Abstract
A role for the Escherichia coli glgX gene in bacterial glycogen synthesis and/or degradation has been inferred from the sequence homology between the glgX gene and the genes encoding isoamylase-type debranching enzymes; however, experimental evidence or definition of the role of the gene has been lacking. Construction of E. coli strains with defined deletions in the glgX gene is reported here. The results show that the GlgX gene encodes an isoamylase-type debranching enzyme with high specificity for hydrolysis of chains consisting of three or four glucose residues. This specificity ensures that GlgX does not generate an extensive futile cycle during glycogen synthesis in which chains with more than four glucose residues are transferred by the branching enzyme. Disruption of glgX leads to overproduction of glycogen containing short external chains. These results suggest that the GlgX protein is predominantly involved in glycogen catabolism by selectively debranching the polysaccharide outer chains that were previously recessed by glycogen phosphorylase.
The synthesis of glycogen in Escherichia coli occurs when carbon is abundant but another nutrient required for growth is limiting. Although glycogen production or accumulation is not required for growth under laboratory culture conditions, the presence of glycogen may increase the viability of E. coli under adverse conditions or in specific ecological niches (reviewed in reference 29). Various E. coli mutants with mutations in glycogen synthesis and catabolism genes have been isolated (4, 7, 10, 28, 30). These mutants fell in three phenotypic classes with respect to the iodine staining phenotype of the bacterial colonies. Class A mutants were devoid of iodine-staining polysaccharide, class B mutants displayed blue staining that revealed the presence of moderately branched polysaccharides, and class C mutants displayed darker staining than the wild type owing to glycogen overproduction (7). These genetic and biochemical studies demonstrated that two activities are absolutely required for the synthesis of α-1,4-glucosyl-glucan, ADP-glucose pyrophosphorylase (EC 2.7.7.17; encoded by glgC) and glycogen synthase (EC 2.4.1.21; encoded by glgA). A third biosynthetic activity, glycogen branching enzyme (EC 2.4.1.18; encoded by glgB), is responsible for the introduction of α-1,6 linkages into the polymer. In E. coli these genes are located in a gene cluster which includes the glgC, glgA, and glgB genes and two other genes thought to be primarily involved in glycogen degradation, glgX and glgP (also known as glgY), encoding a putative glycogen debranching enzyme (EC 3.2.1.-) and glycogen phosphorylase (EC 1.4.1.1), respectively (30). The cluster of genes is expressed as two tandemly arranged operons, one comprised of glgC, glgA, and glgP and the other comprised of glgB and glgX (29). The transcription of these operons is subject to complex regulation (31, 33).
The glgX gene was identified as an open reading frame in plasmid pOP12 (30), which was originally shown to contain the glgC, glgA, and glgB genes (25). Sequence analysis suggested that GlgX was a glucan hydrolase, probably belonging to the isoamylase family of debranching enzymes (31). Evidence supporting this hypothesis was then obtained both by inactivating or overexpressing glgX, although not selectively, and by monitoring the total debranching activity in crude extracts (33). Debranching enzymes are usually classified as either isoamylases or pullulanases. Pullulanases hydrolyze pullulan and amylopectin but have lower activity with glycogen, whereas isoamylases do not hydrolyze pullulan and have high activity with glycogen. However, the isoamylase-type debranching enzyme characterized from E. coli, while unable to hydrolyze pullulan, is not a typical isoamylase as it has only weak activity with amylopectin or glycogen but high activity with β-amylase-limit dextrin and phosphorylase-limit dextrin (GPLD) (15).
Debranching enzymes from a range of bacterial and plant sources have been characterized. In addition to intracellular glg operon-encoded isoamylases homologous to E. coli GlgX, some bacteria produce an extracellular isoamylase that is able to cleave substrates such as glycogen and amylopectin but not pullulan. Examples are the extracellular isoamylases produced by Pseudomonas amyloderamosa (2) and Flavobacterium odoratum (17). In higher plants, three classes of isoamylase genes and one class of pullulanase genes are found. Mutants lacking type 1 isoamylase activity typically have reduced starch contents and accumulate a soluble glycogen-like polysaccharide known as phytoglycogen. Examples of type 1 mutants are the sugary-1 maize mutant, whose mutation is best known as the mutation underpinning sweet corn; the sugary rice mutant, which has a similar phenotype (24); and the sta7 isoamylase-deficient mutant of Chlamydomonas reinhardtii (22), which lacks granular starch but produces a phytoglycogen-like molecule. Analysis of these mutants led to the formulation of a hypothesis which suggests that debranching activities are required to modify the structure of the growing amylopectin molecule in starch to a more ordered structure in order to facilitate its participation in the formation of the crystalline regions of amylopectin which are essential for the integrity of the starch granule (23). In bacterial glycogen synthesis, however, the role of the GlgX-class glycogen-debranching enzyme mutants has not been defined, and so it is not clear whether the enzyme is involved only in glycogen degradation or, as in starch biosynthesis, also acts during synthesis to shape the final structure of the glycogen produced. In this report, we describe the creation by homologous recombination of E. coli strains having defined mutations in glgX, and we describe the accumulation and structure of the glycogen produced by these mutants and relate these properties to the properties of the purified recombinant enzyme.
MATERIALS AND METHODS
Chemicals and reagents.
Amyloglucosidase and the GOPOD reagent were obtained from Megazyme Ltd. (Bray, Ireland). 8-Amino-1,3,6-pyrenetrisulfonic acid (APTS) was purchased from Lambda Fluoreszenztechnologie (Graz, Austria). Columns and buffers for capillary electrophoresis were purchased from Beckman Instruments, Fullerton, Calif.
Bacterial strains, plasmids, and growth conditions.
