Abstract
It is now established that the central nervous system plays an important role in regulating whole body metabolism and energy balance. However, the extent to which sensory systems relay environmental information to modulate metabolic events in peripheral tissues has remained poorly understood. In addition, it has been challenging to map the molecular mechanisms underlying discrete sensory modalities with respect to their role in lipid metabolism. In previous work our lab has identified instructive roles for serotonin signaling as a surrogate for food availability, as well as oxygen sensing, in the control of whole body metabolism. In this study, we now identify a role for a pair of pheromone-sensing neurons in regulating fat metabolism in C. elegans, which has emerged as a tractable and highly informative model to study the neurobiology of metabolism. A genetic screen revealed that GPA-3, a member of the Gα family of G proteins, regulates body fat content in the intestine, the major metabolic organ for C. elegans. Genetic and reconstitution studies revealed that the potent body fat phenotype of gpa-3 null mutants is controlled from a pair of neurons called ADL(L/R). We show that cAMP functions as the second messenger in the ADL neurons, and regulates body fat stores via the neurotransmitter acetylcholine, from downstream neurons. We find that the pheromone ascr#3, which is detected by the ADL neurons, regulates body fat stores in a GPA-3-dependent manner. We define here a third sensory modality, pheromone sensing, as a major regulator of body fat metabolism. The pheromone ascr#3 is an indicator of population density, thus we hypothesize that pheromone sensing provides a salient 'denominator' to evaluate the amount of food available within a population and to accordingly adjust metabolic rate and body fat levels.
Author summary
The central nervous system plays a vital role in regulating whole body metabolism and energy balance. However, the precise cellular, genetic and molecular mechanisms underlying these effects remain a major unsolved mystery. C. elegans has emerged as a tractable and highly informative model to study the neurobiology of metabolism. Previously, we have identified instructive roles for serotonin signaling as a surrogate for food availability, as well as oxygen sensing, in the control of whole body metabolism. In our current study we have identified a role for a pair of pheromone-sensing neurons in regulating fat metabolism in C. elegans. cAMP acts as a second messenger in these neurons, and regulates body fat stores via acetylcholine signaling in the nervous system. We find that the population-density-sensing pheromone detected by these neurons regulates body fat stores. Together, we define a third sensory modality, population density sensing, as a major regulator of body fat metabolism.
Introduction
In relation to fat metabolism and energy balance, the central nervous system plays a more intricate role than historically thought. Initially believed to exert its effects on adiposity predominantly through promoting food intake, several studies have now demonstrated that the underlying neuronal circuits, genetic, molecular and endocrine pathways that regulate body fat reserves in the peripheral metabolic organs are distinct from those that regulate feeding behavior [1–6]. In addition to the pre-eminent role of the mammalian hypothalamus, the sensory nervous system has also been shown to play an important role in regulating whole body metabolism and physiology [7, 8]. Broad sensory dysfunction in humans can be exemplified by ciliopathies such as Bardet-Biedl Syndrome, that leads to profound obesity [9]. In contrast, enhanced sensory environments improve metabolic homeostasis [10]. However, identifying discrete sensory neurons with instructive roles in lipid metabolism has been a challenging undertaking in any system.
In the metazoan Caenorhabditis elegans, the nervous system is well-defined at an anatomic and functional level [11, 12]. The sensory nervous system plays a profoundly important role in regulating whole body physiology and lifespan [13–15]. We and others have shown that the sensory nervous system is an important regulator of systemic lipid metabolism [3, 16, 17]. For example, the presence of food, relayed by serotonergic sensory neurons and amplified by the octopaminergic neurons (octopamine is the invertebrate analog of noradrenaline) is one salient input that regulates the magnitude of fat loss in the intestine [3]. The intestine is the predominant metabolic organ for C. elegans, and expresses all of the genes involved in lipid metabolic processes including fat synthesis, breakdown and its regulation [7, 18, 19]. Furthermore, conserved intestinal fatty acid beta-oxidation has been shown to play a central role in the biosynthesis of the ascarosides, a family of small-molecule pheromones that regulate many aspects of C. elegans physiology and behavior [20, 21]. Thus, metabolic changes in the intestine effectively encapsulate whole body metabolism. The mechanisms governing lipid metabolism are ancient and well-conserved across metazoans [22–28], therefore C. elegans offers an excellent platform to identify new genes and molecular mechanisms underlying neuronal control of fat metabolism, using unbiased approaches.
To systematically examine the role of the sensory nervous system in regulating whole body lipid metabolism, we undertook a screen of the 19 (of 21) viable Gα protein mutants for changes in body fat content [29]. This family of heterotrimeric G proteins is well-known to regulate intracellular signaling cascades in response to changes in the environment, which in turn control many aspects of physiology and behavior [30–32]. An added advantage of this family is that the majority of null mutants are viable, and the anatomical locations of these genes have been well-defined. One gene identified from this screen is the Gα protein, GPA-8, the ortholog of the mammalian gustducin proteins that regulates intracellular cGMP. Previous work from our lab has shown that GPA-8 is expressed in the C. elegans body cavity neurons, and integrates oxygen-sensing with the sensing of internal metabolic state, to drive the rate and extent of fat loss [29, 33]. Thus, we found that environmental oxygen serves as a second physiologically relevant sensory input for the regulation of lipid metabolism.
The most potent 'hit' from our Gα protein screen is called GPA-3, and is a member of the Go/Gi protein family. In this study, we identify the neurons in which GPA-3 functions, define its cellular mechanism of action and the critical downstream neurotransmitter required for its functions in promoting fat loss. In so doing, we define pheromone sensing as a new sensory modality for the regulation of lipid metabolism.
Results and discussion
GPA-3 regulates body fat stores via inducing a conserved triglyceride lipase
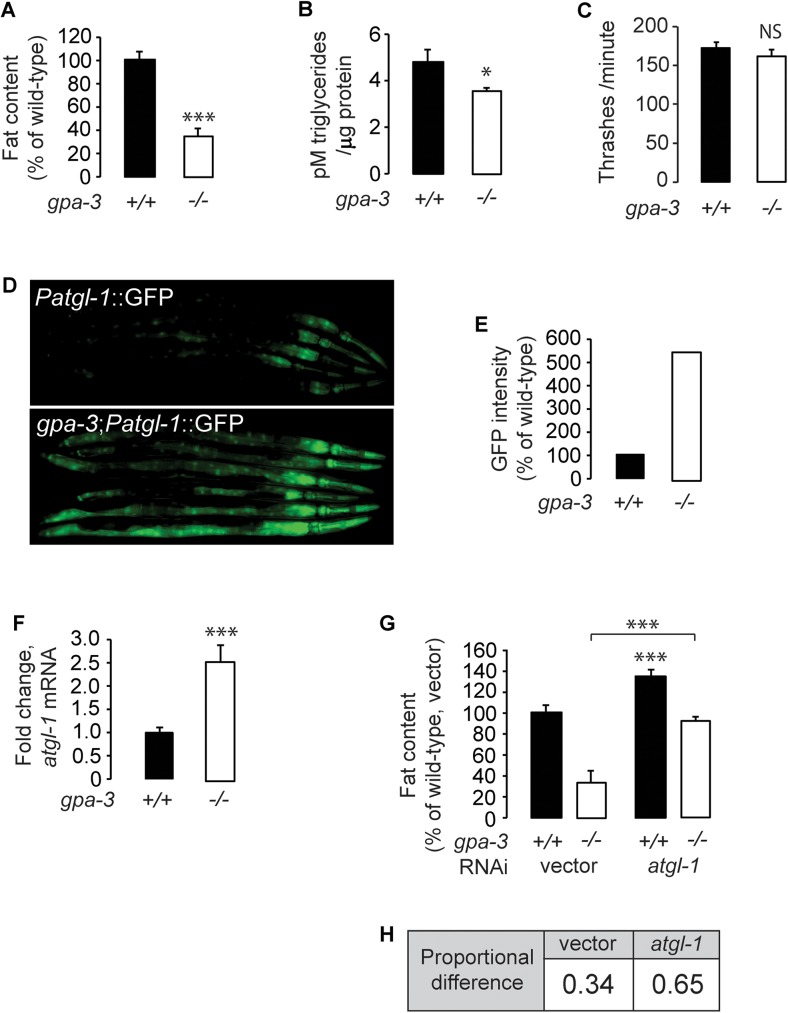
gpa-3(pk35) null mutants (henceforth gpa-3) exhibit a significant decrease in body fat content, as judged by Oil Red O staining of fixed adult animals followed by quantification of lipid droplets in the intestinal cells (Fig 1A and S1A and S1B Fig), and by biochemical extraction of triglycerides from whole adult animals (Fig 1B). The reduced body fat content of gpa-3 mutants could not be explained by differences in locomotor behavior, which is indistinguishable between wild-type and gpa-3 mutants (Fig 1C) [34]. Our previous work had identified a highly conserved lipase called adipocyte triglyceride lipase (ATGL-1) that is rate-limiting for fat loss via the conversion of triglycerides to energy by β-oxidation [3]. Work from other groups has shown that the ATGL-1 protein is stabilized by phosphorylation during fasting thus promoting fat loss [35]. atgl-1 is expressed in the intestine, and is transcriptionally induced in response to neuronal signals that stimulate fat loss. Changes in atgl-1 transcription are tightly correlated with rates of lipolysis [7, 36], thus changes in atgl-1 mRNA reflect physiological shifts in energy utilization. Relative to wild-type animals, gpa-3 mutants have a robust increase in ATGL-1 expression in the intestine (Fig 1D and 1E). Our results indicate that increased fat utilization via induction of triglyceride hydrolysis underlies the reduced body fat of gpa-3 mutants. To corroborate our experiments using the atgl-1 reporter line, we conducted qPCR studies and found an approximately 2.5 fold increase in atgl-1 mRNA in gpa-3 mutants (Fig 1F). Furthermore, RNA-mediated inactivation of ATGL-1 resulted in a nearly 2-fold suppression of fat loss in the gpa-3 mutants (Fig 1G and 1H and S1C Fig). Together, these results show that increased triglyceride hydrolysis is one major mechanism underlying the decreased body fat stores of gpa-3 mutants.
