Abstract
Disruption of the PA2491 gene in a mini-Tn5-tet insertion mutant of a clinical isolate of Pseudomonas aeruginosa increased expression of the mexEF-oprN multidrug efflux genes and decreased production of outer membrane protein OprD, concomitant with enhanced resistance to chloramphenicol, quinolones, and imipenem, which was reminiscent of previously described nfxC mutants. PA2491 encodes a probable oxidoreductase previously shown to be positively regulated by the MexT positive regulator of mexEF-oprN expression (T. Köhler, S. F. Epp, L. K. Curty, and J. C. Pechére, J. Bacteriol. 181:6300-6305, 1999). Spontaneous multidrug-resistant mutants of the P. aeruginosa clinical isolate hyperexpressing mexEF-oprN and showing reduced production of OprD were readily selected in vitro, and all of them were shown to carry mutations in PA2491, highlighting the probable significance of such mutations as determinants of MexEF-OprN-mediated multidrug resistance in vivo.
Three-component multidrug efflux systems of the resistance-nodulation-division (RND) family are prevalent in gram-negative bacteria, in which they contribute significantly to intrinsic and acquired resistance to a range of clinically important antimicrobial agents (i.e., antibiotics and biocides) (48). In Pseudomonas aeruginosa, an opportunistic human pathogen characterized by an innate resistance to multiple antimicrobial agents (19), seven tripartite RND family efflux systems have been described to date (MexAB-OprM [17, 35, 50, 51], MexCD-OprJ [49], MexEF-OprN [32], MexXY-OprM [2, 45, 70], MexJK-OprM [9], and, most recently, MexHI-OpmD [1, 58] and MexVW-OprM [38]), although only the constitutively expressed MexAB-OprM system (37, 51) and the drug-inducible MexXY-OprM system (43) have been shown to contribute to intrinsic resistance. Expression of many of the other systems results from mutations in regulatory, often repressor genes (e.g., mutations in the nfxB [49] and mexL [9] repressor genes in mutants hyperexpressing MexCD-OprJ and MexJK, respectively), and indeed, stable hyperexpression of MexAB-OprM and MexXY also results from mutations in repressor genes (i.e., mexR [25, 52, 54, 64, 73] and mexZ [40, 69], respectively).
The MexEF-OprN system is apparently quiescent in wild-type cells, at least under the usual laboratory growth conditions (32), but it is expressed in nfxC-type multidrug-resistant strains isolated in vitro (14, 32, 42) and in clinics (15, 24). nfxC mutants display resistance to fluoroquinolones, chloramphenicol, trimethoprim, and the carbapenem imipenem (14, 32), although resistance to imipenem results not from MexEF-OprN expression (32) but from the concomitant decrease in outer membrane protein OprD in these mutants (14, 42). OprD is a basic amino acid-peptide channel and a primary route of entry for carbapenems, such as imipenem, in P. aeruginosa (67, 68), and it is often absent in imipenem-resistant strains of P. aeruginosa (4, 31). In addition to its role in the export of and resistance to antimicrobials, MexEF-OprN also promotes resistance (or tolerance) to organic solvents (36), dyes (16), and biocides, such as triclosan (8). As a result of MexEF-OprN hyperexpression, nfxC mutants also express reduced levels of several quorum-sensing (i.e., homoserine lactone)-dependent extracellular virulence factors (33) and are attenuated for virulence (10), apparently as a result of MexEF-OprN-mediated export of a molecule(s) necessary for homoserine lactone production (33).
Unlike the majority of RND-type efflux systems in P. aeruginosa, which are negatively regulated by linked repressor genes, MexEF-OprN expression is positively regulated by the product of the linked mexT gene, a LysR family regulator of mexEF-oprN expression that is also responsible for the decrease in OprD expression seen in nfxC strains (34, 47). Preliminary studies have indicated that MexT control of OprD occurs at the level of transcription (34, 47), although posttranscriptional control of OprD by MexT has also been suggested (34). Expression of mexEF-oprN in P. aeruginosa typically results from mutations in mexT that, surprisingly, revert or suppress inactivating mutations that appear to be common in many so-called wild-type P. aeruginosa PAO strains (33, 41). Still, in strains carrying wild-type mexT, a mutation(s) in other, as-yet-unidentified genes appears to be important for mexEF-oprN hyperexpression (41). In the present report, mutations in PA2491, a gene immediately upstream of mexT, are shown to enhance mexEF-oprN expression and decreased OprD production, providing a multidrug resistance profile indistinguishable from that of previously described nfxC mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cells were cultivated in Luria broth (L-broth) (Miller's Luria broth base [Difco] with 2 g of NaCl per liter of H2O) and on Luria agar (L-agar) (L-broth containing 1.5% [wt/vol] agar) supplemented with antibiotics, as necessary, at 37°C. Plasmid pUCP18 and its derivatives were maintained with 100 μg of ampicillin per ml in Escherichia coli and with 100 to 400 μg of carbenicillin per ml in P. aeruginosa. Plasmid pUT-mini-Tn5-tet was maintained in E. coli with either ampicillin (100 μg per ml) or tetracycline (10 μg/ml), while plasmid pK18MobSacB and its derivatives were maintained in E. coli with 50 μg of kanamycin per ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80d lacZΔM15 endA1 recA1 hsdR17 (rK−mK+) supE44 thi-1 gyrA96 relA1 F− Δ(lacZYA-argF)U169 | 3 |
| S17-1 | thi pro hsdR recA Tra+ | 60 |
| Sm10(λpir) | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu; Kmr λpir | 44 |
| P. aeruginosa strains | ||
| K2153 | Clinical isolate | 61 |
| K2313 | PA2491::mini-Tn5-tet derivative of K2153 | This study |
| K2316 | K2313 ΔmexT | This study |
| K2317 | Spontaneous PA2491 mutant (Tyr127Stop) of K2153 selected with 500 μg of chloramphenicol per ml | This study |
| K2318 | Spontaneous PA2491 mutant (Val333Gly) of K2153 selected with 500 μg of chloramphenicol per ml | This study |
| K2319 | Spontaneous PA2491 mutant (91-bp deletion, ΔT402-G492) of K2153 selected with 500 μg of chloramphenicol per ml | This study |
| K2320 | Spontaneous PA2491 mutant (Ser124Arg) of K2153 selected with 500 μg of chloramphenicol per ml | This study |
| PAO-Geneva | Prototroph (carries wild-type mexT gene) | 41 |
| Plasmids | ||
| pUCP18 | Broad-host-range cloning vector; Apr Cbr | 57 |
| pSN2491 | pUCP18::PA2491 | This study |
| pK18MobSacB | Broad-host-range gene replacement vector; sacB Kmr | 60 |
| pMLS002 | pK18MobSacB::ΔmexT | This study |
| pUT::mini-Tn5-tet | mini-Tn5-tet delivery vector; Apr Tcr | 12 |
Transposon mutagenesis.
