 μg of DEAE fraction; lane 4, 5 μg of Resource-PHE 1 fraction; lane 5, 5 μg of gel filtration fraction; lane 6, 4 μg of Resource-PHE 2 fraction; lane 7, molecular mass marker.
Properties of l-malyl-CoA lyase.
The properties of the purified enzyme are summarized in Table 3. Gel filtration chromatography of the native MCL gave an estimated molecular mass of 195 ± 20 kDa. SDS/PAGE produced one protein band of 35 kDa (Fig. 2). The N-terminal sequence was determined to be SFRTQPP, and only one N-terminal sequence was detected, indicating that the enzyme was pure and is composed of identical subunits. MCL therefore exists as a homohexamer, as is the case for MCL from C. aurantiacus (22) and M. extorquens (18). A search of the database of the almost-completed genome of R. capsulatus revealed a perfect match to the N-terminal sequence of a putative protein with 318 amino acids and a calculated molecular mass of 34 kDa encoded by an open reading frame on contig 2G06-2D11. The deduced amino acid sequence of this ORF had 51% identity to MCL from M. extorquens and was designated mcl1.
TABLE 3.
Molecular and catalytic properties of l-malyl-CoA lyase from R. capsulatus
| Property | Characteristic or value |
|---|---|
| Reaction catalyzed | Acetyl-CoA + glyoxylate ↔ l-malyl-CoA Propionyl-CoA + glyoxylate ↔ erythro-β-methylmalyl-CoA |
| Specific activity (μmol min−1 mg−1) | |
| l-malyl-CoA cleavage | 18 ± 1 |
| erythro-β-Methylmalyl-CoA cleavage | 5.7 ± 0.2 |
| Acetyl-CoA + glyoxylate condensation | 37 ± 1a (25b, 15c) |
| Propionyl-CoA + glyoxylate condensation | 28b |
| Apparent Km value (mM) | |
| Malyl-CoA | 0.015 ± 0.003d |
| erythro-β-Methylmalyl-CoA | 0.021 ± 0.003d |
| Acetyl-CoA | 0.14 ± 0.01a |
| Native molecular mass (kDa) | 195 ± 20 |
| Subunit molecular mass (kDa) | 34 |
| Suggested compostion | α6 |
| N-terminal sequence | SFRTQPP |
| Influence of divalent cations (mM) | Mn2+ (5), 100%; Mg2+ (5), 50%; Co2+ (5), 45%; Zn2+ (1), 33%; Ca2+ (1), 33%; no addition, 33% |
| Inhibitor (mM) | EDTA (0.5), <5% activity |
Determined by coupling malyl-CoA formation to citrate synthase (l-malyl-CoA hydrolysis) with Mg2+.
Activity from HPLC assay with Mn2+.
Activity from HPLC assay with Mg2+.
Determined by following the cleavage reaction.
The initial rate of l-malyl-CoA cleavage was dependent on malyl-CoA concentration in a Michaelis-Menten-type kinetic with an apparent Km value of 15 μM. The MCL-catalyzed reaction is fully reversible and was measured as the glyoxylate-dependent release of CoA from acetyl-CoA in the presence of exogenous citrate synthase, which is known to hydrolyze l-malyl-CoA to malate and CoA (15). The specific activity for l-malyl-CoA formation was 37 U mg−1 with apparent Km values of 0.14 mM for acetyl-CoA and 1.2 mM for glyoxylate. Enzymatic activity was stimulated by the presence of Mg2+, which could be replaced by Co2+, but not by the presence of Zn2+ or Ca2+. Mn2+ was even more effective, with MCL activities being twofold higher than in the presence of Mg2+ or Co2+. EDTA inhibits the enzyme, and activity could be restored by adding an excess of divalent cations.