The E. coli strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains
| E. coli strain | Description | Reference or source |
|---|---|---|
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 8 |
| BWX1 | BW25113 glgX::Kanr | This study |
| BWX2 | BW25113 ΔglgX | This study |
| DH5α | F−endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1Δ(lacIZYA-argF)U169 deoR φ80 dlacΔ(lacZ)M15 | Invitrogen |
| BL21AI | F−ompT hsdSB(rB− mB−) gal dcm araB::T7RNAP-tetA | Invitrogen |
| RR1 | F−hsdS20(rB− mB−) recA+ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44 λ− | 3 |
| MS201 | F−hsdS20(rB− mB−) recA+ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44 λ−glgB::Kanr | This study |
| IK5 | RR1 glgX::Kanr | This study |
Plasmid pMAK705 was generously provided by S. Kushner, and pUC4-Kixx (containing the nptII gene) was purchased from Pharmacia. Luria-Bertani (LB) medium contained 1% tryptone, 0.5% yeast extract, 1% NaCl, and 0.2% glucose. M9 medium contained 0.4% glucose, 0.4% Casamino Acids, 0.1 mM CaCl2, 0.2% MgSO4 · 7H2O, 0.6% Na2HPO4, 0.3% KH2PO4, 0.5% NaCl, and 0.1% NH4Cl. Antibiotic selection plates contained either 50 μg of kanamycin per ml, 30 μg of chloramphenicol per ml, or 100 μg of ampicillin per ml, as required. Homologous recombination experiments were conducted in LB medium by using appropriate temperature conditions. For analysis of glycogen accumulation, 500 ml of M9 medium containing kanamycin, if required, was cultured in a 2-liter baffled flask with vigorous shaking at 37°C. Samples (20 ml) were taken periodically and centrifuged at 5,000 × g for 5 min, and each cell pellet was snap frozen in liquid N2 and stored at −90°C.
Cloning of the glgX gene and construction of glgX-deficient strains.
E. coli cells were made competent and transformed by using the rubidium chloride technique (12) to introduce a plasmid or by electroporation for transformation with PCR products (9).
Two distinct methods were used to inactivate the glgX open reading frame. First, the E. coli glgX gene was cloned from strain MS201, which previously (unpublished data) had an nptII resistance gene inserted into the glgB gene adjacent to glgX by homologous recombination by a standard procedure (11). A genomic library was constructed, and from 150,000 colonies, 3 Kmr colonies were obtained which were shown by DNA sequencing and restriction analysis to contain the entire glgX gene and a region of the adjacent glgB gene containing nptII resistance. This construct was designated pIKX1. Subcloning of this plasmid gave pIKX2, which contained the glgX gene and a 0.35-kb region of the 3′ region of glgB. To create a deletion-insertion glgX gene mutant, pIKX2 was digested with EcoRV to eliminate a 339-bp fragment and religated with a 1.2-kb SmaI fragment from pUC4-Kixx containing an nptII gene. The resulting construct was designated pIKX3. The final cloning step involved the transfer of a 1.78-kb fragment from pIKX3 into the KpnI site of the temperature-sensitive plasmid pMAK705, yielding pMAK-IK3. This plasmid was transformed into E. coli strain RR1, and the procedure of Hamilton et al. (11) was used to resolve strains in which the genomic glgX gene had been replaced by homologous recombination with the kanamycin-disrupted glgX gene from pMAK-IK3.
A second glgX allele was obtained by transformation of the BW25113 wild-type strain with a PCR product obtained from the pKD4 plasmid (8). The amplification product was composed of the kanamycin resistance gene flanked by yeast recombination sites. The primers used contained 45 and 43 nucleotides in 5′ and 3′ regions, respectively, corresponding to homologous regions in glgX. The resulting strain (BWX1) contained a glgX gene with a 1.5-kb deletion and a kanamycin resistance gene inserted at the site of the deletion. BWX1 was transformed with the pCP20 plasmid encoding a yeast recombinase, which allowed eviction of the kanamycin cassette and retention of the BWX2 strain, in which the major part of the glgX open reading frame was deleted.
PCR.
For strain IK5, PCR was used to verify that the homologous recombination procedure had disrupted the target gene. First, PCR amplification of the unique fragment produced by the desired homologous recombination event was performed by using a primer annealing to a region adjacent to the glgX gene but not included in the pMAK-IKX3 construct and a region within the nptII gene. In a second PCR assay we used primers which annealed to the 5′ and 3′ ends of the glgX gene. For BWX1 and BWX2, the primers used to check for replacement of the glgX gene by the kanamycin gene (in BWX1) or the subsequent removal of the kanamycin gene (in BWX2) annealed 200 bp 5′ and 3′ from the disruption region, respectively.
Complementation.
The glgX open reading frame was amplified by performing standard PCR with primers having XbaI restriction sites on each side. The PCR product obtained was then cloned in pUC19 as a translational fusion with lacZ, which allowed induction of glgX by isopropyl-β-d-thiogalactoside (IPTG).
Enzyme purification.
The glgX gene was cloned in frame with a glutathione S-transferase (GST) or six-His N-terminal tag by using the procedure described in the instructions for the Gateway system from Invitrogen. Briefly, the PCR product was cloned in plasmid pDONR201, which allowed recombination with pDEST15 (for the GST tag) or pDEST17 (for the six-His tag). The resulting plasmids were then transferred to the expression host BL21AI. The fusion proteins were expressed by induction with 0.2% arabinose for 3 h at 37°C in BL21AI (Stratagene) cultures containing the plasmid of interest and were purified with commercially available kits for GST-tagged proteins (MagneGST protein purification system; Sigma) or six-His-tagged proteins (Ni-nitrilotriacetic acid agarose; Invitrogen) by following the recommendations of the manufacturers.
Iodine staining.
The iodine staining characteristics of colonies on M9 medium plates were analyzed by exposing the plates to iodine vapor as previously described (7).
Assay of glycogen content.
The cell pellet from 20 ml of culture was suspended in 50 mM Tris-EDTA-acetate buffer (pH 7.8) and centrifuged at 5,000 × g for 5 min. The pellet was resuspended in 200 mM sodium acetate buffer (pH 4.5; 3 ml of buffer/g of cells) and sonicated after addition of 10 μl of 0.1 M Pefabloc per ml. The crude extract was then divided into aliquots for glycogen and protein analyses. The glycogen assay was performed by using a modification of the method proposed in the Megazyme total starch analysis procedure (α-amylase/amyloglucosidase). Amyloglucosidase (10 μl of a 200-U/ml solution) was added to 100 μl of the crude extract and incubated for 30 min at 50°C. After centrifugation for 5 min at 14,000 × g, 1 ml of the glucose oxidase/peroxidase (GOPOD) reagent was added to the supernatant, and the reaction mixture was incubated for 20 min at 50°C prior to measurement of the A510. The glucose concentration was determined by using a standard curve covering the range from 0 to 50 μg of glucose per ml. Control reaction mixtures lacking amyloglucosidase were used for all samples, but they had negligible glucose contents.