The pan-aminoglycoside-resistant P. aeruginosa clinical isolate K2153 was mutagenized with mini-Tn5-tet (12) following mobilization of mini-Tn5-tet-carrying plasmid pUT from E. coli SM10 (λpir) as described previously (7), with some modifications. Briefly, 600 μl of log-phase donor cells and 300 μl of unshaken stationary-phase recipient cells were mixed in a microcentrifuge tube, centrifuged, and resuspended in 100 μl of L-broth before they were spotted in the center of an L-agar plate. Following incubation at 37°C for 2.5 h, bacteria were recovered from the plate in 1 ml of L-broth, and insertion mutants of P. aeruginosa K2153 carrying mini-Tn5-tet in the chromosome were then selected on L-agar plates containing 64 μg of tetracycline per ml (to select mini-Tn5-tet) and 0.5 μg of imipenem per ml (to counterselect donor E. coli). Mini-Tn5-tet insertion mutants showing increased susceptibility to aminoglycosides were subsequently identified by first screening on plates containing paromomycin (512 μg/ml; the MIC for K2153 is 1,024 μg/ml), and the mutants that were not able to grow were examined for increased susceptibility to spectinomycin, kanamycin, and gentamicin (as well as paromomycin) by performing liquid MIC assays (see below). The mini-Tn5-tet-disrupted genes from pan-aminoglycoside-susceptible mutant derivatives of K2153 were recovered following digestion of genomic DNA with PstI (which does not cut within the transposon) and shotgun-cloning into PstI-restricted pK18MobSacB. Transformants of E. coli DH5α carrying pK18MobSacB with inserts of mini-Tn5-tet and flanking chromosomal DNA were selected on Luria-Bertani agar containing tetracycline (10 μg/ml) and kanamycin (50 μg/ml). Chromosomal DNA flanking the mini-Tn5-tet element was sequenced by using primers mini-Tn5-Tc-Right (5′-TGCGTCGCGGTGCATGGAGC-3′) and mini-Tn5-Tc-Left (5′-GACGATGAGCGCATTGTTAG-3′), which are specific to regions 400 to 500 bp upstream of the ends of the element. Once the flanking DNA sequences were obtained, disrupted genes were identified by BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/) searches of the available genome sequence (http://www.pseudomonas.com) (65).
DNA methods.
Standard protocols were used for restriction endonuclease digestion, ligation, transformation of electrocompetent E. coli, plasmid isolation, and agarose gel electrophoresis, as described by Sambrook and Russell (55). Genomic DNA was extracted from P. aeruginosa by using the protocol of Barcak et al. (5). Oligonucleotide synthesis and nucleotide sequencing were carried out by Cortec DNA Services Inc.
Cloning of PA2491 (mexS).
PA2491 was amplified from P. aeruginosa K2153 chromosomal DNA by using primers PA2491-F (5′-CGGCGAATTCCGGATACAGTCACAACCCATGAG-3′; EcoRI site underlined) and PA2491-R (5′-AAGCTTCAAGCTTCGGTCAACGATCTGTGGATCTG-3′; duplicate HindIII sites underlined) in a PCR mixture (50 μl) that included 10 ng of chromosomal DNA, each primer at a concentration of 0.6 μM, 1× ThermoPol buffer (New England Biolabs), each deoxynucleoside triphosphate at a concentration of 0.2 mM, and 2 U of Vent DNA polymerase (New England Biolabs). Amplification of PA2491 was achieved by incubation for 1 min at 95°C, followed by 30 cycles of 0.5 min at 95°C, 0.5 min at 60°C, and 1 min at 72°C and a final 7-min elongation at 72°C. The PA2491 PCR product was subsequently purified with a QIAGEN PCR purification kit, digested with EcoRI and HindIII, and cloned into EcoRI-HindIII-restricted pUCP18 to generate pSN2491, in which the PA2491 coding region was downstream of and in the same orientation as the resident lac promoter of pUCP18. Nucleotide sequencing with universal pUC primers confirmed the absence of mutations in pSN2491-borne PA2491.