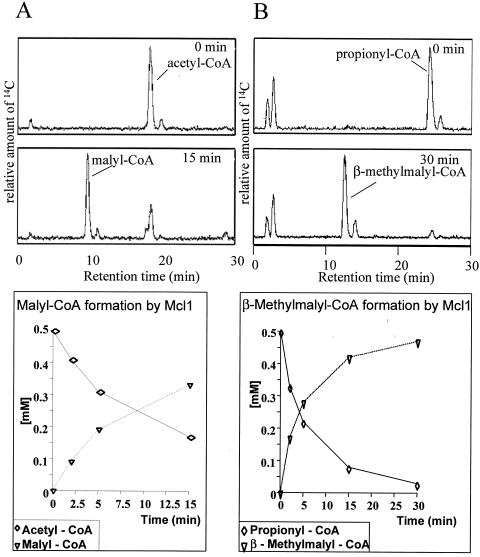
Product formation from glyoxylate and [1-14C] acetyl-CoA was followed by HPLC analysis. Figure 3A shows the time-dependent formation of a new CoA-ester, which coelutes with malyl-CoA. After 15 min, citrate synthase (10 U) was added. This led to a nearly complete hydrolysis of the formed malyl-CoA. Citrate synthase specifically hydrolyzes l-malyl-CoA (15), which was therefore determined to be the product formed from glyoxylate and acetyl-CoA. The specific activities for the formation of malyl-CoA were 15 U mg−1 when Mg2+ was used and 25 U mg−1 when Mn2+ was used in the assay. The specific activities in the HPLC assay, which were low compared to the values obtained from the substrate dependencies, can be explained by the reversibility of the enzyme. Addition of citrate synthase in the coupled photometric assay may draw the equilibrium to l-malyl-CoA synthesis.
FIG. 3.
Identification of products of l-malyl-CoA lyase from R. capsulatus. (Top) HPLC detection of products. (Bottom) Time-dependent formation of products and disappearance of substrates. (A) Formation of malyl-CoA from acetyl-CoA and glyoxylate. (B) Formation of β-methylmalyl-CoA from propionyl-CoA and glyoxylate. Both assays were performed as described in Materials and Methods.
In addition to the reversible cleavage of l-malyl-CoA, the enzyme also catalyzed the reversible cleavage of erythro-β-methylmalyl-CoA to propionyl-CoA and glyoxylate. The rate for methylmalyl-CoA cleavage (5.7 U mg−1) was about one-third of that of malyl-CoA cleavage (18 U mg−1), and the Km values were similar for both substrates. Condensation of glyoxylate with [2-14C] propionyl-CoA resulted in a product which was coeluted with methylmalyl-CoA on a C-18 reversed-phase column (Fig. 3B). The rate of methylmalyl-CoA formation in the presence of Mn2+ was 28 U mg−1, which was similar to the rate of malyl-CoA formation under these conditions.
Indication for the absence of malate synthase activity in cell extracts.
The ratios of the values of the condensation of acteyl-CoA and glyoxylate and the cleavage of malyl-CoA differed between the purified enzyme and cell extracts (Table 4). To exclude the presence of an additional MS, which catalyzes only the first reaction, we compared the rates for the condensation reactions of glyoxylate plus acetyl-CoA and glyoxylate plus propionyl-CoA in cell extracts of acetate-grown R. capsulatus. The activities were determined by HPLC analysis after hydrolyzing the CoA-esters as described in Materials and Methods. The activities were 0.29 U mg−1 for acetyl-CoA and glyoxylate condensation and 0.36 U mg−1 for propionyl-CoA and glyoxylate condensation. The ratio of these two values is similar to those determined for the purified enzyme. Both activities were about twice as high when Mn2+ was substituted for Mg2+ in the assay. The presence of an MS can therefore be excluded. In contrast to malate, β-methylmalate could be seen in cell extracts only after chemical hydrolysis of CoA-esters.
TABLE 4.
