Abstract
CYP26A1 expression is very highly induced by retinoic acid (RA) in the liver, compared to most other tissues, suggesting that a liver-enriched factor may be required for its physiological transcriptional response. HNF4α is a highly conserved liver-specific/enriched member of nuclear receptor superfamily. In this study, we hypothesized that HNF4α and RARs may cooperate in an RA-dependent manner to induce a high level of CYP26A1 expression in liver cells. Partial inhibition of endogenous HNF4α by siRNA reduced the level of RA-induced CYP26A1 mRNA in HepG2 cells. Cotransfection of HNF4α, with or without RARs, demonstrated RA-dependent activation of a human CYP26A1 promoter-luciferase construct. Analysis of a 2.5-kbp putative CYP26A1 promoter sequence identified five potential HNF4α DNA response elements: H1 located in a proximal region overlapping with an RAR element-1 (RARE1 or R1); H2 and H3 in the distal region, close to RARE2 (R2) and RARE3 (R3); and H4 and H5 in intermediary regions. In EMSA and ChIP analyses HNF4α and RARs binding in the proximal and distal CYP26A1 promoter regions was significantly higher in RA-treated cells. Mutational analysis of the individual HNF4α DNA-response elements identified H1 as the major site for HNF4α binding because mutation of H1 inhibited the promoter activity by ~90%, followed by H2 mutation with less than 40% inhibition. Our results indicate that HNF4α coordinates with RARs in an RA-dependent manner to strongly induce CYP26A1 gene expression in the liver, which may explain the high level of response to RA observed in vivo.
Keywords: TRANSCRIPTION FACTOR, HEPATOCYTE, RETINOIC ACID RESPONSE ELEMENT, LIVER-SPECIFIC GENE EXPRESSION
CYP26A1, a member of the cytochrome P450 superfamily, catalyzes the oxidative inactivation of all-trans-retinoic acid (RA), the major hormonal metabolite of vitamin A. RA regulates many physiological processes through its function as a pan-agonist ligand of nuclear retinoic acid receptors (RARα, β, and γ), which partner with retinoid X receptors (RXRα, β, and γ) and bind specifically to retinoic acid response elements (RARE) in target genes [Wei, 2003; Bastien and Rochette-Egly, 2004; Altucci et al., 2007]. The canonical RARE consists of a core of two hexameric motifs of PuG(G/T)TCA(X)nPuG(G/T)TCA, generally oriented as a direct repeat (DR) spaced by two or five nucleotides [Wei, 2003; Bastien and Rochette-Egly, 2004; Altucci et al., 2007]. The intracellular levels of RA are regulated both by its synthesis from retinol, catalyzed by several dehydrogenases, and by catabolism [Napoli, 2012], for which the CYP26 family, including CYP26A1, is considered to play a major role [Ross and Zolfaghari, 2011].
Not only is RA the specific substrate for CYP26A1 but also a potent inducer of CYP26A1 gene expression [Pennimpede et al., 2010; Ross and Zolfaghari, 2011]. Results from analysis of the functional promoter region of the CYP26A1 gene have identified at least four RAREs, one a DR5 in the proximal promoter region, and two other DR5 elements plus one half-site located in a cluster more distal from the CYP26A1 transcription start site [Loudig et al., 2000; Loudig et al., 2005; Zhang et al., 2010]. The induction of CYP26A1 gene expression exhibits tissue specificity as it is more highly induced by RA in the liver than in several other tissues [Wang et al., 2002], while in some cell types it is not induced at all [Ross and Zolfaghari, 2011]. In the liver, CYP26A1 appears to be exclusively expressed in hepatocytes [Ross et al., 2011]. Previously, we reported that treatment of vitamin A-deficient rats with RA resulted in a rapid increase in CYP26A1 mRNA levels in the liver, reaching 2000-fold over the control level, which is normally low, within few hours [Wang et al., 2002]. In addition, hepatic expression of CYP26A1 mRNA was highly correlated with the intake of dietary vitamin A and with liver retinol concentrations, an well-accepted indicator of whole-body vitamin A status [Yamamoto et al., 2000], and that increased CYP26A1 expression after RA treatment was due to activation of the transcriptional process [Zolfaghari et al., 2007; Zolfaghari and Ross, 2009].
The potent induction of CYP26A1 expression by RA in the liver suggests that its physiological transcriptional response may require a liver-enriched factor. One candidate is hepatocyte nuclear factor4-alpha (HNF4α), or NR2A1 [Huang et al., 2014], which is a highly conserved member of the nuclear receptor superfamily of ligand-dependent transcriptional factors [Bolotin et al., 2010]. HNF4α is indispensible during embryonic development as mice lacking HNF4α died before completion of gastrulation [Li et al., 2000; Hayhurst et al., 2001]. In tissues of the adult human, HNF4α is expressed at high levels in liver, small intestine, and kidney and at lower levels in pancreas and colon [Drewes et al., 1996]. HNF4α is a key regulator of numerous hepatocyte genes that play important roles in lipid and glucose metabolism, and in the catabolism of xenobiotics and drugs particularly by cytochrome P450 enzymes [Jover et al., 2001; Holloway et al., 2008; Hwang-Verslues and Sladek, 2010]. Similar to other members of the nuclear receptor superfamily HFN4α contains a DNA binding domain and a ligand binding domain, HNF4α has been shown to bind exclusively to the sequence NNNNCAAAGTCCA [Fang et al., 2012]. Linoleic acid, an essential fatty acid, was recently identified as a reversible physiological ligand for HNF4α; however, ligand occupancy was not shown to have a significant effect on HNF4α transcriptional activity [Yuan et al., 2009]. HNF4α is associated with several human diseases including diabetes, hemophilia, hepatitis, atherosclerosis, and inflammatory bowel diseases [Bolotin et al., 2010].
Whereas the null mutation of the HNF4α gene is embryonic lethal [Li et al., 2000], the creation of a conditional liver-specific knockout of HNF4α has allowed for development through early adulthood [Hayhurst et al., 2001]. Using this model in a gene array analysis [Holloway et al., 2008], CYP26A1 mRNA levels were decreased by more than 50% in the liver of both male and female HNF4α-deficient mice as compared to wild type (WT) mice. In addition, a protein binding microarray analysis using a human liver cell line identified potential DNA binding regions for HNF4α in the putative promoter region of CYP26A1 gene [Hwang-Verslues and Sladek, 2010]. Based on these previous results, our overall objectives in the present study were to investigate whether HNF4α is a regulator of the expression of the CYP26A1 gene in liver cells. Using HepG2 human hepatocarcinoma cells as a hepatocyte model [Knowles et al., 1980], we found that RA up-regulates the expression of the HNF4α gene with kinetics similar to that for RA induction of the CYP26A1 gene, and that HNF4α, with an apparent increase in binding, enhances the promoter activity of the CYP26A1 gene in RA-treated HepG2 cells.
MATERIALS AND METHODS
MATERIALS
HepG2 cells, and human embryonic kidney HEK293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) and 0.5% penicillin-streptomycin at 37 °C in a 5% CO2-air incubator. The cells were plated and used at 80% confluency.
RA was purchased from Sigma–Aldrich Chemical Company and prepared as stocks at 1 and 10 mM in ethanol. Rabbit polyclonal IgG antibodies against RARα, RARβ, RXRα, Pol II (H-224), and mouse monoclonal antibody against RARγ, mouse monoclonal IgG2a and goat polyclonal IgG antibodies against HNF4α, and a goat polyclonal IgG goat anti-mouse IgG2a-HRP were all from Santa Cruz Biotechnology (Santa Cruz, CA). SuperSignal® West Pico Reagent containing an enhanced chemoluminescent substrate for detection of horseradish peroxidase as well as HNF4α siRNA together with non-targeting siRNA and DharmaFECT Transfection Reagents were from Thermo Scientific (Rockford, IL). pGL3-Basic-luc, pRLTK, and pGEMT-Easy plasmid vectors were purchased from Promega (Madison, WI) and pcDNA3.1 plasmid vector for expression from Invitrogen (Carsbad, CA). Mouse RARα, β, and γ and RXRα clones, each in pSG5 expression plasmid vector, were provided by Dr. Pierre Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire, CNRS/INSERM/ULP/Collège de France, Illkirch-Cedex, Strasbourg, France). MMLV Reverse transcriptase, oligo dT nucleotide, and T4 polynucleotide kinase were from Promega, High Fidelity Tag DNA polymerase, Lipofectamine 2000 transfection reagent and Trizol reagent were all from Invitrogen Biotechnology. Reagents for RT-qPCR was purchased from BioRad (Hercules, CA), plasmid DNA Purification Kits were from Qiagen (Valencia, CA), and DNA gel extraction kit was from Omega Bio-Tek (Thermo Scientific). Protease inhibitor cocktail and G25 column were purchased from Roche (Indianapolis, IN) and X-ray film was from VWR International (University Park, PA). All the restriction enzymes were obtained from New England Biolab (Igswich, MA).