Fig 1. GPA-3 regulates body fat stores via regulating a conserved triglyceride lipase.
(A) Wild-type animals and gpa-3(pk35) mutants were fixed and stained with Oil Red O. Fat content was quantified for gpa-3 mutants and is expressed as a percentage of wild-type animals ± SEM (n = 20). ***, p<0.001 by Student’s t-test. See also S1A and S1B Fig. (B) Extracted lipids were quantified by liquid chromatography/mass spectrometry, data were normalized to protein, and quantified using the Pierce BCA Protein Assay kit. gpa-3 mutants have a significant reduction in triglycerides compared to wild-type animals. *, p<0.05 by Student’s t-test. (C) Thrashing rate was measured for wild-type animals and gpa-3 mutants. Young adults were individually introduced to M9 buffer and allowed to swim freely for 1 minute to become accustomed to the environment. The number of thrashes was then measured for the next 1 minute. gpa-3 mutants showed similar motor function to wild-type animals. Data is expressed as number of thrashes per minute ± SEM (n = 15). NS, not significant by Student’s t-test. (D) Representative images are shown of wild-type animals and gpa-3 mutants bearing an integrated atgl-1::GFP transgene. (E) The fluorescence intensity of atgl-1 expression was quantified for 6 randomly selected worms for each genotype and is expressed as a percentage of wild-type animals (F) atgl-1 mRNA levels were measured by quantitative PCR in gpa-3 mutants and is expressed as fold change relative to wild-type animals ± SEM (n = 3 biological replicates). ***, p<0.001 by Student’s t-test. (G) Wild-type animals and gpa-3 mutants were grown on vector or atgl-1 RNAi containing bacteria and then fixed and stained with Oil Red O. Fat content was quantified for each genotype and condition and is expressed as a percentage of wild-type animals grown on vector RNAi ± SEM (n = 20). Loss of atgl-1 in gpa-3 mutants led to a nearly 2-fold increase in fat content relative to wild-type. ***, p<0.001 by Student’s t-test. (H) Data from 1G showing proportional difference in fat content when ATGL-1 is inactivated in wild type animals and gpa-3 mutants. See also S1C Fig.
GPA-3 regulates body fat via cAMP-mediated signaling from amphid sensory neurons
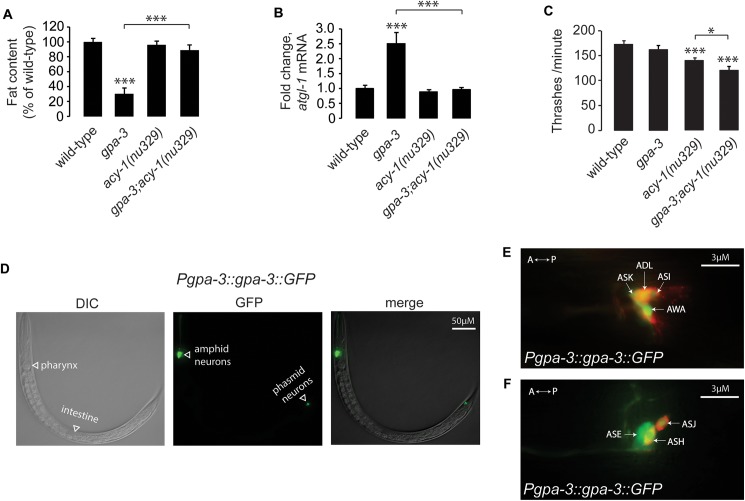
GPA-3 is orthologous to the mammalian cAMP-regulating Gαo/i class [31], sharing 73% similarity (7e-139). Gαo/i family members are known to regulate intracellular cAMP via inhibition of adenylyl cyclases. The C. elegans cAMP adenylyl cyclase ACY-1 is expressed in neurons, and viable loss-of-function (nu239) mutants are available [37]. Although the acy-1(nu329) mutants did not show an appreciable difference in body fat (Fig 2A and S2C Fig), removal of acy-1 in the gpa-3 mutants resulted in a near-complete suppression of the gpa-3 body fat phenotype. To provide a second, molecular readout of the fat loss, we measured atgl-1 mRNA by qPCR, and found that the gpa-3-mediated induction of atgl-1 in the gpa-3 mutants was also suppressed in the gpa-3;acy-1 double mutants (Fig 2B). Thus, ACY-1 function is required downstream of GPA-3 in the regulation of body fat via the induction of ATGL-1-mediated lipolysis. acy-1 mutants have a mild locomotor defect and gpa-3;acy-1 double mutants have a statistically significant additive effect (Fig 2C). However because gpa-3 mutants themselves do not have a locomotor phenotype, these effects are non-specific to the gpa-3 fat regulatory pathway. To determine the direction of the effect of increased cAMP on body fat, we exogenously administered a non-hydrolyzable analog of cAMP called 8-Bromo-cAMP (8-Br-cAMP), which led to a dose-dependent decrease in body fat stores (S2A Fig). Together, our results indicate that GPA-3 controls fat utilization through inhibition of the adenylyl cyclase ACY-1, and the resultant regulation of cAMP concentrations (S2B Fig).
Fig 2. GPA-3 regulates body fat via cAMP-mediated signaling from amphid sensory neurons.
(A) Animals were fixed and stained with Oil Red O. Fat content was quantified for each genotype and is expressed as a percentage of wild-type animals ± SEM (n = 20). ***, p<0.001 by one-way ANOVA. See also S2C Fig. (B) atgl-1 mRNA levels were measured by quantitative PCR in gpa-3, acy-1(nu329) and gpa-3;acy-1 mutants and is expressed as fold change relative to wild-type animals ± SEM (n = 3 biological replicates). ***, p<0.001 by one-way ANOVA. (C) Thrashing rate was measured for each genotype. Although gpa-3 mutants show similar motor function to wild-type animals, acy-1(nu329) and gpa-3;acy-1 mutants. Data is expressed as number of thrashes per minute ± SEM (n = 15). *, p<0.05; ***, p<0.001 by one-way ANOVA. (D) Representative images showing the expression of GPF in transgenic animals bearing a transgene driving gpa-3 expression with the endogenous promoter. DIC (left panel), GFP (central panel) and a merged representation of these 2 images (right panel). GPA-3 is solely expressed in the nervous system and not in the intestine, where its metabolic phenotype manifests. (E, F). Using DiI staining (red), a method used to identify C. elegans sensory neurons, we found that GPA-3 is expressed in 9 bilaterally symmetric pairs of amphid sensory neurons with ciliated endings that are directly exposed to the environment. A, anterior; P, posterior.
Examination of our gpa-3-expressing transgenic lines showed that GPA-3 is solely expressed in the nervous system and not in the intestine, where its metabolic phenotype manifests (Fig 2D). GPA-3 is expressed in 9 bilaterally symmetric pairs of amphid sensory neurons with ciliated endings that are directly exposed to the environment: ADF, ADL, ASE, ASG, ASH, ASI, ASJ, ASK, and sporadically in AWA (Fig 2E and 2F), confirming previous observations [38, 39]. The localization of GPA-3 to the amphid sensory neurons suggests that the subset of neurons from which GPA-3 regulates body fat either directly or indirectly regulate a long-range neuroendocrine factor that acts in the intestine to elicit fat loss.
