Abstract
While chromatin characteristics in interphase are widely studied, characteristics of mitotic chromatin and their inheritance through mitosis are still poorly understood. During mitosis chromatin undergoes dramatic changes: Transcription stalls, chromatin binding factors leave the chromatin, histone modifications change and chromatin becomes highly condensed. Many key insights into mitotic chromosome state and conformation have come from extensive microscopy studies over the last century. Over the last decade the development of 3C-based techniques has enabled the study of higher order chromosome organization during mitosis in a genome-wide manner. During mitosis chromosomes lose their cell type specific and locus-dependent chromatin organization that characterizes interphase chromatin and fold into randomly positioned loop arrays. Upon exit of mitosis cells are capable of quickly rearranging the chromosome conformation to form the cell type specific interphase organization again. The information that enables this rearrangement after mitotic exit is thought to be encoded at least in part in mitotic bookmarks, e.g. histone modifications and variants, histone remodelers, chromatin factors and non-coding RNA. Here we give an overview of the chromosomal organization and epigenetic characteristics of the interphase and mitotic chromatin in vertebrates. Second, we describe different ways in which mitotic bookmarking enables epigenetic memory of the features of the interphase chromatin through mitosis. And third, we explore the role of epigenetic modifications and mitotic bookmarking in cell differentiation.
Keywords: chromatin organization, cell cycle, mitosis, mitotic bookmarking, epigenetics, histone modification, epigenetic memory
Introduction
A major question in cell biology is how cell type identity is maintained through mitosis. Imaging studies have been instrumental in studying mitotic chromosomes in live cells and after purification. These pioneering studies, mostly by the Laemmli group, led to fundamental insights into the architecture of mitotic chromosomes (Earnshaw & Laemmli, 1983; Maeshima & Laemmli, 2003; Marsden & Laemmli, 1979). More recently, high-throughput genomic methods have been used to gain deeper and more detailed insights into the folding of chromatin inside mitotic chromosomes and the local characteristics of the chromatin fiber such the presence of open sites, patterns of histone modifications and the binding of other factors (Hsiung et al., 2015; Hsiung et al., 2016; Naumova et al., 2013).
Decades of genetic and epigenetic studies have revealed many features of chromosome structure and how these could be involved in transcriptional control in the interphase cell. Over the last decade emerging high-throughput sequencing techniques like chromosome conformation capture (3C) based techniques, assays for transposase-accessible chromatin using sequencing (ATAC-seq), DamID and chromatin immunoprecipitation sequencing (ChIP-seq) enable the study of chromosome conformation, nuclear organization, chromatin state, its function and its regulators (Buenrostro, Giresi, Zaba, Chang, & Greenleaf, 2013; Dekker, Rippe, Dekker, & Kleckner, 2002; Lieberman-Aiden et al., 2009; Orlando, 2000; van Steensel, Delrow, & Henikoff, 2001). Large-scale consortia like the Encode project and NIH epigenome roadmap provide comprehensive overviews of cell type specific profiles of histone modifications, nuclear organization and DNA binding factors in non-synchronous, mostly interphase, cells (The ENCODE Project Consortium, 2012; Consortium Roadmap Epigenomics et al., 2015). These cell type specific features establish regulatory control of the genome and its effects on the phenotype of a cell. However, the characteristics of vertebrate chromatin change dramatically during mitosis. Chromosome conformation transforms from a cell type specific to a universal condensed organization, many chromatin factors and the transcription machinery are thought to no longer bind to the DNA, nuclear envelope and therefore lamina interactions disintegrate and new histone modifications specific for mitosis are deposited. After mitosis, chromatin returns to its uncondensed cell type specific shape, chromatin factors are bound again, the nuclear envelope and lamina interactions are restored and the histone modification pattern specific for interphase is reestablished (Hsiung et al., 2015; Martinez-Balbas, Dey, Rabindran, Ozato, & Wu, 1995; Naumova et al., 2013; Prescott & Bender, 1962). However, for many of these changes in the vertebrate mitotic chromatin it is unknown how, with which function and in which order they occur and how the interphase chromatin state is re-formed upon mitotic exit.
Mitosis has been an area of interest for over a century since condensation of chromosomes was first observed by microscopy. For many decades the main focus was the study of the mitotic chromatin through different microscopy techniques like FISH and immunofluorescence to localize chromatin proteins (Levsky & Singer, 2003). Because of the clear morphological features of mitotic cells, it is relatively easy to single out cells for study using microscopy. A downside of studies using these techniques is the limitations of the scale of the experiment, since it is not possible to do genome-wide experiments (probing the position of all loci) using microscopy. The development of high-throughput sequencing techniques have opened new ways to study mitotic chromosomes. These methods enable genome-wide detection of chromatin state, and the mapping of chromatin structure to specific sequences. However, these methods have their own set of limitations. Most particularly, these methods do not analyze single cells, but determine population-averaged features. For this, they typically require large numbers of cells, which means that for cell cycle studies one has to carefully synchronize large cell cultures in the cell cycle phase of interest (Banfalvi, 2011; Vassilev, 2006; Vassilev et al., 2006; Xeros, 1962). When doing such population based studies, one needs to obtain samples of a homogenous population. Although synchronization protocols have been optimized over the years, it is good to keep in mind that it remains difficult to obtain a fully synchronized population and that heterogeneity in the population can be the cause of inconsistencies between different studies and contamination with unsynchronized cells can reduce the quality of the obtained data.
Here we review and discuss chromosome conformation in interphase and mitosis and explore how epigenetic information can be contained within the local and global organization of chromatin. While we focus on vertebrate chromosomes, there is wealth of data on these phenomena in plants as well. We refer the reader to several key publications for those studies (Eichten, Schmitz, & Springer, 2014; Saze, 2008; Weimer, Demidov, Lermontova, Beeckman, & Van Damme, 2016).
Chromatin folding in Interphase and Metaphase
The fact that chromosomes do not simply consist of floating linear strands of DNA has been known since their discovery. In fact, chromosomes were first observed because of their dramatic condensation during mitosis, which allowed their visualization by microscopes of that time, described by Walther Flemming in the late 1800s (Flemming, 1882; Paweletz, 2001). For decades chromosomes and chromatin were studied by microscopy and techniques like X-ray crystallography. In the era of molecular biology and the development of sequencing, the research focus shifted towards unraveling the human genome by sequence and the concept of chromosome structure and conformation became less studied. However, it is clear other factors beyond DNA sequence contain instructions for the cell. The structural and physical organization of chromosomes inside the nucleus is an important carrier of information, which is important in many processes such as gene expression regulation and is in part specific for cell type identity (Bickmore & Van Steensel, 2013; Dekker, Marti-Renom, & Mirny, 2013; Dekker & Mirny, 2016).
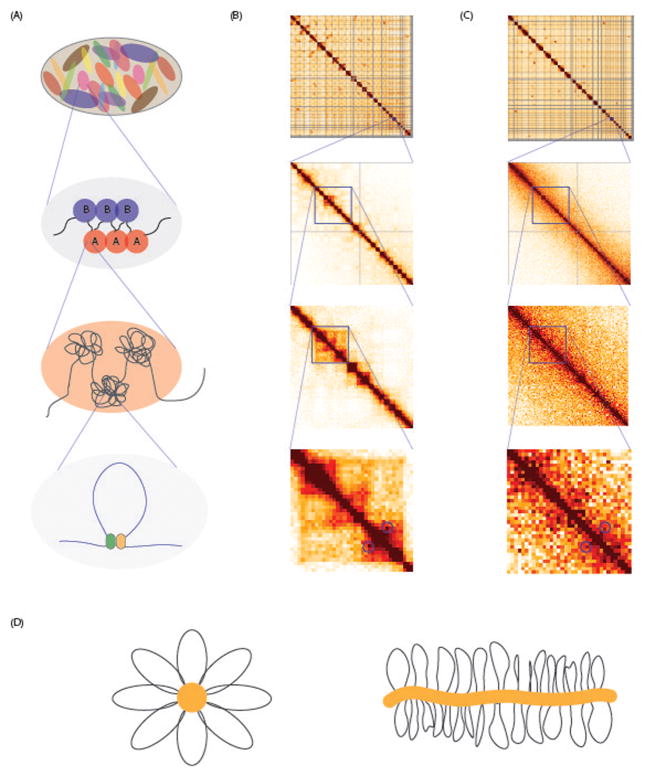
Eukaryotic chromatin is organized on different levels which are represented in figure 2a as cartoons and as observations of these organization levels in Hi-C heatmaps of interphase HeLa cells represented in figure 2b (previously published data in a study by Naumova et al (Naumova et al., 2013)). As interphase chromosomes are too large to freely diffuse inside the nucleus, they occupy their own territories (figure 2a–b, first panel). These individual chromosome territories were already observed in the 1990s and have been confirmed with microscopy and chromosome conformation capture techniques (Cremer et al., 1993; Lieberman-Aiden et al., 2009). Chromosomes can interact with neighboring chromosomal territories by looping part of one chromosome into another chromosome territory (Branco & Pombo, 2006). As a result of chromosome territories interchromosomal interactions are much less frequent than interactions between loci located on the same chromosome. The organization of chromosome territories within the nucleus is highly conserved between cell types and even across species (Tanabe et al., 2002).
Figure 2.
(A) Cartoon representation of chromosome territories (panel 1), compartments (panel 2), TADS (panel 3) and chromosome loops (panel 4). (B) Representation of the different chromosome organization levels in Hi-C heatmaps interphase of HeLa cell. Panel 1 shows chromosome territories in a heatmap showing all chromosomes. Panel 2 shows multiple compartments in a zoom in to the right arm of chromosome 18. Panel 3 shows a zoom in to 3.1–49.6 Mb of chromosome 18 representing multiple TADs. Panel 4 represents a possible looping interaction in a zoom in to chromosome 18 34.5–39.8 Mb. (C) Hi-C heatmaps of the same regions shown in (B) but for metaphase HeLa cells. Hi-C data shown was published in Naumova et al. (2013). (D) Model of the bottle brush polymer conformation of the mitotic chromosome suggested by Naumova et al. (2013) and Goloborodko et al (2016). A color version of this figure is available online.
The next layer of interphase chromosomal conformation is the organization in distinct sub-nuclear compartments where active and inactive regions, both from the same chromosome and occasionally from different chromosomes, cluster together (figure 2a–b, second panel) (Lieberman-Aiden et al., 2009; Wang et al., 2016). These regions are categorized as A or B compartments; A compartments are described as sub-nuclear neighborhoods of active and transcribed loci and B compartments mostly contain inactive regions. The compartmentalization correlates with other well-studied characteristics of the chromatin, such as gene expression, genome accessibility and histone marks: A-compartments are highly correlated with open, active, euchromatic regions and B compartments mostly contain loci of closed, inactive, heterochromatin regions. The organization in compartments leads to spatial separation of active loci from inactive loci, preventing interactions between active and inactive regions which may help prevent heterochromatin spreading. Compartmentalization is cell type specific (Lieberman-Aiden et al., 2009; Rao et al., 2014), likely because gene expression and chromatin modifications are cell type specific. Some studies show that the organization in active and inactive compartments is correlated with an increased concentration of factors involved in the regulation of these regions. An example of such an enrichment can be found in so-called transcription factories; a concept introduced by Iborra et al (Iborra, Pombo, Jackson, & Cook, 1996). Transcription factories are regions in the nucleus, where active genes and the transcription machinery are concentrated. This mechanism of enrichment would fit with an organization into A compartments. However, it is still to be determined whether compartmentalization in A and B compartments is cause or consequence of these proposed mechanisms.