The full-length (FL) promoter of CYP26A1 gene was cloned from a human CYP26A1 genomic clone containing the extended 5′- and 3′-flanking regions in FOSMID vector (obtained from Children’s Hospital Oakland-BACPAC Resources, Oakland, CA) and the deleted CYP26A1 promoter clones in pGL3-Basic-luc vector were reported previously [Zhang et al., 2010]. [γ-32P]ATP was purchased from GE Healthcare (UK). For preparation of all other reagents in our laboratory, we followed the protocols reported in Sambrook and Russell [2001].
RNA EXTRACTION AND ANALYSIS
Total RNA was extracted from the cells using Trizol reagent, re-precipitated in sodium acetate, and then dissolved in DEPC-treated autoclaved dH2O for analysis. The RNA samples were quantified by Nano-Drop spectrophotometry and analyzed for CYP26A1 and HNF4α mRNA transcript levels with 18 S rRNA as an internal control by reverse transcriptase real-time PCR (qPCR) using primer pair 5′-GCTGCCTCTCTAACCTGCAC-3′ as sense and 5′-TGCTTTAGTGCCTGCATGTC-3′ as antisense for CYP26A1; two pairs of primers, 5′-AGAATGTGCAGGTGTTGACG-3′ as sense and CTCGAGGCACCGTAGTGTTT, as antisense in pair 1, 5′-GAGCTGCAGATCGATGACAA-3′ as sense and5′-TACTGGCGGTCGTTGATGTA-3′ as antisense in pair 2 for HNF4α; and 5′-CGCGGTTCTATTTTGTTGGT-3′ as sense and 5′-AGTCGGCATCGTTTATGGTC-3′ as antisense for 18 S rRNA. The PCR program was set to run first at 94 °C for 3 min for activation of the polymerase and then 40 cycles of 20 s at 94 °C for melting, 30 s at 60 °C for annealing, 30 s at 72 °C for extension and 1 s at 75 °C for reading.
RNA INTERFERENCE
HepG2 cells were incubated in triplicate in 24-well plates with 5% FBS medium without antibiotics and then transfected with either human HNF4α siRNA or non-targeting siRNA, as the control, overnight according to the protocol furnished by Thermo Scientific as the manufacturer. The cells were washed with PBS and incubated with either ethanol as the vehicle or RA for 4 h. The cells were washed with PBS and then subjected to either extraction for total RNA or total proteins.
CLONING AND PREPARATION OF THE HUMAN HNF4A EXPRESSION VECTOR AND CYP26A1 PROMOTER CONSTRUCTS
Poly A+ RNA (0.5 μg) from human liver samples [Zolfaghari and Ross, 2004] was first reverse transcribed using MMLV reverse transcriptase and oligo dT from Promega in a 25 μl reaction mixture. Following dilution to 100 μl with deionized water, 5 μl was used to amplify the open reading frame of HNF4α cDNA with 5′-TTGGATCCGCCACCATGCGACTCTCCAAAACCC-3′ and 5′-TTTCTAGACTAGATAACTTCCTGCTTGG-3′ (underlined letters indicate the restriction sites for cloning) sense and antisense primers respectively, using High Fidelity Taq DNA polymerase from Invitrogen following the manufacturer protocol for 30 cycles. The PCR product was run on agarose gel by electrophoresis and the DNA band was cut and extracted from the gel using DNA gel extraction kit. The DNA was first double digested with Bam HI/XbaI and cloned into the pcDNA3.1(+) expression vector. The human HNF4α expression vector was first transformed into JM109 bacteria for screening and amplification and then purified using Plasmid Midi Kit from Qiagen. The DNA expression plasmid was subjected to sequencing for confirmation in the Nucleic Acid Facility, Pennsylvania State University.
A 2.5 kbp fragment of DNA spanning from −2498 bp to −1 upstream of the ATG codon of the human CYP26A1 gene was cloned by PCR from human FOSMID clone containing human CYP26A1 gene using 5′-TTGCTAGCTGTGACTCCAGTCTAGTCC-3′ and 5′-TTCTCGAGGCGCGCCGCGACCT-3′ as forward and reverse primers, respectively and High Fidelity Taq DNA polymerase as described above. The DNA fragment as the full length CYP26A1 promoter DNA was then isolated, double digested with NheI/XhoI, and then cloned into pGL3-Basic-Luciferase. The DNA deleted CYP26A1 promoter clones in pGL3-Basic-Luc including E1, E2, E3, E4, E5, and E6, were all from previous study [Zhang et al., 2010]. The CYP26A1 promoter clones with individual mutated RAR regions in pGL3-Basic-Luc including R1, R2, R3, and R4 were all previously reported [Zhang et al., 2010]. The CYP26A1 mutant promoter constructs in HNF4α nucleotide sequences (H1–H5) were generated by PCR following the protocol described previously [Zolfaghari and Ross, 2009]. Primers used for mutant constructs were: pair 1: 5′-TTGCTAGCTGTGACTCCAGTCTAGTCC-3′ (forward), 5′-AAGTTTAAACTTAAATTTTTACTCGCCAAGCTTTCGTGGG-3′ (reverse); pair 2: 5′-AAGTTTAAACTTTTAACCTGGTAATTTATTGACCCC-3′ (forward), 5′-AAATCCCGGGTCCGCCCC-3′ (reverse); pair 3: 5′-TTGCTAGCTGTGACTCCAGTCTAGTCC-3′ (forward), 5′-AAGTTTAAACTTTTAAAAATTCCTGCACGCTGGAAATTGCC-3′ (reverse); pair 4: 5′-AAGTTTAAACTTTTAAGAGCTGAACTTGGCGAGGTGG-3′ (forward), 5′-AAATCCCGGGTCCGCCCC-3′ (reverse); pair 5: 5′-GGGGCGGACCCGGGATTTC-3′ (forward), 5′-AAGTTTAAACTTAAATTTTTAATCAGCTGCTACTGAGCTGC-3′ (reverse); pair 6: 5′-AAGTTTAAACTTTTAATTCCCAGTACGGATCCCTTC-3′ (forward), 5′-AGCCGGAATTCCAAGCTCTAG-3′ (reverse); pair 7: 5′-GGGGCGGACCCGGGATTTC-3′ (forward), 5′-AAGTTTAAACTTAAATTTTTAAGCCTCGGGGCATCGGGCGC-3′ (reverse); pair 8: 5′-AAGTTTAAACTTTTAACTTCTGCGCGATCCTTCGGA-3′ (forward), 5′-TTCTCGAGGCGCGCCGCGACCT-3′ (reverse).
TRANSIENT TRANSFECTION AND LUCIFERASE ASSAYS
HepG2 cells or HEK293T cells grown routinely in T-75 flask in DMEM medium with 10% fetal bovine serum were trypsinized and transferred into 12-well plates 1–2 day before transfection. When the cells in the wells were about 80% confluent, the pGL3-Basic-Luc plasmid constructs together with the expression vectors were added into the same medium containing 5% fetal bovine serum with no antibiotics using Lipofectamine 2000 following the protocol recommended for transfection. pRLTK plasmid containing the Renilla-Luc gene was cotransfected to provide an internal control for transfection efficiency. After overnight transfection the cells were washed with PBS and then incubated with full-growth medium containing 10% fetal bovine serum and 0.1–1 μM all-trans-RA in ethanol at the final concentration of 0.001% ethanol. After incubation at 37 °C for 24 h the cells were washed with PBS, and then lysed to assay for firefly-Luc and Renilla-Luc activities using the DRL Luc assay system from Promega in Luminator 20. Promoter activity was defined as the ratio of firefly-Luc to Renilla-Luc activity. Each reported activity is an average of at least 3–4 wells.