GPA-3 functions in ADL amphid sensory neurons to regulate intestinal fat utilization
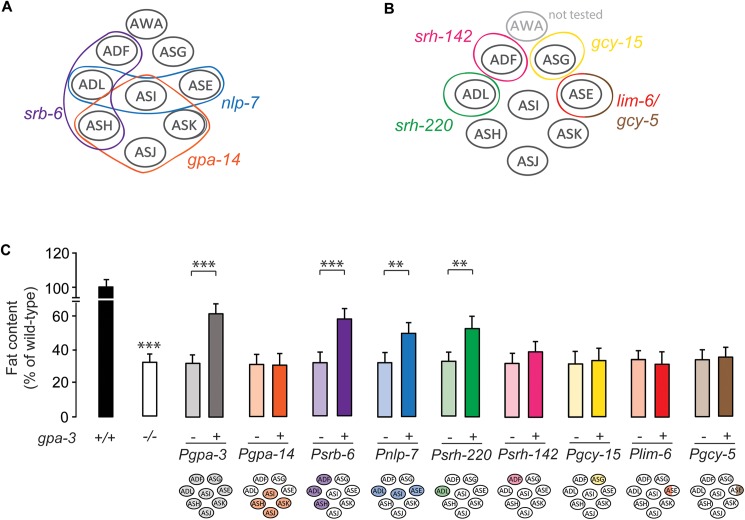
To identify the neurons in which GPA-3 acts to regulate fat stores in the intestine, we generated expression constructs to drive gpa-3 cDNA in subsets of amphid neurons in which gpa-3 is normally expressed. The transgenic rescue strategy is given in Fig 3A and 3B. In gpa-3 null mutants, restoration of gpa-3 cDNA using either 5kb or 7kb of endogenous gpa-3 upstream regulatory regions, gave significant restoration of intestinal fat content (Fig 3C and S3A Fig), confirming that GPA-3 functions in sensory neurons to regulate intestinal fat stores. However, we noted that in the gpa-3 transgenic animals, neither the 5kb nor the 7kb promoter were sufficient to confer a complete restoration of body fat stores (S3A Fig), which prompted us to examine the feeding behavior of gpa-3 mutants. Relative to wild-type animals, gpa-3 mutants displayed ~15–20% decrease in food intake (S3B Fig). However, we found that re-expression of gpa-3 under either 5kb or 7kb promoters did not restore food intake to wild-type levels (S3B Fig).
Fig 3. GPA-3 functions in ADL amphid sensory neurons to regulate intestinal fat utilization.
(A-B) Model depicting the transgenic rescue strategy utilized to restore gpa-3 cDNA, first in subsets of amphid neurons (A), and then in individual neuron types (B). (C) gpa-3 mutants bearing gpa-3 expression using the indicated promoter were fixed and stained with Oil Red O. Relative to non-transgenic controls (-, light bars), transgenic animals (+, dark bars) bearing the gpa-3 transgene in ADL neurons restored body fat content. Data are expressed as a percentage of body fat in wild-type animals ± SEM (n = 12–16). **, p<0.01; ***, p<0.001 by one-way ANOVA. See also S3A Fig.
We next wanted to test whether acy-1 mutants suppressed the decreased food intake of gpa-3 mutants, and accordingly measured food intake in the relevant mutants. We found that acy-1 mutants also displayed decreased food intake to a similar extent as the gpa-3 mutants. However, the gpa-3;acy-1 double mutants resembled either single mutant alone (S3C Fig). Thus, unlike the suppression of GPA-3-mediated fat loss (Fig 2A) or its induction of atgl-1 (Fig 2B), acy-1 mutants do not suppress GPA-3-mediated food intake.
We wanted to further examine the role of GPA-3 in specific subsets of neurons. Accordingly, we devised a transgenic rescue strategy that allowed us to include or exclude a role for GPA-3 in subsets of neurons in which it is expressed (Fig 3A). In gpa-3 null mutants, restoration of gpa-3 cDNA expression using the srb-6 (ADL, ADF, ASH) and nlp-7 (ADL, ASI, ASE) promoters, but not the gpa-14 (ASH, ASI, ASK, ASJ) promoter significantly restored intestinal fat content (Fig 3C). This combinatorial strategy first eliminated a role for GPA-3 in ASI, ASH, ASK and ASJ neurons and second, identified a potential role for the ADL neurons because it is the only neuron pair that overlaps between the two rescuing promoters, srb-6 and nlp-7. We next drove gpa-3 cDNA expression in the individual neurons ADL, ASG and ASE using neuron-specific promoters (Fig 3B; the AWA neurons were not tested). Restoration of gpa-3 in the ADL neurons alone significantly restored body fat stores in the gpa-3 null mutants (Fig 3C). Thus, GPA-3 function in the ADL neurons regulates body fat stores. Although the transgenic rescue strategy revealed a clear role for GPA-3 in controlling body fat in subsets of neurons, in no case were we able to restore the feeding phenotype of gpa-3 mutants, including the endogenous promoter that was sufficient to restore fat stores (Fig 3C and S3A and S3B and S3D Fig). These results suggest the possibility that background effects unrelated to the gpa-3 gene contribute to the reduced feeding phenotype in these mutants, despite the gpa-3 mutant having been outcrossed 7 times. Another possibility is that although unusual in C. elegans [40], additional regulatory elements further upstream from the chosen 7kb gpa-3 promoter region may play a role in controlling gpa-3 expression. However, our data also suggest that the reduced food intake only accounts for a small percentage of the net change in body fat stores, because expression of gpa-3 in the ADL neurons significantly restores body fat stores without altering food intake (Fig 3C). Together our data suggest that gpa-3 functions in the ADL neurons to regulate fat content, independent of changes in food intake or locomotion.
Enhanced cAMP production in ADL neurons decreases intestinal fat
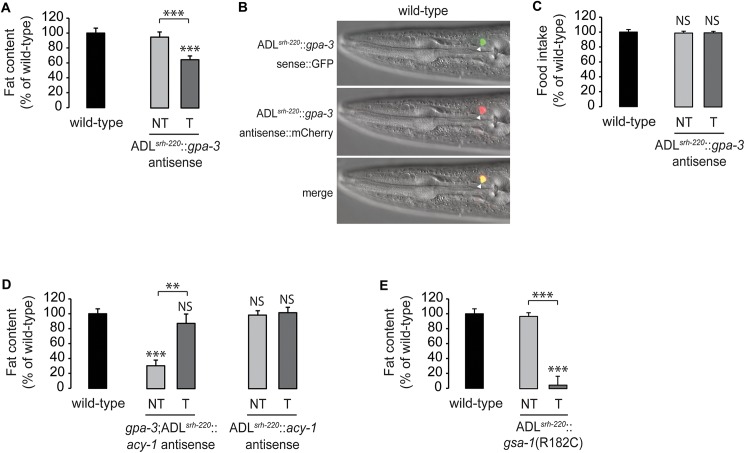
To determine the necessity of GPA-3 in the ADL neurons for the regulation of body fat, we conducted antisense mediated inhibition experiments [41] using an ADL-specific promoter. Inactivation of gpa-3 in the ADL neurons lowered fat content to 65% of that seen in wild-type animals (Fig 4A and 4B and S4C Fig). Notably, eliminating gpa-3 in the ADL neurons in an otherwise wild-type background did not alter food intake (Fig 4C), reinforcing our observations that gpa-3-mediated regulation of body fat via the ADL neurons occurs independently of feeding. Together with the transgenic rescue experiments, we find that GPA-3 expression in ADL neurons is necessary and sufficient to maintain body fat stores.
Fig 4. Enhanced cAMP production in ADL neurons decreases intestinal fat.
(A) Wild-type animals bearing antisense-mediated inactivation of gpa-3 expression in ADL neurons using the srh-220 promoter were fixed and stained with Oil Red O. Relative to non-transgenic controls (NT, light gray bar), transgenic animals (T, dark gray bar) bearing gpa-3 antisense in ADL neurons had a significant decrease in body fat. Data are expressed as a percentage of body fat in wild-type animals ± SEM (n = 20). ***, p<0.001 by one-way ANOVA. See also S4A Fig. (B) Representative images showing the expression of gpa-3 sense::GFP (upper panel), gpa-3 antisense::mCherry (central panel) and their co-localization in ADL (merged, lower panel). (C) Food intake in wild-type animals bearing antisense-mediated inactivation of gpa-3 expression in ADL neurons. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant by one-way ANOVA. (D) Wild-type animals and gpa-3 mutants bearing antisense-mediated inactivation of acy-1 expression in ADL neurons using the srh-220 promoter were fixed and stained with Oil Red O. Relative to non-transgenic controls in the gpa-3 background (NT, light gray bar), transgenic animals (T, dark gray bar) bearing acy-1 antisense in ADL neurons restored body fat content to wild-type. In wild-type animals, there was no difference in fat content of non-transgenic (NT, light gray bar) and (T, dark gray bar) transgenic animals. Data are expressed as a percentage of body fat in wild-type animals ± SEM (n = 20). NS, not significant; **, p<0.01; ***, p<0.001 by one-way ANOVA. See also S4B Fig. (E) Wild-type animals bearing gsa-1(R182C), a dominant, gain-of-function mutation of C. elegans Gαs, in ADL neurons were fixed and stained with Oil Red O. Relative to non-transgenic controls (NT, light gray bar), transgenic animals (T, dark gray bar) bearing gsa-1(R182C) in ADL neurons had a significant decrease in body fat. Data are expressed as a percentage of body fat in wild-type animals ± SEM (n = 14–20). ***, p<0.001 by one-way ANOVA. See also S4C Fig.