Construction of a ΔmexT mutant of P. aeruginosa.
To introduce an in-frame deletion of the mexT gene into P. aeruginosa, a deletion construct was first prepared in plasmid pK18MobSacB by cloning PCR-amplified 1-kb DNA fragments corresponding to the regions upstream and downstream of the mexT sequences to be deleted. Sequences 5′ to the deletion were amplified by using primers MexTUp-F (5′-GAATTCGAATTCGTAGAACTGCAGGTGCTTC-3′; tandem EcoRI sites underlined) and MexTUp-R (5′-GGATCCGGATCCCAGGTCGTTTCGGTTCATG-3′; tandem BamHI sites underlined). Sequences 3′ to the deletion were amplified by using primers MexTDown-F (5′-GGATCCGGATCCCGAACTGTCGATGGCTTGG-3′; tandem BamHI sites underlined) and MexTDown-R (5′-AAGCTTAAGCTTGACGTACTTGAGGAACACG-3′; tandem HindIII sites underlined). The 50-μl PCR mixture was formulated as described above, and PCR was performed by using the same parameters as those outlined for amplification of PA2491. The PCR products (purified with a QIAGEN PCR purification kit) were subsequently digested with EcoRI and BamHI or BamHI and HindIII, as appropriate, and separately cloned into appropriately restricted pK18MobSacB, yielding plasmids pMSMexTUp (5′ upstream fragment) and pMSMexTDown (3′ downstream fragment). Following nucleotide sequencing to confirm the absence of mutations in the cloned DNAs, the 3′ downstream fragment was excised from pMSMexTDown by digestion with BamHI and HindIII and was cloned into EcoRI-BamHI-restricted pMSMexTUp, yielding pMLS002. The latter plasmid carried an in-frame deletion of the mexT gene, as confirmed by nucleotide sequencing. The pMLS002 vector was then introduced into E. coli S17-1 and mobilized into P. aeruginosa strain K2313 as described previously (61), and transconjugants carrying the plasmid in the chromosome were selected on L-agar plates containing kanamycin (1,000 μg/ml) and chloramphenicol (15 μg/ml; to counterselect E. coli). Kanamycin-resistant colonies were streaked onto L-agar plates containing 10% (wt/vol) sucrose, and sucrose-resistant colonies were screened for the presence of the mexT deletion by PCR with primers MexT-F (5′-CATGAACCGAAACGACCTG-3′) and MexT-R (5′-GACGTACTTGAGGAACACG-3′) and the parameters described above.
Amplification and sequencing of mexT.
The mexT gene of P. aeruginosa PAO-Geneva was amplified by PCR by using primers mexT-F1 (5′-GGTTGTGACTGTATCCGCCC-3′) and mexT-R1 (5′-TGGAATAAGCCGCACACCC-3′) in a reaction mixture that was formulated as described previously (72) and subjected to heating at 94°C for 3 min, followed by 30 cycles of 0.5 min at 94°C, 0.5 min at 58°C, and 1 min at 72°C and then incubation for 10 min at 72°C. However, amplification of mexT from clinical isolate K2153 with these PCR primers repeatedly failed to generate a fragment of the expected size that could be sequenced. Thus, the gene was amplified from K2153 in two overlapping parts, with primers mexT-F1 and mexT-R2 (5′-CCAGCAGGTTCGGCATCAATAG-3′) amplifying the 5′ portion of the gene and primers mexT-F2 (5′-ATGAGTCGCGCCAGCGAG-3′) and mexT-R3 (5′-AGGAGAAGTGGGATGACTGT-3′) amplifying the 3′ portion of the gene. The reaction mixtures were formulated and treated as described above, except that the annealing temperatures used were 57°C (mexT-F1-mexT-R2 reaction) and 53°C (mexT-F2-metT-R3 reaction). PCR products were purified and sequenced by using the corresponding PCR primers.
Antimicrobial susceptibility testing.
The antimicrobial susceptibilities of strains were assessed in microtiter trays by using a twofold serial dilution technique (27). Briefly, 50-μl aliquots of log-phase cells grown in L-broth were added to an equal volume of L-broth containing serial twofold dilutions of antibiotic to obtain a final cell concentration of 2.75 × 105 cells/ml. Following incubation at 37°C for 18 h, growth was assessed visually, and the MIC was defined as the lowest concentration of antibiotic that inhibited visible growth.
RT-PCR.
RNA isolation from overnight cultures of P. aeruginosa and subsequent reverse transcription (RT)-PCR to assess expression of rpsL, mexE, and mexT were carried out as described previously (61) by using the primer pairs rpsLF and rpsLR (61), mexEF (5′-GTCATCGAACAACCGCTG-3′) and mexER (5′GTCGAAGTAGGCGTAGACC-3′), and mexTF (5′-CATGAACCGAAACGACCTG-3′) and mexTR (5′-CGAAGATTTCCTGGGCTCG-3′), respectively. Strains carrying pUCP18 and its derivatives were cultured in the presence of carbenicillin.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
P. aeruginosa strains were screened for OprD production by using an OprD-specific antiserum (23) following Western immunoblotting of electrophoretically separated cell envelopes by using established procedures (62).
Selection of spontaneous PA2491 mutants.