NUCLEAR EXTRACT PREPARATION
Nuclear extracts from HepG2 cells treated with either ethanol as the vehicle or all-trans-RA for 6 h at 37 °C were prepared as previously described [Zolfaghari and Ross, 2009]. Cell pellets was homogenized in a hypotonic buffer containing10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM PMSF, 0.5 mM DTT, 1 mM sodium orthovanadate, 0.5% Nonidet P-40 using a Dounce tissue homogenizer. After centrifugation at 2500 × g at 4 °C for 10 min, the supernatant was aspirated and then pellets were washed once with hypotonic buffer containing no detergent. Then a hypertonic buffer containing 20 mM HEPES, pH 7.9, 10% glycerol, 1.5 mM MgCl2, 400 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT, 1 mM sodium orthovanadate was added to extract nuclear protein. After 30 min of incubation on ice, the mixture was centrifuged at 15,000 × g for 30 min. The supernatant was collected as the nuclear extract, aliquoted, and stored at −80 °C.
ELECTROPHORETIC MOBILITY SHIFT ASSAY (EMSA)
The annealed double-stranded oligonucleotide fragments previously used for RARE1 (H1) [Zhang et al., 2010] and RARE2 were first 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase, purified using a G25 column, and counted in a liquid scintillation spectrometer. The [32P]-end labeled DNA probe (5 × 104 cpm) was incubated with the nuclear extract from HepG2 cells for 30 min at room temperature. For EMSA antibody supershift assays, nuclear extracts were incubated with antibody to HNF4α for 20 min prior to addition of labeled probe. Incubation mixtures were electrophoresed on 5% native polyacrylamide gels using 0.5× TBE electrophoresis buffer. Gels were then vacuum dried and then subjected to autoradiography.
WESTERN BLOT ANALYSIS
After washing in PBS, HepG2 cells were lysed in RIPA buffer and subjected to total protein measurement. Twenty micrograms of protein along with rainbow molecular weight markers (Bio-Rad) were separated in a 10% gel by SDS-PAGE and electroblotted onto a Pure Nitrocellulose membrane (Bio-Rad). After blocking in 5% non-fat dry milk in washing buffer (1× PBS containing 0.05% Tween 20) for 2 h at room temperature, the membrane was incubated with the primary antibody (1:500) in blocking buffer at 4 °C overnight. The membrane was washed three times with washing buffer, each for 10 min, and then incubated with goat anti-mouse IgG2a as the secondary antibody (1:2000) conjugated with horseradish peroxidase for 1 h at room temperature in 1% non-fat dry milk in washing buffer and then washed as described above. Horseradish peroxidase activity was analyzed through visualization by SuperSignal West Pico Chemiluminescent substrate solutions (Thermo Biotechnology, Rockford, IL) followed by exposure to Hyperfilm ECL films (GE Healthcare). The density and area of the protein bands on the developed film were scanned and quantified in Photoshop (Adobe, San Jose, CA). For loading controls, the membranes were stained for proteins with Ponceau solution.
CHROMATIN IMMUNOPRECIPITATION (CHIP) ASSAY
HepG2 cells grown in full growth medium containing 10% FBS in 100 mm plate were incubated with either ethanol as the vehicle or 1 μM all-trans-RA for less than 1 h at 37 °C and then treated by adding formaldehyde directly to the tissue culture medium to a final concentration of 1% and then incubated for 10 min at room temperature. Cross-linking reactions were stopped by the addition of PBS-glycine to a final concentration of 0.125 M. Cells were washed twice with ice-cold PBS, scraped, and centrifuged at 1000 rpm for 10 min at 4 °C. Cells were then resuspended in cell lysis buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5% Nonidet P-40) containing protease inhibitors cocktail and 1 mM PMSF and kept on ice for 10 min. Cells were homogenized using a Dounce homogenizer (B pestle) for 15 strokes, and the resultant homogenates were centrifuged at 2000 rpm for 5 min at 4 °C to pellet the nuclei. The pellets were resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA and 1% SDS) containing protease inhibitors and PMSF and incubated on ice for 20 min. The nuclear lysates were sonicated on ice to an average chromatin length of 500–1000 kb and then centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatants were then subjected to ChIP assays as described [Hollingshead et al., 2008]. Purified antibodies specific to RARs, RNA Pol-II, and HNF4α were used for immunoprecipitation. The pulled-down DNA was subjected to real-time PCR with 5′-GGGTCACAGGCGGGTCAG-3′ and 5′-GGCTGTAAACACGGCGAACC-3′ as the primer pair for distal region and 5′-GAGCTCAGCACACCTTGGAT-3′ and 5′-CCCACGT-TACCTTGGTCAGA-3′ as the primer pair for proximal region of human promoter using SYBR Green/Fluorescein PCR Master Mix as described [Zhang et al., 2010].
STATISTICAL ANALYSIS
All data shown are the mean ± standard deviation or standard error of the mean of at least triplicate in each experiment. The data were analyzed by either one- or two-way ANOVA followed by Fisher’s least significant difference test using Prism6 Statistical Software (GraphPad). A P value <0.05 was considered statistically significant.
RESULTS
RA INCREASES THE EXPRESSION OF HNF4α IN HEPG2 CELLS
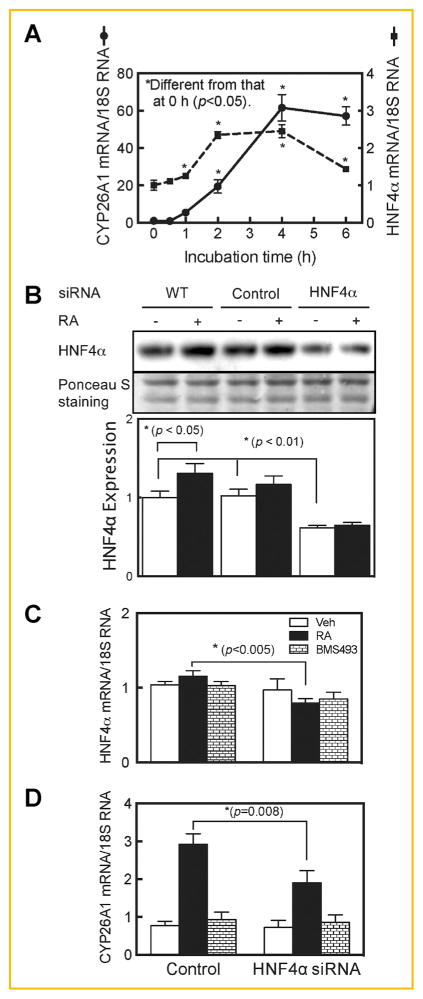
When HepG2 cells were cultured in a full growth medium with 10% FBS they expressed CYP26A1 mRNA at a very low level, similar to that reported for CYP26A1 in the liver of vitamin A adequate rats [Wang et al., 2002; Zhang et al., 2010]. However, upon treatment of the cells with RA for up to 6 h, CYP26A1 mRNA was increased by 20-fold after 2 h (P <0.05) and peaked at more than 60-fold over baseline by 4 h (P <0.0001) (Fig. 1A). During this time, the expression of HNF4α mRNA also peaked, with a maximum of >2 fold after 2–4 h (P <0.05) (Fig. 1A). Moreover, cells treated with RA for 4 h exhibited a greater than 30% increase (P <0.05) in HNF4α protein as determined by western blot analysis (Fig. 1B, lane 1 compared to lane 2). Using two different pairs of primers for analysis of HNF4α by RT-qPCR, the basal expression levels of HNF4α mRNA in HepG2 cells were much higher than those of CYP26A1 mRNA (data not shown). Previously, similar kinetic responses have been reported for HNF4α expression in HepG2 cells treated with 9-cis-RA [Hatzis and Talianidis, 2001; Mosialou et al., 2010].
Fig. 1.