Our genetic epistasis experiments (Fig 2A and 2B and S2A Fig) suggested that gpa-3 negatively regulates acy-1 to control intracellular cAMP. We next wanted to determine the extent to which this signaling pathway functions in the ADL neurons. Accordingly, we inactivated acy-1 solely in the ADL neurons in the gpa-3 mutant background using antisense inhibition. Relative to non-transgenic controls, inactivation of acy-1 selectively in the ADL neurons led to a significant suppression of the decreased body fat of the gpa-3 mutants, resulting in body fat content similar to wild-type levels (Fig 4D and S4B Fig). As seen with global acy-1 loss (Fig 2A), inactivation of acy-1 specifically in the ADL neurons also did not appreciably alter fat stores (Fig 4D and S4B Fig). Together, these experiments reveal a role for GPA-3 as a negative regulator of ACY-1 and intracellular cAMP in the ADL neurons, for the control of body fat stores. In C. elegans, the stimulatory Gαs that activates adenylyl cyclase to increase intracellular cAMP [42] is called GSA-1. To test the prediction that enhanced cAMP production in ADL neurons decreases body fat, we selectively expressed gsa-1(R182C), a dominant, gain-of-function mutation of C. elegans Gαs [43] in the ADL neurons, which resulted in a near-complete loss of intestinal fat (Fig 4E and S4C Fig). Thus, enhanced cAMP signaling in the ADL neurons stimulates fat loss in the intestine, and the cAMP second messenger in ADL neurons is instructive for the control of fat stores in the intestine.
Cholinergic signaling drives GPA-3-mediated fat loss
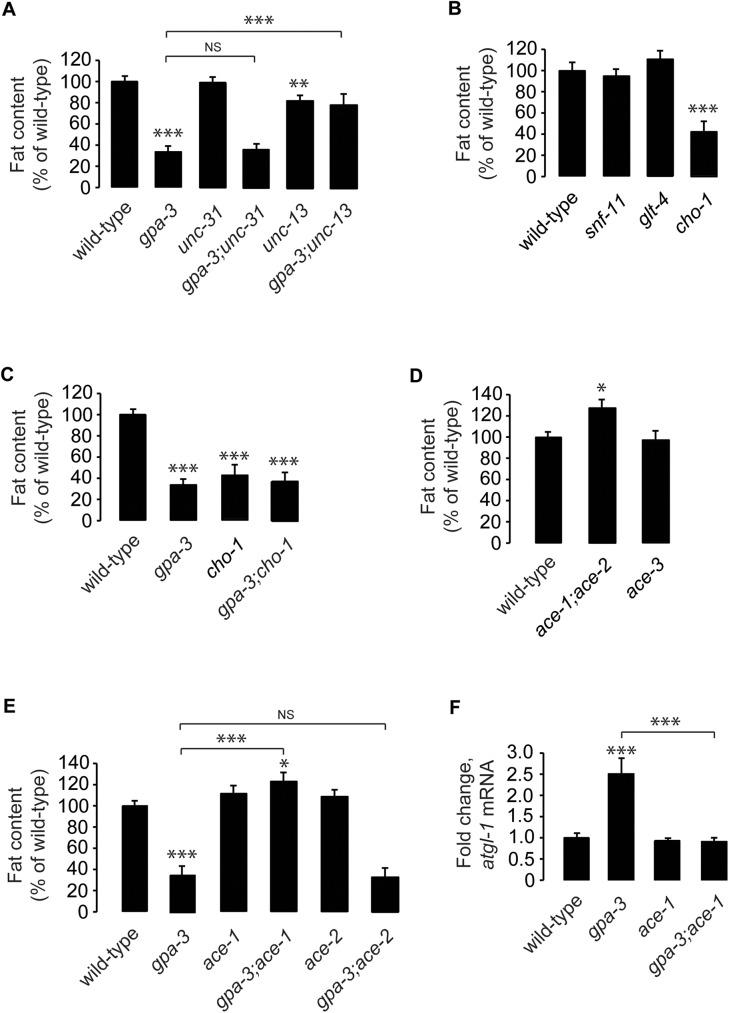
Information from the ADL neurons to the intestine could be relayed either directly via the release of a neuroendocrine factor, or indirectly via modifying the properties of other neurons. These possibilities can be distinguished in the following way: long-range neuropeptides and neuromodulators are localized to dense core vesicles, which require the conserved Calcium-dependent Activator Protein for Secretion (CAPS, UNC-31 in C. elegans) for fusion with the plasma membrane [44–46]. On the other hand, the canonical neurotransmitters (acetylcholine, ACh; γ-amino butyric acid, GABA; and glutamate) are localized to small clear synaptic vesicles, which require a protein called UNC-13 (MUNC-13 in mammals) for fusion at the synapse [47, 48]. Thus, loss of UNC-31 function blocks the release of neuropeptides and biogenic amines from neurons [46], and loss of UNC-13 function blocks release of the canonical neurotransmitters [48]. We generated gpa-3;unc-31(e928) and gpa-3;unc-13(n2813) mutants and measured the body fat of the respective single and double mutants. Interestingly, we found that loss of unc-13 resulted in complete suppression of the fat loss seen in the gpa-3 mutants (Fig 5A and S5A Fig). This result suggested that rather than neuropeptides and biogenic amines, the canonical neurotransmitters acetylcholine, GABA or glutamate are required for the effects of GPA-3 signaling.
Fig 5. Cholinergic signaling drives GPA-3-mediated fat loss.
(A-E) Animals were fixed and stained with Oil Red O. Fat content was quantified for each genotype as indicated, and is expressed as a percentage of wild-type animals ± SEM (n = 14–20). NS, not significant; *, p<0.05; **, p<0.01; ***, p<0.001 by one-way ANOVA. See also S5A–S5E Fig. (F) atgl-1 mRNA levels were measured by quantitative PCR in gpa-3, ace-1 and gpa-3;ace-1 mutants and is expressed as fold change relative to wild-type animals ± SEM (n = 3 biological replicates). ***, p<0.001 by one-way ANOVA.
To determine which of the three canonical neurotransmitter pathways are required downstream of GPA-3, we first examined mutants of the presynaptic re-uptake transporters for GABA (snf-11), glutamate (glt-4) and ACh (cho-1). Loss of the re-uptake transporters of the conventional neurotransmitters would disrupt their steady-state levels in the synaptic cleft, and thus indicate a potential role in regulating body fat stores. snf-1(ok156) and glt-4(bz69) mutants did not appreciably alter body fat stores, whereas cho-1(ok1069) mutants had approximately 40% of the body fat of wild-type animals (Fig 5B and S5B Fig), resembling gpa-3 null mutants. ACh synthesis and breakdown occur via mechanisms distinct from the other neurotransmitters: after release into the synapse, unbound ACh is cleaved to form acetyl-CoA and choline by the enzyme acetylcholinesterase within the synaptic cleft itself. Choline is then taken up into the pre-synaptic neuron by the CHO-1 re-uptake transporter, and this step is a rate-limiting source of choline for presynaptic ACh synthesis. Thus, cho-1 mutants are defective in the re-uptake of synaptic choline and are deficient in ACh [49, 50]. gpa-3;cho-1 mutants display similar fat content as either single mutant alone (Fig 5C and S5C Fig). To determine the extent to which changes in ACh signaling regulate body fat stores, we examined the available mutants in the acetylcholinesterase genes, ace-1, ace-2 and ace-3, which have increased synaptic ACh [51–53]. Relative to wild-type animals, ace-1;ace-2 double mutants had a significant increase in body fat stores, whereas ace-3 mutants did not show an appreciable difference (Fig 5D and S5D Fig). These results suggested that alterations in synaptic ACh result in changes in body fat stores.