Debranching enzyme assay procedures. (i) Glucan solubilization assay.
An extract from RR1 and IK5 cells (0.2 mg) was incubated with 1 mg of polysaccharides in 100 mM acetate buffer (pH 5.0) containing 5 mM dithiothreitol (DTT) at 37°C for 1 h. At the end of this incubation, 3 volumes of ethanol was added, and the mixture was centrifuged for 5 min. The supernatant (0.2 ml) was freeze-dried, and the glucan content was determined with a starch assay kit (Sigma). The freeze-dried material was resuspended in 0.2 ml of 33 mM citrate buffer (pH 4.6), added to an equal volume of starch assay reagent (containing amyloglucosidase), and incubated at 60°C for 15 min. After 1 min of centrifugation, the amount of glucose in 0.1 ml of supernatant was assayed with a second reagent (0.5 ml) containing hexokinase/glucose-6-phosphate dehydrogenase, ATP, and NADP and incubated at room temperature for 15 min. The absorbance at 340 nm was measured, and the amount of glucose produced was estimated by using a glucose standard curve.
(ii) Branch linkage assay.
The debranching activity of GlgX was determined by measuring the amount of reducing ends produced by using the dinitrosalicylic acid colorimetric method (19). An extract of E. coli cells (0.4 mg of proteins) was incubated at 37°C with 1 mg of substrate in 100 mM acetate buffer (pH 5.0) containing 5 mM DTT in a 300-μl (total volume) mixture for 1 h. Samples were then boiled for 2 min, and each supernatant was diluted to 450 μl with water. Then 450 μl of a dinitrosalicylic acid solution (1% dinitrosalicylic acid, 0.2% phenol, 0.05% sodium sulfite, 1% sodium hydroxide) was added, and the mixture was incubated at 90°C for 15 min. A potassium sodium tartrate solution (150 μl of a 40% [wt/vol] solution) was added to stabilize the red-brown color that developed. After the preparation cooled to room temperature, the absorbance at 575 nm was recorded. Maltotriose was used as the standard.
1H-nuclear magnetic resonance (NMR) spectroscopy.
Extracts of wild-type (RR1) and mutant (IK5) E. coli cells (1.6 mg of protein) were incubated with 4 mg of phosphorylase-limit dextrin in a 1-ml mixture containing 50 mM acetate buffer (pH 5) and 2 mM DTT at 37°C for 4 h. The reaction was stopped by boiling. After centrifugation for 5 min, the supernatant was lyophilized, and then it was dissolved in D2O (300 μl) twice and lyophilized again. The final lyophilized product was dissolved in 99.996% D2O, and the spectra were recorded at 40°C with a Varian INOVA 600 spectrometer.
Debranching of P-limit dextrin by commercial isoamylase (Megazyme) was determined as described above but at pH 4.5 and in the absence of DTT by using 15 U of enzyme.
Analysis of the glycogen structure.
Fluorophore-assisted carbohydrate electrophoresis was used to analyze the degree of polymerization (DP) of oligosaccharides released from glycogen by enzymatic digestion.
(i) Isoamylase debranching.
Extracted glycogen (5 mg) was resuspended in 400 μl of 0.25 M NaOH and boiled for 5 min. Glacial acetic acid (12.8 μl), 1.6 ml of 0.05 M of sodium acetate buffer (pH 4), and 10 μl of isoamylase (200 U/ml) were added. The mixture was incubated at 37°C for 2 h and boiled for 20 min to denature the enzyme.
(ii) Reductive amination.
The charged fluorophore APTS was attached to reducing ends via a reductive amination reaction. After centrifugation of the sample (5 min at 12,000 × g), buffer salts were removed from the supernatant by treating a 1-ml aliquot with 0.2 g of a mixed-bed ion-exchange resin for 20 min. Aliquots (10 μl) of the supernatant were dried in vacuo. The reductive amination reaction was carried out by addition of 5 μl of reducing agent (6.3 mg of NaCNBr in 100 μl of water) and 5 μl of 0.2 M APTS. The mixture was incubated overnight at 37°C. The reaction was stopped by adding 90 μl of 6 M urea in 0.04 M boric acid-0.04 M Tris (pH 8.6).
(iii) Separation.
Separation was performed with a P/ACE 5010 instrument with the cathode on the injection side (reversed polarity) and was monitored with a laser-induced fluorescence detector (Beckman Instruments) with an argon ion laser as the excitation source. The capillary used was a 50-μm-diameter eCAP neutral coated capillary (obtained from an eCAP N-linked oligosaccharide profiling kit [Beckman Instruments] along with the carbohydrate separation gel buffer) cut to a length of 47 cm (40 cm to the detector), and the sample was introduced by pressure injection of the diluted reductive amination reaction mixture (typically 2 to 5 s at 0.5 lb/in2). The separating medium consisted of carbohydrate separation gel buffer, and separation was achieved by using an applied voltage of 23.5 kV (current, 14 μA) at 25°C.
Protein analysis.
Protein was analyzed by a Coomassie blue protein assay obtained from Pierce Chemical Co. Ltd. by using bovine serum albumin as the standard. Standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis and Coomassie blue staining were used to monitor the purification of both GST-GlgX and His6-GlgX. These proteins were purified with a MagneGST protein purification system obtained from Promega (Madison, Wis.) or an Ni-nitrilotriacetic acid purification system obtained from Invitrogen (Paisley, United Kingdom).
RESULTS
Construction of a glgX-deficient E. coli strain by homologous recombination.
One aim of this study was to characterize an E. coli strain with specific inactivation of the glgX gene. Two different methods to create a defined deletion-insertion lesion in the glgX gene by homologous recombination were used. This was done to compare different isogenic pairs in different genetic backgrounds. Indeed, we observed that glycogen accumulation levels were very sensitive to the genetic backgrounds of the strains used. The first mutant strain, strain IK5, was obtained by homologous recombination with pMAK-IK3 by using a previously described procedure (11).
