Abstract
Pseudomonas aeruginosa possesses two aerotaxis transducers, Aer and Aer-2. A deletion-insertion mutation of alternative sigma factor RpoS eliminated Aer-2-mediated aerotaxis but not Aer-mediated aerotaxis. Transcriptional analysis revealed that cheY2, cheA2, cheW2, and aer-2 were expressed in an RpoS-dependent manner as a single transcript.
An aerotactic response in Pseudomonas aeruginosa PAO1 (10) has previously been characterized as the movement of a cell toward oxygen (18). In P. aeruginosa PAO1, chemotaxis proteins such as CheA, CheB, CheR, CheW, and CheY are required and two methyl-accepting chemotaxis proteins, Aer and Aer-2, function as independent sensor-transducers for aerotaxis (11). In Escherichia coli, two methyl-accepting chemotaxis proteins, Aer and Tsr, independently sense and transduce aerotactic signals (2, 17). Recently, it was demonstrated that E. coli Aer is a methylation-independent transducer (3) and the PAS domain of E. coli Aer requires the C-terminal HAMP domain for native-fold formation and flavin adenine dinucleotide binding (9, 21).
The aerotactic responses of P. aeruginosa cells were induced during the transition from exponential to stationary growth phase (12). In the previous study, we demonstrated that aer is transcriptionally regulated by the anaerobic regulator ANR (12). ANR is involved in the anaerobic induction of various enzymatic systems, including those required for arginine fermentation, cyanogenesis, and denitrification (1, 7, 8, 23). ANR activates target promoters by binding to ANR boxes, the consensus sequences shared by the ANR-dependent promoters (1, 8). The aer promoter contains two ANR boxes at −42.5 and −93.5 bp upstream of the transcriptional start site of aer, and both of them are essential for expression of the aer gene (12). The anr mutation eliminated Aer-mediated aerotaxis, but not Aer-2-mediated aerotaxis, suggesting that aer-2 expression is regulated by a factor other than ANR. In the present study, we report that the alternative sigma factor RpoS is required for Aer-2-mediated aerotaxis and the transcription of aer-2 is dependent on RpoS.
The sigma factor RpoS is known to have a role in regulating the expression of stationary-phase genes in a wide range of bacteria, including P. aeruginosa (13, 19). To assess the possibility that RpoS is involved in the stationary induction of Aer-2-mediated aerotaxis, the rpoS gene was disrupted by inserting a tet (conferring tetracycline resistance) cassette (22) into the wild-type gene in the P. aeruginosa PAO1 genome as described previously (14). The resulting rpoS mutant, designated PAO-CH1, was fully motile and grew as well as the parent strain, PAO1. PAO-CH1 was examined for the ability to exhibit aerotaxis.
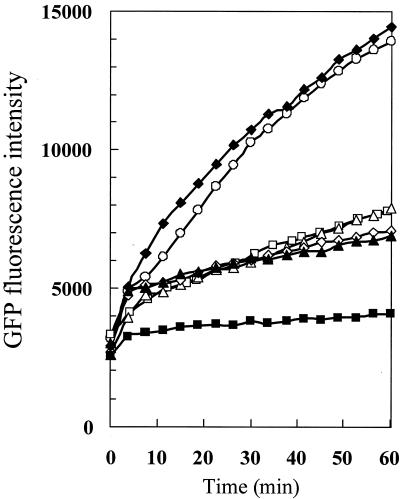
Aerotactic responses of P. aeruginosa were assessed by the chemotaxis well chamber method (18). Stationary-phase cells of the rpoS mutant PAO-CH1 harboring the green fluorescent protein expression vector pMRP9-1 (16) exhibited decreased but significant aerotaxis. The entire rpoS gene (19) was cloned into pMRP9-1 to construct pCSH9-11. Plasmid pCSH9-11 complemented the mutation of PAO-CH1 (Fig. 1), showing that the mutation phenotype was not due to polar effects. The intensity of aerotaxis by PAO-CH1 was as strong as those of the aer and aer-2 single mutants (11) (Fig. 1). These results suggest the possibility that RpoS regulates only one of the aerotaxis transducer genes. To confirm this possibility, we constructed the aer rpoS and aer-2 rpoS double mutants by inserting a kan (conferring kanamycin resistance) cassette into the wild-type aer and aer-2 genes in the PAO-CH1 genome, respectively. The aer rpoS and aer-2 rpoS double mutants were designated PAO-CH2 and PAO-CH3, respectively. Aerotaxis assays revealed that PAO-CH2 failed to exhibit aerotaxis, whereas PAO-CH3 showed the same level of aerotactic responses as the aer-2 single mutant (Fig. 1). These results demonstrated that Aer-2-mediated aerotaxis, but not Aer-mediated aerotaxis, requires RpoS.