To determine the relationship between gpa-3 signaling and ACh, and to identify the key acetylcholinesterase responsible for the effects of ACh on body fat, we crossed gpa-3 mutants with the ace-1;ace-2 mutants to generate each mutant combination, as well as the ace-1 and ace-2 single mutants. We found that the gpa-3;ace-1 mutants fully suppressed the reduced body fat of gpa-3 single mutants (Fig 5E and S5E Fig), whereas the gpa-3;ace-2 double mutants did not, and resembled the gpa-3 single mutants alone (Fig 5E and S5E Fig). ace-1 mutants also suppressed the transcriptional induction of atgl-1 seen in gpa-3 mutants (Fig 5F). Thus, ACE-1 is required downstream of GPA-3 in the regulation of body fat, suggesting that the GPA-3-mediated fat regulatory signal is transmitted from the ADL neurons via the cholinergic pathway.
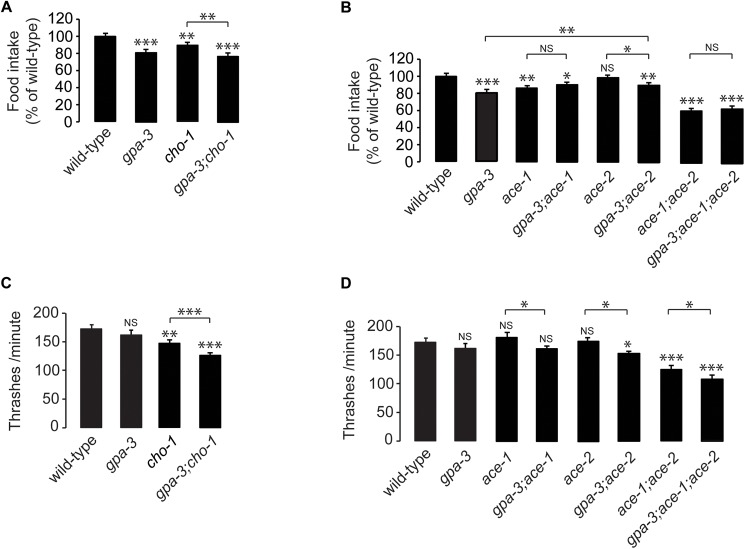
We measured food intake and locomotion of the mutants in the cholinergic pathway, with and without gpa-3 (Fig 6). As described in S3B Fig, gpa-3 mutants had an ~15–20% decrease in food intake (Fig 6A and 6B). Cholinergic signaling has been known to alter rhythmic behaviors [54, 55], and as expected, cho-1 mutants also have a significant reduction in food intake, albeit to a lesser extent than the gpa-3 mutants themselves. gpa-3;cho-1 double mutants resemble the gpa-3 single mutants with respect to feeding deficits (Fig 6A). We next examined the ace genes with and without gpa-3 with respect to food intake. ace-1 mutants have decreased food intake similar to the gpa-3 mutants, and gpa-3;ace-1 double mutants do not suppress the gpa-3 phenotype. Rather, they resemble either single mutant alone (Fig 6B). This is in contrast to the suppression of GPA-3-mediated fat loss, as judged by fat levels as well as by measuring the induction of atgl-1 by GPA-3 (Fig 5E and 5F). ace-2 mutants have a negligible effect on food intake, and gpa-3;ace-2 mutants resemble gpa-3 mutants alone. Taken together, the ace-1-mediated suppression of fat loss of the gpa-3 mutants is specific, and is not accompanied by suppression of food intake (Fig 6B). Similar results were observed with locomotion (Fig 6C and 6D); additionally, gpa-3 mutants and ace-1 mutants do not show appreciable differences in locomotion. Thus, the fat phenotype of gpa-3 mutants occurs as a selective consequence of a shift towards fat mobilization.
Fig 6. Feeding and locomotion data for mutants of genes required for GPA-3-mediated fat regulation.
(A-B) Food intake was measured for each genotype as indicated and is expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; *, p<0.05; **, p<0.01; ***, p<0.001 by one-way ANOVA. (C-D) Thrashing rate was measured for each genotype as indicated and is expressed as number of thrashes per minute ± SEM (n = 15). NS, not significant; *, p<0.05; **, p<0.01; ***, p<0.001 by one-way ANOVA.
Pheromone signaling regulates body fat stores via GPA-3 signaling
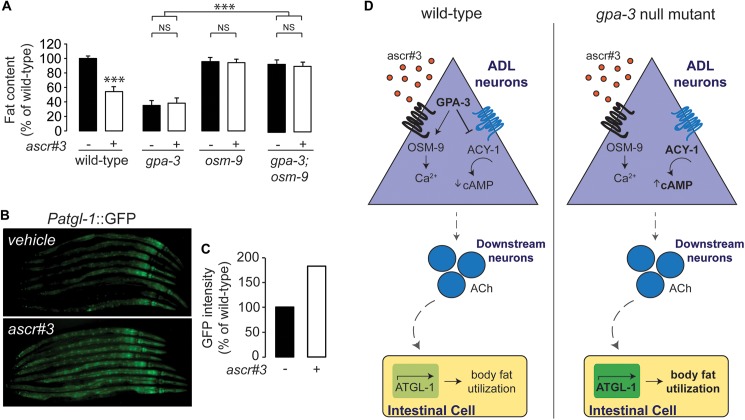
The ADL neurons mediate avoidance behavior from aversive stimuli [56–58] and are also shown to modulate social feeding behavior in response to high O2 levels [59]. These effects are mediated predominantly through the TRPV channel, OSM-9 [59, 60]. We found that osm-9 mutants had wild-type body fat levels, and fully suppressed the reduced body fat of gpa-3 mutants (Fig 7A and S6 Fig). These results suggested that an aversive function encoded by ADL neurons was related to the GPA-3-mediated fat phenotype. ADL neurons detect an ascaroside pheromone called ascr#3 (also called C9) and initiate an aversive response in N2 wild-type animals that is abrogated in osm-9 mutants [61]. Pheromone signaling in C. elegans was originally shown to control developmental fate decisions [62–64]. However, a recent body of evidence has shown that a chemically-diverse family of ascaroside-based pheromones function individually and in combination, to elicit behaviors that collectively transmit population structure and population density information [20, 27, 65]. We wondered whether ascr#3, the ascaroside detected by the ADL neurons would alter body fat stores. Administration of ascr#3 at a dose known to elicit Ca2+ transients in ADL neurons [66] led to a robust decrease in body fat stores (Fig 7A and S6 Fig). gpa-3 and osm-9 single mutants, and gpa-3;osm-9 double mutants, did not display a further reduction in body fat upon ascr#3 administration, suggesting that activation of ADL neurons by ascr#3 decreases body fat stores via GPA-3-dependent signaling (Fig 7A and S6 Fig). As expected, administration of ascr#3 also robustly induced atgl-1 expression in the intestine (Fig 7B and 7C).
Fig 7. Pheromone signaling regulates body fat stores via GPA-3 signaling.
(A) Animals were transferred at L4 to plates containing either ddH2O vehicle or 80nM ascaroside, ascr#3, then fixed and stained with Oil Red O. Fat content was quantified for each condition and is expressed as a percentage of vehicle-treated wild-type animals ± SEM (n = 20). NS, not significant; ***, p<0.001 by two-way ANOVA. (B) Representative images are shown of wild-type animals bearing an integrated atgl-1::GFP transgene exposed either to ascr#3 or vehicle. (C) The fluorescence intensity of atgl-1 expression was quantified for 8 randomly selected worms for each condition and is expressed as a percentage of wild-type animals exposed to vehicle. (D) Model depicting the ascr#3-mediated regulation of cAMP signaling, via the G protein, GPA-3, in the ADL neurons which controls acetylcholine release in to-be-defined cholinergic neurons, and in turn regulates a stimulatory signal to control body fat stores via the rate-limiting ATGL-1 lipase in the intestine. Under normal conditions, in wild-type animals, environmental levels of the ascaroside ascr#3 regulate the extent to which GPA-3 in the ADL neurons inhibits the downstream adenylyl cyclase ACY-1 and, therefore, controlling levels of cAMP in the ADL neurons. This, in turn, controls the level of acetylcholine released from small, clear vesicles in downstream neurons, and initiates a signal to the intestinal cells to regulate the activity of ATGL-1 (left panel). In gpa-3 mutants, irrespective of ascr#3 levels, the GPA-3 mediated inhibition of ACY-1 is lost, causing more cAMP to be produced in the ADL neurons. This, in turn, causes more acetylcholine to be released in downstream neurons, ultimately up-regulating ATGL-1 in the intestinal cells, and a constitutive loss of body fat due to increased fat utilization (right panel).