One hundred microliters of a 1/10 dilution of an overnight L-broth culture of P. aeruginosa K2153 was plated onto L-agar plates containing 500 μg of chloramphenicol per ml. After 48 h of incubation at 37°C, 50 representative chloramphenicol-resistant colonies were screened for a resistance profile characteristic of a MexEF-OprN-hyperexpressing strain (i.e., growth on elevated levels of nalidixic acid [512 μg/ml], norfloxacin [4 μg/ml], and imipenem [4 μg/ml]). Strikingly, all spontaneous chloramphenicol-resistant K2153 derivatives showed reduced susceptibility to nalidixic acid, norfloxacin, and imipenem, and seven of these derivatives were randomly selected for further study. To assess the presence of PA2491 mutations in these organisms, the PA2491 gene was amplified from the chromosomes of strain K2153 and these mutants by using primers PA2491-F and PA2491-R as described above and was sequenced with the same primers.
RESULTS
Mutations in PA2491 promote mexEF-oprN expression and multidrug resistance.
Clinical P. aeruginosa isolate K2153 displays a pan-aminoglycoside-resistant phenotype owing, at least in part, to enhanced expression of the mexXY multidrug efflux genes (61). In an attempt to identify the gene(s) required for MexXY-dependent aminoglycoside resistance in this strain, K2153 was mutagenized with mini-Tn5-tet, mutants showing enhanced aminoglycoside susceptibility were recovered, and the disrupted genes were identified. One such mutant, K2313, showed increased susceptibility to several aminoglycosides but also showed a marked increase in resistance to chloramphenicol, quinolones, and imipenem (Table 2), a resistance profile strikingly reminiscent of nfxC mutants that overproduce MexEF-OprN (32). By using RT-PCR, expression of mexE (as a measure of mexEF-oprN expression) was examined in K2313 and compared to the expression in the parent strain, K2153. As shown in Fig. 1A, lane 2, the mexE levels in K2313 were clearly greater than the levels in K2153, in which mexE was barely detectable (Fig. 1A, lane 1). Recovery and sequencing of chromosomal DNA flanking the transposon insert in K2313 resulted in identification of PA2491 as the disrupted gene in this mutant. Intriguingly, PA2491 occurs upstream of and is apparently transcribed divergently from mexT, and it is predicted to encode a polypeptide with substantial homology to Zn2+-dependent oxidoreductases/dehydrogenases (see COG0604, COG1064, COG2130, COG1063, COG1062, and pfam0017 at http://ww.ncbi.nlm.nih.gov), including amino acid dehydrogenases associated with amino acid metabolism (COG1063). Introduction of the PA2491 gene cloned from K2153 (on plasmid pSN2491) into this strain reversed both the enhanced mexE (mexEF-oprN) expression (Fig. 1A, compare lanes 3 and 4) and multidrug resistance (Table 2), which is consistent with the hypothesis that PA2491 disruption in K2313 is responsible for the multidrug resistance and efflux gene expression seen in K2313.
TABLE 2.
Influence of PA2491 mutations on antibiotic susceptibility of P. aeruginosa
| Strain | PA2491 statusa | MIC (μg/ml) of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Paromomycin | Streptomycin | Spectinomycin | Gentamicin | Kanamycin | Chloramphenicol | Imipenem | Nalidixic acid | Norfloxacin | ||
| K2153 | WT | 1,024 | 128 | 1,024 | 8 | 256 | 128 | 4 | 256 | <1 |
| K2313 | Null | 128 | 16 | 256 | 4 | 64 | 1,024 | 8 | 2,048 | 8 |
| K2313/pUCP18 | Null | —d | — | 256 | 4 | 64 | 1,024 | 8 | 2,048 | 8 |
| K2313/pSN2491 | WT | — | — | 1,024 | 8 | 256 | 256 | 4 | 512 | 4 |
| K2316 | Null | — | — | 1,024 | 8 | 256 | 64 | 4 | 256 | <1 |
| K2317 | Tyr127Stopb | — | — | 512 | 4 | 64 | 2,048 | 8 | 2,048 | 8 |
| K2318 | Val333Gly | — | — | 512 | 4 | 64 | 2,048 | 8 | 2,048 | 4 |
| K2319 | Truncatedc | — | — | 512 | 4 | 64 | 2,048 | 8 | 2,048 | 8 |
| K2320 | Ser124Arg | — | — | 512 | 4 | 64 | 2,048 | 8 | 2,048 | 8 |
The PA2491 product is wild type (WT), absent (null), truncated, or mutated (mutation indicated).
This mutation was found in four of seven chloramphenicol-selected multidrug-resistant derivatives of K2153.
Truncated owing to a 91-bp deletion in the PA2491 gene.
—, not determined.
FIG. 1.
Expression of mexE (A), mexT (B), and rpsL (C) in P. aeruginosa as assessed by RT-PCR. Lane 1, strain K2153 (PA2491+); lane 2, K2313 (PA2491−); lane 3, K2313/pUCP18 (PA2491−); lane 4, K2313/pSN2491 (PA2491+); lane 5, K2316 (K2313 ΔmexT); lane 6, K2153 (PA2491+); lane 7, K2317 (PA2491−, Tyr127Stop); lane 8, K2318 (PA2491−, Val333Gly); lane 9, K2319 (PA2491−, 91-bp deletion); lane 10, K2320 (PA2491−, Ser124Arg) (genotypes are indicated in parentheses). The PCR portions of the reactions were carried out for 28 cycles (top panel) and 29 cycles (bottom panel) for mexE (A), for 29 cycles (top panel) and 30 cycles (bottom panel) for mexT (B), and for 18 cycles (top panel) and 19 cycles (bottom panel) for rpsL (C). The rpsL reaction served as an internal control that ensured that equal amounts of RNA were employed in all of the RT-PCRs.