FIG. 1.
Aerotactic responses by wild-type and mutant strains of P. aeruginosa. Changes in the green fluorescent protein (GFP) fluorescence intensity of the upper well were measured by a fluorescence spectrophotometer. Symbols: ○, PAO1(pMRP9-1) (wild type); ▵, TLPC01(pMRP9-1) (aer mutant) (11); □, TLPG01(pMRP9-1) (aer-2 mutant) (11); ⋄, PAO-CH1(pMRP9-1) (rpoS mutant); ▴, PAO-CH3(pMRP9-1) (aer-2 rpoS mutant); ▪, PAO-CH2(pMRP9-1) (aer rpoS mutant); ♦, PAO-CH1(pCSH9-11).
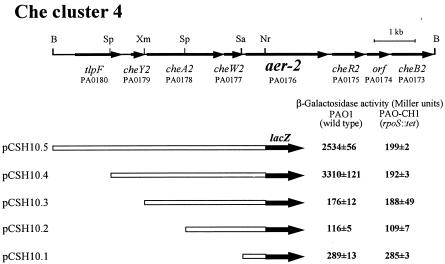
The aer-2 gene is located near an mcp-like gene (tlpF) and a complete set of chemotaxis-like genes (cheY2, cheA2, cheW2, cheR2, and cheB2) encoding homologues of CheY, CheA, CheW, CheR, and CheB (11) (Fig. 2). These genes are juxtaposed and have the same transcriptional polarity. tlpF, cheY2, cheA2, and cheW2 are located upstream of aer-2. Their coding regions overlap or are separated by short intergenic regions (27 to 197 bp), suggesting that these genes are expressed as a single transcript. To locate the promoter of the transcript containing aer-2, we constructed promoter fusions in the broad-host-range transcriptional fusion vector pQF50 (5). Regions upstream of aer-2 were isolated and inserted individually upstream from the promoterless lacZ gene in pQF50 (Fig. 2). Each of five constructs was transformed into PAO1 and PAO-CH1. β-Galactosidase activities were then measured in stationary-phase cells of transformants at 37°C (15). High levels of β-galactosidase activity were detected with PAO1(pCSH10.5 [carrying tlpF cheY2A2W2 aer-2::lacZ]) and PAO1(pCSH10.4 [carrying cheY2A2W2 aer-2::lacZ]) (Fig. 2). In PAO1 harboring pCSH10.3 (carrying cheA2W2 aer-2::lacZ), pCSH10.2 (carrying cheW2 aer-2::lacZ), and pCSH10.1 (carrying aer-2::lacZ), β-galactosidase levels were about 150 U, which was similar to the basal levels seen in PAO1 harboring the control plasmid pQF50. These results suggest that cheY2, cheA2, cheW2, and aer-2 are expressed as a single transcript and the transcript starts from the region upstream of cheY2. The lacZ fusion pCSH10.4 gave a basal level of β-galactosidase activity in PAO-CH1, demonstrating that RpoS is required for transcription of the cheY2A2W2 and aer-2 genes.
FIG. 2.
Physical map of Che cluster 4 of P. aeruginosa PAO1 (11) and regions cloned in the promoter probe vector pQF50. Specific restriction sites used to isolate each fragment are shown on the map. Restriction sites: B, BamHI; Nr, NruI; Sa, Sau3AI; Sp, SphI; Xm, XmnI. The locations and orientations of tlpF, cheY2, cheA2, cheW2, aer-2, cheR2, PA0174, and cheB2 are indicated by horizontal arrows. Gene identification numbers used in the P. aeruginosa genome sequencing project (http://www.pseudomonas.com/) are indicated below the gene names. Open bars are P. aeruginosa chromosomal DNA fragments subcloned into pQF50. The lacZ gene is shown by the black arrow. β-Galactosidase activities were determined in P. aeruginosa wild-type PAO1 and its rpoS mutant, PAO-CH1, containing the aer-2::lacZ transcriptional fusion plasmids shown. β-Galactosidase activity is shown along with the standard deviation (mean of four independent experiments).