We propose a model in which pheromone-mediated regulation of cAMP signaling in the ADL neurons controls acetylcholine release in to-be-defined cholinergic neurons, which in turn regulates a fat-stimulatory signal to control body fat stores via the rate-limiting ATGL-1 lipase in the intestine (Fig 7D). Under normal conditions, in wild-type animals, population-density-dependent levels of the ascaroside ascr#3 regulates the extent to which GPA-3 in the ADL neurons inhibits the downstream adenylyl cyclase ACY-1 thus controlling cAMP levels in the ADL neurons. This, in turn, controls the level of acetylcholine released from small, clear vesicles in cholinergic neurons, and initiates a signal to the intestinal cells to regulate the activity of ATGL-1 (Fig 7D, left panel). In gpa-3 mutants, irrespective of ascr#3 levels, the GPA-3 mediated inhibition of ACY-1 is lost, causing more cAMP to be produced in the ADL neurons. This, in turn, causes more acetylcholine to be released in downstream neurons, ultimately up-regulating ATGL-1 in the intestinal cells, and a constitutive loss of body fat due to increased fat utilization (Fig 7D, right panel). Receptors for GPA-3 have been identified in the context of the dauer developmental decision [67]. Two G protein coupled receptors, srbc-64 and srbc-66 require GPA-3 signaling from the ASK neurons to mediate the dauer decision in response to the dauer pheromone and the ascaroside C6. srbc-64 and -66 are not reported to be expressed in the ADL neurons, nor known to be responsive to ascr#3, and therefore likely function via distinct mechanisms.
Our previous work has suggested that food and oxygen are salient environmental cues that regulate body fat stores via modulation of neuronal circuit function [3, 29]. Based on the studies presented here, we now propose pheromone sensing as a third sensory modality that regulates body fat stores. As an animal encounters a new patch of food, it must adjust its metabolism to reflect its environment. A patch of food that contains other worms must necessarily be shared, whereas a patch of food without worms reflects a relatively greater amount of food. We speculate that ascr#3/GPA-3 signaling from the ADL neurons provides C. elegans a mechanism to discriminate between these distinct environments, and accordingly modulate its metabolism. Our experiments provide the first insights into the molecular mechanisms by which pheromone sensing from the nervous system regulates peripheral lipid metabolism. In future studies, it will be interesting to determine the extent to which these discrete sensory inputs intersect to coordinate body fat metabolism.
Materials and methods
Animal maintenance and strains
C. elegans was cultured as described [68]. N2 Bristol, obtained from the Caenorhabditis Genetic Center (CGC) was used as the wild-type reference strain. The mutant and transgenic strains used are listed in S1 Table. Animals were synchronized for experiments by hypochlorite treatment, after which hatched L1 larvae were seeded on plates with the appropriate bacteria. All experiments were performed on day 1 adults.
Cloning and transgenic strain construction
Promoters and genes were cloned using standard PCR techniques from N2 Bristol worm lysates or cDNA and cloned using Gateway Technology (Life Technologies). Promoter lengths were determined based on functional rescue and are available upon request. All rescue plasmids were generated using polycistronic GFP. Transgenic rescue strains were constructed by microinjection into the C. elegans germline followed by visual selection of transgenic animals under fluorescence. For the microinjections, 5–10 ng/μl of the desired plasmid was injected with 25 ng/μl of an unc-122::GFP or myo-2::mCherry coinjection marker and 65–70 ng/μl of an empty vector to maintain a total injection mix concentration of 100 ng/μl. In each case, 10–20 stable transgenic lines were generated. Two lines were selected for experimentation based on consistency of expression and transmission rate.
Triglyceride extraction and quantitation
Triglycerides were extracted from wild-type and mutant C. elegans as described [3]. Extracted lipids were quantified by liquid chromatography/mass spectrometry on an HP 1100 MSDTM, using a neutral lipid Pheromex Luna C5 column, following the methodology from Nomura and colleagues [69]. Data were normalized to protein, quantified by the Pierce BCA Protein Assay kit.
Oil Red O staining
Oil Red O staining was performed as described [3]. For Oil Red O experiments in which animals were treated with a non-hydrolyzable cAMP analogue, animals were added to plates containing either M9 vehicle or 20, 200, or 500μM 8-Bromoadenosine 3′,5′-cyclic monophosphate (Sigma Aldrich). For Oil Red O experiments in which animals were treated with ascaroside ascr#3, animals were added to plates containing either ddH2O vehicle or 80nM ascr#3. Within a single experiment, roughly 3500 animals were fixed and stained, 100 animals were visually inspected on slides, following which 15–20 animals were imaged for each genotype/condition. All experiments were repeated at least 3 times. Wild-type and gpa-3 mutants were included as controls for each experiment.
Thrashing assay
Thrashing rate was measured as previously described [70]. For each animal, a movement where the head and/or tail swung to the other side was counted as one thrash. 15–20 animals were assessed for each phenotype.
RNAi
RNAi experiments were conducted as previously described [3]. Plates were seeded with HT115 bacteria containing vector or the relevant RNAi clone four days prior to seeding larvae.
Image acquisition and quantitation
Black and white images of Oil Red O stained animals and fluorescent images were captured using a 10X objective on a Zeiss Axio Imager microscope. Lipid droplet staining in the first four pairs of intestinal cells was quantified as described [3]. We have found that quantification of the anterior intestine reliably captures fat content of the entire intestine. For all atgl-1::GFP images, an equal number of worms were chosen blindly and lined up side by side. Fluorescence intensity for all chosen worms was quantified for each condition. Images were quantified using ImageJ software (NIH).
Quantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). Genomic DNA was removed using an RNase-free DNase kit (QIAGEN). cDNA was prepared using a iScript Reverse Transcription Supermix for RT-qPCR kit (BioRad) according to the manufacturer’s instructions. Quantitative PCR was performed using the SsoAdvanced Universal SYBR® Green Supermix according to the manufacturer’s instructions. Data were normalized to actin mRNA. Primer sequences are available upon request.
DiI staining
Animals of mixed developmental stages were incubated in a 1:200 dilution of DiI stain (Life Technologies) for 3 hours on a rotating rack. After staining, the animals were seeded onto a plate containing an OP50 bacterial lawn and allowed to dry for approximately 30 minutes. Fluorescent images of animals in the L2-L3 larval stages were captured using a 100X objective on a Zeiss Axio Imager microscope.
Food intake
Food intake was measured by counting pharyngeal pumping, as previously described [71]. For each animal, the rhythmic contractions of the pharyngeal bulb were counted over a 10 s period under a Zeiss M2 Bio Discovery microscope. For each genotype, 10 animals were counted per condition and the experiment was repeated at least three times.
Statistics
Wild-type animals were included as controls for every experiment. Error bars represent SEM. Student’s t-test, one-way ANOVA, and two-way ANOVA were used as indicated in the figure legends.
Supporting information
(A) Images of wild-type animals and gpa-3 mutants fixed and stained with Oil Red O. Animals are oriented facing upwards, and the head and intestinal cells are as marked. Oil Red O stained droplets are indicated. For each genotype, images depict the full range of the observed phenotype. (B) The integrated density of the lipid droplets is used to quantify body fat stores, as described in the Materials and Methods. Graph represents the integrated density values of individual wild-type animals and gpa-3 mutants. ***, p<0.001 by Student’s t-test. (C) Representative images of wild-type animals and gpa-3 mutants exposed to vector control or atgl-1 RNAi fixed and stained with Oil Red O. The model depicts the section of anterior intestine being represented for each genotype and condition.
(TIF)
(A) Animals were transferred at L4 to plates containing either M9 vehicle or 20, 200, or 500μM 8-Bromoadenosine 3′,5′-cyclic monophosphate (8-Br-cAMP), then fixed and stained with Oil Red O. Fat content was quantified for each condition and is expressed as a percentage of vehicle-treated wild-type animals ± SEM (n = 12). ***, p<0.001 by one-way ANOVA. (B) Model depicting epistatic relationship between the Go/I protein GPA-3 and the adenylyl cyclase ACY-1 for the regulation of cAMP. (C) Representative images of wild-type animals, gpa-3, acy-1(nu329), and gpa-3;acy-1 mutants fixed and stained with Oil Red O.
(TIF)
(A) gpa-3 mutants bearing gpa-3 expression using a 5kb or a 7kb endogenous promoter were fixed and stained with Oil Red O, as indicated. Relative to non-transgenic controls (-, light gray bars), transgenic animals (+, dark gray bars) bearing the gpa-3 transgene restored body fat content to the same extent, whether driven by the 5kb or 7kb promoter. Data are expressed as a percentage of body fat in wild-type animals ± SEM (n = 12–16). ***, p<0.001 by one-way ANOVA. (B) Food intake for gpa-3 mutants bearing gpa-3 expression using a 5kb or a 7kb endogenous promoter was measured. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; ***, p<0.001 by one-way ANOVA. (C) Food intake for wild-type animals, gpa-3, acy-1(nu329), and gpa-3;acy-1 mutants was measured. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; ***, p<0.001 by one-way ANOVA. (D) Food intake for gpa-3 mutants bearing gpa-3 expression using the indicated promoter was measured. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; ***, p<0.001 by one-way ANOVA.