The second mutant strain, strain BWX1, was obtained by direct transformation with a PCR product of the BW25113 wild-type strain containing the pKD46 plasmid by using an established protocol (8). The resulting strain, designated BWX2, had a 1.5-kb deletion of the glgX gene and a scar due to the kanamycin resistance gene excision. This second technique that allowed removal of the kanamycin gene reduced the risk of a polar effect of the mutation on neighboring genes. Confirmation that the desired homologous recombination events had occurred was obtained by PCRs.
Iodine-staining properties of glgX-deficient strain.
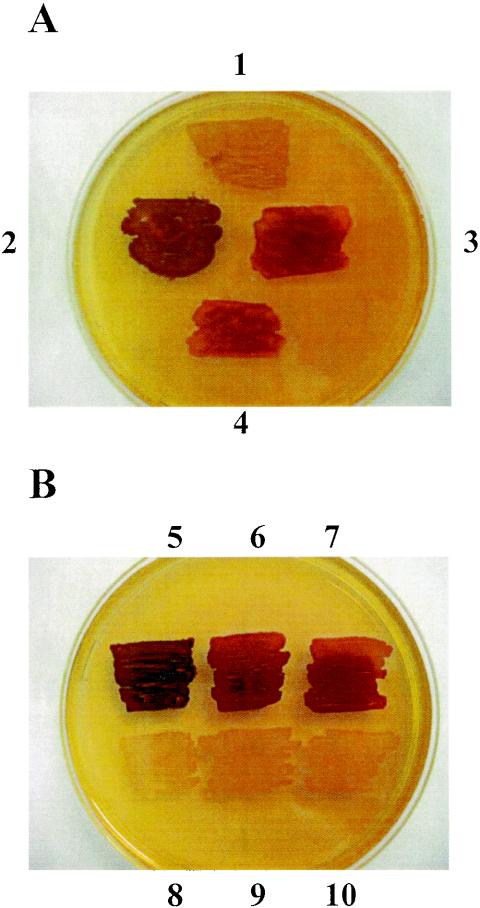
Iodine staining is frequently used as a diagnostic tool to determine the amount and structure of starch or glycogen produced by microorganisms or unicellular algae. Cells were grown on minimal medium (M9 medium) supplemented with glucose to stimulate glycogen synthesis. The light red-brown color observed for E. coli RR1 was typical of the color reaction of E. coli cells containing a wild-type glycogen biosynthetic pathway (Fig. 1). The BW25113 wild-type cells were indistinguishable from RR1 cells by iodine staining (data not shown). IK5 cells consistently stained a distinctly different darker brown color than RR1 cells (Fig. 1A), and the BWX1 and BWX2 mutant strains had a darker brown phenotype than BW25113. IK5 stained a darker color than BWX1 or BWX2, possibly due to the different genetic backgrounds in which the mutations were generated.
FIG. 1.
Iodine staining. Cell patches were grown overnight on M9 medium plates supplemented with glucose. The plates were then flooded with an iodine solution (4% I2, 0.4% KI). (A) Cell patches 1 to 4 correspond to RR1, IK5, BWX1, and the transduced strain containing the IK5 mutation in the BW25113 genetic background, respectively. (B) Three different glgX mutant strains (IK5, BWX1, and BWX2 containing an empty pUC19) are at the top (cell patches 5, 6, and 7, respectively), and the same strains carrying the complementation plasmid (pX) are at the bottom (cell patches 8, 9, and 10, respectively). The M9 medium plate used for this test was supplemented with 100 μg of ampicillin per ml and 5 mM IPTG.
To check this hypothesis, transduction with phage P1vir was carried out by using the method of Miller (20). The glgX::kan mutation in IK5 was transferred by P1 phage into strain BW25113. As shown in Fig. 1A, the transduced strain containing the IK5 mutation in BW25113 yielded the same iodine staining phenotype that the BWX1 or BWX2 mutant cells had. To check if the iodine staining phenotype was due to the glgX mutation, the three mutant strains (IK5, BWX1, and BWX2) were transformed with the pX plasmid containing a translational fusion between the eight first amino acids of lacZ and the glgX gene in pUC19. The resulting ampicillin-resistant clones were used for iodine staining on M9 medium containing 5 mM IPTG. Both the mutants and the wild type displayed the same light iodine staining, suggesting that there was complementation of the mutant defects (Fig. 1B). No effect of the pX plasmid was observed without addition of IPTG. These data showed that the glgX deficiency in E. coli was responsible for the darker phenotype observed in the iodine staining test.
Isoamylase activities of wild-type, glgX-deficient, and complemented strains.
The isoamylase activities of the wild-type and glgX-deficient strains were measured by using mid-log-phase cells grown in LB medium. Two assay methods were used to determine the amount and nature of the debranching activity present in IK5 and RR1.
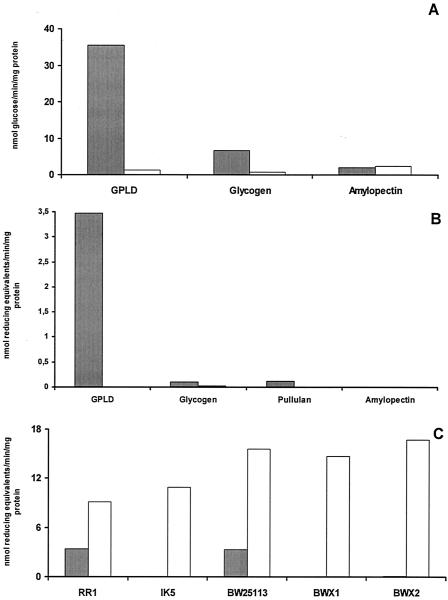
Figure 2A shows the results when the activities of IK5 and RR1 were assayed with a variety of substrates by using a glucan solubilization assay. In this assay, the release of ethanol-soluble maltooligosaccharides was monitored after incubation of the substrate with enzyme. Figure 2B shows the results of quantification of reducing sugars released from various substrates by extracts of IK5 and RR1. Both assays demonstrated that the major debranching enzyme activity of E. coli was absent from IK5 cells and confirmed that the spectrum of activity of the enzyme in the RR1 cells was consistent with previous reports (15), which showed that the E. coli debranching enzyme has a much higher activity with GPLD as the substrate than with glycogen or amylopectin (Fig. 2). The glgX-deficient strain, IK5, did not contain levels of debranching enzyme above the baseline level with the phosphorylase-limit dextrin of glycogen, glycogen, or amylopectin. Both the BWX1 and BWX2 mutant strains also lacked measurable debranching enzyme activity in these assays (Fig. 2C). The debranching activity was also monitored with crude extracts of the mutant or the wild-type reference strain transformed with plasmid pX. In this case, the culture conditions were the same, except that 100 μg of ampicillin per ml was added. All the strains expressing GlgX at high levels exhibited three to four times more activity with GPLD than untransformed bacterial strains exhibited (Fig. 2C). These high levels of the debranching enzyme were consistent with the yellow phenotype observed after iodine staining (Fig. 1B). These data, together with complementary studies (33), support the conclusion that the E. coli isoamylase activity characterized in detail by Jeanningros et al. (15) is encoded by the glgX gene. Evidence that the activity observed was catalyzed by a debranching enzyme and not by another exo- or endoamylase was provided by 1H-NMR (Fig. 3).