HNF4α is regulated by RA and HNF4α silencing reduces the expression of CYP26A1 in human hepatoma HepG2 cells. RA induced the expression of CYP26A1 and HNF4α genes fast in HepG2 cells (A). HepG2 cells in 6-well plates were incubated with 1 μM all-trans-RA for 0–6 h and then collected for total RNA extraction for analysis of CYP26A1, HNF4α, and 18 S rRNA, as the control, by Reverse-Transcriptase qPCR. Each point represents the mean of 3 wells ± SEM. Inhibition of HNF4α by siRNA reduced expression of CYP26A1 protein (B) and mRNA (C and D) in HepG2 cells. Cells were treated with either HNF4α or non-targeting siRNA, as the control for 24 h, then further incubated with either vehicle, or 5 nM RA or 1 μM BMS493 as an RARα antagonist, together with RA for 4 h and then washed and collected for protein analysis by western blotting using HNF4α specific monoclonal antibody and for RNA analysis by real time RT-PCR. Ponceau S staining was used as a loading control for protein analysis and 18S rRNA, as the control for RNA analysis. Each point in bar graphs represents the mean of 3 wells ± SEM.
To determine the possible involvement of endogenous HNF4α in CYP26A1 gene expression, HepG2 cells were incubated with HNF4α specific siRNA for 24 h and then further treated with RA at a low concentration (5 nM) for 4 h, after which the cells were washed and subjected to RNA analysis by RT-qPCR and protein by western blot analysis. HNF4α gene silencing resulted in reduction of both HNF4α mRNA by about 30% (Fig. 1C) and HNF4α protein by more than 40% in RA-treated cells (Fig. 1B). As a result of reducing HNF4α expression, CYP26A1 mRNA was reduced by about 35%, as compared to HepG2 cells treated with non-targeted siRNA as the control (Fig. 1D). However, comparatively, this reduction was not of the same magnitude as that observed in cells treated with BMS493, an antagonist of RARα, which almost completely knocked down the expression of CYP26A1 mRNA induced by RA (Fig. 1D).
CYP26A1 GENE EXPRESSION IS SENSITIVE TO RA TREATMENT IN HNF4α-CONTAINING HEPG2 CELLS BUT NOT IN HEK293T CELLS LACKING HNF4α
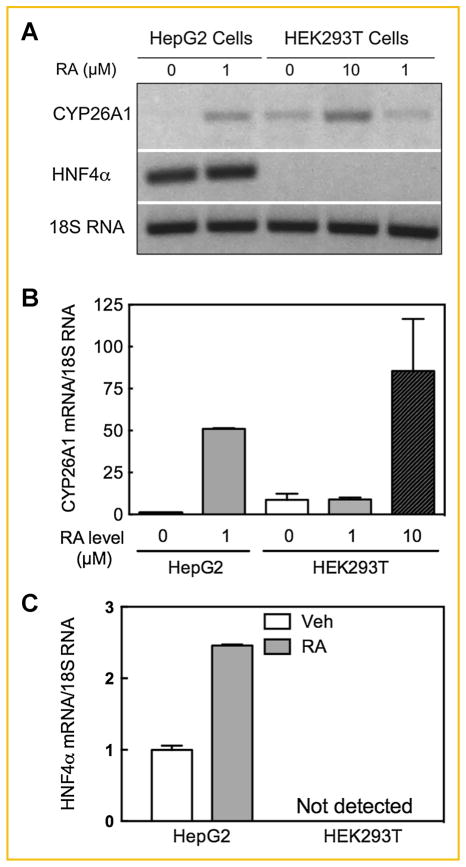
Since HEK293T cells lack any endogenous expression of HNF4α [Jiang et al., 2003] we evaluated the functional contribution of HNF4α in the activity of the CYP26A1 promoter in HepG2 cells as well as HEK293T cells as a model of non-hepatic cells for comparison. HepG2 cells and HEK293T cells were each incubated with RA for 4 h. CYP26A1 is endogenously expressed in HEK293T cells but only at a very low level in HepG2 cells in the absence of RA (Fig. 2A,B). Upon addition of 1 μM RA to both cell types, CYP26A1 was induced by more than 50 fold in HepG2 cells as compared to none in HEK293T cells. Even 10 μM RA induced CYP26A1 by not more than 7 fold in HEK293T cells. For HNF4α, basal expression was constitutively high in HepG2 cells and up-regulated by RA, whereas HNF4α was not detected in HEK293T cells in either the absence of RA, in agreement with Jiang et al. [2003] or in the presence of RA (Fig. 2A,C).
Fig. 2.
CYP26A1 gene expression is sensitive to RA treatment in HNF4α-containing HepG2 cells. HepG2 or HEK293T cells were grown in 6-well plates and treated with either ethanol as the vehicle control or RA in triplicate for 4 h and then collected for RNA analysis by electrophoresis in ethidium bromide treated agarose gel (A) and real time RT-PCR (B and C) as described in the Materials and Methods section. The relative expression of CYP26A1 or HNF4α mRNA/18S rRNA level, as the control, was set as 1 in HepG2 cells treated with the vehicle in bar graphs. Each point represents the mean of 3 wells ± SEM.
HNF4A INDUCES THE PROMOTER ACTIVITY OF THE CYP26A1 GENE
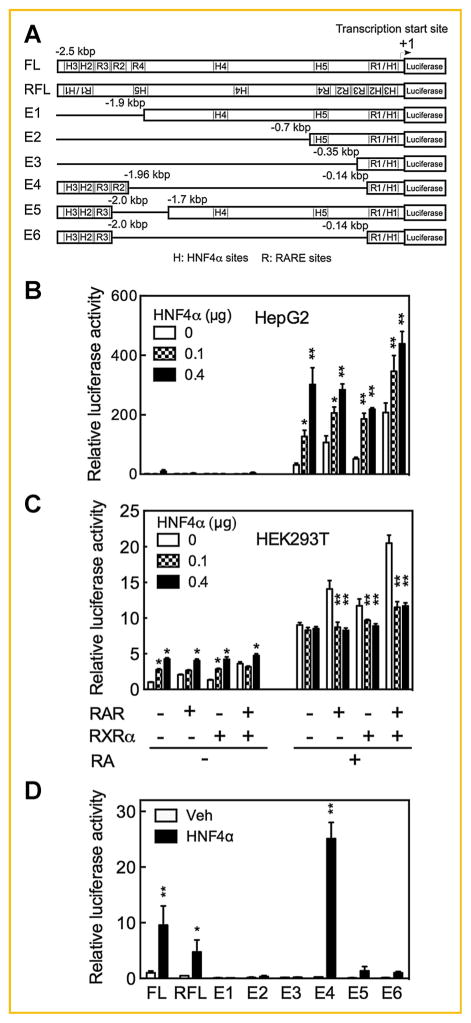
As illustrated in Fig. 3A, the promoter region of the CYP26A1 gene contains at least four previously defined RARE elements: a proximal DR5 (R1) and a cluster of two distal DR5 (R2 and R3) together with a single half DR (R4) [Loudig et al., 2000; Loudig et al., 2005; Zhang et al., 2010]. We have shown that all four elements of these RARE are essential for the full expression of the CYP26A1 gene in HepG2 cells [Zhang et al., 2010]. In order to learn where HNF4α binds in the DNA promoter region of CYP26A1 gene, we first physically analyzed the sequence of the full-length (FL) promoter using the MatInspector computer program (www.genomatix.de). In addition to the four RAREs above, five potential HNF4α binding sites were identified within this 2.5 kbp promoter region as illustrated in Fig. 3A for the FL promoter. One of these sites, designated H1, lies within the R1 sequence in the proximal region. Two of the elements, H2 and H3, are located in the distal region close to the RARE cluster, and another two elements, H4 and H5, are located in the middle region of the promoter (Fig. 3A, FL sequence).
Fig. 3.
HNF4α enhances CYP26A1 promoter activity in HepG2 cells but not in HEK293T cells treated with RA. Cells grown in 24-well plates were cotransfected in triplicate with pGL3-Basic-luc-hCYP26A1 promoter containing full-length human CYP26A1 promoter (FL) (A) and HNF4α at 0, 0.1, and 0.4 μg without or with retinoid receptors and then treated with either vehicle or RA at 1 μM for HepG2 cells and 10 μM for HEK293T cells for 24 h after which the cells were collected for luciferase assay. HNF4α dose dependently increased the CYP26A1 promoter activity in HepG2 cells (B) but not in HEK293T cells treated with RA (C). Nucleotide deletion analysis of CYP26A1 promoter indicates that HNF4α response element(s) might be present around or within the retinoic acid response elements (D). *P <0.05; **P <0.001.