The third layer of chromosomal organization in interphase is the organization of DNA into topologically associated domains, TADs (figure 2a and b, third panel) (Dixon et al., 2012; Nora et al., 2012). TADs are defined as contiguous genomic regions, typically several hundred kilobases in size that show elevated levels of self-interactions. TADs are separated from each other by boundaries that prevent interactions between loci located in adjacent TADs (Fudenberg et al., 2016; Giorgetti et al., 2014; Sanborn et al., 2015). TAD boundaries are enriched for certain protein binding sites. It is still under debate which proteins are involved in defining TAD boundaries and which genetic features define a TAD region, however some characteristics of TAD boundaries are well described, such as the CTCF binding motif (Dixon et al., 2012). These motifs are bound by structure-mediating proteins like CTCF and cohesin, which can act as an insulator between TADs (Phillips-Cremins & Corces, 2013; Rao et al., 2014; Tang et al., 2015; Wit et al., 2015; Zuin et al., 2014). Since CTCF binding motifs are genetically defined, TAD boundaries are regarded as universal features of all cell types (Dixon et al., 2012). Cell-type invariant TADs that can be located far apart on the same chromosome or on different chromosomes cluster together forming the cell type-specific A- and B-type compartments described above, depending on their chromatin and expression state in a given cell type. Therefore TADs have been proposed to be basic building blocks of chromosome and nuclear organization (Gibcus & Dekker, 2013).
At the scale of up to tens of Kb, chromatin interactions are organized in DNA loops (figure 2a–b fourth panel). Looping between two loci on a 10 to 100 kb scale, enables direct interaction between for example an enhancer and a promoter (Doyle, Fudenberg, Imakaev, & Mirny, 2014; Rao et al., 2014; Sanyal, Lajoie, Jain, & Dekker, 2012, Tang et al., 2015). Many looping interactions appear to occur between loci located within the same TAD or insulated domain (Hnisz et al., 2016; Jin et al., 2015; Smith, Lajoie, Jain, & Dekker, 2016). Even though TAD organization is often conserved among cell types, looping interactions can be cell type specific, but do rarely seem to cross TAD boundaries (Smith, Lajoie, Jain, & Dekker, 2016). The presence of a looping interaction can alter gene expression in a cell type specific manner by bringing promoters in close proximity to promoters (Deng et al., 2014; Sanyal et al., 2012; Tolhuis, Palstra, Splinter, Grosveld, & De Laat, 2002; Vernimmen, De Gobbi, Sloane-Stanley, Wood, & Higgs, 2007). Looping interactions have been suggested to function as a fine-tuning mechanism of (transcriptional) regulation. It has been shown for several diseases, like cancer and polydactyly, how alteration in looping interactions is associated with the disease phenotype (Hnisz et al., 2016; Lupianez et al., 2015; Zhao et al., 2009). To summarize, interphase chromatin architecture is established by clustering of sets of TADs into cell type specific A and B compartments. The cell type specific regulation of a certain locus within a TAD is then enabled by forming loops of for example regulating loci with an effector locus.
This partly cell type specific conformation of interphase chromosomes changes dramatically during mitosis. Observations using microscopy techniques already proved the dramatic changes many decades ago. The distribution of the DNA in the nucleus changes from amorphous territories to elongated rode shaped structures, with the characteristic banding pattern upon staining (Speicher & Carter, 2005; Uchida & Lin, 1971). It was shown that these mitotic bands along the chromosomes are the same between different cell types (Craig & Bickmore, 1993). Both the dramatic change in shape and the cell type indifferent band patterns already suggested loss of the higher organization in compartments and TADs known for interphase chromosomes, as the rod shape and the individualization of the chromosomes do not allow for interaction within and between compartments. Using 5C and Hi-C-techniques Naumova et al. observed these dramatic changes in a genome wide manner, represented in figure 2c. Clearly, cells are capable of reestablishing the same chromosomal organization in early G1 phase as was present before mitosis (Naumova et al., 2013). A recent study of Hsuing et al measured enhancer promoter interactions in interphase and mitosis using Capture-C, a technique to capture interactions anchored at hundreds of loci at the same time (C. Hsiung et al., 2016; Hughes et al., 2014). For the promoter enhancer pairs measured in this study, it was found that they are specific to interphase, and show to have largely reduced interaction frequencies in mitotic cells. This suggests that chromosomal organization in loops between functional elements is also impaired in mitosis, and that these interactions must be re-established in the next G1 phase.
Folding of the mitotic chromosome
Although the folding characteristics of interphase chromatin appear almost completely lost during mitosis, this does not mean mitotic chromatin has no higher order organization. The mitotic chromosome of vertebrates condenses 2–3 times in volume compared to interphase (Vagnarelli, 2012). The prevailing model for mitotic chromosome architecture that is supported by pioneering microscopy studies by Laemmli and co-workers, more recent 5C and Hi-C analyses and polymer modeling is that chromosomes fold as longitudinally compressed arrays or stochastically positioned consecutive chromatin loops (Earnshaw & Laemmli, 1983; Naumova et al., 2013). Furthermore, during the condensation process, sister chromatids are separated and individualized during pro- and prometaphase to accommodate proper division over the two new daughter cells (Liang et al., 2015; Llères, James, Swift, Norman, & Lamond, 2009; Nagasaka, Hossain, Roberti, Ellenberg, & Hirota, 2016).
The main machineries that drive mitotic chromosome morphogenesis are condensin complexes, including condensin I and II (Hirano & Mitchison, 1994; Kimura & Hirano, 1997; Ono et al., 2003). It was found that depletion of subunits of the condensin complexes delays the condensation process and progression of mitosis (Ono et al., 2003; T Hirota, Gerlich, Koch, Ellenberg, & Peters, 2004). Although the exact working mechanism is not yet completely understood, the prevailing model is that condensin functions as a ring-like structure that keeps two strands of DNA from the same chromosome together (Cuylen, Metz, Hruby, & Haering, 2013). Condensins are abundant chromosomal protein complexes that are located along the central axis of the mitotic chromosome, consistent with its proposed role to organize the chromosome as a long array of chromatin loops. Whereas condensin II is present in the nucleus throughout the cell cycle and mainly exerts its mitotic function in prophase, condensin I mostly gains access to the chromatin upon nuclear envelope breakdown in prometaphase (Hirota et al., 2004).
A model introduced by Nasmyth proposes that protein complexes like condensins are capable of forming loops that progressively become larger until further progression is blocked by other chromatin bound factors (e.g. other condensins) (Nasmyth, 2003). This idea is further elaborated in loop extrusion models, proposed in earlier form by Arthur Riggs over 25 years ago (Riggs, 1990), and recently developed to describe chromatin interaction data obtained with Hi-C (Dekker, 2014; Fudenberg et al., 2016; Goloborodko, Imakaev, Marko, & Mirny, 2016; Sanborn et al., 2015). Computational modeling and experimental studies applied extrusion models for understanding formation of interphase chromatin domains (Alipour & Marko, 2012; Dekker and Mirny, 2016; Fudenberg et al., 2016; Sanborn et al., 2015; Yang Zhang, Isbaner, & Heermann, 2013). Naumova et al. proposed that a loop extrusion process could lead to formation of the mitotic chromatin loop array (Naumova et al., 2013). One of the most recent studies directly tested the loop extrusion model for mitotic chromosome formation (Goloborodko, Imakaev, Marko, & Mirny, 2016). During the mitotic condensation process condensin complexes concentrate onto the chromatid axis and as interphase-specific boundary elements like CTCF dissociate from the mitotic chromatin, loop extrusion can occur unimpeded, which causes the DNA to progressively condense. Using this model, Goloborodko et al were able to computationally predict the condensation process into prophase-like chromosomes that show the same characteristics as was observed in experimental studies (Earnshaw & Laemmli, 1983; Naumova et al., 2013). Loop-extrusion leads to arrays of stochastically positioned consecutive loops. General chromatin attraction leads to further loop condensation and stacking of loops on top of each other. The physical characteristics of DNA predict steric repulsion between loops which results in a bottle-brush like organization of the DNA with loops arranged as rosettes around a more centrally located axis of loop bases, represented in figure 2d. This is a structure very reminiscent of the radial loop model for mitotic chromosomes proposed many years ago by Laemmli and co-workers (Earnshaw & Laemmli, 1983; Rattner & Lin, 1985). This loop repulsion also mediates separation of the sister chromatids after disentanglement is established, which will be described later in this section. Although the loop extrusion model can explain many features of the condensing chromatin, it does not explain the higher order rod-like organization chromosomes in mitosis. The forces involved in the loop-extrusion model would cause a complete condensation and collapse of the DNA into a ball shape, caused by the chromosomes sticking to each other, if there would not be an external agent that functions as a surfactant coating the mitotic chromosomes to prevent multiple mitotic chromosomes sticking together (Ohta, Wood, Bukowski-Wills, Rappsilber, & Earnshaw, 2011). A recent study suggested that the positively charged protein Ki-67 could act as such an agent to keep the mitotic chromosomes separated (Cuylen et al., 2016). Ki-67 is associated with nucleoli in interphase cells, however it has been known for many years that Ki-67 is associated with the outside of the mitotic chromosome (Scholzen et al., 2002). This makes Ki-67 a good candidate to be a key mediator in the individualization of mitotic chromosomes.
Another key player that regulates the organization of the mitotic chromosome is the cohesin complex, which holds sister chromatids together until late mitosis when the sister chromatids are separated and the DNA is divided between the new daughter cells (Foti et al., 2015; Haering, Löwe, Hochwagen, & Nasmyth, 2002; Ivanov & Nasmyth, 2005; Tanaka, Cosma, Wirth, & Nasmyth, 1999). Sister chromatids are tightly intertwined after DNA replication in S-phase up till prophase. This implies that the initial stages of condensation occur while the sister chromatids are entangled and interacting. However, sister chromatids need to be separated to enable proper segregation, equal distribution of chromosomes over the daughter cells and to prevent DNA breaks caused by the strong forces on the DNA by the mitotic spindle. It has been known that cohesin removal from the mitotic chromosome has two distinct pathways (Waizenegger, Hauf, Meinke, & Peters, 2000). Cohesin is first removed from the chromosome arms during prophase; however localization of cohesin at the centromeres is maintained. This enables coupled progression of the condensation and sister separation of the chromosome arms, while they are still attached at the centromeres necessary for metaphase plate alignment. Cohesin complexes located at the centromeres are removed after completion of the condensation process and formation of the mitotic spindle in metaphase to anaphase transition (Waizenegger et al., 2000). However, the question how the topological entanglement of the sister chromatids is resolved after removal from cohesin in prophase, remains to be addressed. A recent study from Liang et al (Liang et al., 2015) showed how topoisomerase IIα functions to disentangle the sister chromatids. In addition to this, Goloborodko et al showed in their computational study how addition of topoisomerase II-activity to the loop extrusion model leads to separation of the sister chromatids. The function of topoisomerase in sister segregation was later also confirmed using a labeling technique to visualize the chromatids separately by super resolution microscopy (Nagasaka, Hossain, Roberti, Ellenberg, & Hirota, 2016). It was shown that sister chromatid separation is initiated in early prophase, however further condensation is halted when topoisomerase IIα is inhibited.