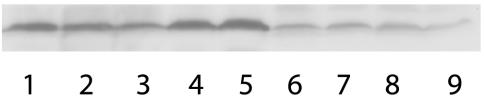
nfxC strains expressing mexEF-oprN, including strains with documented mexT mutations, display reduced OprD production that is, in fact, responsible for the imipenem resistance of these mutants (32, 34). It was of interest, therefore, to see if the imipenem resistance resulting from the PA2491 disruption in K2313 was similarly due to decreased OprD production. By using antibodies specific to OprD, OprD levels were assessed in the PA2491 mutant K2313 and its K2153 parent strain, and the results clearly show that there was a modest but definite decline in OprD levels in K2313 (Fig. 2, lanes 1 and 2). Moreover, the cloned PA2491 gene on plasmid pSN2491 increased OprD levels in K2313 (Fig. 2, compare lanes 3 and 4), in parallel with a reduction in the imipenem MIC to wild-type levels as well (Table 2). As in previously documented nfxC strains, a mutation in PA2491 simultaneously increased mexEF-oprN expression and reduced OprD production, providing the characteristic multidrug resistance of this mutant.
FIG. 2.
Immunoblot detection of OprD. Lane 1, clinical isolate K2153 (PA2491+); lane 2, K2313 (PA2491−); lane 3, K2313/pUCP18 (PA2491−); lane 4, K2313/pSN2491 (PA2491+); lane 5, K2316 (K2313 ΔmexT); lane 6, K2317 (PA2491−, Tyr127Stop); lane 7, K2318 (PA2491−, Val333Gly); lane 8, K2319 (PA2491−, 91-bp deletion); lane 9, K2320 (PA2491−, Ser124Arg) (genotypes are indicated in parentheses).
MexT is required for mexEF-oprN expression and multidrug resistance in PA2491 mutants.
One possible explanation for the increased expression of mexEF-oprN and the corresponding multidrug resistance of PA2491::mini-Tn5-tet strain K2313 is that the transposon insertion in PA2491 creates or provides a promoter that drives downstream mexT expression, with enhanced levels of MexT responsible for the increase in mexEF-oprN expression. To assess this possibility, the levels of expression of mexT were examined in the parent strain K2153 and in K2313 by using RT-PCR. As shown in Fig. 1B, lanes 1 and 2, no increase in mexT expression resulted from transposon disruption of PA2491 in K2313, and complementation of this mutant with the cloned PA2491 gene (on plasmid pSN2491) similarly had no effect on mexT expression (Fig. 1B, compare lanes 3 and 4) despite its negative impact on mexEF-oprN expression and resistance (see above). Thus, the impact of a PA2491 mutation on mexEF-oprN expression is not related to changes in MexT levels. MexT is, however, required for mexEF-oprN expression and the resultant multidrug resistance of K2313, since an in-frame deletion of mexT in this strain compromised mexEF-oprN expression (Fig. 1A, compare lanes 2 and 5) and resistance (Table 2, compare K2316 and K2313). This also indicates that unlike the mexT genes of many so-called wild-type strains, the mexT gene of clinical isolate K2153 must encode a functional activator. Consistent with this, sequencing of the mexT gene from this strain revealed the presence of only six silent mutations (C263T, T383C, G391C, A475G, G490C, and T511C) compared to the wild-type mexT gene of P. aeruginosa PAO-Geneva (whose sequence in this study differed from that published previously by a single C76G transition that replaced a leucine with a valine in the MexT protein). As expected, elimination of mexT in the K2316 derivative of K2313 also increased OprD levels (Fig. 2, compare lanes 2 and 5) concomitant with a decrease in the imipenem MIC (Table 2), indicating that the OprD decrease that resulted from the PA2491 disruption in K2313, like the mexEF-oprN increase, was MexT dependent.
Isolation of spontaneous PA2491 mutants.
Previous studies have shown that chloramphenicol readily selects NfxC-type mexEF-oprN-hyperexpressing multidrug-resistant mutants in the lab (26, 33). Thus, an overnight culture of K2153 was plated onto chloramphenicol-containing plates, and resistant isolates obtained after 48 h of growth were screened for resistance to quinolones, chloramphenicol, and imipenem, which is typical of nfxC mutants. All 50 chloramphenicol-resistant derivatives of K2153 chosen at random showed elevated resistance to nalidixic acid, norfloxacin, chloramphenicol, and imipenem (Table 2), and seven representative derivatives were screened for mutations in PA2491. While the PA2491 gene of K2153 carried multiple (fourteen) mutations relative to the PA2491 gene of the sequenced P. aeruginosa strain (www.pseudomonas.com), all but one mutation (a G-to-A transition yielding an Asp244Asn substitution in the PA2491 protein) were silent. All seven of the chloramphenicol-selected multidrug-resistant mutants carried additional mutations in PA2491, including four unique mutations, Tyr127Stop (e.g., K2317), Val333Gly (e.g., K2318), Ser124Arg (e.g., K2320), and a 91-bp deletion from T402 to G492 in the gene (e.g., K2319). Moreover, these mutations were associated with substantially increased mexE expression (Fig. 1A, compare lane 6 with lanes 7 to 10) and reduced OprD production (Fig. 2, compare lane 1 with lanes 6 to 9), confirming that spontaneous nfxC-like multidrug-resistant PA2491 mutants are readily selected, at least in vitro.