Malyl-CoA lyase activities from various sources
| Source | Activity (μmol min−1 mg−1)
|
|||
|---|---|---|---|---|
| Lytic cleavage of:
|
Glyoxylate condensation with:
|
|||
| l-Malyl-CoA | β-methylmalyl-CoA | acetyl-CoA | propionyl-CoA | |
| Cell extract of acetate-grown R. capsulatus | 0.052 | NDe | 0.20a (0.29b) | ND (0.36b) |
| Purified malyl-CoA lyase from R. capsulatus | 18 | 5.7 | 37a (25c) | ND (28c) |
| Purified malyl-CoA lyase from C. aurantiacusd | 2.3 | 6.5 | 0.48 | 0.5 |
Determined by coupling malyl-CoA formation to exogenous citrate synthase (l-malyl-CoA hydrolysis) with Mg2+.
Activity from HPLC assay with Mn2+.
Activity from HPLC assay with Mg2+.
Condensation reactions determined discontinuously as acetyl-CoA-dependent consumption of glyoxylate (22).
ND, not determined.
Argument against citrate synthase as malyl-CoA-hydrolyzing enzyme.
Citrate synthase activity in cell extracts of acetate-grown R. capsulatus was 300 to 400 nmol min−1 mg−1, depending on the cell batch. This is up to 10 times the rate measured for malyl-CoA cleavage. To determine whether citrate synthase was able to support the release of CoA from malyl-CoA formed by malyl-CoA lyase under physiological conditions, we added exogenous citrate synthase in a 20:1 ratio (based on the respective activities in cell extract) to purified malyl-CoA lyase and determined the glyoxylate-dependent release of CoA from acetyl-CoA as described for the coupled assay in Materials and Methods. The release of CoA under these conditions was less than 0.5% of the expected rate. Citrate synthase activity therefore appears not to be sufficient to explain the malyl-CoA-hydrolyzing enzyme activity. Hence, another malyl-CoA-thioester-hydrolyzing activity has to be postulated.
DISCUSSION
Isocitrate lyase-negative, malyl-CoA lyase-positive acetate utilizers.
R. capsulatus has the genetic potential for a functioning glyoxylate cycle, and this pathway has been postulated to operate in acetate assimilation, because both key enzymes of the cycle (ICL and MS) were reported in extracts of acetate-grown cells (27, 31, 46). However, Albers and Gottschalk failed to detect ICL activity (1). Under our growth and test conditions, ICL activity was also not detected, and therefore, a functional glyoxylate cycle was ruled out. The reason for this discrepancy is not known. This, then, would group R. capsulatus phenotypically with other ICL-negative organisms, which use an unknown pathway for acetate assimilation (1, 3, 8, 10, 19, 23, 25, 27).
Acetate-grown R. capsulatus probably also lacked MS activity. The apparent MS activity detected in cell extracts is due to the combined action of a malyl-CoA lyase forming malyl-CoA from acetyl-CoA and glyoxylate and a malyl-CoA-hydrolyzing enzyme. The nature of the enzyme responsible for malyl-CoA hydrolysis is not known. The absence of a “true” MS activity (forming malate directly from acetyl-CoA and glyoxylate) has been reported previously for ICL-negative R. sphaeroides and M. extorquens (11, 20, 44, 45). We could confirm this observation for the ICL-negative R. rubrum. In contrast, the apparent MS activity in the ICL-positive R. palustris was due to a true MS. We therefore propose that a unifying feature of ICL-negative bacteria is the presence of MCL rather than MS during growth on acetate. MCL may be involved in either malyl-CoA (β-methylmalyl-CoA) formation or cleavage.
Malyl-CoA lyase from R. capsulatus.
MCL isolated from acetate-grown R. capsulatus has properties (Table 3) similar to those of l-malyl-CoA/β-methylmalyl-CoA lyase from C. aurantiacus (22) and MCL from M. extorquens (18) with respect to molecular weight and subunit composition. The R. capsulatus enzyme differs, however, in its preference of Mn2+ over Mg2+ as the stimulating cation. All three enzymes catalyze not only the reversible cleavage of l-malyl-CoA but also the reversible cleavage of β-methylmalyl-CoA to propionyl-CoA and glyoxylate. The β-methylmalyl-CoA condensation activity for the enzyme of M. extorquens, however, is low and was not studied in detail (18). The R. capsulatus and the C. aurantiacus enzymes differ also with respect to the preference for condensation or cleavage reaction (Table 4). The R. capsulatus enzyme seems better adapted to catalyzing the condensation reaction, while the C. aurantiacus enzyme seems better adapted to catalyzing the cleavage reaction.