Chromatin condensation not only occurs at the level of chromatin loops and whole chromosomes, at the nucleosomal level changes are observed during mitosis as well. As many chromatin binding factors are thought to migrate off the chromatin during mitosis, the nucleosomes can rearrange their relative positioning. Although it is unknown how the activity and movement of ring-like complexes like condensin, as proposed for loop extrusion, are affected by the presence nucleosomes on the DNA, it has been shown that nucleosomes are evicted from the chromatin in order for condensin to load and start forming loops (Toselli-Mollereau et al., 2016). One can imagine that the forces on the chromatin fiber during the condensation process might cause the nucleosomes to redistribute along the chromatin. Using an electron microscopy technique called EM-assisted nucleosome interaction capture (EMANIC), Grigoryev et al were able to capture hierarchical looping of nucleosome chains that are ordered in a zig-zag like fashion (Grigoryev, Bascom, Buckwalter, Schubert, & Woodcock, 2015). The zigzag stacking of nucleosomes is also present in interphase chromatin, however the higher order organization of loops of nucleosomes appears to be unique for mitosis (Grigoryev, 2012). This emphasizes the importance of nucleosomes, their positioning and their modifications in mitosis, which will be more elaborately discussed below.
Chromatin accessibility in mitosis
Mitotic chromatin is highly condensed and appears to have universal organization as detected by Hi-C. This does not mean that there are no differences in levels of local condensation and chromatin accessibility along the mitotic chromosome, as described for the interphase chromatin. A study by Hsuing et al observed variability in the accessibility of the mitotic chromatin using DNA hypersensitivity assay (Hsiung et al., 2015). Hsuing and colleagues showed that certain elements like promoters are maintained accessible during mitosis, where other elements such as enhancers are only accessible in interphase but not during mitosis. This is interesting, because this suggests there are chromatin features present in the interphase chromatin that are maintaining these promoter sites accessible during mitosis. The accessibility of promoter during mitosis sites also implies that the motifs, although temporarily less stable bound by factors,, maintain their open conformation and are accessible for these factors upon mitotic exit. This was surprising as the 3D organization of the mitotic chromatin does not seem to suggest differences across the mitotic chromatin (Naumova et al., 2013). Furthermore, again seemingly contradictory with the data on the 3D organization of the mitotic chromatin, this locus specific accessibility can be different between cell types and therefore enable epigenetic memory of chromatin accessibility after mitosis. The mechanism that enables the maintenance of chromatin accessibility in mitosis however still remains to be resolved and might be different for individual loci.
Mitotic Bookmarking and Epigenetic Memory
Mitotic chromosomes have their own, temporary, chromosomal organization with unique characteristics. Interestingly, this organization is highly similar between different cell types, whereas interphase chromosomal organization is on some levels highly cell type specific. Even though most transcription activity ceases and at least a subset of proteins dissociate from the chromosomes during mitosis, cells are capable of rearranging their chromosomes back into cell type specific conformations in early G1 (Martinez-Balbas et al., 1995; Naumova et al., 2013; Prescott & Bender, 1962). This suggest that the information to rearrange chromosomes into their interphase chromosomal organization after condensation is contained in the mitotic chromosomes, in the soluble fraction, or both even though it is temporary overruled by machineries in mitosis that enable chromosome condensation and segregation. One way cell type-specific information can be maintained could be through mitotic bookmarking of cell type specific gene regulatory elements by patterns of local histone modifications and other epigenetic marks. These mitotic bookmarks can be histone modifications and variants, DNA methylation, noncoding RNA and less frequently specific transcription factors and histone reader complexes that remain bound during mitosis (F. Wang & Higgins, 2013). There are two distinct functions of mitotic bookmarks. First, there are mitotic bookmarks that exert a function during mitosis, for example histone phosphorylation that is implicated in chromatin condensation (Wilkins et al., 2014). In addition to that, there are mitotic bookmarks that are important for epigenetic memory, which enables proper open or closed chromatin re-formation in early G1, like the phospho/methyl-switch H3K9me3-S10ph and the histone acetyl transferase Brd4 (Devaiah et al., 2016; Jeong, Cho, Park, Ko, & Kang, 2010). The latter bookmarks may act at single sites, like enhancers and promoters, or at larger domains, for example larger heterochromatic lamina associated domains. Below we describe different forms of mitotic bookmarking, their function and mechanism and give some examples of bookmarks that are passed on through mitosis.
Histone modifications and variants
During S-phase the cell replicates not just the DNA but also the local chromatin state. The appropriate set of histone modifications has to be added onto the new histones assembled on the newly replicated DNA. Although new nucleosomes are incorporated directly after DNA duplication, the histone modifications are added in the time between S-phase up till early G1. A recent study by Alabert et al showed that not all histone modifications are added to the new nucleosomes at the same time (Alabert et al., 2015; Lin, Yuan, Han, Marchione, & Garcia, 2016).
Analysis of histone modifications as bookmarks in mitosis has been limited and sometimes contradictory because of the limitations of proper antibodies used to study the marks using immunofluorescence microscopy or ChIP-seq. Although mass spectrometry techniques have been used to study histone modifications in an antibody free and unbiased way, these techniques are not without limits either. Mass spectrometry can measure the mass change caused by the histone modifications. Furthermore the co-localization of modifications on the same histone tail can be studied using mass spectrometry. However, a downside of mass spectrometry is that it cannot detect the location of the studied histones on the DNA, so the information of where these histone modifications reside on the mitotic chromatin is lost. In addition to that, like all population studies, mass spectrometry relies on the level of homogeneity of the synchronized population that is studied. A recently developed technique by the Bernstein lab combines the power of high resolution imaging with antibody detection of histone modifications followed by sequencing (Shema et al., 2016). Although this technique still relies on antibodies for detection, it can detect combinations of bivalent histone modifications at specific genomic locations.
Even though analysis of histone modifications can be challenging, it will be important to understand the pathways of mitotic bookmarks and the mechanisms by which modifications affect the dynamics and characteristics of mitotic chromosomes, and the transmission of gene regulatory programs that determine cell type identity. Although we will not be able to describe all studies, we will highlight recent and important studies of histone modifications and their modifiers in mitosis.
Histone Phosphorylation
One of the most pronounced histone modifications in mitosis is histone phosphorylation. Many serine and threonine residues on the histone 3 tail become phosphorylated during mitosis, which was already observed in the 1970s (Bradbury, Inglis, Matthews, & Sarner, 1973). Since many of these histone phosphorylation residues exert their function solely during mitosis and are dephosphorylated upon mitotic exit, this is an example of a mitotic bookmark that has a function during mitosis and may be involved marking gene regulatory elements specifically during mitosis when factors that bind them in interphase are not bound, or in organizing the mitotic chromosome itself. One well-studied histone phosphorylation event is histone 3 serine 10 phosphorylation (H3S10ph). H3S10ph is important for chromosome condensation, most likely through recruitment of regulatory and structural proteins, but the precise mechanisms through which this modification affects chromosome conformation are not fully understood (Antonin & Neumann, 2016; Johansen & Johansen, 2006; Wilkins et al., 2014). The kinase that is the main histone writer of this modification, Aurora B, is shown to be colocalized with other histone kinases like Haspin, which phosphorylates a second histone residues H3T3ph (Wang et al., 2010, 2012). Although Haspin and Aurora B can act on individual chromosome arms, centromeric histone H3 phosphorylation is regulated by a positive feedback loop of these kinases. Recruitment of Aurora B and Haspin and the subsequent hyper phosphorylation of the centromere, enables the recruitment of the chromosomal passenger complex (CPC) (Wang et al., 2011; Yamagishi, Honda, Tanno, & Watanabe, 2010). The CPC is required for attachment of the mitotic spindle and kinetochores to the mitotic chromatin, which is necessary for proper sister chromatid segregation and completion of cytokinesis (Gurden, Anderhub, Faisal, & Linardopoulos, 2016; Wang et al., 2012).
Phospho/Methyl Switches
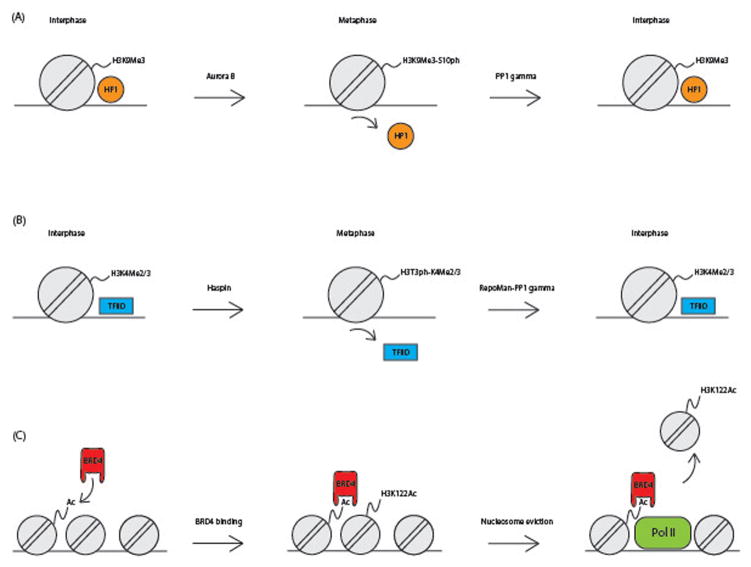
H3S10ph and H3T3ph not only exert their function during mitosis to guide recruitment of the CPC to centromeres, but also have a function in maintaining epigenetic memory along the chromosomal arms during mitosis. Several residues that can be phosphorylated are located next to a lysine, that can be mono, di or tri-methylated (Martin & Zhang, 2005). For instance H3S10 is located immediately adjacent to H3K9. Many of these lysines are modified and bound by regulating factors in interphase. However when the neighboring serine or threonine becomes phosphorylated in mitosis, these regulating factors can no longer bind the modified lysine residue. The temporary phosphorylation of the neighboring residue switches the lysine off as a regulating histone modification, a so-called phospho/methyl switch (Fischle, Wang, & Allis, 2003). An example of such a phospho/methyl switch is H3K9me3S10pho, where the function of tri-methylated H3 lysine 9 is affected by H3S10 phosphorylation in mitosis (figure 3a) (Toru Hirota, Lipp, Toh, & Peters, 2005; Jeong et al., 2010). The positioning of H3S10ph next to histone 3 lysine 9 di- and tri-methylation functions like a temporary shield from the chromatin binding factors. During interphase H3K9me3 acts like a heterochromatic mark, which binds to the heterochromatin protein 1 (HP1). HP1 and H3K9me3 together enable heterochromatic spreading and compaction of the heterochromatin (Lachner, O’Carroll, Rea, Mechtler, & Jenuwein, 2001). However, when the neighboring residue H3S10 is phosphorylated during mitosis, HP1 can no longer bind to H3K9me3. This enables the heterochromatin mark to be temporary overruled by the mitotic machinery, inactivating its role in interphase chromosome architecture and allows the chromatin to be condensed in a locus-independent mitosis-specific way. Then, upon mitotic exit, the H3S10ph is dephosphorylated by the PP1gamma complex and heterochromatin protein 1 (HP1) can bind H3K9me2/3 again which mediates proper reestablishment of the heterochromatin, and thus may contribute to the re-establishment of the interphase – specific spatial chromosome conformation (Qian, Beullens, Lesage, & Bollen, 2013).