Distribution of PA2491-mexT pairs in bacteria.
Despite the presence of numerous oxidoreductase homologues of PA2491 and homologues of MexT and MexEF-OprN in gram-negative bacteria, very few instances of linked PA2491-mexT-like genes were found in a search of the GenBank databases. Of five such instances (all in the Pseudomonadaceae and Burkholderiaceae), only two were linked to efflux systems of the RND family (in Pseudomonas putida and Azotobacter vinelandii) (Table 3). Still, a PA2491-mexT-like pair in Pseudomonas syringae serovar Tomato encoding predicted products with very high homology to the PA2491 gene product and MexT (Table 3) was observed adjacent to a probable membrane protein gene that was, however, disrupted by an insertion-like element, and so we could not rule out the possibility that this pair was once linked to an RND-type efflux gene.
TABLE 3.
PA2491-mexT pairs in other bacteriaa
| Organism | PA2491 homologue | MexT homologue | MexE homologue | MexF homologue | OprN homologue |
|---|---|---|---|---|---|
| Pseudomonas syringae pv. Tomato | AAO56529 (57/74)b | AAO56530 (85/91) | —c | —c | —c |
| Pseudomonas putida | CAD59443 (55/75) | TtgS (85/90) | TtgDd (34/62) | TtgEd (38/73) | TtgFd (31/60) |
| Azotobacter vinelandii | ZP_00090326 (60/76) | ZP_000900325 (77/83) | ZP_00090324 (68/77) | ZP_00090323 (78/86) | —e |
| Burkholderia cepacia | ZP_00218119 (31/59) | ZP_00218118 (25/61) | — f | — f | — f |
| Ralstonia eutropha | ZP_00170295 (29/42) | ZP_00170294 (21/52) | — f | — f | — f |
Microbial genome sequences available from the GenBank database were screened for genes that both encoded protein homologues of PA2491 and occurred immediately adjacent to genes encoding MexT/LysR-like proteins. These sequences were then further screened for linked efflux genes encoding RND family efflux systems (e.g., MexEF-OprN), and the PA2491, MexT, and MexEF-OprN homologues discovered are indicated.
The numbers in parentheses are the percentage of identity/percentage of similarity to PA2491, MexT, MexE, MexF, or OprN.
Insertion-like sequences were reported adjacent to the P. syringae PA2491-mexT homologues, suggestive of a rearrangement of the original neighboring sequences.
The ttgDEF operon encoding TtgDEF is separated by two genes or open reading frames from the genes encoding the PA2491-MexT homologues CAD59443 and TtgS.
A gene encoding a probable outer membrane-OprN component was absent.
No efflux genes were linked to genes encoding the corresponding PA2491-MexT homologues.
DISCUSSION
MexEF-OprN is typically quiescent in wild-type P. aeruginosa, and its expression and MexEF-OprN-dependent multidrug resistance typically occur following mutation (32) (e.g., in the mexT gene that encodes a positive regulator of mexEF-oprN expression [34, 47]). Such mutations provide for both mexEF-oprN expression and the concomitant decline in OprD levels that characterizes these so-called nfxC mutants (32), although, surprisingly, all of the documented nfxC mutations actually revert or suppress inactivating mexT mutations that appear to be commonplace in wild-type P. aeruginosa PAO strains (41). While examples of nfxC-like multidrug-resistant mutants derived from P. aeruginosa strains harboring a wild-type mexT gene are known (34, 41), the nature of the mutation(s) responsible is unknown, although a mexS designation has been proposed for them (41). In the present paper, we report the identity of a possible mexS candidate, PA2491, whose disruption in a mini-Tn5-tet insertion mutant and in spontaneous mutants provides multidrug resistance owing to a concomitant increase in MexEF-OprN and a decrease in OprD. As such, it represents the first example of a mutation in a nonregulatory locus that is responsible for increased expression of a multidrug efflux system in P. aeruginosa and one of only a few such examples in bacteria (e.g., certain auxotrophic mutations in E. coli have been linked to increased expression of the AcrAB components of theAcrAB-TolC multidrug efflux system in this organism [20]).
PA2491 is a previously identified oxidoreductase/dehydrogenase homologue, designated Qrh by Köhler et al. (34), who reported that it, like mexEF-oprN, was positively regulated by MexT. Coregulation of an oxidoreductase and an efflux system might be explained by their having a common role in, for example, detoxification of cellular metabolites, reminiscent of glutathione-mediated detoxification and export of exogenous and endogenous toxic compounds in yeast (59). Moreover, it is likely that such metabolites, as substrates for PA2491 and MexEF-OprN, are the effector molecules for the MexT regulator, to which they bind, and stimulate MexT-dependent activation of PA2491 and mexEF-oprN expression. Still, the fact that loss of PA2491 activity specifically activates mexEF-oprN expression suggests that the constitutively expressed (34) PA2491 gene has the primary role in detoxification and that only under circumstances in which the enzyme is unable to keep up with metabolite production, due either to mutational loss of PA2491 or perhaps to an excess of metabolite production, is MexEF-OprN recruited. The finding that enhanced mexEF-oprN expression in previously described nfxC mutants and our newly described PA2491 mutants is coupled with reduced production of a basic amino acid-peptide porin, OprD 22), suggests that such metabolites might well be derived from basic amino acids and peptides, whose reduced uptake in such mutants would clearly limit the production of downstream metabolites that might be the substrates for MexEF-OprN and PA2491. Certainly, there is precedence for bacterial efflux of amino acids (6, 11, 39, 71) and their metabolites (11), indicating that under certain circumstances the accumulation of these compounds within the cell is detrimental to cell health.