Based on the N-terminal sequence of the purified protein, a gene on contig 2G06-2D11 of R. capsulatus (designated mcl1) was identified, with a deduced amino acid sequence which showed significant sequence similarities to MCL from M. extorquens (51% sequence identity, GenBank accession number AAB58884) and MCL from C. aurantiacus (33% sequence identity, GenBank accession number ZP_00017884). These proteins belong to a large family, which also includes MS of type A and G and the β subunit of citrate/citramalate lyases (Pfam 01274) (7, 13). Amino acids identified to be important in catalysis for MSs type G (24, 38) are conserved in MCL of R. capsulatus, pointing to a similar mechanism of both enzymes. Apart from the substrate specificity (MS does not accept propionyl-CoA as a substrate), the main difference between MS and MCL is the reversibility of the latter, forming the corresponding CoA-ester of malate. The nature of the mechanistic differences responsible for this feature cannot be deduced from sequence comparison, but it is highly probable that the differences occur in the phase of acetyl-CoA binding and enolization.
Comparative genomic analysis.
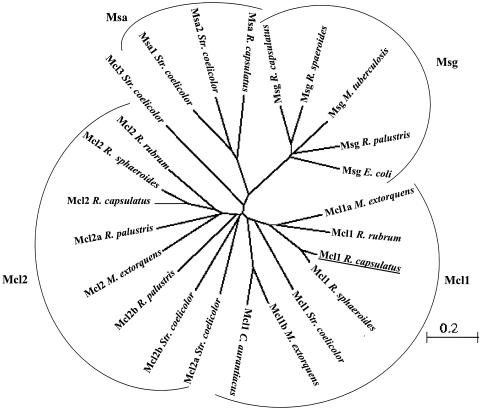
The genomes of several ICL-negative bacteria are completely sequenced (R. sphaeroides and S. coelicolor, both deposited at NCBI) or almost finished (R. capsulatus and M. extorquens [both deposited at Integrated Genomics] and R. rubrum [deposited at NCBI]) and were analyzed for the presence of genes encoding MCL-like proteins (Table 5). Figure 4 shows a phylogenetic analysis of amino acid sequences of various MCL-like proteins. MSs of type A and G clearly are outgrouping in the alignment. Among the MCL, two main subgroups can be observed. One subgroup contains all proteins, which have been shown to exhibit MCL activity (designated mcl1); no protein of the second subgroup (designated mcl2) has been characterized, and the substrates for those enzymes are unknown.
FIG. 4.
Phylogenetic tree of homologues of malyl-CoA lyase from R. capsulatus (underlined) based on amino acid sequences. The scale gives the distance equivalent to an exchange of 20/100 amino acid. Accession numbers are as follows: for M. extorquens, malyl-CoA lyase (Mcl1a) AAB58884 and RMQ06958, RMQ05381 (Mcl1b), and RMQ07283 (Mcl2); for C. aurantiacus, malyl-CoA/methylmalyl-CoA lyase ZP_00017884 (Mcl1); for S. coelicolor, NP_630554 (Mcl1), NP_626293 (Mcl2a), NP_639627 (Mcl2b), NP_625374 (Mcl3), NP_630343 (Msa1), and NP_625279 (Msa2); for R. capsulatus, RRC00706 (Msa), RRC01959 (Msg), RRC00315 (Mcl1), and RRC02787 (Mcl2); for R. rubrum, ZP_00014117 (Mcl1) and ZP_00015247 (Mcl2); for R. sphaeroides, ZP_00005846 (Mcl1), ZP_00007562 (Mcl2), and ZP_00006289 (Msg); for R. palustris, ZP_00007562 (Mcl2a), ZP_00009331 (Mcl2b), and NP_949552 (Msg); for E. coli, NP_417450 (Msg); for Mycobacterium tuberculosis, NP_216353 (Msg). Sequences for R. capsulatus and M. extorquens were provided by Integrated Genomics. All other sequences were provided by NCBI (http://www.ncbi.nlm.nih.gov).