Figure 3.
Examples of epigenetic bookmarks in mitosis and interphase. (A) The heterochromatic mark H3K9 trimethylation is shielded by H3S10 phosphorylation caused by the Aurora B kinase during mitosis, which causes heterochromatin protein 1 (HP1) to temporarily dissociated from the mitotic chromatin. When H3S10 is dephosphorylated by the PP1γ complex, HP1 binding is restored. (B) The histone mark H3K4 di and tri methylation becomes shielded during mitotis by the H3T3 phosphorylation mark regulated by the kinase Haspin. This causes the euchromatic regulatory protein TFIID to dissociate from the chromatin. Upon mitotic exit H3T3 is dephosphorylated by the RepoMan-PP1 γ complex which restores TFIID binding to the interphase chromatin. (C) The histone acetyl transferase BRD4 can bind to histone acetylation marks in interphase and mitosis. When bound to the chromatin BRD4 can then acetylate H3K122Ac, which results in nucleosome eviction. Local nucleosome eviction enables reorganization of the nucleosome distribution and binding of big complexes like the transcription machinery. A color version of this figure is available online.
There are also phospho/methyl switches at euchromatic marks that are temporarily affected by a neighboring phosphorylated residue. An example of this is H3T3ph/H3K4me3 (Varier et al., 2010), which is represented in figure 3b. H3 lysine 4 tri-methylation is a mark known to be associated with active promoter sites (Hon, Hawkins, & Ren, 2009). H3K4me3 on the interphase chromatin has high affinity for the transcription factor TFIID, which is known to recruit the transcription machinery (Orphanides, Lagrange, & Reinberg, 1996; Vermeulen et al., 2007). In mitotic chromatin however, the H3K4me3 neighboring residue H3T3 becomes phosphorylated by Haspin (Eswaran et al., 2009). TFIID binding to H3K4me3 has been shown in a collaborative study by the labs of Jonathan Higgins and Marc Timmers to be severely reduced in mitosis as a result of H3T3 phosphorylation (Varier et al., 2010). Upon mitotic exit, H3T3 becomes dephosphorylated by the phosphorylase complex RepoMan PP1 gamma (Prévost et al., 2013; Qian, Lesage, Beullens, Van Eynde, & Bollen, 2011). Once the H3T3 residue is no longer phosphorylated, TFIID can bind again to H3K4me3 and RNA polII can bind to the promoter site to start transcription. The importance of maintenance of H3K4me3 as mitotic bookmark is emphasized by the fact that H3K4me3 is copied on to the new nucleosome early after DNA replication. This is in contrast with H3K4me1, a mark for enhancer regions, which is not shielded by H3T3ph and is shown to be added later in the cell cycle (Lin et al., 2016; Varier et al., 2010). This result, combined with observations by Hsiung et al that showed that chromatin in promoter regions, typically marked by H3K4me2/3, remains accessible, whereas many enhancer regions close, or become less accessible, during mitosis (Hsiung et al., 2015), suggests that bookmarking sites are correlated with maintenance of accessibility at these sites through mitosis.
Phospho/methyl-switches provide an elegant system to temporarily overrule the regulatory and structural effects of cis-elements by dissociating the trans factors that in interphase mediate their activity including long-range interactions with other elements, while maintaining their positional information so that in the next G1 the same set of regulatory elements can be re-activated. Although a lot is still unknown about the exact working mechanisms and whether the kinases and phosphorylases are specifically targeted to these sites, the phospho/methyl switch is one of the best described and understood mechanisms of mitotic bookmarking.
Histone Acetylation
Most histone acetyltransferases and histone deacetylases migrate off the mitotic chromatin (Kruhlak et al., 2001). This implies that many acetylation marks once added after S-phase and G2 are stable throughout mitosis. Although phosphorylation marks are not known to shield acetylation marks like they do for some methylation marks, there are pathways that may help the reestablishment of factors binding to these acetylation marks and the associated chromatin after mitosis. An example is the chromatin binding protein bromodomain 4 protein (BRD4), that is known to bind either H3K14 or H4K5 or K12 acetylated residues (Chiang, 2009; Dey, Nishiyama, Karpova, McNally, & Ozato, 2009). Its working mechanism in interphase is represented in figure 3c. Brd4 binds acetylated histones, but also has histone acetyltransferase activity itself, which preferentially acetylates the H3 lysine 122 residue and several lysine residues on the tail of H3 and H4 (Devaiah et al., 2016). H3K122 is located in the core of the nucleosome where the nucleosome is bound to the DNA and acetylation of this residue causes nucleosome clearance of the chromatin (Tropberger et al., 2013). The removal of nucleosomes results in local chromatin decompaction and exposes free DNA to which protein complexes like the transcription machinery can bind (Devaiah et al., 2016). Although this process is only described in interphase and it is unknown if the histone acetyltransferase activity of Brd4 is present in mitosis, it is known that Brd4 is one of the few HATs that remain bound to mitotic chromatin (Dey, Chitsaz, Abbasi, Misteli, & Ozato, 2003; Dey et al., 2009). Furthermore, the sites where it remains bound to the mitotic chromatin are associated to post-mitotic transcription and Brd4 overexpression was shown to accelerate the reactivation of transcription upon mitotic exit (Dey et al., 2009; Zhao, Nakamura, Fu, Lazar, & Spector, 2011). This suggests that Brd4 and its associated histone acetylation marks function as bookmarks during mitosis and enable reestablishment of post-mitotic transcription in a timely matter. One can imagine that loss of chromatin accessibility of certain regions as shown by Hsiung et al can be reestablished after mitosis by enzymes like Brd4 (Hsiung et al., 2015).
Another histone acetylation mark associated with reactivation of post-mitotic transcription is H3K27Ac. In a recent paper by Hsiung et al. polII binding upon mitotic exit was measured using ChIP-seq to study prevalence, location and order of spikes in transcription after mitotic exit (Hsiung et al., 2016). An interesting observation was that the histone modification H3K27Ac could best predict which sites were going to be transcriptionally active after mitosis. The regulatory pathways of H3K27Ac deposition and inheritance, and how this modification is involved in a spike in post-mitotic transcription still remain to be resolved. It will be very interesting to better understand the interplay of histone modification and re-initiation of transcription upon mitotic exit.
Histone Variants
Not much is known about the function of histone variants in mitosis and when histone variants are incorporated after DNA replication. During S-phase only the canonical histone variants H3.1 and H3.2 are incorporated in the newly synthesized DNA, which implies that histone variants that are functioning as mitotic bookmarks should be incorporated before the end of mitosis (Campos et al., 2010). Which remodelers are regulating this replacement and when in the cell cycle this occurs is not known. The best studied histone variant with a known function in mitosis is the centromeric variant H3, CENP-A in humans (Ahmad & Henikoff, 2001; Sánchez & Losada, 2011). CENP-A is localized at centromeres and has been shown to help the localization of the mitotic kinetochores, which enables proper DNA segregation and cytokinesis. Defects in the CENP-A localization have been shown to affect proper DNA segregation in mitosis (Maheshwari et al., 2015). A recent study by Roulland et al showed the importance of the flexible tails of CENP-A in mitosis. When the flexible tail of CENP-A is switched with the more rigid tail of the canonical H3, several kinetochore proteins dislocate and severe mitotic and cytokinetic defects like aneuploidy are observed (Roulland et al., 2016). Although DNA sequences in centromeres have been shown not to be conserved, the centromeric H3 histone variant shows high levels of similarity across species (Cheeseman et al., 2004). This suggests that DNA sequence does not determine sites of CENP-A incorporation, but is likely epigenetically determined, e.g. by the presence of old CENP-A. CENP-A incorporation is uncoupled from DNA-replication and is mediated by a dedicated histone chaperone HJURP (Bassett et al., 2012; Dunleavy et al., 2009; Foltz et al., 2009; McKinley & Cheeseman, 2015).
Histone variant H3.3 is another histone 3 variant known to be inherited through mitosis. H3.3 can function as a mark for promoters of transcriptionally active genes (Chow et al., 2005). H3.3-containing nucleosomes appear less stable than the canonical H3 containing nucleosomes, which enables nucleosome clearance and remodeling, and therefore initiates transcription (Ahmad & Henikoff, 2002). Furthermore, H3.3 is associated with many active histone modifications, such as histone acetylation and methylation (e.g. H3K4me3), which attracts histone modifiers that can spread the histone modifications to neighboring histones, both H3.3 and canonical H3 histones. In contrast to other H3 variants, H3.3 can be incorporated to the chromatin independent of DNA replication, as its remodeler is expressed in G1 and G2 as well as S-phase (Campos et al., 2010; Chow et al., 2005; Ray-Gallet et al., 2002; Tagami, Ray-Gallet, Almouzni, & Nakatani, 2004).
Linker Histone H1
Hyper phosphorylation of the histone variant H1 is also a hallmark of mitosis (Boggs, Allis, & Chinault, 2000; Bradbury, Inglis, & Matthews, 1974). Histone H1 accumulates phosphorylation marks during the cell cycle, starting at no or low levels of phosphorylation in G1 to the highest levels of phosphorylation in M-phase (Bleher & Martin, 1999; Halmer & Gruss, 1996). Some histone H1 variants have been shown to become more phosphorylated than other variants, however all H1 variants gain phosphorylation marks in mitosis (Clausell, Happel, Hale, Doenecke, & Beato, 2009; Gréen, Lönn, Peterson, Ollinger, & Rundquist, 2010). It has been suggested that H1 and its phosphorylated forms are key mediators of chromatin structure and chromosome condensation (Ohsumi, Katagiri, & Kishimoto, 1993; Thoma, Koller, & Klug, 1979; Thoma & Koller, 1977). Maresca et al immuno-depleted H1 in Xenopus laevis egg extracts (Maresca, Freedman, & Heald, 2005) and found chromatin does not condense properly and chromosomes have elongated arms. Furthermore, the chromatids showed misalignment on the metaphase plate, which lead to defects in sister segregation. Interestingly, positioning of the kinetochore proteins was unaffected in the H1 depleted chromatin. It has been suggested that the centromeric H3 variant CENP-A might not need histone H1 to interact with the linker DNA, as CENP-A contains a conserved domain that shows highly similarity with motifs present on the H1 tails. The results of the recent study by Roulland et al confirms these suggestions as they show a low binding affinity between CENP-A and H1 as a result of the flexible histone tails of CENP-A (Roulland et al., 2016).