Next, to examine whether the CYP26A1 promoter is regulated by HNF4α in HepG2 cells and in HEK293T cells, we used the FL promoter of CYP26A1 containing all four RARE and five putative HNF4α response elements, constructed in pGL3-Basic-Luc as a reporter gene. Using both HepG2 and HEK293T cells, the FL reporter construct together with pRLTK plasmid containing Renilla luciferase were contransfected overnight with either 0, 0.1, and 0.4 μg of human HNF4α DNA in a pcDNA3.1 expression vector, both without or with expression vectors for RARs, RXRα, or both receptors. Following these transfections, the HepG2 cells or HEK293T cells were incubated with either vehicle or RA for 24 h, after which luciferase activity was determined. The promoter activity of the CYP26A1 gene was almost undetectable in vehicle-treated HepG2 cells (Fig. 3B). However, upon treatment of the transfected cells with RA the activity of the CYP26A1 promoter increased by more than 30 fold, similar to the results obtained for analysis of endogenous CYP26A1 mRNA expression after RA treatment (see Fig. 1A and Fig. 2A,B). In contrast, the promoter activity of the CYP26A1 gene was relatively high in HEK293T cells in the absence of RA; however, treatment of these cells with RA increased the promoter activity by less than 10 fold, similar to the results observed for endogenous mRNA analysis of CYP26A1 in HEK293T cells (see Fig. 2A,B). It is noteworthy that HNF4α did not have any significant effect on the promoter activity of the CYP26A1 gene in HepG2 cells in the absence of RA. However, expression of HNF4α dose dependently increased the promoter activity of the CYP26A1 gene in RA-treated cells, either without or with cotransfection of RARs (Fig. 3B). HNF4α alone increased the promoter activity by 4 fold (P <0.05) at 0.1 μg HNF4α DNA and by 10 fold at 0.4 μg DNA (P <0.001) (Fig. 3B). Transfection of either RARs or RXRα separately did not have any significant effect on the induction of CYP26A1 promoter activity by HNF4α. However, the combination of RARs/RXRα increased the inductive effect of HNF4α on the promoter activity of CYP26A1 gene significantly (P <0.001) (Fig. 3B). In contrast, in HEK293T cells transfection with HNF4α DNA increased the promoter activity of the CYP26A1 gene only in vehicle-treated cells, although only by 2–3 fold, but not in cells treated with RA (Fig. 3C). In fact, HNF4α significantly (P <0.001) suppressed the inductive effect of RARs, RXRα, or both in combination, on the promoter activity of the CYP26A1 gene in HEK293T cells following treatment with RA (Fig. 3C).
MUTATION BY NUCLEOTIDE DELETION INDICATES THAT HNF4α RESPONSE ELEMENT MAY OVERLAP WITH THE RA RESPONSE ELEMENTS
Previously, we constructed several different clones of the promoter of CYP26A1 gene carrying different deletions in pGL3-Basic-Luc vector to test the role of the individual RARE in the promoter activity of the CYP26A1 gene [Zhang et al., 2010]. Those clones include, in addition to FL, reverse-oriented FL and clones with eliminations designated E1 through E6 (shown in Fig. 3A). The individual deleted clones as well as the WT FL clone, each along with pRLTK as an internal control, were cotransfected without or with HNF4α expression vector into HepG2 cells, which following transfection were treated with RA for 24 h. Again, the luciferase activity of the promoter constructs was much higher in the transfected cells after RA treatment (data not shown). The orientation of the FL promoter construct in the promoterless pGL3-Basic-Luc vector appears to be critical for full luciferase activity since the FL reverse oriented promoter (shown in Fig. 3A) resulted in less activity than the WT FL promoter either without or with expression of HNF4α (Fig. 3D). The distal regions of the CYP26A1 promoter including R2 through R4 as well as H2 and H3 in coordination with proximal region containing R1 and H1 were essential for full activity of the CYP26A1 promoter in response to RA and HNF4α; however, the intermediate region containing H4 and H5 was dispensable in response to RA and as well as to HNF4α (Fig. 3D). Constructs referred to as E1, E2, and E3 which are lacking the distal region had no significant activity either in the absence or presence of HNF4α. Elimination of R2 and R4 in the distal region, as in the constructs E5 and E6, resulted in complete loss of activity in response to RA and about 90% inhibition of the activity in response to HNF4α. Elimination of the DNA intermediate region including the half site R4 as well as H4 and H5 (construct E4), which brings the distal and proximal regions closer together, reduced the CYP26A1 promoter activity in HepG2 cells treated with RA but increased it 2.5 times as compared to the WT CYP26A1 promoter in response to HNF4α in RA-treated cells (Fig. 3D). Thus, whereas the proximal promoter regions in concert with the distal regions are essential for full activity, the intermediate region, oppositely, was inhibitory to the CYP26A1 promoter activity in response to HNF4α in cells treated with RA. Based on these results, the potential response element(s) that bind HNF4α may be associated with, i.e., overlap, the RARE regions.
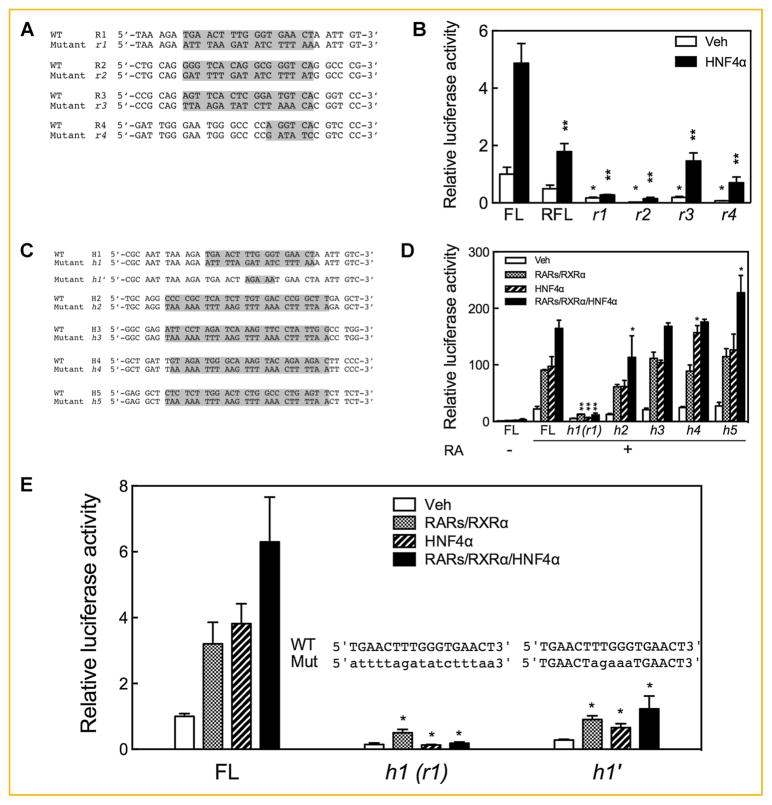
MUTATION IN ANY INDIVIDUAL RARE RESULTED IN SIGNIFICANT SUPPRESSION OF THE PROMOTER ACTIVITY OF THE CYP26A1 GENE IN RESPONSE TO HNF4α
To evaluate the role of the individual RARE and HNF4α response elements in the promoter activity of the CYP26A1 gene, we mutated the individual sites (illustrated in Fig. 4A) by nucleotide substitution [Zhang et al., 2010] in the FL construct and used this construct for cotransfection without or with the HNF4α expression vector in HepG2 cells, which after transfection were treated with RA for 24 h. Mutation of each of the individual RARE sites resulted in the complete loss of CYP26A1 promoter activity in response to RA (Fig. 4B). By comparison, in response to HNF4α the mutation of r1 or r2, shown in Fig. 4A, resulted in the complete loss of CYP26A1 promoter activity (P <0.0001), while the FL promoter constructs with mutations r3 or r4, also shown in Fig. 4A, retained some residual activity, although less than that of the WT FL control (P <0.05) (Fig. 4B).