To identify the 5′ end of the mRNA containing cheY2A2W2 and are-2, we carried out rapid amplification of cDNA ends (RACE) as described previously (6, 12). cheY2 sequence-specific primer Y2SPI (5′-GCGTCACTCGAGCAGTTTC-3′) was used for reverse transcription of the total RNA from PAO1. A nested PCR using forward primers T17ADP (5′-GAGTCGACTCGAGAATTCTTTTTTTTTTTTTTTTT-3′) and ADP (5′-GAGTCGACTCGAGAATTC-3′) and cheY2 sequence-specific primers Y2SPII (5′-TTCATCGCGTCGCTCGATTC-3′) and Y2SPIII (5′-ATTCGGTGGTCAGCATGAGG-3′) enabled amplification of a RACE product (data not shown) that was subsequently isolated and subcloned into pUC118 (20). DNA sequencing indicated that the 5′ ends were located at an A nucleotide at position −101 and a G nucleotide at position −100 (relative to the cheY2 start codon) (Fig. 3). A potential RpoS −10 region (CTTTACT) was located at positions −13 to −7 upstream of the transcription start point (the A nucleotide) (Fig. 3), with six of the seven bases being identical to the consensus sequence (CTATACT) (4).
FIG. 3.
Promoter region of P. aeruginosa cheY2. The transcriptional start sites, which were determined by 5′ RACE and DNA sequencing, are indicated by the angled arrow. Numbering is relative to the transcriptional start site (the A nucleotide). The putative translation start site, ribosome-binding site (RBS), and −35 promoter sequence are underlined. Double underlining indicates a putative RpoS −10 region.
In summary, P. aeruginosa possesses two aerotaxis transducers, Aer and Aer-2. aer expression is dependent on the anaerobic regulator ANR, which is converted to its active form under low oxygen supply. aer-2 is transcribed together with cheY2A2W2 (and probably with cheR2B2). The stationary-phase sigma factor RpoS is required for transcription of the operon.
REFERENCES
- 1.Arai, H., M. Mizutani, and Y. Igarashi. 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149:29-36. [DOI] [PubMed] [Google Scholar]
- 2.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikov, S. I., A. C. Miller, K. K. Gosink, and J. S. Parkinson. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus sequence structure for σS-dependent promoters. Mol. Microbiol. 21:657-659. [DOI] [PubMed] [Google Scholar]
- 5.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frohman, M. A., M. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galimand, M., M. Gamper, A. Zimmermann, and D. Haas. 1991. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, D., M. Gamper, and A. Zimmermann. 1992. Anaerobic control in Pseudomonas aeruginosa, p. 177-187. In E. Galli, S. Silver, and B. Witholt (ed.), Pseudomonas: molecular biology and biotechnology. American Society for Microbiology, Washington, D.C.
- 9.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong, C. S., M. Shitashiro, A. Kuroda, T. Ikeda, N. Takiguchi, H. Ohtake, and J. Kato. 2004. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 231:247-252. [DOI] [PubMed] [Google Scholar]
- 12.Hong, C. S., A. Kuroda, T. Ikeda, N. Takiguchi, H. Ohtake, and J. Kato. 2004. The aerotaxis transducer gene aer, but not aer-2, is transcriptionally regulated by the anaerobic regulator ANR in Pseudomonas aeruginosa. J. Biosci. Bioeng. 97:184-190. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen, F., M. Bally, V. Chapomherve, G. Michel, A. Lazdunski, P. Williams, and G. S. Stewart. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835-844. [DOI] [PubMed] [Google Scholar]
- 14.Kato, J., T. Nakamura, A. Kuroda, and H. Ohtake. 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63:151-161. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Parsek, M. R., and E. P. Greenberg. 1999. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol. 31:43-55. [DOI] [PubMed] [Google Scholar]
- 17.Rebbapragata, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shitashiro, M., T. Fukumura, J. Kato, A. Kuroda, T. Ikeda, N. Takiguchi, and H. Ohtake. 2003. Evaluation of bacterial aerotaxis for its potential use in detecting the toxicity of chemicals to microorganisms. J. Biotechnol. 101:11-18. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, K., and H. Takahashi. 1994. Cloning, analysis and expression of an rpoS homolog gene from Pseudomonas aeruginosa PAO1. Gene 150:81-85. [DOI] [PubMed] [Google Scholar]
- 20.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 21.Watts, K. J., Q. Ma, M. S. Johnson, and B. L. Taylor. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, H., J. Kato, A. Kuroda, T. Ikeda, N. Takiguchi, and H. Ohtake. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 182:3400-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann, A., C. Reimmann, M. Galimand, and D. Haas. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulator gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5:1483-1490. [DOI] [PubMed] [Google Scholar]