FIG. 2.
Debranching activity of E. coli extracts with various substrates. Crude extracts of RR1 (solid bars) and IK5 (open bars) were prepared from log-phase cells grown in LB medium. The phosphorylase-limit dextrin of glycogen (GPLD) was produced by exhaustive incubation of glycogen with excess phosphorylase in the presence of phosphate buffer until no further release in G1P from glycogen in the presence of Pi and active enzyme was obtained. (A) Activity determined by a glucan solubilization assay procedure. The activities are expressed in nanomoles of glucose equivalents liberated per minute per milligram of protein. (B) Debranching activity measured by the branch linkage assay. The assay conditions used are described in Materials and Methods. The activities are expressed in nanomoles of equivalent reducing ends produced per minute per milligram of protein. (C) Debranching activity on GPLD assayed with wild-type strains RR1 and BW25113 and the IK5, BWX1, and BWX2 glgX-deficient strains. The solid bars indicate the activities of the original strains (in nanomoles of glucose equivalents liberated per minute per milligram of protein), and the open bars indicate the activities of the same strains containing the pX plasmid and grown in the presence of 100 μg of ampicillin per ml.
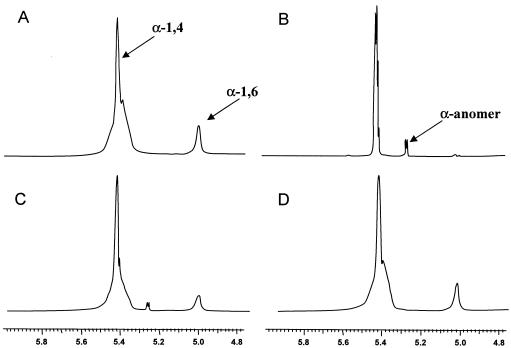
FIG. 3.
Analysis of GlgX activity by 1H-NMR spectroscopy. The proton NMR spectrum of phosphorylase-limit dextrin (A) is compared to the spectra of the phosphorylase-limit dextrin cleavage products induced by wild-type (C) and mutant (D) extracts and the phosphorylase-limit dextrin cleavage products generated by pure commercial isoamylase (B). The chemical shifts for the protons of α-(1,4) linkages and α-(1,6) linkages are at 5.4 and 5.0 ppm, respectively. The chemical shift of the α-anomer of the reducing ends produced is at 5.27 ppm.
Assay of isoamylase activity by 1H-NMR.
The substrate utilized was the phosphorylase-limit dextrin of glycogen (Fig. 3A), in which the chemical shifts assigned to the α-1,4 (5.42 ppm) and α-1,6 (5.0 ppm) linkages could be clearly observed. Figure 3B shows the spectrum for the product of incubation of the substrate with isoamylase, which resulted in almost complete removal of the α-1,6 chemical shift and the appearance of a chemical shift corresponding to the α-anomer of the released reducing sugar at 5.27 ppm.
Figure 3C shows the spectrum for the product of incubation of the substrate with extract from E. coli RR1 cells, which revealed the decrease in the chemical shift for the α-1,6 linkage and the appearance of the chemical shift at 5.27 ppm. There was no evidence of any debranching activity when the substrate was incubated with IK5 cell extract (Fig. 3D). Table 2 shows the integrated data quantifying these observations. Complete debranching of the phosphorylase-limit dextrin substrate by RRI extract was not observed because the GlgX enzyme was apparently unable to efficiently hydrolyze the α-1,6 linkages of side chains longer than four glucosyl residues, and its action was therefore confined to the outer chains of the limit dextrin, leaving the core structure intact.
TABLE 2.
Debranching activity measured by 1H-NMR spectroscopy
| Source of debranching enzyme | Ratio of branching (%) | % of reducing ends |
|---|---|---|
| RR1 | 9.7 | 1.02 |
| IK5 | 9.1 | 0.00 |
| Isoamylase | 10.8 | 10.30 |
| No enzyme | 10.1 | 0.00 |
Accumulation of glycogen.
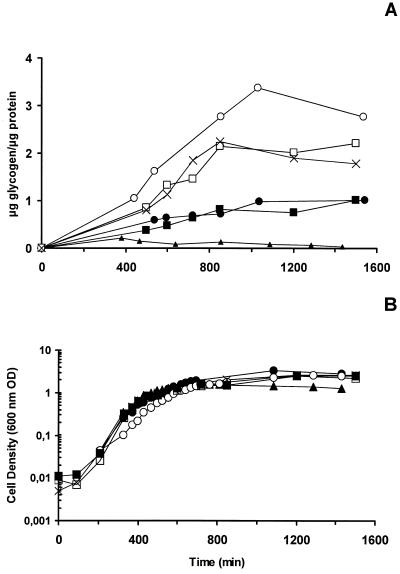
The amount of glycogen accumulated by IK5, BWX1, or BWX2 was two to three times greater than the amounts of glycogen accumulated by the parental RR1 and BW25113 strains, and this difference was observed throughout the E. coli growth curve (Fig. 4), supporting the hypothesis that GlgX plays a role in glycogen degradation. A twofold overacccumulation of glycogen was also observed for the dark-brown iodine staining class C mutants deficient for allosteric regulation of ADP-glucose pyrophosphorylase reported previously (7), while MS201, the glgB-deficient strain, had a very small quantity of glucan. These results are consistent with a major role for the branching enzyme and GlgX in glycogen synthesis and degradation, respectively.