(TIF)
(A-C) Representative images of all genotypes fixed and stained with Oil Red O.
(TIF)
(A-E) Representative images of all genotypes fixed and stained with Oil Red O.
(TIF)
Representative images of all genotypes and conditions fixed and stained with Oil Red O.
(TIF)
(TIF)
Acknowledgments
We thank Prof. Benjamin Cravatt (The Scripps Research Institute, La Jolla) for assistance with mass spectrometry, Prof. Randy Blakely (Florida Atlantic University Brain Institute, Jupiter) and Prof. James Rand (University of Oklahoma Health Sciences Center, Oklahoma) for strains, Dr. Emily Witham (Janssen Pharmaceutical Companies of Johnson and Johnson, La Jolla) and Harkaranveer Ratanpal (Liberty University College of Osteopathic Medicine, Virginia) for assistance with construct design. We also thank Dr. Lavinia Palamiuc, Dr. Nicole Littlejohn, and other members of the Srinivasan lab for critical comments on the manuscript. We thank the CGC for providing strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by research grants to SS from the National Institute of Diabetes and Digestive and Kidney Diseases (https://www.niddk.nih.gov/) (DK077427 and DK095804). Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kong D, Tong Q, Ye C, Koda S, Fuller PM, Krashes MJ, et al. GABAergic RIP-Cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell. 2012;151(3):645–57. doi: 10.1016/j.cell.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Li T, Yang D, Ma R, Moran T, Smith W. Synphilin-1 alters metabolic homeostasis in a novel Drosophila obesity model. International journal of obesity. 2012;36(12):1529–36. doi: 10.1038/ijo.2012.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noble T, Stieglitz J, Srinivasan S. An Integrated Serotonin and Octopamine Neuronal Circuit Directs the Release of an Endocrine Signal to Control C. elegans Body Fat. Cell Metabolism. 2013;18(5):672–84. doi: 10.1016/j.cmet.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu M, Sarruf DA, Talukdar S, Sharma S, Li P, Bandyopadhyay G, et al. Brain PPARγ Promotes Obesity and is Required for the Insulin–Sensitizing Effect of Thiazolidinediones. Nature medicine. 2011;17(5):618–22. doi: 10.1038/nm.2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterson Michael J, Horvath Tamas L. Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell Metabolism. 2015;22(6):962–70. doi: 10.1016/j.cmet.2015.09.026 [DOI] [PubMed] [Google Scholar]
- 6.Toda C, Kim JD, Impellizzeri D, Cuzzocrea S, Liu Z-W, Diano S. UCP2 Regulates Mitochondrial Fission and Ventromedial Nucleus Control of Glucose Responsiveness. Cell. 2016;164(5):872–83. doi: 10.1016/j.cell.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan S. Regulation of Body Fat in Caenorhabditis elegans. Annual Review of Physiology. 2015;77(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linford NJ, Kuo T-H, Chan TP, Pletcher SD. Sensory Perception and Aging in Model Systems: From the Outside In. Annual Review of Cell and Developmental Biology. 2011;27(1):759–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen H-j, Beck JS, et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nature genetics. 2002;31(4):435–8. doi: 10.1038/ng935 [DOI] [PubMed] [Google Scholar]
- 10.Cao L, Choi EY, Liu X, Martin A, Wang C, Xu X, et al. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell metabolism. 2011;14(3):324–38. doi: 10.1016/j.cmet.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaslaver A, Liani I, Shtangel O, Ginzburg S, Yee L, Sternberg PW. Hierarchical sparse coding in the sensory system of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(4):1185–9. doi: 10.1073/pnas.1423656112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsch J, Ventimiglia D, Bargmann CI, Albrecht DR. High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2013;110(45):E4266–E73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402(6763):804–9. doi: 10.1038/45544 [DOI] [PubMed] [Google Scholar]
- 14.Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of Bacterial Secondary Metabolites Modulates Neuroendocrine Signaling and Behavior of C. elegans. Cell. 2014;159(2):267–80. doi: 10.1016/j.cell.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, et al. Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell. 2016;166(6):1553–63.e10. doi: 10.1016/j.cell.2016.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greer ER, Pérez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and Molecular Dissection of a C. elegans Sensory Circuit that Regulates Fat and Feeding. Cell Metabolism. 2008;8(2):118–31. doi: 10.1016/j.cmet.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neal SJ, Park J, DiTirro D, Yoon J, Shibuya M, Choi W, et al. A Forward Genetic Screen for Molecules Involved in Pheromone-Induced Dauer Formation in Caenorhabditis elegans. G3 (Bethesda, Md). 2016;6(5):1475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(9):5854–9. doi: 10.1073/pnas.092064799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brock TJ, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176(2):865–75. doi: 10.1534/genetics.107.071860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, et al. Comparative Metabolomics Reveals Biogenesis of Ascarosides, a Modular Library of Small-Molecule Signals in C. elegans. Journal of the American Chemical Society. 2012;134(3):1817–24. doi: 10.1021/ja210202y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroeder FC. Modular assembly of primary metabolic building blocks: a chemical language in C. elegans. Chemistry & biology. 2015;22(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Riordan VB, Burnell AM. Intermediary metabolism in the dauer larva of the nematode Caenorhabditis elegans—1. Glycolysis, gluconeogenesis, oxidative phosphorylation and the tricarboxylic acid cycle. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry. 1989;92(2):233–8. [Google Scholar]
- 23.Holt SJ, Riddle DL. SAGE surveys C. elegans carbohydrate metabolism: evidence for an anaerobic shift in the long-lived dauer larva. Mechanisms of Ageing and Development. 2003;124(7):779–800. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130(8):1621–34. [DOI] [PubMed] [Google Scholar]
- 25.van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS biology. 2005;3(2):e53 doi: 10.1371/journal.pbio.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKay RM, McKay JP, Avery L, Graff JM. C. elegans: A Model for Exploring the Genetics of Fat Storage. Developmental Cell. 2003;4(1):131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454(7208):1115–8. doi: 10.1038/nature07168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiavi A, Torgovnick A, Kell A, Megalou E, Castelein N, Guccini I, et al. Autophagy induction extends lifespan and reduces lipid content in response to frataxin silencing in C. elegans. Experimental gerontology. 2013;48(2):191–201. doi: 10.1016/j.exger.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witham E, Comunian C, Ratanpal H, Skora S, Zimmer M, Srinivasan S. C. elegans body cavity neurons are homeostatic sensors that integrate fluctuations in oxygen availability and internal nutrient reserves. Cell reports. 2016;14(7):1641–54. doi: 10.1016/j.celrep.2016.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roayaie K, Crump JG, Sagasti A, Bargmann CI. The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20(1):55–67. [DOI] [PubMed] [Google Scholar]
- 31.Jansen G, Thijssen KL, Werner P, van derHorst M, Hazendonk E, Plasterk RHA. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21(4):414–9. doi: 10.1038/7753 [DOI] [PubMed] [Google Scholar]
- 32.Reynolds NK, Schade MA, Miller KG. Convergent, RIC-8-dependent Gα signaling pathways in the Caenorhabditis elegans synaptic signaling network. Genetics. 2005;169(2):651–70. doi: 10.1534/genetics.104.031286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringstad N. A Controlled Burn: Sensing Oxygen to Tune Fat Metabolism. Cell Rep. 2016;14(7):1569–70. doi: 10.1016/j.celrep.2016.02.015 [DOI] [PubMed] [Google Scholar]
- 34.Yemini E, Jucikas T, Grundy LJ, Brown AE, Schafer WR. A database of Caenorhabditis elegans behavioral phenotypes. Nature methods. 2013;10(9):877–9. doi: 10.1038/nmeth.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Kong J, Jang JY, Han JS, Ji Y, Lee J, et al. Lipid droplet protein LID-1 mediates ATGL-1-dependent lipolysis during fasting in Caenorhabditis elegans. Molecular and cellular biology. 2014;34(22):4165–76. doi: 10.1128/MCB.00722-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakrabarti P, V. Kandror K. Adipose Triglyceride Lipase: A New Target in the Regulation of Lipolysis by Insulin. Current Diabetes Reviews. 2011;7(4):270–7. [DOI] [PubMed] [Google Scholar]