Interestingly, the K2313 PA2491::mini-Tn5-tet insertion mutant was originally selected because of its increased susceptibility to aminoglycosides compared to its MexXY-expressing parent strain, K2153. In this instance, however, it is probable that overexpression of MexEF-OprN in K2313 indirectly affected aminoglycoside resistance as a result of a negative effect on MexXY expression. MexEF-OprN-overproducing nfxC mutants are also known to be aminoglycoside and β-lactam hypersusceptible (14), apparently owing to decreased expression of MexXY and MexAB-OprM, respectively. In fact, there have been several reports of increased expression of one RND-type multidrug efflux system compromising expression of another system in P. aeruginosa (18, 21, 37, 63).
Whatever the intended functions(s) of PA2491 and MexEF-OprN, the apparently limited distribution of linked genes encoding PA2491-MexT-MexEF-OprN homologues (present in only a few Pseudomonadaceae) suggests either that they provide a function unique to these organisms or that they represent a novel approach to a more common operation that is addressed by other means in other organisms. It is interesting that the highly similar PA2491-MexT counterparts in P. putida, CAD59443 (GenBank) and TtgS, are encoded by genes linked (two open reading frames away) to an RND family efflux operon, ttgDEF, which was previously shown to respond and provide tolerance to organic solvents only (46, 53), although it is unclear at present whether TtgS has any role in regulating ttgDEF expression or whether CAD59443 mutants overproduce TtgDEF. Since solvents induce stress response genes in Pseudomonas and other organisms (30, 56, 66), it is also not clear whether solvents are the primary inducers of this system or whether the changes that they induce in the cell ultimately result in accumulation of substances (related to the MexEF-OprN substrates?) that are the true inducers and intended substrates of TtgDEF. Given the substantial similarity in amino acid sequence and genetic organization of the PA2491-MexT-MexEF-OprN and CAD59443-TtgS-TtgDEF systems, one might expect these systems to be functionally related, and indeed, MexEF-OpN (36), like TtgDEF and, in fact, many RND family efflux systems (48), has been shown to contribute to solvent tolerance. Whether these systems respond to and export related metabolites remains to be seen.
A recent transcriptome analysis of P. aeruginosa after 12 h of interaction with airway epithelial cells revealed substantial (10- to 15-fold) increases in expression of both mexEF-oprN and PA2491, apparently in parallel with increased damage of the epithelial cells (and release of cell contents?), likely resulting from prolonged interaction with this organism (13). Possibly epithelial cell contents generate intracellular pools of metabolite substrates for MexEF-OprN and PA2491, either following uptake of epithelial cell contents (as, e.g., nutrients) or as a result of physiological changes promoted by exposure to these cell contents (a stress response?). In any case, these data suggest that P. aeruginosa may well encounter circumstances in vivo where PA2491 and MexEF-OprN are needed. Also, the demonstration that mutants overexpressing MexEF-OprN were readily recovered from an experimental model of rat pneumonia in the absence of antibiotic selection (28) indicates that there is some advantage to MexEF-OprN expression in vivo, independent of antimicrobial export. The recent observation that mutational loss of the VsqR quorum-sensing and virulence regulator compromises mexEF-oprN expression in cells under oxidative stress (i.e., stress due to H2O2) suggests that this system may normally be induced in response to this stress (29). mexEF has also been shown to be induced by a synthetic analogue of a naturally occurring macroalga (seaweed) furanone compound known to adversely affect quorum-sensing-dependent gene expression in P. aeruginosa (20a). Nonetheless, mexEF has never been shown to be regulated by quorum sensing, suggesting that the furanone effect on this efflux system is related to some other effect of this compound on P. aeruginosa unrelated to its quorum-sensing antagonist properties.
While these studies and our demonstration in this study that PA2491 mutations enhance mexEF-oprN expression clearly indicate that MexEF-OprN has a role other than antimicrobial export and resistance, the intended function of this efflux system and the identity of its natural substrates remain a mystery. Still, intended transcriptome studies of PA2491 (and nfxC) mutants may provide some clues.
Acknowledgments
We thank H. Schweizer and L. Tomalty for providing strains and R. E. W. Hancock for providing the anti-OprD antiserum.
This work was supported by an operating grant from the Canadian Cystic Fibrosis Foundation. M.L.S. was supported by a studentship from the Natural Science and Engineering Research Council.