FIG. 4.
Glycogen accumulation by IK5 (○), MS201 (▴), RR1 (•), BW25113 (▪), BWX1 (×), and BWX2 (□). Strains were grown in M9 medium containing 4 g of glucose per liter (and 50 μg of kanamycin per ml for IK5 and BWX1). (A) Ratio of glycogen to protein accumulation over time. (B) Cell density, as measured by A600 (600 nm OD).
Glycogen structure.
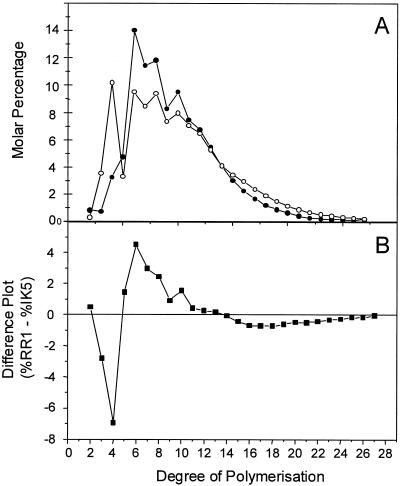
While iodine staining provided a rapid indication of the structure of glycogen or starch, it did not provide the detailed information on structure required to define the role of the GlgX debranching enzyme. Chain length analysis by the fluorophore-assisted carbohydrate electrophoresis technique (21, 26) provided an accurate method for determining the impact of a specific mutation on glycogen structure. This technique involved debranching the extracted glycogen with a highly purified isoamylase and then labeling the reducing ends of the oligosaccharide population by reductive amination with the charged fluorophore APTS. The labeled oligosaccharides were then separated and detected by capillary electrophoresis. Comparison of the glycogens produced by the wild-type RR1 strain and IK5 showed that the major differences between these glycogens were in the chain lengths with DP between 3 and 7.
When this analytical system was used, significant differences in the glycogens produced in the presence and in the absence of the glgX gene were observed; on a molar basis, the glycogen of IK5 had significantly more short chains than the wild-type glycogen (Fig. 5A). A plot of the normalized distribution of the chain lengths of IK5 subtracted from the chain lengths of RR1 is shown in Fig. 5B; 9.6% of the chains in RR1 had a DP of 3 to 5, whereas 17.4% of the chains in IK5 fell in this range.
FIG. 5.
Chain length distribution of oligosaccharides determined by capillary electrophoresis of the isoamylase-debranched glycogen produced by RR1 (•) and IK5 (○). (A) Chain length distributions expressed as the percentage of total chains, expressed on a molar basis. (B) Difference plot generated by subtracting the molar percentage for IK5 at each chain length from the corresponding molar percentage for RR1 at the corresponding chain length.
GlgX purification.
The GlgX protein was tagged at its N terminus with GST or six-His, which allowed purification of the enzyme. Each construct was introduced into BL21AI cells, and production of the proteins of interest was induced for 3 h by using 0.2% arabinose in LB medium supplemented with 100 μg of ampicillin per ml at 37°C. The production of both tagged proteins was highly enhanced by arabinose. Most of the GST-tagged enzyme was found in the soluble phase. Around 10% of the six-His-tagged GlgX was in the soluble extract, which allowed purification of the enzyme from this fraction. The remaining six-His-tagged GlgX was in inclusion bodies. The purified enzymes were checked on a 7.5% acrylamide sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel stained with Coomassie brilliant blue. The GST-tagged GlgX appeared as a single band at 110 kDa (data not shown). A few low-molecular-weight contaminating proteins were always purified with the six-His-tagged GlgX, which appeared as a faint band at 76 kDa due to the small amount of protein in the soluble phase used for purification. Protein samples from both expression systems were assayed with GPLD as described above, and this allowed us to estimate the yields and purification factors for each enzyme. GST-GlgX was purified with a yield of 2% and a purification factor of 30-fold, and the six-His-tagged protein was purified 51-fold with a yield of 9% compared with the activities determined for the soluble fraction obtained from the induced crude extracts used for purification. The low yields observed may have been due to the instability of the enzymes after purification.
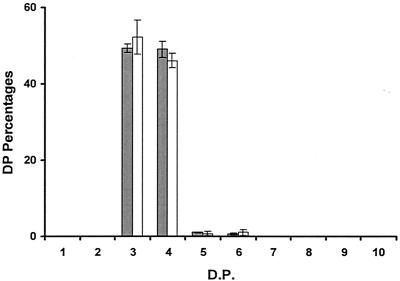
Both purified proteins were immediately used in incubation assays with rabbit liver glycogen (Sigma, St. Louis, Mo.) by using the glucan solubilization assay procedure described previously, but the debranched oligosaccharides obtained were labeled and analyzed by capillary electrophoresis. Both purified GST- and six-His-tagged GlgX debranched oligosaccharides with degrees of polymerization of three or four glucose residues with high efficiency. These two purified enzymes hydrolyzed the α-1,6 linkages of longer chains with very low efficiency (Fig. 6). Oligosaccharides with DP greater than six glucose residues were not debranched by the purified enzymes. Some chains with DP of five or six were observed, but they did not account for more than 1% of the total debranched oligosaccharides. The same activity pattern was observed for both purified tagged proteins. The debranching activity encoded by the glgX gene of E. coli therefore defines an isoamylase type of debranching enzyme with high specificity for hydrolysis of branched maltotriose or maltotetraose.
FIG. 6.
Products of GlgX incubation with glycogen: detection of the oligosaccharides produced by incubation of purified GST-GlgX (shaded bars) and purified six-His-tagged GlgX (open bars) with rabbit liver glycogen by capillary electrophoresis. The results are expressed as percentages of the DP liberated. The x axis indicates the degree of polymerization. The results were obtained with two different enzyme preparations for each tagged protein.
DISCUSSION
Although previous studies had shown that the glgX gene was not absolutely required for synthesis of linear α-1,4-glucosyl-glucan (30), no E. coli strain specifically lacking the glgX gene alone was available prior to this study. Our results clearly indicate that the absence of the glgX gene product leads to glycogen overproduction, which suggests that the gene has an important function in storage polysaccharide catabolism.