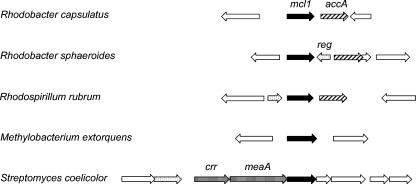
The mcl1 gene of R. capsulatus is located upstream of a gene designated accA; the deduced amino acid sequence of AccA shows significant sequence similarity to the α-subunit of the carboxytransferase component of several biotin-dependent acyl-CoA carboxylases. Together with the β-subunit (AccD), AccA catalyses the transfer of the biotin-bound bicarbonate onto the acyl-CoA substrate. A similar genomic arrangement is found for mcl1 homologs of R. sphaeroides and R. rubrum, with the exception of an additional gene, possibly encoding a regulator protein, as part of the gene cluster (Fig. 5). Propionyl-CoA carboxylase has been implicated in acetate assimilation by R. rubrum (5) and M. extorquens (9); in addition, a butyryl-CoA carboxylase is part of the glyoxylate regeneration pathway proposed for M. extorquens (8, 29).
FIG. 5.
Genetic environment of mcl1 for R. capsulatus, R. sphaeroides, R. rubrum, M. extorquens, and S. coelicolor. Only genes implicated in acetate assimilation and putative regulators are annotated. Filled arrow, mcl1 malyl-CoA lyase; hatched pattern, accA subunit of acyl-CoA carboxylase; dotted pattern, reg putative regulators; striped pattern, ccr crotonyl-CoA reductase or meaA potential mutase.
ICL-negative S. coelicolor also contains an mcl1 homolog in its genome, which is downstream of genes encoding an alcohol dehydrogenase (adhA), a crotonyl-CoA reductase (crr), and a coenzyme B12-dependent mutase (meaA). All three of these genes and their products have been shown to be involved in assimilation of C1 or C2 compounds by Streptomyces collinus and M. extorquens (9, 19, 29, 30, 39, 41). The mcl1 homolog of M. extorquens encodes a protein, which is upregulated in methanol- but not succinate-grown cells; its function, however, is unknown (34). An mcl1 homolog is not present in the complete genome of R. palustris.
Proposed role of malyl-CoA lyase.
The following arguments suggest that l-malyl-CoA/β-methylmalyl-CoA lyase, encoded by mcl1, is part of the unknown acetate assimilation pathway in ICL-negative bacteria. (i) MCL activity is present in cell extracts of ICL-negative R. capsulatus, R. rubrum, and R. sphaeroides (44, 45; this work). M. extorquens expresses two distinct MCL during growth on C1 compounds, one of which has been shown to be required for glyoxylate regeneration from acetyl-CoA in order to sustain the serine cycle for C1 metabolism (4, 8, 37). The second MCL may be involved primarily in a pathway for assimilation of acetyl-CoA and polyhydroxyalkanoate utilization. R. palustris, which is ICL positive and uses the glyoxylate cycle for acetate assimilation, does not express MCL activity. (ii) MCL activity in cell extracts of R. rubrum and R. capsulatus is up-regulated severalfold in acetate-grown cells but not in cells which were grown with sugars as the carbon source. (iii) The protein responsible for MCL activity in cell extracts of acetate-grown R. capsulatus is encoded by mcl1. Homologs to mcl1 are present in all known genomes of ICL-negative bacteria but not in the complete genome of the ICL-positive R. palustris. (iv) The