Fig. 4.
Mutation in any of the retinoic acid response elements or of the HNF4α response elements abrogates the activity of CYP26A1 promoter in response to HNF4α. HepG2 cells were cotransfected with FL CYP26A1 promoter in pGL3-Basic-luc mutated by nucleotide substitution in individual RAREs (r1 to r4) (A and B) or individual HNF4α response elements (h1, h2, h3, h4, h5, and h1′) (C, D, and E) with HNF4α expression vector and then treated with 0.1 μM RA for 24 h after which the cells were assayed for luciferase activity. Data from each bar represent the mean of n = 3 wells ± SEM. *P <0.05; **P <0.001.
MUTATION IN INDIVIDUAL HNF4α RESPONSE ELEMENTS INDICATES THAT THE FUNCTIONAL HNF4α RESPONSE ELEMENT MAY RESIDE WITHIN THE R1 SITE OF THE CYP26A1 PROMOTER
The five putative HNF4α response elements present in the FL promoter region of the CYP26A1 gene (see Fig. 3A), along with their mutations, designated h1 to h5, are shown in Fig. 4C. While the mutation designated r1 almost completely eliminated activity of the FL promoter in response to both RA and HNF4α, mutation of only h2 but not h3 in the distal region of the CYP26A1 FL promoter caused no more than a 40% reduction of activity in response to RA and HNF4α (Fig. 4D). Mutations in the distal region, either in h3 or individual mutations in h4 and h5, did not significantly reduce FL promoter activity in response to RA and HNF4α (Fig. 4D). In fact, mutation h5 slightly increased the activity of the FL promoter in response to HNF4α. These results imply that H1 located within the R1 element in the proximal region of the CYP26A1 promoter is indispensable not only for RA but also for HNF4α responsiveness.
Since the mutation in h1 (overlapping r1) includes nucleotides for both the DR5 hexamer of the RARE (R1) and the five nucleotides between the two hexamers, which are identified as the core of the HNF4α binding site CAAA [Fang et al., 2012], we made another mutated FL promoter construct, designated h1′ (Fig. 4C) which contained mutation only in the five intervening nucleotides between the DR5 hexameric sequences but not within the hexamer repeats themselves. Assuming that the five intervening nucleotides are nonspecific for the RARE (R1), we expected that this construct would have full activity in response to RA and its receptors but not to HNF4α, for which CAAA is critical. To our surprise, mutation in the five intervening nucleotides (h1′) but not in the hexamer repeat sequence r1 (or h1) still caused a significant reduction in FL promoter in response not only to HNF4α but also, to a lesser extent, to RARs/RXRα. This reduction in activity was, however, less severe in the h1′ mutation than in the r1 (h1) mutation (Fig. 4E). This result suggests that the intervening nucleotides can be a factor in retinoid receptor binding as well as being important for HNF4α.
HNF4α BINDS WITH TO BOTH PROXIMAL AND DISTAL REGIONS IN THE PROMOTER OF CYP26A1 GENE IN RESPONSE TO RA IN HEPG2 CELLS
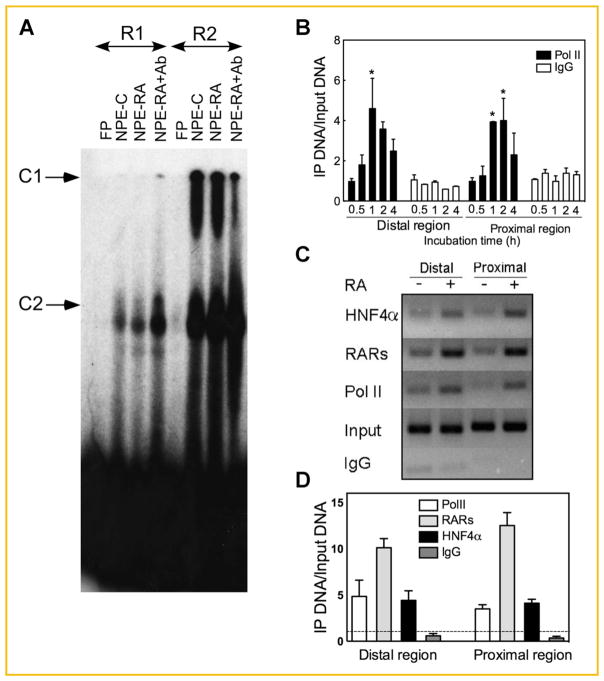
Previous results from protein binding array analysis indicated that HNF4α binds in the DNA region spanning from −2000 to +10000 of the CYP26A1 gene, but the exact binding site or sites have not been specified [Hwang-Verslues and Sladek, 2010]. Among the H binding sites we found that the proximally located H1 within the R1 site plays the most important role in the activation of the CYP26A1 gene (Fig. 4D,E). On the other hand, among four other RARE elements, the R2 site in the distal region has been shown to be the most responsive not only to RA ([Zhang et al., 2010] and Fig. 4B) but also to HNF4α. To determine whether HNF4α binds to the sites located in these proximal and distal regions of the CYP26A1 gene promoter we used the same DNA oligonucleotides that were used previously [Zhang et al., 2010] and shown to be involved RAR binding in both the proximal and distal regions. Nuclear protein extracts from HepG2 cells that had been treated with either vehicle or RA were incubated with [32P]-labeled oligonucleotides as the probes and incubated in either the absence or presence of HNF4α antibody, then separated by native polyacrylamide gel electrophoresis. EMSA results showed that the nuclear proteins from RA-treated HepG2 cells formed more intense complexes with all individual oligonucleotide probes than those from vehicle-treated control cells (Fig. 5A). Protein binding was previously shown to be specific since unlabeled oligonucleotides of the same sequence competed with the labeled probes, and, in addition, mutant oligonucleotide probes did not displace the binding [Zhang et al., 2010]. The R2 oligonucleotide probe formed large complexes (C1), which could not pass through the gel. Such complexes (C1) were not observed in the other sites. However, upon incubation with HNF4α antibody the oligonucleotide probe spanning the R1 site formed C1 complexes in addition to a C2 complex (Fig. 5A). The C1 complexes were diminished when the R2 oligonucleotide probe was incubated with HNF4α antibody. Based on these results, HNF4α is involved in binding to R1 in the proximal region as well as in the distal region R2 site (Fig. 5A) but not in the R3 and R4 sites (data not shown).
Fig. 5.
RA increases the binding of HNF4α to both the proximal and distal regions of the CYP26A1 gene promoter in HepG2 cells. (A) R1 and R2 may interact with HNF4α in the nuclear extract from HepG2 cells. Nuclear protein extracts from HepG2 cells treated with either vehicle or all-trans-RA were incubated with individual [32P]-labeled RARE oligonucleotide (see Materials and Methods section for the oligonucleotide sequences) as probe in the absence or presence of HNF4α antibody and then run on native polyacrylamide gel by electrophoresis. The gel was dried under vacuum and then exposed to X-ray film as described in the Materials and Methods section. (B) ChIP assay indicates that Pol II binding increased (*P <0.05 compared to 0 h) in both proximal and distal regions of CYP26A1 promoter in HepG2 cells within 1–2 h after treated with RA. HepG2 cells grown in 100 mm plates were treated with 1 μM RA for 4 h and prepared for ChIP assay using either anti human Pol II antibody or anti human CD1d antibody as an irrelevant IgG control. (C and D) RA increases the binding of HNF4α to both the proximal and distal regions in the promoter of CYP26A1 gene in HepG2 cells. HepG2 cells were incubated with either ethanol as the vehicle or 1 μM RA for 1 h and then collected for ChIP assay. The DNA samples pulled down by individual antibodies were then subjected to PCR using primer pairs designed for the proximal and distal regions of CYP26A1 promoter. RA treatment of HepG2 cells increased the binding of HNF4α to both proximal and distal regions in the promoter of CYP26A1 gene similar to RARs and Pol II used as the controls. Non-immune IgG was used as the negative control. The dashed line in panel D indicates the relative binding in the cells treated with vehicle. The data represent the mean ± SEM, n = 3.