DNA methylation
DNA methylation is a layer of epigenetic regulation that is closest to the genetic information in the DNA and was one of the first epigenetic marks to be discovered (Holliday & Pugh, 1975; Holliday, 1987). Methylated cytosines can function as a chromatin silencing mark and enable imprinting of gene silencing (Curradi, Izzo, Badaracco, & Landsberger, 2002). In contrast to histone modifications and histone variants, DNA methylation of the newly synthesized strand is established immediately after the replication fork has passed, using the old strands as template (Wigler, Levy, & Perucho, 1981). This enables correct copying of cytosine methylation and prevents loss during multiple cell divisions. This makes DNA methylation unique among the other mitotic bookmarks, since the copying of the other bookmarks are delayed and spread out over G2 and M-phase. DNA binding of factors can be both positively and negatively affected by DNA methylation (Jones, 2012; Maurano et al., 2015; Schübeler, 2015), which can influence the chromatin accessibility state and long-range chromatin interactions. How these phenomena are altered or modulated in mitotic chromosomes to facilitate the folding of the chromosomes as linear loop arrays is not known yet. It will be interesting to study the interplay between DNA methylation and other mitotic bookmarks that mark repressive regions in the chromatin like histone 3 lysine 9 tri-methylation.
Non-coding RNA
During recent years roles for non-coding RNAs (ncRNAs) in epigenetic regulation of the genome have been uncovered. Most of these studies are done in non-synchronously cycling cells and little is known about roles of ncRNA and formation of mitotic chromosomes. It was generally assumed that RNAs do not play a big role in the mitotic chromatin, since the transcription machinery stalls and migrates off the mitotic chromatin (Prescott & Bender, 1962). This implies that there is no production of new RNAs during mitosis and RNA molecules that are retained on the mitotic chromatin have to be produced before mitosis initiation. A recent study however showed the presence of large group of ncRNAs during mitosis, which were coined mitotic chromatin associated RNAs, mCARs (Meng et al., 2016). These authors used a novel technique called 5′-tag sequencing to detect a large set of new ncRNAs detected that either bind mitotic chromatin (mCARs) or interphase chromatin (iCARs). A large fraction of the detected mCARs consisted of small nucleolar RNA molecules (snoRNAs) and long non-coding RNAs (lncRNAs). Furthermore, it was shown that the mCARs are highly conserved across species in a wide range of vertebrate species (Meng et al., 2016), which suggests a conserved function of these mCARs in mitosis. In an attempt to study the localization of these mCARs on mitotic chromatin, it was observed that there are two classes of mCARs which located either on the condensed chromosomal exterior or on the interior of the condensed chromosome. However, it remains to be resolved what the role of mCARs is during mitosis, whether their identity and location are cell type-specific, and whether they can function as a mitotic bookmarks.
Transcription Factors
It was suggested for many years that most transcription factors and other interphase chromatin binding factors migrate from the chromatin upon mitotic entry and would remain in the cytosol until after DNA decondensation (Chen et al., 2005). However several studies have identified chromatin binding factors that remain on the mitotic chromatin, like the previously described histone acetyl transferase BRD4 and transcription factors like FOXA1, MYC and RUNX2, which have been shown to be important in cell fate determination (Caravaca et al., 2013; Devaiah et al., 2016; Topham et al., 2015; Young et al., 2007). FOXA1 was shown to specifically remain bound to highly transcribed regions in mitosis and promotes initiation of transcription of these regions after mitosis (Caravaca et al., 2013). Although it is not clear why some transcription factors remain on the mitotic chromosome and others migrate away from the chromatin, it is clear that some transcription factors play an important role in mitotic bookmarking and transcriptional reactivation after mitosis (Zaret, 2014). A recent paper by Teves et al. even suggests that many transcription factors remain associated to mitotic chromosomes, although in a more dynamic fashion compared to interphase (Teves et al., 2016). Teves et al. show that earlier studies showing transcription factors migrating of mitotic chromatin might be caused by fixation artefacts. Future studies will have to explore which transcription factors indeed remain bound to the mitotic chromosome and which function these factors exert during mitosis.
Lamina interactions
One of the major morphological changes of the vertebrate cell that occurs during mitosis is the breakdown of the nuclear envelope. During interphase the proteins that coat the inside of the nuclear membrane, the nuclear lamina, play roles in providing support and directing higher order chromosome folding and nuclear organization (Chubb, Boyle, Perry, & Bickmore, 2002; van Steensel & Dekker, 2010). Using an elegant system to track individual lamina interactions in single cells over time, it was shown that lamina interacting regions are not necessarily the same after mitotic exit and that the lamina interacting regions reassemble over the nuclear envelope. However the regions that interact with the nuclear lamina after mitosis are always associated with other regions that are associated with transcription repression and regions assigned as LADs (lamina interacting domains) in population studies (Kind et al., 2013). This implies that information regarding lamina interactions are not maintained by a mitotic bookmark throughout mitosis, however the epigenetic marks that label these regions as repressed and candidates for lamina interactions are maintained throughout mitosis. One of the histone modifications that could act as such a mark is H3K9me2, as sites of hyper methylated nucleosomes are typically found in regions containing LADs (Kind et al., 2013).
Meiotic bookmarking
Although not elaborately described in this review, it is known that epigenetic marks are not only inherited through mitotic cell divisions, but can also be transmitted through meiosis and passed on to next generations. Recent studies in Drosophila showed how epigenetic marks in the parental epigenome are inherited into the following generation (Öst et al., 2014). This was surprising considering that spermatocytes do not have nucleosomes (or only a small number at specific sites), but use other DNA binding proteins to enable compact folding and protection against DNA damage causing factors to which the spermatozoa are exposed (Banerjee, Smallwood, & Hultén, 1995; Brunner, Nanni, & Mansuy, 2014). Even more so, it has been shown that upon fertilization, the gametes undergo epigenome reprogramming (Gill, Erkek, & Peters, 2012). However, studies like those performed by Ost et al showed that specific marks are able to escape this epigenetic reprogramming and can be passed on to epigenome of offspring. Further research is needed to explore the mechanisms by which meiotic epigenomic bookmarking is established and maintained.
Mitotic bookmarking in differentiation
As described in the paragraphs above, there are many different mechanisms that can establish mitotic bookmarking and epigenetic inheritance. However, one can imagine that the epigenetic marks need to change when cells undergo differentiation. During differentiation the chromatin characteristics of cells changes in many ways. Chromatin organization is changed at the scale of positioning and modification of individual nucleosomes at cell type specific gene regulatory elements (The ENCODE Project Consortium et al., 2012; Thurman et al., 2012), at the level of long-range interactions between such elements and the formation chromatin domains such as A- and B- compartments (Beagan et al., 2016; Sanyal et al., 2012; Wijchers et al., 2016). Correspondingly, gene expression programs are affected (Djebali et al., 2012). It is unlikely that large-scale changes in chromosome conformation are made during G1 as chromosomes are too big to move around in the interphase nucleus, which would be necessary to change the structures on the level of compartments (Dekker & Mirny, 2016; Rosa & Everaers, 2008). During mitotic exit and early G1 chromosomes decondense and the nucleus reforms, providing the cell a window of opportunity to spatially rearrange its genome in accordance with changes in cell type. This concept is interesting in the context of cell type differentiation. During differentiation there are two scenarios, one where the cell divides into two equal daughter cells with a distinct cell type from the original cell. The second scenario is one where the two daughter cells have different states, e.g. one daughter cell is initiated to become a different cell type and the other daughter cell will remain the same type as the mother cell, a stem cell. In both scenarios epigenetic features change before, during or after mitosis, suggesting roles for bookmarking processes. In addition, in the second scenario, where the daughter cells acquire each a different state, epigenetic changes in bookmarks are specific to each of the sister chromatids. This adds another layer of complexity to the mechanisms of epigenetic inheritance and modification: how do cells change their mitotic bookmarks in order to initiate correct differentiation and how do cells initiate and control differences in epigenetic states of otherwise identical sister chromatids?
Several histone modifications have been described to change during cell type differentiation and repression of enzymes that deposit these modifications represses the differentiation process and the activation of differentiation-dependent genes. Examples of these are H3K4me3, which has been shown to change during differentiation of human embryonic stem cells, and H3K36me3, which was shown to be required for differentiation of mouse embryonic stem cells to endoderm (Grandy et al., 2016; Yuanliang Zhang et al., 2014).
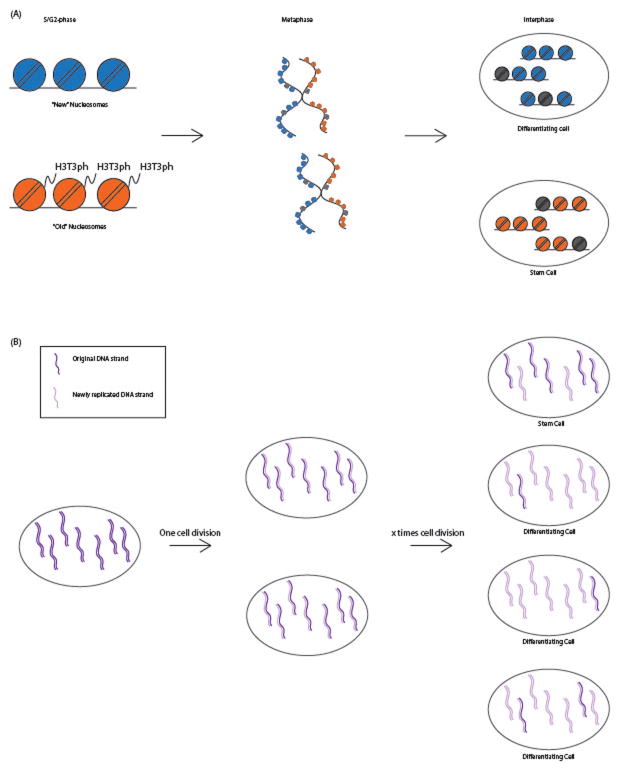
Several techniques have been developed to study how sister chromatids separate from each other. One of these techniques is Strand-seq (Falconer et al., 2010, 2012). Using BrdU labelling the sister chromatids can be separately observed and followed over stem cell differentiation. A hypothesis called the Immortal Strand Hypothesis has been introduced many years ago that proposes that stem cells will try to maintain the original copy of the chromosome to prevent accumulation of DNA replication mistakes (Cairns, 1975). Although some studies have contradicted this hypothesis, it is clear that at least some cells are retaining certain chromosomes in the daughter stem cells specifically (Conboy, Karasov, & Rando, 2007; Karpowicz et al., 2005; Potten, Owen, & Booth, 2002; Shinin, Gayraud-Morel, Gomès, & Tajbakhsh, 2006; G. H. Smith, 2005). Using Strand-seq, the distribution of strands was followed over multiple cell cycles after labelling with BrdU for 1 cycle. The immortal strand hypothesis suggests that for some chromosomes the labeled copy of the chromosome will be unequally distributed into on daughter cell, the stem cell (represented as a flowchart in figure 4a).