Evidence from sequence comparisons (30) and a preliminary study of the glgX gene product (33) suggested that glgX was likely to encode a functional debranching enzyme and that this enzyme was likely to be the enzyme studied in detail by Jeanningros et al. (15). The absence of isoamylase-type debranching enzyme from the glgX-deficient mutant and the substrate specificity of the debranching enzyme present in wild-type cells (which showed higher reaction rates with phosphorylase-limit dextrin than with glycogen) provide strong evidence which suggests that glgX encodes the enzyme characterized by Jeanningros et al. (15). To definitively establish this, we expressed a recombinant glgX gene product and purified it to homogeneity. The pure protein displayed the same specificity for debranching chains consisting of three or four glucose residues that the enzyme studied by Jeanningros et al. displayed.
It has been suggested previously that the role of GlgX may be in the catabolism of glycogen in bacteria (15, 33). The results presented here demonstrate that disruption of the glgX gene alters the accumulation pattern and structure of the glycogen produced in E. coli throughout the growth of a culture. A role in glycogen degradation is supported by the accumulation phenotype. It is thought that glycogen is first acted on by glycogen phosphorylase (encoded by glgP), yielding glycogen-containing short external chains consisting of approximately four glucosyl units (32). This phosphorylase-limit dextrin is then acted upon by the GlgX isoamylase, yielding a population of linear maltooligosaccharides with a distribution of DP centered around maltotetraose. This population of maltooligosaccharides is thought to be disproportionated by amylomaltase (encoded by malQ), providing a substrate that is depolymerized by the malP-encoded maltodextrin phosphorylase, yielding glucose-1-phosphate (G1P) (32).
The results presented here are consistent with the mechanism that was first proposed by Palmer et al. (27) and was expanded upon by Krebs and Preiss (16). Briefly, in the absence of the glgX-encoded isoamylase, glycogen with short outer chains, resulting from the action of the GlgP glycogen phosphorylase, accumulates, inducing a shift in chain length distribution toward lower DP. Analysis of the glycogen in the glgX-deficient strain IK5 showed that the level of external chains in glycogen is increased severalfold, which is consistent with the lack of glgX activity and with its specificity for removing short external chains of glycogen. In addition, because neither glycogen phosphorylase nor amylomaltase can act on phosphorylase-limit dextrins, glycogen degradation is impaired in the absence of GlgX, and glycogen accumulates. While the role of GlgX is clearly predominantly to facilitate degradation, the data presented here also indicate that the enzyme remains active during glycogen synthesis, shaping the structure of the glycogen produced by reducing the frequency of short external chains. This activity may facilitate the production of larger glycogen molecules as the presence of very short chains has been predicted to lead to allosteric crowding and inhibition of synthesis (18).
The complex regulation of genes in the E. coli glycogen operons described previously (13, 29, 33) indicates that there is complex transcriptional control of the expression of the genes for glycogen biosynthesis and catabolism. However, the impact of the glgX mutation in induction of both glycogen accumulation and an altered glycogen structure suggests that glycogen biosynthesis and degradation occur concomitantly in an E. coli culture and that these pathways may both be active and in balance in an individual cell. It has been demonstrated that a key enzyme for glycogen degradation, glycogen phosphorylase (GlgP), is present throughout growth when there is net glycogen accumulation (5, 6). The substrate specificities and regulatory properties of the synthesis and degradation enzymes suggest that both synthesis and degradation pathways may be present in the same cell but kept in balance through the specificities of the respective enzyme systems and the availability of carbon flux. During synthesis, the pattern of action of E. coli branching enzyme, transferring chains larger than six glucosyl residues, produces external chains that are longer than those that can be acted on efficiently by GlgX, ensuring that there is not an extensive futile cycling between branching and debranching enzyme activities. Glycogen phosphorylase-mediated phosphorolysis of external chains should be limited relative to glycogen synthase-mediated extension of external chains due to the availability of ADP-glucose (promoting glycogen synthesis via ADP-glucose pyrophosphorylase and inhibiting glycogen degradation through inhibition of glycogen phosphorylase [6]), high G1P levels, and low free Pi and AMP levels in the cell (AMP is a positive allosteric effector of glycogen phosphorylase in E. coli [5]). Therefore, the narrow specificity of GlgX, which cleaves only glycogen chains with external chains with DP of four or less, allows this activity to coexist in the cell with net glycogen synthesis and is consistent with the hypothesis that the two genes of the glgB-glgX operon are tandemly expressed, as suggested by Romeo et al. (30). Isoamylases with broad specificity, such as plant isoamylases and extracellular bacterial isoamylases, would be inconsistent with net glycogen synthesis as they would induce a futile cycle; thus, they would need to be extremely tightly controlled at the transcriptional or translational level to allow net glycogen synthesis. The data presented in this report suggest that even during periods of net glycogen synthesis, components of the glycogen degradation pathway are active, but they also suggest that it is only when growth ceases, ADP-glucose becomes limiting, G1P levels fall, and Pi accumulates that there are significant rates of net glycogen degradation.
Debranching enzymes belong to family 13 of the glycosylhydrolases, which are defined by the presence of a conserved domain (domain A) that assumes a barrel shape whose sides are a succession of eight β sheets and α helices. The loops connecting adjoining α helices and β sheets are numbered according to their positions within the polypeptide chain.
Differences in the length of the loop 4 region of isoamylases have been suggested to underpin differences in substrate specificity between isoamylases (1, 14). Both the plant isoamylase type 1 genes and P. amyloderamosa genes have a longer loop 4 region than GlgX; it has been suggested that this region is required to accommodate the longer side chains of amylopectin and glycogen compared to the short side chains of phosphorylase-limit dextrins preferentially hydrolyzed by GlgX. It has recently been shown that plant type 3 isoamylases have loop 4 regions whose lengths are similar to those of E. coli GlgX and which utilize phosphorylase-limit dextrins preferentially over substrates with longer external side chains (14). All of the recently reported isoamylase-like sequences with the exception of plant isoamylase type 1 and the P. amyloderamosa family of secreted isoamylases have predicted loop 4 regions whose lengths are similar to those of E. coli GlgX. Confirmation that a short loop 4 region is indicative of substrate specificity favoring phosphorylase-limit dextrins over substrates with longer external chains requires additional evidence.