RA TREATMENT INCREASES POL II, RAR, AND HNF4α BINDING TO THE PROXIMAL AND DISTAL REGIONS OF THE CYP26A1 PROMOTER IN CHROMATIN OF INTACT HEPG2 CELLS
To confirm the direct involvement of HNF4α in binding to the CYP26A1 gene in intact chromatin, a ChIP assay was employed to evaluate the extent of in vivo occupancy of HNF4α to the DNA in the promoter region of the CYP26A1 gene. Since CYP26A1 mRNA expression was increased to its maximum level by 4 h with treatment of RA in HepG2 cells (Fig. 1A), the cells in this experiment were treated with 1 μM RA for up to 4 h and then evaluated for the extent of recruitment of the DNA-dependent RNA polymerase Pol II to the promoter region of the CYP26A1 gene in the intact chromatin. Fig. 5B shows that both DNA in the proximal as well as distal regions around RAR response elements are involved in the binding of Pol II. RA treatment after 1–2 h increased the binding of the Pol II by the maximum levels of 4–5 fold (P <0.05) both in the proximal and distal regions of the CYP26A1 promoter in the intact chromatin in HepG2 cells. In a separate experiment, a 1-h treatment with RA increased not only the binding of Pol II but also RARs by about 10–12 fold and HNF4α by about 3–4 fold in both regions including the one proximal to the transcription start site as well as the distal region ~2 kbp upstream from the transcription start (Fig. 5C,D).
DISCUSSION
In this report we have studied the FL promoter of the CYP26A1 gene and demonstrated that HNF4α, a liver-enriched nuclear receptor, can activate the promoter of the CYP26A1 gene in HepG2 cells, a model for hepatocytes [Knowles et al., 1980], in an RA-dependent manner. It was known from previous work that the CYP26A1 gene is expressed at very low levels in HepG2 cells but is highly inducible by RA whereas, in contrast, the CYP26A1 gene is constitutively expressed in HEK293T cells, a non-hepatic cell, but is minimally induced by RA [Zhang et al., 2010]. In the present study, we found, first, that whereas HNF4α is not expressed at all in HEK293T cells it is highly expressed in HepG2 cells, and its mRNA and protein expression is induced after treatment of the cells with RA. Although the fold increase for HFN4α was less than that for CYP26A1, this may be because the basal level of HNF4α expression is much higher than CYP26A1 expression. Thus the absolute change in HNF4α expression could be quite substantial. Moreover and conversely, transfection of HepG2 cells with HNF4α siRNA resulted in suppression of CYP26A1 mRNA in cells treated with a low concentration of RA. Although the suppressive effect of HNF4α siRNA on CYP26A1 mRNA expression was not as substantial as that of an RARα antagonist, BMS493, its effective level was still significant as compared to the non-targeted siRNA control. This could be due to incomplete suppression of HNF4a expression, as a 60% residual level of HNF4α remained in HepG2 cells. However, the ability to detect a reduction in CYP26A1 expression concomitant with a modest reduction in HNF4a may suggest that natural physiological changes in HNF4α expression could help to regulate the expression of CYP26A1 in hepatocytes.
Secondly, HNF4α dose-dependently induced the FL promoter activity of the CYP26A1 gene in RA-treated HepG2 cells, both without or with cotransfection with RARs, but not significantly in vehicle-treated cells. In fact, similar to the endogenous gene, which had almost no activity, the transfected promoter had negligible activity, both without or with retinoid receptors, in the absence of RA in HepG2 cells. Although HNF4α activated the promoter of CYP26A1 gene in HEK293T cells treated with vehicle it suppressed the induction activity of CYP26A1 promoter by nuclear retinoid receptors in the cells treated with RA. Third, using mutation by either nucleotide elimination or substitution the results show that potential HNF4α response element(s) are present within the RARE regions of the CYP26A1 gene because, (i) elimination of non-RARE regions resulted in an increase in promoter activity of the CYP26A1 gene by more than three times in response to HNF4α, as compared to the FL promoter; and (ii) mutation in any of the four RARE present in the FL promoter resulted in a significant reduction in the CYP26A1 promoter activity in response to HNF4α in RA-treated HepG2 cells. In fact, mutation by nucleotide substitution in either r1 in the proximal region or especially r2 in the distal region of the FL promoter resulted in almost complete inactivation of the promoter in response to both HNF4α and RA in HepG2 cells. This indicates a requirement for both regions for high induction of CYP26A1 by RA, although the potential HNF4α response element among five putative HNF4α response elements was found within R1 in the proximal region of the promoter. In addition, to our surprise, the specific mutation in the core region of HNF4α located within R1 resulted in almost complete inhibition of promoter activity in response to both RARs/RXRα as well as HNF4α although the hexameric sequences of the DR5 which constitutes the R1 region remained intact. This may indicate that cooperative binding of HNF4α with those retinoid receptors to proximal region of the promoter is essential for complete activation of CYP26A1 gene. It also suggests that the nucleotide sequence required for HNF4α to cooperate with RARs in the proximal region of the CYP26A1 gene is TGAACTTTGGGTGAACT (with TGGG complementary, in reverse, to the HNF4α binding sequence CAAA), which is more rigidly specified than that of a typical DR5 RARE. This sequence is completely conserved at least among the human, mouse, and rat genomes [Zhang et al., 2010].
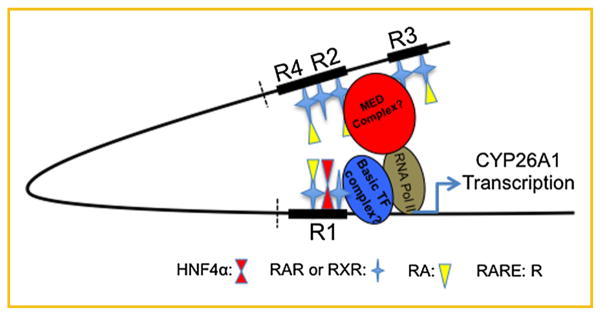
Fourth, these results are supported by those obtained from EMSA as well as ChIP assays. Whereas tighter bounds were formed between nucleotide fragments containing individual RARE elements with nuclear extract proteins from the cells treated with RA as compared to those of the cells treated with vehicle, HNF4α was shown to be involved in binding only with R1 and R2 in forming complexes in the cells treated with RA. In ChIP assay experiments we showed that HNF4α, like Pol II and RARs, bound to both proximal and distal regions of the CYP26A1 in intact chromatin in HepG2 cells treated with RA. Similar results to our previous report we found that RARs as well as PolII as the basic transcription component bound to both proximal and distal regions but bound faster in the cells treated with RA as compared to those in vehicle control cells. Based on the results from a ChIP-loop assay study, Bruck et al. [2009] speculated that RARα may mediate bridge formation that results in positioning of the distal region close to the proximal region of CYP26A1 promoter, in the induction of gene expression in human MCF7 cells in response to RA. Based on their speculation, the results of the present study, and those of our previous report [Zhang et al., 2010], we suggest that upon treatment of hepatocytes with RA, HNF4α may participate in acceleration of bridge formation between distal and proximal RAREs (R) in the promoter of CYP26A1 gene, as shown in the model in Fig. 6. It may be that the absence of HNF4α reduces the occurrence of this proposed bridge formation by retinoid receptors in response to RA, as the induction of CYP26A1 expression by RA was lower in HEK293T cells lacking HNF4α (Fig. 2). CYP26A1 mRNA expression has been reported to be active in several cell lines [Ross and Zolfaghari, 2011]. For example, it is expressed at substantial levels in HEK293T, a human embryonic cell line, but induced to a lesser extent (fold change) in those cells as compared to HepG2 cells [Zhang et al., 2010]. Previously, we also showed that RARs as well as Pol II were bound only in the proximal but not in the distal region of the CYP26A1 promoter within intact chromatin in HEK293T cells, as compared to both proximal and distal regions in HepG2 cells upon treatment with RA [Zhang et al., 2010]. Based on those results we speculated that the extent of the bending (“bridging”) of the CYP26A1 promoter could be much lower in the intact chromatin in HEK293T cells as compared to in HepG2 cells upon treatment with RA. In fact, in this report we found that HNF4α could not activate the promoter of the CYP26A1 gene but rather reduced the activity of the gene both in the absence or presence of RARs/RXRα. Based on the results in this study we speculate that HNF4α may be considered as an important factor in forming a bridge between the RARE1 and RARE2 loci of the CYP261 promoter as has been proposed by Bruck et al. [2009] in the cells including HepG2 cells, but not specifically implicating HNF4α in this process. Our study implicates HNF4α as a potentially important contributor to CYP26A1 activation in the liver, possibly through this bridging mechanism.