ratio of the rates of β-methylmalyl-CoA and malyl-CoA (malate) formation in cell extracts is similar to that found with the purified enzyme. If an additional MS were present, the rate of malate formation should be higher. (v) In the case of S. coelicolor, mcl1 is part of a gene cluster required for acetate assimilation. For R. capsulatus, R. sphaeroides, and R. rubrum, a gene next to mcl1 may encode a subunit of a protein, propionyl-CoA carboxylase, which has been implicated in acetate assimilation in R. rubrum (5). (vi) MCL from R. capsulatus catalyzes not only the reversible condensation of acetyl-CoA and glyoxylate to malyl-CoA but also the reversible cleavage of β-methylmalyl-CoA into propionyl-CoA and glyoxylate. Both reactions are part of the proposed citramalate cycle for acetate assimilation for R. rubrum (25). MCL from C. aurantiacus was proposed to fulfill two functions, l-malyl-CoA cleavage and β-methylmalyl-CoA synthesis in a reversal of the citramalate cycle (22). This pathway functions in glyoxylate assimilation during autotrophic CO2 fixation in C. aurantiacus (21). (vii) Cell extracts of R. capsulatus did not hydrolyze β-methylmalyl-CoA to β-methylmalate and CoA, in contrast to malyl-CoA. This is also in agreement with the proposed citramalate cycle which requires only β-methylmalyl-CoA cleavage to propionyl-CoA and glyoxylate. In contrast, malyl-CoA formation is pulled forward by subsequent hydrolysis of malyl-CoA to malate and CoA.
We propose a dual role of malyl-CoA lyase (Mcl1) in acetate assimilation. First, the enzyme is thought to cleave β-methylmalyl-CoA to propionyl-CoA and glyoxylate. β-Methylmalyl-CoA is a proposed intermediate in the unknown acetate assimilation pathway, and the source of it (e.g., citramalate) remains to be identified. Propionyl-CoA is subsequently carboxylated, leading finally to succinyl-CoA. Second, the enzyme combines glyoxylate with acetyl-CoA, forming malyl-CoA, which is cleaved to malate by a second unidentified enzyme, resulting in unidirectional acetyl-CoA assimilation into a C4 compound.
TABLE 5.
Comparative genomic analysis for the presence of genes encoding Mcl-like proteins msa, msg, mcl1 and mcl2a
| Organism | msa | msg | mcl1 | mcl2 |
|---|---|---|---|---|
| R. capsulatus | + | + | + | + |
| R. sphaeroides | − | + | + | + |
| R. rubrum | − | − | + | + |
| M. extorquens | − | − | + | + |
| S. coelicolor | + | − | + | + |
| R. palustris (ICL positive) | + | + | − | + |
+, Gene present in genome; −, gene not present in genome.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany, the Fonds der Chemischen Industrie, Frankfurt, Germany, and the Graduiertenkolleg “Biochemie der Enzyme” (fellowship awarded to M.M.).
REFERENCES
- 1.Albers, H., and G. Gottschalk. 1976. Acetate metabolism in Rhodopseudomonas gelatinosa and several other rhodospirillaceae. Arch. Microbiol. 111:45-49. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L., and R. C. Fuller. 1967. Photosynthesis in Rhodospirillum rubrum. II. Photoheterotrophic carbon dioxide fixation. Plant Physiol. 42:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, England.