REFERENCES
- 1.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 2.Aires, J. R., T. Köhler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 4.Ballestero, S., A. Fernandez-Rodriguez, R. Villaverde, H. Escobar, J. C. Perez-Diaz, and F. Baquero. 1996. Carbapenem resistance in Pseudomonas aeruginosa from cystic fibrosis patients. J. Antimicrob. Chemother. 38:39-45. [DOI] [PubMed] [Google Scholar]
- 5.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 6.Bellmann, A., M. Vrljic, M. Patek, H. Sahm, R. Kramer, and L. Eggeling. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765-1774. [DOI] [PubMed] [Google Scholar]
- 7.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC type multidrug resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 8.Chuanchuen, R., K. Beinlich, T. T. Hoang, A. Becher, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Gebbie, M. Benghezal, C. Van Delden, L. K. Curty, and T. Köhler. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dassler, T., T. Maier, C. Winterhalter, and A. Bock. 2000. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 36:1101-1112. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisk, A., J. R. Schurr, G. Wang, D. C. Bertucci, L. Marrero, S. H. Hwang, D. J. Hassett, and M. J. Schurr. 2004. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect. Immun. 72:5433-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda, H., M. Hosaka, K. Hirai, and S. Iyobe. 1990. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob. Agents Chemother. 34:1757-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda, H., M. Hosaka, S. Iyobe, N. Gotoh, T. Nishino, and K. Hirai. 1995. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:790-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germ, M., E. Yoshihara, H. Yoneyama, and T. Nakae. 1999. Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 261:452-455. [DOI] [PubMed] [Google Scholar]
- 17.Gotoh, N., H. Tsujimoto, K. Poole, J.-I. Yamagishi, and T. Nishino. 1995. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob. Agents Chemother. 39:2567-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotoh, N., H. Tsujimoto, M. Tsuda, K. Okamoto, A. Nomura, T. Wada, M. Nakahashi, and T. Nishino. 1998. Charaterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock, R. E. W., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updates 3:247-255. [DOI] [PubMed] [Google Scholar]
- 20.Helling, R. B., B. K. Janes, H. Kimball, T. Tran, M. Bundesmann, P. Check, D. Phelan, and C. Miller. 2002. Toxic waste disposal in Escherichia coli. J. Bacteriol. 184:3699-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirai, K., S. Suzue, T. Irikura, S. Iyobe, and S. Mitsuhashi. 1987. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 31:582-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, H., and R. E. Hancock. 1993. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J. Bacteriol. 175:7793-7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, H., D. Jeanteur, F. Pattus, and R. E. Hancock. 1995. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol. Microbiol. 16:931-941. [DOI] [PubMed] [Google Scholar]
- 24.Jalal, S., O. Ciofu, N. Hoiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 26.Jalal, S., G. Wretlind, N. Gotoh, and B. Wretlind. 1999. Rapid identification of mutations in a multidrug efflux pump in Pseudomonas aeruginosa. APMIS 107:1109-1116. [DOI] [PubMed] [Google Scholar]
- 27.Jo, J. T., F. S. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Join-Lambert, O. F., M. Michea-Hamzehpour, T. Kohler, F. Chau, F. Faurisson, S. Dautrey, C. Vissuzaine, C. Carbon, and J. C. Pechére. 2001. Differential selection of multidrug efflux mutants by trovafloxacin and ciprofloxacin in an experimental model of Pseudomonas aeruginosa acute pneumonia in rats. Antimicrob. Agents Chemother. 45:571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juhas, M., L. Wiehlmann, B. Huber, D. Jordan, J. Lauber, P. Salunkhe, A. S. Limpert, F. von Gotz, I. Steinmetz, L. Eberl, and B. Tummler. 2004. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150:831-841. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, H., M. Yamamoto, and R. Aono. 1998. Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiology 144:353-359. [DOI] [PubMed] [Google Scholar]
- 31.Kohler, T., M. Michea-Hamzehpour, S. F. Epp, and J. C. Pechére. 1999. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob. Agents Chemother. 43:424-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J.-C. Pechére. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 33.Kohler, T., C. Van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechére. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Köhler, T., S. F. Epp, L. K. Curty, and J.-C. Pechére. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, X.-Z., L. Zhang, and K. Poole. 1998. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J. Bacteriol. 180:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, X. Z., N. Barre, and K. Poole. 2000. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 46:885-893. [DOI] [PubMed] [Google Scholar]
- 38.Li, Y., T. Mima, Y. Komori, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 52:572-575. [DOI] [PubMed] [Google Scholar]
- 39.Livshits, V. A., N. P. Zakataeva, V. V. Aleshin, and M. V. Vitushkina. 2003. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 154:123-135. [DOI] [PubMed] [Google Scholar]
- 40.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maseda, H., K. Saito, A. Nakajima, and T. Nakae. 2000. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107-112. [DOI] [PubMed] [Google Scholar]
- 42.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mine, T., Y. Morita, A. Kataoka, T. Mitzushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosqueda, G., and J. L. Ramos. 2000. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J. Bacteriol. 182:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochs, M. M., M. P. McCusker, M. Bains, and R. E. Hancock. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poole, K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 49.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J.-I. Yamagishi, X.-Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 50.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 51.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Santos, P. M., D. Benndorf, and I. Sa-Correia. 2004. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 4:2640-2652. [DOI] [PubMed] [Google Scholar]
- 57.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 58.Sekiya, H., T. Mima, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Functional cloning and characterization of a multidrug efflux pump, MexHI-OpmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47:2990-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma, K. G., R. Kaur, and A. K. Bachhawat. 2003. The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch. Microbiol. 180:108-117. [DOI] [PubMed] [Google Scholar]
- 60.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 61.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srikumar, R., X.-Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 66.Tomas, C. A., J. Beamish, and E. T. Papoutsakis. 2004. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J. Bacteriol. 186:2006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680-15684. [PubMed] [Google Scholar]
- 69.Vogne, C., J. R. Aires, C. Bailly, D. Hocquet, and P. Plesiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yen, M. R., Y. H. Tseng, P. Simic, H. Sahm, L. Eggeling, and M. H. Saier, Jr. 2002. The ubiquitous ThrE family of putative transmembrane amino acid efflux transporters. Res. Microbiol. 153:19-25. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, L., X.-Z. Li, and K. Poole. 2001. The SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziha-Zarifi, I., C. Llanes, T. Koehler, J.-C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]