Acknowledgments
We acknowledge M. Eb of the University of Science and Technology, Lille, France, and Rudi Appels, CSIRO Plant Industry, for their assistance in arranging a student exchange visit for I.S.K.
We acknowledge funding support from the CRC for Plant Science.
REFERENCES
- 1.Abe, J.-I., C. Ushijima, and S. Hizukuri. 1999. Expression of the isoamylase gene of Flavobacterium odoratum KU in Escherichia coli and identification of essential residues of the enzyme by site-directed mutagenesis. Appl. Environ. Microbiol. 65:4163-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amemura, A., R. Chakraborty, M. Fujita, T. Noumi, and M. Futai. 1988. Cloning and nucleotide sequence of the isoamylase gene from Pseudomonas amyloderamosa SB-15. J. Biol. Chem. 263:9271-9275. [PubMed] [Google Scholar]
- 3.Bolivar, F., R. L. Rodriguez, D. J. Greene, M. C. Betlach, H. L. Heynecker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Cattaneo, J., M. Damotte, N. Sigal, F. Sanchez-Medina, and J. Puig. 1969. Genetic studies of Escherichia coli K12 mutants with alterations in glyconeogenesis and properties of an altered adenosine diphosphate glucose pyrophosphorylase. Biochem. Biophys. Res. Commun. 34:694-701. [DOI] [PubMed] [Google Scholar]
- 5.Chen, G. S., and I. H. Segal. 1968. Escherichia coli polyglucose phosphorylases. Arch. Biochem. Biophys. 127:164-174. [DOI] [PubMed] [Google Scholar]
- 6.Chen, G. S., and I. H. Segal. 1968. Purification and properties of glycogen phosphorylase from Escherichia coli. Arch. Biochem. Biophys. 127:175-186. [DOI] [PubMed] [Google Scholar]
- 7.Damotte, M., J. Cattaneo, N. Sigal, and J. Puig. 1968. Mutants of E. coli K12 altered in their ability to store glycogen. Biochem. Biophys. Res. Commun. 32:916-920. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govons, S., R. Vinopal, J. Ingraham, and J. Preiss. 1969. Isolation of mutants in Escherichia coli B altering their ability to synthesize glycogen. J. Bacteriol. 97:970-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton, C. M., M. Aldea, P. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hengge-Aronis, R., and D. Fischer. 1992. Identification and analysis of glgS, a novel growth-phase regulated and rpoS dependent gene involved in glycogen synthesis in E. coli. Mol. Microbiol. 6:1877-1886. [DOI] [PubMed] [Google Scholar]
- 14.Hussain, H., A. Mant, R. Seale, S. Zeeman, E. Hinchcliffe, A. Edwards, C. Hylton, S. Bornemann, A. M. Smith, C. Martin, and R. Bustos. 2003. Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15:133-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeanningros, R., N. Creuzet-Sigal, C. Frixon, and J. Cattaneo. 1976. Purification and properties of debranching enzyme from E. coli. Biochim. Biophys. Acta 438:186-199. [DOI] [PubMed] [Google Scholar]
- 16.Krebs, E. G., and J. Preiss. 1975. Regulatory mechanisms in glycogen metabolism, p. 337-389. In W. J. Whelan (ed.) Biochemistry of carbohydrates, vol. 1. MTP International Review of Science. University Park Press, Baltimore, Md.
- 17.Krohn, B. M., G. F. Barry, and G. M. Kishore. 1997. An isoamylase with neutral pH optimum from a Flavobacterium species: cloning, characterization and expression of the iam gene. Mol. Gen. Genet. 254:469-478. [DOI] [PubMed] [Google Scholar]
- 18.Melendez-Hevia, E., T. G. Waddell, and E. D. Shelton. 1996. Optimization of molecular design in the evolution of metabolism: the glycogen molecule. Biochem. J. 295:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 20.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Morell, M. K., M. S. Samuel, and M. G. O'Shea. 1998. Analysis of starch structure using fluorophore-assisted carbohydrate electrophoresis. Electrophoresis 19:2603-2611. [DOI] [PubMed] [Google Scholar]
- 22.Mouille, G., M.-L. Maddelein, N. Libbessart, P. Tagala, A. Decq, B. Delrue, and S. Ball. 1996. Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8:1353-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers, A. M., M. K. Morell, M. G. James, and S. G. Ball. 2000. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122:989-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura, Y. 1996. Some properties of starch debranching enzymes and their possible role in amylopectin biosynthesis. Plant Sci. 121:1-18. [Google Scholar]
- 25.Okita, T. W., R. L. Rodriguez, and J. Preiss. 1981. Biosynthesis of bacterial glycogen. Cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J. Biol. Chem. 256:6944-6952. [PubMed] [Google Scholar]
- 26.O'Shea, M. G., and M. K. Morell. 1996. High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis 17:681-686. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, T. N., G. Wober, and W. J. Whelan. 1973. The pathway of exogenous and endogenous carbohydrate utilization in Escherichia coli: a dual function for the enzymes of the maltose operon. Eur. J. Biochem. 39:601-612. [DOI] [PubMed] [Google Scholar]
- 28.Preiss, J., A. Sabraw, and E. Greenberg. 1971. An ADP-glucose pyrophosphorylase with lower apparent affinities for substrate and effector molecules in an Escherichia coli B mutant deficient in glycogen synthesis. Biochem. Biophys. Res. Commun. 42:180-186. [DOI] [PubMed] [Google Scholar]
- 29.Preiss, J., and T. Romeo. 1994. Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 47:329-399. [DOI] [PubMed] [Google Scholar]
- 30.Romeo, T., A. Kumar, and J. Preiss. 1988. Analysis of the E. coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene 70:363-376. [DOI] [PubMed] [Google Scholar]
- 31.Romeo, T., J. Black, and J. Preiss. 1990. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vivo effects of the catabolite repression and stringent response systems in glg gene expression. Curr. Microbiol. 21:131-137. [Google Scholar]
- 32.Schinzel, R., and B. Nidetzky. 1999. Bacterial α-glucan phosphorylases. FEMS Microbiol. Lett. 171:73-79. [DOI] [PubMed] [Google Scholar]
- 33.Yang, H., M. Y. Liu, and T. Romeo. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the csrA gene product. J. Bacteriol. 178:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]