Fig. 6.
HNF4α may enhance the bridge formation by RAR in the promoter of CYP26A1 gene in intact chromatin in HepG2 cell treated with RA. Based on the results of this study with those of our previous report [Zhang et al., 2010], and of bridge formation mediated by retinoic acid receptors in the chromatin as proposed by Bruck et al. [Bruck et al., 2009], we speculate that upon treatment of hepatocytes with RA, HNF4α may participate in acceleration of bridge formation between distal and proximal RAREs (R) in the promoter of the CYP26A1 gene. Absence of HNF4α may lower the occurrence of this speculated bridge formation by retinoid receptors in response to RA as lower induction of CYP26A1 expression by RA has been observed in the cells lacking HNF4α. Elimination of the bracketed region in the promoter highly increases the promoter activity of the CYP26A1 gene by HNF4α in HepG2 cells in response to RA as shown in Fig. 3D.
In summary, we have found that high induction of CYP26A1 gene expression by RA in hepatocytes is enhanced dose-dependently by the presence of HNF4α, a liver specific nuclear transcription factor that is highly expressed in hepatocytes. Indeed the increased CYP26A1 promoter activity by HNF4α required the presence of RA. Among the five HNF4α binding sites identified, the site located within the RARE proximal to the transcription start site was shown to be the HNF4α binding site that is essential for the increased activity of CYP26A1 in RA-treated hepatocytes. However, because the relative DNA binding of HNF4α was increased in the proximal region as well as in distal regions of the promoter of CYP26A1 gene in hepatocytes treated with RA, as shown by ChIP assays, our evidence suggests that HNF4α may function cooperatively with RARs in forming a bridge, as illustrated in Fig. 6, which may potentially bring the distal site close to the proximal transcription start site, and thereby enhance the promoter activity of the CYP26A1 gene in hepatocytes upon treatment with RA.
Acknowledgments
This work was supported by NIH grant CA90214 and funds from the Dorothy Foehr Huck endowment, Pennsylvania State University. We thank Dr. Yao Zhang for the CYP26A1 promoter constructs used in this study.
Abbreviations
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- HNF4α
hepatocyte nuclear factor-4 alpha
- RA
all-trans-retinoic acid
- RAR
retinoic acid receptor
- RARE
retinoic acid response element
- RNA PolII
RNA polymerase II
- RXR
retinoid X receptor
Footnotes
Disclosures: All authors state that they have no conflicts of interest.
Materials disclosure: Information about the materials and methods used is available from Dr. Ross (acr6@psu.edu) or Dr. Zolfaghari (rxz7@psu.edu). No permanent new materials or models were generated.
Authors’ contribution: Study design, conduct, data interpretation and manuscript preparation: RZ and ACR. Study design, data and manuscript review: ACR. Both authors take responsibility for the integrity of the data analysis.
References
- Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Bolotin E, Schnabl JM, Sladek FM. Homo sapiens hepatocyte nuclear factor 4, alpha. 2010 http://www.cisreg.ca/cgi-bin/tfe/articles.pl?tfid=140.
- Bruck N, Vitoux D, Ferry C, Duong VAB, de Thé H, Rochette-Egly C. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 2009;28:34–47. doi: 10.1038/emboj.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes T, Senkel S, Holewa B, Ryffel GU. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol. 1996;16:925–931. doi: 10.1128/mcb.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Mane-Padros D, Bolotin E, Jiang T, Sladek FM. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucl Acids Res. 2012;40:5343–5356. doi: 10.1093/nar/gks190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis P, Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol Cell Biol. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead HE, Borland MG, Billin AN, Willson TM, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) and inhibition of cyclooxygenase 2 (COX2) attenuate colon carcinogenesis through independent signaling mechanisms. Carcinogenesis. 2008;29:169–176. doi: 10.1093/carcin/bgm209. [DOI] [PubMed] [Google Scholar]
- Holloway MG, Miles GD, Dombkowski AA, Waxman DJ. Liver-specific hepatocyte nuclear factor-4alpha deficiency: greater impact on gene expression in male than in female mouse liver. Mol Endocrinol. 2008;22:1274–1286. doi: 10.1210/me.2007-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Chandra V, Rastinejad F. Retinoic acid actions through mammalian nuclear receptors. Chem Rev. 2014;114:233–254. doi: 10.1021/cr400161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang-Verslues WW, Sladek FM. HNF4α-role in drug metabolism and potential drug target? Curr Opin Pharmacol. 2010;10:698–705. doi: 10.1016/j.coph.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Tanaka T, Iwanari H, Hotta H, Yamashita H, Kumakura J, Watanabe Y, Uchiyama Y, Aburatani H, Hamakubo T, Kodama T, Naito M. Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4alpha (HNF4alpha) isoforms in human and rats. Nucl Recept. 2003;1:5. doi: 10.1186/1478-1336-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, Castell JV. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology. 2001;33:668–675. doi: 10.1053/jhep.2001.22176. [DOI] [PubMed] [Google Scholar]
- Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J. 2005;392:241–248. doi: 10.1042/BJ20050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialou I, Zannis VI, Kardassis D. Regulation of human apolipoprotein m gene expression by orphan and ligand-dependent nuclear receptors. J Biol Chem. 2010;285:30719–30730. doi: 10.1074/jbc.M110.131771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821:152–167. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res A Clin Mol Teratol. 2010;88:883–894. doi: 10.1002/bdra.20709. [DOI] [PubMed] [Google Scholar]
- Ross AC, Cifelli CJ, Zolfaghari R, Li N-Q. Multiple cytochrome P-450 genes are concomitantly regulated by vitamin A under steady-state conditions and by retinoic acid during hepatic first-pass metabolism. Physiol Genomics. 2011;43:57–67. doi: 10.1152/physiolgenomics.00182.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu Rev Nutr. 2011;31:65–87. doi: 10.1146/annurev-nutr-072610-145127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Wang Y, Zolfaghari R, Ross AC. Cloning of rat cytochrome P450RAI (CYP26) cDNA and regulation of its gene expression by all-trans-retinoic acid in vivo. Arch Biochem Biophys. 2002;401:235–243. doi: 10.1016/S0003-9861(02)00043-7. [DOI] [PubMed] [Google Scholar]
- Wei LN. Retinoid receptors and their coregulators. Annu Rev Pharmacol Toxicol. 2003;43:47–72. doi: 10.1146/annurev.pharmtox.43.100901.140301. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Zolfaghari R, Ross AC. Regulation of CYP26 (cytochrome P450RAI) mRNA expression and retinoic acid metabolism by retinoids and dietary vitamin A in liver of mice and rats. FASEB J. 2000;14:2119–2127. doi: 10.1096/fj.00-0061com. [DOI] [PubMed] [Google Scholar]
- Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zolfaghari R, Ross AC. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene. 2010;464:32–43. doi: 10.1016/j.gene.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari R, Cifelli CJ, Lieu SO, Chen Q, Li NQ, Ross AC. Lipopolysaccharide opposes the induction of CYP26A1 and CYP26B1 gene expression by retinoic acid in the rat liver in vivo. Am J Physiol Gastrointest Liver Physiol. 2007;292:1029–1036. doi: 10.1152/ajpgi.00494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. Cloning, gene organization and identification of an alternative splicing process in lecithin:retinol acyltransferase cDNA from human liver. Gene. 2004;341:181–188. doi: 10.1016/j.gene.2004.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. An essential set of basic DNA response elements is required for receptor-dependent transcription of the lecithin:retinol acyltransferase (Lrat) gene. Arch Biochem Biophys. 2009;489:1–9. doi: 10.1016/j.abb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]