Figure 4.
Mitotic bookmarks in cell differentiation. (A) Representation of the silent sister hypothesis as suggested by Falconer et al (2013). During stem cell differentiation it is believed that the daughter cells destined to be the stem cell will retain the original DNA strands of some chromosomes, where the differentiation daughter cells will mainly contain the newly replicated DNA strands. (B) Nucleosomes on one sister chromatid are specifically labelled with the H3T3 phosphorylation mark. This enables the cell to retain the sister chromatid containing to the stem cell and the other sister chromatid will be passed on to the differentiation daughter cell. A color version of this figure is available online.
In order to understand how histones and there modifications can segregate to the differentiating daughter cell, it is important to understand that the H3/H4 tetramer of nucleosomes are always incorporated together in the nucleosomes as the parental H3/H4 tertramer stays together during DNA replication. This implies that nucleosomes can be assigned as new and old nucleosomes after DNA replication (Xu et al., 2010). A paper by Tran et al in 2013 already showed that new and old nucleosomes are not always equally represented on each sister chromatid, as they show that in stem cell differentiation of the male germline of Drosophila melanogaster preexisting nucleosomes preferentially segregate to the germline stem cell, whereas the new nucleosomes segregate to the differentiating daughter cell (Tran, Lim, Xie, & Chen, 2013). Furthermore, recent studies in Drosophila and mammalian cell lines observed differences in histone modifications between these old and new nucleosomes (Alabert et al., 2015; Lin et al., 2016; Tran, Lim, & Xie, 2012). It was shown that H3T3 phosphorylation is preferentially more represented on newly synthesized nucleosomes in the male Drosophila germline, represented in figure 4b, and when H3T3ph is impaired, proper differentiation of the germline was affected (Xie et al., 2015). It will be very interesting to explore the pathways and mechanisms that are used to initiate epigenetic changes in cellular phenotype, how differences between sister chromatids are established and proper sister segregation is controlled.
Concluding remarks
We are only starting to understand the mechanisms by which epigenetic information contained within the vertebrate chromatin is transmitted through mitosis and how this occurs in the context of a mitotic chromosome conformation that is dramatically different from interphase. One important question that remains unanswered is how molecular details of epigenetic bookmarks are read in early G1 and enable re-establishment of cell type specific chromatin organization. Insights into these processes promise not only to lead to mechanistic understanding of mitotic inheritance of cell type specific chromatin state, they will also reveal how the spatial organization of interphase chromosomes is determined in general by the action of cis-acting elements along the chromatin fiber. This will also lead to a better understanding of what epigenetic mechanisms underlie processes in which cell type identity is changed, for example in stem cell differentiation or in diseases that result in cancer development and aging.
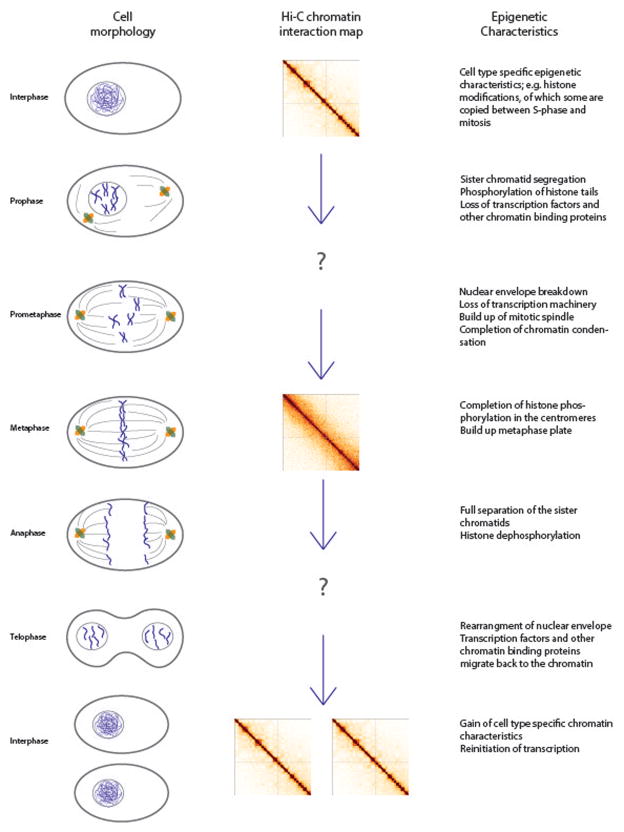
Figure 1.
Overview of cell morphological changes, changes in the chromatin organization and changes known epigenetic characteristics during the different phases of mitosis. Hi-C data shown is in HeLa cells and was previously published in Naumova et al (2013). A color version of this figure is available online.
Footnotes
Declaration of Interest
Work in our laboratory is supported by the National Human Genome Research Institute (R01 HG003143, U54 HG007010, U01 HG007910), the National Cancer Institute (U54 CA193419), the NIH Common Fund (U54 DK107980, U01 DA 040588), the National Institute of General Medical Sciences (R01 GM 112720), and the National Institute of Allergy and Infectious Diseases (U01 R01 AI 117839). The authors declare that they have no competing interests. J.D. is an investigator of the Howard Hughes Medical Institute.
References
- Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. Journal of Cell Biology. 2001;153(1):101–109. doi: 10.1083/jcb.153.1.101. http://doi.org/10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. The Histone Variant H3.3 Marks Active Chromatin by Replication-Independent Nucleosome Assembly ment are not clear. A study in Tetrahymena concluded that no protein difference between histone H3 variants was required for replacement histone deposition and Molecular. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen O, … Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes and Development. 2015;29(6):585–590. doi: 10.1101/gad.256354.114. http://doi.org/10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Research. 2012;40(22):11202–11212. doi: 10.1093/nar/gks925. http://doi.org/10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Neumann H. Chromosome condensation and decondensation during mitosis. Current Opinion in Cell Biology. 2016;40:15–22. doi: 10.1016/j.ceb.2016.01.013. http://doi.org/10.1016/j.ceb.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Smallwood a, Hultén M. ATP-dependent reorganization of human sperm nuclear chromatin. Journal of Cell Science. 1995;108(Pt 2):755–765. doi: 10.1242/jcs.108.2.755. [DOI] [PubMed] [Google Scholar]
- Banfalvi G. Overview of Cell Synchronization. 2011:1–23. doi: 10.1007/978-1-61779-182-6_1. http://doi.org/10.1007/978-1-61779-182-6_1. [DOI] [PubMed]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, … Black BE. HJURP Uses Distinct CENP-A Surfaces to Recognize and to Stabilize CENP-A/Histone H4 for Centromere Assembly. Developmental Cell. 2012;22(4):749–762. doi: 10.1016/j.devcel.2012.02.001. http://doi.org/10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan JA, Gilgenast TG, Kim J, Plona Z, Norton HK, Hu G, … Phillips-Cremins JE. Local Genome Topology Can Exhibit an Incompletely Rewired 3D-Folding State during Somatic Cell Reprogramming. Cell Stem Cell. 2016;18(5):611–24. doi: 10.1016/j.stem.2016.04.004. http://doi.org/10.1016/j.stem.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, Van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. doi: 10.1016/j.cell.2013.02.001. http://doi.org/10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Bleher R, Martin R. Nucleo-cytoplasmic translocation of histone H1 during the HeLa cell cycle. Chromosoma. 1999;108(5):308–316. doi: 10.1007/s004120050382. http://doi.org/10.1007/s004120050382. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Allis CD, Chinault AC. Immunofluorescent studies of human chromosomes with antibodies against phosphorylated H1 histone. Chromosoma. 2000;108(8):485–90. doi: 10.1007/s004120050400. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10794570. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, … Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biology. 2005;3(5):e157. doi: 10.1371/journal.pbio.0030157. http://doi.org/10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EM, Inglis RJ, Matthews HR. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature. 1974;247:257–261. doi: 10.1038/247257a0. [DOI] [PubMed] [Google Scholar]
- Bradbury EM, Inglis RJ, Matthews HR, Sarner N. Phosphorylation of very-lysine-rich histone in Physarum polycephalum. Correlation with chromosome condensation. European Journal of Biochemistry / FEBS. 1973;33(1):131–9. doi: 10.1111/j.1432-1033.1973.tb02664.x. http://doi.org/10.1111/j.1432-1033.1973.tb02664.x. [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biology. 2006;4(5):780–788. doi: 10.1371/journal.pbio.0040138. http://doi.org/10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AM, Nanni P, Mansuy IM. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics & Chromatin. 2014;7(1):2. doi: 10.1186/1756-8935-7-2. http://doi.org/10.1186/1756-8935-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013;10(12):1213–8. doi: 10.1038/nmeth.2688. http://doi.org/10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:cp1. doi: 10.1038/255197a0. http://doi.org/10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WHW, … Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nature Structural and Molecular Biology. 2010;17(11):1343–1351. doi: 10.1038/nsmb.1911. http://doi.org/10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes & Development. 2013;27(3):251–60. doi: 10.1101/gad.206458.112. http://doi.org/10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes and Development. 2004;18(18):2255–2268. doi: 10.1101/gad.1234104. http://doi.org/10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural protiens. Journal of Cell Biology. 2005;168(1):41–54. doi: 10.1083/jcb.200407182. http://doi.org/10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biology Reports. 2009;1(December):98. doi: 10.3410/B1-98. http://doi.org/10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, … Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Reports. 2005;6(4):354–60. doi: 10.1038/sj.embor.7400366. http://doi.org/10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Current Biology. 2002;12(6):439–445. doi: 10.1016/s0960-9822(02)00695-4. http://doi.org/10.1016/S0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PloS One. 2009;4(10):e0007243. doi: 10.1371/journal.pone.0007243. http://doi.org/10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biology. 2007;5(5):1120–1126. doi: 10.1371/journal.pbio.0050102. http://doi.org/10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium Roadmap Epigenomics. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, … Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. http://doi.org/10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Bickmore WA. Chromosome bands--flavours to savour. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 1993;15(5):349–54. doi: 10.1002/bies.950150510. http://doi.org/10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, … Lichter P. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symposia on Quantitative Biology. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Molecular and Cellular Biology. 2002;22(9):3157–73. doi: 10.1128/MCB.22.9.3157-3173.2002. http://doi.org/10.1128/MCB.22.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, Muller-Reichert T, Neumann B, Poser I, … Gerlich DW. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;44:1–19. doi: 10.1038/nature18610. http://doi.org/10.1038/nature18610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Hruby A, Haering CH. Entrapment of Chromosomes by Condensin Rings Prevents Their Breakage during Cytokinesis. Developmental Cell. 2013;27(4):469–478. doi: 10.1016/j.devcel.2013.10.018. http://doi.org/10.1016/j.devcel.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics & Chromatin. 2014;7(1):1–12. doi: 10.1186/1756-8935-7-25. http://doi.org/10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom Ma, Mirny La. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature Reviews Genetics. 2013;14(6):390–403. doi: 10.1038/nrg3454. http://doi.org/10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164(6):1110–1121. doi: 10.1016/j.cell.2016.02.007. http://doi.org/10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science (New York, NY) 2002;295(5558):1306–11. doi: 10.1126/science.1067799. http://doi.org/10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, … Blobel GA. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158(4):849–860. doi: 10.1016/j.cell.2014.05.050. http://doi.org/10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, … Singer DS. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nature Structural & Molecular Biology. 2016;23(August 2015):1–12. doi: 10.1038/nsmb.3228. http://doi.org/10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(15):8758–8763. doi: 10.1073/pnas.1433065100. http://doi.org/10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Molecular Biology of the Cell. 2009;20(23):4899–909. doi: 10.1091/mbc.E09-05-0380. http://doi.org/10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, … Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80. doi: 10.1038/nature11082. http://doi.org/10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis Ca, Merkel A, Dobin A, Lassmann T, Mortazavi A, … Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. http://doi.org/10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny La. Chromatin Loops as Allosteric Modulators of Enhancer-Promoter Interactions. PLoS Computational Biology. 2014;10(10):e1003867. doi: 10.1371/journal.pcbi.1003867. http://doi.org/10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Almouzni-Pettinotti G. HJURP Is a Cell-Cycle-Dependent Maintenance and Deposition Factor of CENP-A at Centromeres. Cell. 2009;137(3):485–497. doi: 10.1016/j.cell.2009.02.040. http://doi.org/10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Laemmli UK. Architecture of metaphase chromosomes and chromosome scaffolds. Journal of Cell Biology. 1983;96(1):84–93. doi: 10.1083/jcb.96.1.84. http://doi.org/10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Schmitz RJ, Springer NM. Epigenetics: Beyond Chromatin Modifications and Complex Genetic Regulation. Plant Physiology. 2014;165(3):933–947. doi: 10.1104/pp.113.234211. http://doi.org/10.1104/pp.113.234211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, … Knapp S. Structure and functional characterization of the atypical human kinase haspin 2VUW 3IQ7 3DLZ inhibitor iodotubercidin & AMP. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(48):20198–20203. doi: 10.1073/pnas.0901989106. http://doi.org/10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Chavez Ea, Henderson A, Poon SSS, McKinney S, Brown L, … Lansdorp PM. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463(7277):93–97. doi: 10.1038/nature08644. http://doi.org/10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Hills M, Naumann U, Poon SSS, Chavez Ea, Sanders AD, … Lansdorp PM. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nature Methods. 2012;9(11) doi: 10.1038/nmeth.2206. http://doi.org/10.1038/nmeth.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425(6957):475–9. doi: 10.1038/nature02017. http://doi.org/10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Flemming W. Zellsubstanz, kern und zelltheilung. Leipzig: F.C.W. Vogel; 1882. [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, … Cleveland DW. Centromere-Specific Assembly of CENP-A Nucleosomes Is Mediated by HJURP. Cell. 2009;137(3):472–484. doi: 10.1016/j.cell.2009.02.039. http://doi.org/10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, … Buonomo SB. Nuclear architecture organized by Rif1 underpins the replication-timing program. Molecular Cell. 2015:1–14. doi: 10.1016/j.molcel.2015.12.001. http://doi.org/10.1016/j.molcel.2015.12.001. [DOI] [PMC free article] [PubMed]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Reports. 2016;(15):1–12. doi: 10.1016/j.celrep.2016.04.085. http://doi.org/10.1101/024620. [DOI] [PMC free article] [PubMed]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Molecular Cell. 2013;49(5):773–82. doi: 10.1016/j.molcel.2013.02.011. http://doi.org/10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill ME, Erkek S, Peters AHFM. Parental epigenetic control of embryogenesis: A balance between inheritance and reprogramming? Current Opinion in Cell Biology. 2012;24(3):387–396. doi: 10.1016/j.ceb.2012.03.002. http://doi.org/10.1016/j.ceb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, … Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157(4):950–63. doi: 10.1016/j.cell.2014.03.025. http://doi.org/10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 2016:1–20. doi: 10.7554/eLife.14864. http://doi.org/10.1101/038281. [DOI] [PMC free article] [PubMed]
- Grandy RA, Whitfield TW, Wu H, Fitzgerald MP, VanOudenhove JJ, Zaidi SK, … Stein GS. Genome-wide studies reveal that H3K4me3 modification in bivalent genes is dynamically regulated during the pluripotent cell cycle and stabilized upon differentiation. Molecular and Cellular Biology. 2016;36(4):615–627. doi: 10.1128/MCB.00877-15. http://doi.org/10.1128/MCB.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gréen A, Lönn A, Peterson KH, Ollinger K, Rundquist I. Translocation of histone H1 subtypes between chromatin and cytoplasm during mitosis in normal human fibroblasts. Cytometry Part A: The Journal of the International Society for Analytical Cytology. 2010;77(5):478–84. doi: 10.1002/cyto.a.20851. http://doi.org/10.1002/cyto.a.20851. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA. Nucleosome spacing and chromatin higher-order folding. Nucleus (Austin, Tex) 2012;3(6):493–9. doi: 10.4161/nucl.22168. http://doi.org/10.4161/nucl.22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. PNAS. 2015 doi: 10.1073/pnas.1518280113. http://doi.org/10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed]
- Gurden MD, Anderhub SJ, Faisal A, Linardopoulos S. Aurora B prevents premature removal of spindle assembly checkpoint proteins from the kinetochore: A key role for Aurora B in mitosis. Oncotarget. 2016 doi: 10.18632/oncotarget.10657. http://doi.org/10.18632/oncotarget.10657. [DOI] [PMC free article] [PubMed]
- Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Molecular Cell. 2002;9(4):773–788. doi: 10.1016/s1097-2765(02)00515-4. http://doi.org/10.1016/S1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Halmer L, Gruss C. Effects of cell cycle dependent histone H1 phosphorylation on chromatin structure and chromatin replication. Nucleic Acids Research. 1996;24(8):1420–1427. doi: 10.1093/nar/24.8.1420. http://doi.org/10.1093/nar/24.8.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79(3):449–458. doi: 10.1016/0092-8674(94)90254-2. http://doi.org/10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci. 2004;117(Pt 26):6435–6445. doi: 10.1242/jcs.01604. http://doi.org/jcs.01604 [pii]\r. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438(7071):1176–80. doi: 10.1038/nature04254. http://doi.org/10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton A, Bak RO, Li CH, … Young RA. Activation of proto-oncogenes by disruption of chromosome neighborhoods, 9024. 2016 doi: 10.1126/science.aad9024. http://doi.org/10.1126/science.aad9024. [DOI] [PMC free article] [PubMed]
- Holliday R. Epigenetic Defects. Science. 1987;238(11):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science (New York, NY) 1975;187(4173):226–232. http://doi.org/10.1126/science.1111098. [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Human Molecular Genetics. 2009;18(R2):12–14. doi: 10.1093/hmg/ddp409. http://doi.org/10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung C, Bartman CR, Huang P, Ginart P, Stonestrom AJ, Keller CA, … Blobel GA. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes & Development. 2016;30:1423–1439. doi: 10.1101/gad.280859.116. http://doi.org/10.1101/gad.280859.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung CC, Morrissey CS, Udugama M, Frank CL, Keller Ca, Baek S, … Gerd a. Genome accessibility is widely preserved and locally modulated during mitosis. 2015:1–29. doi: 10.1101/gr.180646.114. http://doi.org/10.1101/gr.180646.114. [DOI] [PMC free article] [PubMed]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, … Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature Genetics. 2014;46(2):205–12. doi: 10.1038/ng.2871. http://doi.org/10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo a, Jackson Da, Cook PR. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. Journal of Cell Science. 1996;109(Pt 6):1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122(6):849–860. doi: 10.1016/j.cell.2005.07.018. http://doi.org/10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Jeong YS, Cho S, Park JS, Ko Y, Kang Y-K. Phosphorylation of serine-10 of histone H3 shields modified lysine-9 selectively during mitosis. Genes to Cells: Devoted to Molecular & Cellular Mechanisms. 2010:15. doi: 10.1111/j.1365-2443.2009.01375.x. http://doi.org/10.1111/j.1365-2443.2009.01375.x. [DOI] [PubMed]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, … Zhao K. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528(7580):142–146. doi: 10.1038/nature15740. http://doi.org/10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Research: An International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology. 2006;14(4):393–404. doi: 10.1007/s10577-006-1063-4. http://doi.org/10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484–92. doi: 10.1038/nrg3230. http://doi.org/10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Karpowicz P, Morshead C, Kam A, Jervis E, Ramuns J, Cheng V, Van Der Kooy D. Support for the immortal strand hypothesis: Neural stem cells partition DNA asymmetrically in vitro. Journal of Cell Biology. 2005;170(5):721–732. doi: 10.1083/jcb.200502073. http://doi.org/10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell. 1997;90(4):625–634. doi: 10.1016/s0092-8674(00)80524-3. http://doi.org/10.1016/S0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, De Vries SS, Janssen H, … Van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153(1):178–192. doi: 10.1016/j.cell.2013.02.028. http://doi.org/10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, … Bazett-Jones DP. Regulation of Global Acetylation in Mitosis through Loss of Histone Acetyltransferases and Deacetylases from Chromatin. Journal of Biological Chemistry. 2001;276(41):38307–38319. doi: 10.1074/jbc.M100290200. http://doi.org/10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–20. doi: 10.1038/35065132. http://doi.org/10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. Journal of Cell Science. 2003;116(Pt 14):2833–8. doi: 10.1242/jcs.00633. http://doi.org/10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Liang Z, Zickler D, Prentiss M, Chang FS, Witz G, Maeshima K, Kleckner N. Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles. Cell. 2015;161(5):1124–1137. doi: 10.1016/j.cell.2015.04.030. http://doi.org/10.1016/j.cell.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, … Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, NY) 2009;326(5950):289–93. doi: 10.1126/science.1181369. http://doi.org/10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Yuan Z-F, Han Y, Marchione DM, Garcia BA. Preferential phosphorylation on old histones during early mitosis in human cells. Journal of Biological Chemistry. 2016;291(29) doi: 10.1074/jbc.M116.726067. jbc.M116.726067. http://doi.org/10.1074/jbc.M116.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llères D, James J, Swift S, Norman DG, Lamond AI. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. Journal of Cell Biology. 2009;187(4):481–496. doi: 10.1083/jcb.200907029. http://doi.org/10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, … Mundlos S. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–1025. doi: 10.1016/j.cell.2015.04.004. http://doi.org/10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Laemmli UK. A Two-step scaffolding model for mitotic chromosome assembly. Developmental Cell. 2003;4(4):467–480. doi: 10.1016/s1534-5807(03)00092-3. http://doi.org/10.1016/S1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Maheshwari S, Tan EH, West A, Franklin FCH, Comai L, Chan SWL. Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids. PLoS Genetics. 2015;11(1):1–20. doi: 10.1371/journal.pgen.1004970. http://doi.org/10.1371/journal.pgen.1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. The Journal of Cell Biology. 2005;169(6):859–69. doi: 10.1083/jcb.200503031. http://doi.org/10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden MPF, Laemmli UK. Metaphase chromosome structure: Evidence for a radial loop model. Cell. 1979;17(4):849–858. doi: 10.1016/0092-8674(79)90325-8. http://doi.org/10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature Rev. 2005;6(11):838–49. doi: 10.1038/nrm1761. http://doi.org/10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83(1):29–38. doi: 10.1016/0092-8674(95)90231-7. http://doi.org/10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, Stamatoyannopoulos JA. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Reports. 2015;12(7):1184–1195. doi: 10.1016/j.celrep.2015.07.024. http://doi.org/10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nature Reviews Molecular Cell Biology. 2015 doi: 10.1038/nrm.2015.5. http://doi.org/10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed]