- 4.Arps, P. J., G. F. Fulton, E. C. Minnich, and M. E. Lidstrom. 1993. Genetics of serine pathway enzymes in Methylobacterium extorquens AM1: phosphoenolpyruvate carboxylase and malyl coenzyme A lyase. J. Bacteriol. 175:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, I. A., L. V. Filatova, and R. N. Ivanovsky. 2002. Inhibition of acetate and propionate assimilation by itaconate via propionyl-CoA carboxylase in isocitrate lyase-negative purple bacterium Rhodospirillum rubrum. FEMS Microbiol. Lett. 216:49-54. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Buckel, W., and A. Bobi. 1976. The enzyme complex citramalate lyase from Clostridium tetanomorphum. Eur. J. Biochem. 64:255-262. [DOI] [PubMed] [Google Scholar]
- 8.Chistoserdova, L., S.-W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chistoserdova, L. V., and M. E. Lidstrom. 1996. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extroquens AM1. Microbiology 142:1459-1468. [DOI] [PubMed] [Google Scholar]
- 10.Claassen, P. A. M., and A. J. B. Zehnder. 1986. Isocitrate lyase activity in Thiobacillus versutus grown anaerobically on acetate and nitrate. J. Gen. Microbiol. 132:3179-3185. [Google Scholar]
- 11.Cox, R. B., and J. R. Quayle. 1976. Synthesis and hydrolysis of malyl-coenzyme A by Pseudomonas AM1: an apparent malate synthase activity. J. Gen. Microbiol. 95:121-133. [DOI] [PubMed] [Google Scholar]
- 12.Cutinelli, C., G. Ehrensvärd, L. Reio, E. Saluste, and R. Stjernholm. 1951. Acetic acid metabolism in Rhodospirillum rubrum under anaerobic condition. II. Arkiv. fuer Kemi. 3:315-322. [Google Scholar]
- 13.Dimroth, P., and H. Eggerer. 1975. Isolation of subunits of citrate lyase and characterization of their function in the enzyme complex. Proc. Natl. Acad. Sci. USA 72:3458-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggerer, H., and C. H. Grünewälder. 1964. Zum Mechanismus der biologischen Umwandlung von Citronensäure. IV. Synthese von Malyl-Coenzym A und seiner Diastereomeren. Justus Liebigs Ann. Chem. 667:200-208. [Google Scholar]
- 15.Eggerer, H., U. Remberger, and C. H. Grünewälder. 1964. Zum Mechanismus der biologischen Umwandlung von Citronensäure. V. Citrat-synthase, eine Hydrolase für Malyl-coenzym A. Biochem. Z. 339:436-453. [PubMed] [Google Scholar]
- 16.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, S. C., and E. E. Dekker. 1984. Malyl-CoA formation in the NAD+-, CoASH-, and α-ketoglutarate dehydrogenase-dependent oxidation of 2-keto-4-hydroxyglutarate. J. Biol. Chem. 259:10012-10019. [PubMed] [Google Scholar]
- 18.Hacking, A. J., and J. R. Quayle. 1974. Purification and properties of malyl-coenzyme A lyase from Pseudomonas AM1. Biochem. J. 139:399-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, L., and K. A. Reynolds. 1997. A novel alternate anaplerotic pathway to the glyoxylate cycle in streptomycetes. J. Bacteriol. 179:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersh, L. B. 1973. Malyl coenzyme A lyase. Mechanism of action deduced by kinetic analysis. J. Biol. Chem. 249:5208-5212. [PubMed] [Google Scholar]
- 21.Herter, S., J. Farfsing, N. Gad'on, C. Rieder, W. Eisenreich, A. Bacher, and G. Fuchs. 2001. Autotrophic CO2 fixation by Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J. Bacteriol. 183:4305-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herter, S., A. Busch, and G. Fuchs. 2002. l-Malyl-coenzyme A lyase/β-methylmalyl-coenzyme A lyase from Chloroflexus aurantiacus, a bifunctional enzyme involved in autotrophic CO2 fixation. J. Bacteriol. 184:5999-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoare, D. S. 1963. The photo-assimilation of acetate by Rhodospirillum rubrum. Biochem. J. 87:284-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard, B. R., J. A. Endrizzi, and S. J. Remington. 2000. Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 Å resolution: mechanistic implications. Biochemistry 39:3156-3168. [DOI] [PubMed] [Google Scholar]