- 37.Saifee O, Metz LB, Nonet ML, Crowder CM. A gain-of-function mutation in adenylate cyclase confers isoflurane resistance in Caenorhabditis elegans. The Journal of the American Society of Anesthesiologists. 2011;115(6):1162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwaal RR, Mendel JE, Sternberg PW, Plasterk RH. Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans Dauer-inducing pheromone. Genetics. 1997;145(3):715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lans H, Rademakers S, Jansen G. A network of stimulatory and inhibitory Gα-subunits regulates olfaction in Caenorhabditis elegans. Genetics. 2004;167(4):1677–87. doi: 10.1534/genetics.103.024786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinke V, Krause M, Okkema P. Transcriptional regulation of gene expression in C. elegans. WormBook: the online review of C elegans biology. 2013:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esposito G, Di Schiavi E, Bergamasco C, Bazzicalupo P. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene. 2007;395(1–2):170–6. doi: 10.1016/j.gene.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 42.Korswagen HC, Park J-H, Ohshima Y, Plasterk R. An activating mutation in a Caenorhabditis elegans Gs protein induces neural degeneration. Genes & development. 1997;11(12):1493–503. [DOI] [PubMed] [Google Scholar]
- 43.Schade MA, Reynolds NK, Dollins CM, Miller KG. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the Gαs pathway and define a third major branch of the synaptic signaling network. Genetics. 2005;169(2):631–49. doi: 10.1534/genetics.104.032334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walent JH, Porter BW, Martin TFJ. A novel 145 kd brain cytosolic protein reconstitutes Ca2+-regulated secretion in permeable neuroendocrine cells. Cell. 1992;70(5):765–75. [DOI] [PubMed] [Google Scholar]
- 45.Berwin B, Floor E, Martin TFJ. CAPS (Mammalian UNC-31) Protein Localizes to Membranes Involved in Dense-Core Vesicle Exocytosis. Neuron. 1998;21(1):137–45. [DOI] [PubMed] [Google Scholar]
- 46.Lin X-G, Ming M, Chen M-R, Niu W-P, Zhang Y-D, Liu B, et al. UNC-31/CAPS docks and primes dense core vesicles in C. elegans neurons. Biochemical and biophysical research communications. 2010;397(3):526–31. doi: 10.1016/j.bbrc.2010.05.148 [DOI] [PubMed] [Google Scholar]
- 47.Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nature neuroscience. 1999;2(11):959–64. doi: 10.1038/14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madison JM, Nurrish S, Kaplan JM. UNC-13 Interaction with Syntaxin Is Required for Synaptic Transmission. Current Biology. 2005;15(24):2236–42. doi: 10.1016/j.cub.2005.10.049 [DOI] [PubMed] [Google Scholar]
- 49.Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, et al. A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rand JB. Acetylcholine. 2007.
- 51.Johnson CD, Duckett JG, Culotti JG, Herman RK, Meneely PM, Russell RL. An acetylcholinesterase-deficient mutant of the nematode Caenorhabditis elegans. Genetics. 1981;97(2):261–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Culotti JG, Von Ehrenstein G, Culotti MR, Russell RL. A second class of acetylcholinesterase-deficient mutants of the nematode Caenorhabditis elegans. Genetics. 1981;97(2):281–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson CD, Rand JB, Herman RK, Stern BD, Russell RL. The acetylcholinesterase genes of C. elegans: identification of a third gene (ace-3) and mosaic mapping of a synthetic lethal phenotype. Neuron. 1988;1(2):165–73. [DOI] [PubMed] [Google Scholar]
- 54.McKay JP, Raizen DM, Gottschalk A, Schafer WR, Avery L. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics. 2004;166(1):161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avery L, You Y-J. C. elegans feeding. WormBook: the online review of C elegans biology. 2012:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83(2):207–18. [DOI] [PubMed] [Google Scholar]
- 57.Sambongi Y, Nagae T, Liu Y, Yoshimizu T, Takeda K, Wada Y, et al. Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport. 1999;10(4):753–7. [DOI] [PubMed] [Google Scholar]
- 58.Hilliard MA, Bergamasco C, Arbucci S, Plasterk RH, Bazzicalupo P. Worms taste bitter: ASH neurons, QUI‐1, GPA‐3 and ODR‐3 mediate quinine avoidance in Caenorhabditis elegans. The EMBO journal. 2004;23(5):1101–11. doi: 10.1038/sj.emboj.7600107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O 2 responses and aggregation in C. elegans. Current Biology. 2006;16(7):649–59. doi: 10.1016/j.cub.2006.03.023 [DOI] [PubMed] [Google Scholar]
- 60.de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. 2002;419(6910):899–903. doi: 10.1038/nature01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jang H, Kim K, Neal Scott J, Macosko E, Kim D, Butcher Rebecca A, et al. Neuromodulatory State and Sex Specify Alternative Behaviors through Antagonistic Synaptic Pathways in C. elegans. Neuron. 2012;75(4):585–92. doi: 10.1016/j.neuron.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golden JW, Riddle DL. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science. 1982;218(4572):578–80. [DOI] [PubMed] [Google Scholar]
- 63.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes & development. 2008;22(16):2149–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, Mangelsdorf DJ, et al. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Developmental cell. 2006;10(4):473–82. doi: 10.1016/j.devcel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 65.Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, Schroeder FC, et al. Ascaroside signaling is widely conserved among nematodes. Current Biology. 2012;22(9):772–80. doi: 10.1016/j.cub.2012.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, et al. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron. 2012;75(4):585–92. doi: 10.1016/j.neuron.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, et al. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science (New York, NY). 2009;326(5955):994–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng S-W, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140(1):49–61. doi: 10.1016/j.cell.2009.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nawa M, Kage-Nakadai E, Aiso S, Okamoto K, Mitani S, Matsuoka M. Reduced expression of BTBD10, an Akt activator, leads to motor neuron death. Cell Death & Differentiation. 2012;19(8):1398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srinivasan S, Sadegh L, Elle IC, Christensen AG, Faergeman NJ, Ashrafi K. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell metabolism. 2008;7(6):533–44. doi: 10.1016/j.cmet.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Images of wild-type animals and gpa-3 mutants fixed and stained with Oil Red O. Animals are oriented facing upwards, and the head and intestinal cells are as marked. Oil Red O stained droplets are indicated. For each genotype, images depict the full range of the observed phenotype. (B) The integrated density of the lipid droplets is used to quantify body fat stores, as described in the Materials and Methods. Graph represents the integrated density values of individual wild-type animals and gpa-3 mutants. ***, p<0.001 by Student’s t-test. (C) Representative images of wild-type animals and gpa-3 mutants exposed to vector control or atgl-1 RNAi fixed and stained with Oil Red O. The model depicts the section of anterior intestine being represented for each genotype and condition.
(TIF)
(A) Animals were transferred at L4 to plates containing either M9 vehicle or 20, 200, or 500μM 8-Bromoadenosine 3′,5′-cyclic monophosphate (8-Br-cAMP), then fixed and stained with Oil Red O. Fat content was quantified for each condition and is expressed as a percentage of vehicle-treated wild-type animals ± SEM (n = 12). ***, p<0.001 by one-way ANOVA. (B) Model depicting epistatic relationship between the Go/I protein GPA-3 and the adenylyl cyclase ACY-1 for the regulation of cAMP. (C) Representative images of wild-type animals, gpa-3, acy-1(nu329), and gpa-3;acy-1 mutants fixed and stained with Oil Red O.
(TIF)
(A) gpa-3 mutants bearing gpa-3 expression using a 5kb or a 7kb endogenous promoter were fixed and stained with Oil Red O, as indicated. Relative to non-transgenic controls (-, light gray bars), transgenic animals (+, dark gray bars) bearing the gpa-3 transgene restored body fat content to the same extent, whether driven by the 5kb or 7kb promoter. Data are expressed as a percentage of body fat in wild-type animals ± SEM (n = 12–16). ***, p<0.001 by one-way ANOVA. (B) Food intake for gpa-3 mutants bearing gpa-3 expression using a 5kb or a 7kb endogenous promoter was measured. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; ***, p<0.001 by one-way ANOVA. (C) Food intake for wild-type animals, gpa-3, acy-1(nu329), and gpa-3;acy-1 mutants was measured. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; ***, p<0.001 by one-way ANOVA. (D) Food intake for gpa-3 mutants bearing gpa-3 expression using the indicated promoter was measured. Data are expressed as a percentage of wild-type animals ± SEM (n = 10). NS, not significant; ***, p<0.001 by one-way ANOVA.
(TIF)
(A-C) Representative images of all genotypes fixed and stained with Oil Red O.
(TIF)
(A-E) Representative images of all genotypes fixed and stained with Oil Red O.
(TIF)
Representative images of all genotypes and conditions fixed and stained with Oil Red O.
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.