- 25.Ivanovsky, R. N., E. N. Krasilnikova, and I. A. Berg. 1997. A proposed citramalate cycle for acetate assimilation in the purple non-sulfur bacterium Rhodospirillum rubrum. FEMS Microbiol. Lett. 153:399-404. [Google Scholar]
- 26.Kornberg, H. L., and H. A. Krebs. 1957. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179:988-991. [DOI] [PubMed] [Google Scholar]
- 27.Kornberg, H. L., and J. Lascelles. 1960. The formation of isocitratase by the athiorhodaceae. J. Gen. Microbiol. 23:511-517. [DOI] [PubMed] [Google Scholar]
- 28.Kornberg, H. L., P. J. R. Phizackerley, and J. R. Sadler. 1959. Synthesis of cell constituents from acetate by Escherichia coli. Biochem. J. 72:32-33. [Google Scholar]
- 29.Korotkova, N., L. Chistoserdova, V. Kuksa, and M. E. Lidstrom. 2002. Glyoxylate regeneration pathway in the methylotroph Methylbacterium extorquens AM1. J. Bacteriol. 184:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korotkova, N., and M. E. Lidstrom. 2004. MeaB is a component of the methylmalyonyl-CoA mutase complex required for protection of the enzyme from inactivation. J. Biol. Chem. 279:13652-13658. [DOI] [PubMed] [Google Scholar]
- 31.Kortstee, G. J. J. 1981. The second part of the icl-serine pathway, p. 211-219. In H. Dalton (ed.), Microbial growth on C1 compounds. Heyden, London, England.
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Large, P. J., D. Peel, and J. R. Quayle. 1961. Microbial growth on C1 compounds. 2. Synthesis of cell constituents by methanol- and formate-grown Pseudomonas AM1, and methanol-grown Hyphomicrobium vulgare. Biochem. J. 81:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laukel, M., M. Rossignol, G. Borderies, U. Völker, and J. A. Vorholt. 2004. Comparison of the proteome of Methylobacterium extorquens AM1 grown under methylotrophic and nonmethylotrophic conditions. Proteomics 4:1247-1264. [DOI] [PubMed] [Google Scholar]
- 35.Oh, M.-K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 36.Pfennig, N. 1978. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped vitamin B12-requiring member of the family rhodospirillaceae. Int. J. Bacteriol. 28:283-288. [Google Scholar]
- 37.Salem, A. R., A. J. Hacking, and J. R. Quayle. 1973. Cleavage of malyl-coenzyme A into acetyl-coenzyme A and glyoxylate by Pseudomonas AM1 and other C1-unit-utilizing bacteria. Biochem. J. 136:89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, C. V., C.-C. Huang, A. Miczak, D. G. Russell, J. C. Sacchettini, and K. Höner zu Bentrup. 2003. Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis. J. Biol. Chem. 278:1735-1743. [DOI] [PubMed] [Google Scholar]
- 39.Smith, L. M., W. G. Meijer, L. Dijkhuizen, and P. M. Goodwin. 1996. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology 142:675-684. [DOI] [PubMed] [Google Scholar]
- 40.Srere, P. A., H. Brazil, and L. Gonen. 1963. The citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem. Scand. 17:129-134. [Google Scholar]
- 41.Stone, S., and D. M. Goodwin. 1988. Characterization and complementation of mutants of Methylobacterium AM1 which are defective in C-1 assimilation. J. Gen. Microbiol. 135:227-235. [Google Scholar]
- 42.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 43.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promotor system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuboi, S., and G. Kikuchi. 1962. Enzymic cleavage of malate to glyoxylate and acetyl-coenzyme A. Biochim. Biophys. Acta 62:188-190. [DOI] [PubMed] [Google Scholar]
- 45.Tuboi, S., and G. Kikuchi. 1965. Enzymic cleavage of malyl-coenzyme A into acetyl-coenzyme A and glyoxylic acid. Biochim. Biophys. Acta 96:148-153. [DOI] [PubMed] [Google Scholar]
- 46.Willison, J. C. 1988. Pyruvate and acetate metabolism in the photosynthetic bacterium Rhodobacter capsulatus. J. Gen. Microbiol. 134:2429-2439. [Google Scholar]
- 47.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low-background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]