- Meng Y, Yi X, Li X, Hu C, Wang J, Bai L, … Shao Z. The non-coding RNA composition of the mitotic chromosome by 5′-tag sequencing. Nucleic Acids Research. 2016 doi: 10.1093/nar/gkw195. gkw195. http://doi.org/10.1093/nar/gkw195. [DOI] [PMC free article] [PubMed]
- Nagasaka K, Hossain MJ, Roberti MJ, Ellenberg J, Hirota T. Sister chromatid resolution is an intrinsic part of chromosome organization in prophase. Nature Cell Biology. 2016 Apr; doi: 10.1038/ncb3353. http://doi.org/10.1038/ncb3353. [DOI] [PubMed]
- Nasmyth K. Disseminating the Genome: Joining, Resolving, and Separating Sister Chromatids During Mitosis and Meiosis. 2003:673–745. doi: 10.1146/annurev.genet.35.102401.091334. Http://Dx.Doi.Org/10.1146/Annurev.Genet.35.102401.091334. [DOI] [PubMed]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny La, Dekker J. Organization of the mitotic chromosome. Science (New York, NY) 2013;342(6161):948–53. doi: 10.1126/science.1236083. http://doi.org/10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, … Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–5. doi: 10.1038/nature11049. http://doi.org/10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K, Katagiri C, Kishimoto T. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science. 1993;262(5142):2033–2035. doi: 10.1126/science.8266099. http://doi.org/10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- Ohta S, Wood L, Bukowski-Wills JC, Rappsilber J, Earnshaw WC. Building mitotic chromosomes. Current Opinion in Cell Biology. 2011;23(1):114–121. doi: 10.1016/j.ceb.2010.09.009. http://doi.org/10.1016/j.ceb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115(1):109–121. doi: 10.1016/s0092-8674(03)00724-4. http://doi.org/10.1016/S0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends in Biochemical Sciences. 2000;25(3):99–104. doi: 10.1016/s0968-0004(99)01535-2. http://doi.org/10.1016/S0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Development. 1996;10(21):2657–83. doi: 10.1101/gad.10.21.2657. http://doi.org/10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Öst A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, … Pospisilik JA. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell. 2014;159(6):1352–1364. doi: 10.1016/j.cell.2014.11.005. http://doi.org/10.1016/j.cell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Paweletz N. Walther Flemming: pioneer of mitosis research. Nature Reviews Molecular Cell Biology. 2001;2(1):72–75. doi: 10.1038/35048077. http://doi.org/10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Corces VG. Chromatin insulators: linking genome organization to cellular function. Molecular Cell. 2013;50(4):461–74. doi: 10.1016/j.molcel.2013.04.018. http://doi.org/10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. Journal of Cell Science. 2002;115(Pt 11):2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- Prescott DM, Bender M. DNA synthesis and mitosis in cultures of human peripheral leukocytes. Experimental Cell Research. 1962;27:221–9. doi: 10.1016/0014-4827(62)90225-2. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/13867092. [DOI] [PubMed] [Google Scholar]
- Prévost M, Chamousset D, Nasa I, Freele E, Morrice N, Moorhead G, Trinkle-Mulcahy L. Quantitative fragmentome mapping reveals novel, domain-specific partners for the modular protein RepoMan (recruits PP1 onto mitotic chromatin at anaphase) Molecular & Cellular Proteomics: MCP. 2013;12(5):1468–86. doi: 10.1074/mcp.M112.023291. http://doi.org/10.1074/mcp.M112.023291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Beullens M, Lesage B, Bollen M. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold Repo-Man. Current Biology: CB. 2013;23(12):1136–43. doi: 10.1016/j.cub.2013.05.017. http://doi.org/10.1016/j.cub.2013.05.017. [DOI] [PubMed] [Google Scholar]
- Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Current Biology: CB. 2011;21(9):766–73. doi: 10.1016/j.cub.2011.03.047. http://doi.org/10.1016/j.cub.2011.03.047. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, … Aiden EL. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. http://doi.org/10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Lin CC. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985;42(1):291–296. doi: 10.1016/s0092-8674(85)80124-0. http://doi.org/10.1016/S0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EMD, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Molecular Cell. 2002;9(5):1091–1100. doi: 10.1016/s1097-2765(02)00526-9. http://doi.org/10.1016/S1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Riggs AD. DNA Methylation and Late Replication Probably Aid Cell Memory, and Type 1 DNA Reeling Could Aid Chromosome Folding and Enhancer Function. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 1990;326(1235):285–297. doi: 10.1098/rstb.1990.0012. http://doi.org/DOI:10.1098/rstb.1990.0012. [DOI] [PubMed] [Google Scholar]
- Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Computational Biology. 2008;4(8) doi: 10.1371/journal.pcbi.1000153. http://doi.org/10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulland Y, Ouararhni K, Naidenov M, Ramos L, Shuaib M, Syed SH, … Dimitrov S. The Flexible Ends of CENP-A Nucleosome Are Required for Mitotic Fidelity. Molecular Cell. 2016;63(4):674–685. doi: 10.1016/j.molcel.2016.06.023. http://doi.org/10.1016/j.molcel.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, … Aiden EL. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1518552112. http://doi.org/10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed]
- Sánchez P, Losada A. New clues to understand how CENP-A maintains centromere identity. Cell Division. 2011;6(1):11. doi: 10.1186/1747-1028-6-11. http://doi.org/10.1186/1747-1028-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–13. doi: 10.1038/nature11279. http://doi.org/10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H. Epigenetic memory transmission through mitosis and meiosis in plants. Seminars in Cell and Developmental Biology. 2008;19(6):527–536. doi: 10.1016/j.semcdb.2008.07.017. http://doi.org/10.1016/j.semcdb.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Endl E, Wohlenberg C, van der Sar S, Cowell IG, Gerdes J, Singh PB. The Ki-67 protein interacts with members of the heterochromatin protein 1 (HP1) family: a potential role in the regulation of higher-order chromatin structure. The Journal of Pathology. 2002;196(2):135–144. doi: 10.1002/path.1016. http://doi.org/10.1002/path.1016. [DOI] [PubMed] [Google Scholar]
- Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. http://doi.org/10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- Shema E, Jones D, Shoresh N, Donohue L, Ram O, BE Single-molecule decoding of combinatorially modified nucleosomes. Science. 2016;352(6286):717–721. doi: 10.1126/science.aad7701. http://doi.org/10.1126/science.aad7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biology. 2006;8(7):677–687. doi: 10.1038/ncb1425. http://doi.org/10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. American Journal of Human Genetics. 2016;98(1):185–201. doi: 10.1016/j.ajhg.2015.12.002. http://doi.org/10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development (Cambridge, England) 2005;132(4):681–7. doi: 10.1242/dev.01609. http://doi.org/10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nature Reviews Genetics. 2005;6(10):782–92. doi: 10.1038/nrg1692. http://doi.org/10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nature Biotechnology. 2010;28(10):1089–1095. doi: 10.1038/nbt.1680. http://doi.org/10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 Complexes Mediate Nucleosome Assembly Pathways Dependent or Independent of DNA Synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. http://doi.org/10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- Tanabe H, Müller S, Neusser M, von Hase J, Calcagno E, Cremer M, … Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4424–9. doi: 10.1073/pnas.072618599. http://doi.org/10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of Cohesin Association Sites at Centromeres and along Chromosome Arms. Cell. 1999;98(6):847–858. doi: 10.1016/s0092-8674(00)81518-4. http://doi.org/10.1016/S0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, … Ruan Y. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163(7):1–17. doi: 10.1016/j.cell.2015.11.024. http://doi.org/10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. A Dynamic Mode of Mitotic Bookmarking by Transcription Factors. eLife. 2016:66464. doi: 10.7554/eLife.22280. http://doi.org/10.1101/066464. [DOI] [PMC free article] [PubMed]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. http://doi.org/10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Koller TH. Influence of histone H1 on chromatin structure. Cell. 1977;12(2):101–107. doi: 10.1016/0092-8674(77)90188-x. http://doi.org/10.1016/0006-291X(80)91264-4. [DOI] [PubMed] [Google Scholar]
- Thoma F, Koller TH, Klug A. Involvement of Histone H1 in the Organization of the Nucleosome and of the Salt-Dependent Superstructures of Chromatin. 1979 Nov;83 doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R, Rynes E, Humbert R, Vierstra J, Maurano M, Haugen E, … Stamatoyannopoulos J. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. http://doi.org/10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, De Laat W. Looping and interaction between hypersensitive sites in the active??-globin locus. Molecular Cell. 2002;10(6):1453–1465. doi: 10.1016/s1097-2765(02)00781-5. http://doi.org/10.1016/S1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Topham C, Tighe A, Ly P, Bennett A, Sloss O, Nelson L, … Taylor SS. MYC Is a Major Determinant of Mitotic Cell Fate. Cancer Cell. 2015;28(1):129–140. doi: 10.1016/j.ccell.2015.06.001. http://doi.org/10.1016/j.ccell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toselli-Mollereau E, Robellet X, Fauque L, Lemaire S, Schiklenk C, Klein C, … Bernard P. Nucleosome eviction in mitosis assists condensin loading and chromosome condensation. EMBO Journal. 2016;35(14):1–17. doi: 10.15252/embj.201592849. http://doi.org/10.15252/embj.201592849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Lim C, Xie J. Asymmetric Division of Drosophila. Science. 2012;1309(November):2010–2013. doi: 10.1126/science.1226028. http://doi.org/10.1126/science.1226028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Lim C, Xie J, Chen X. Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions. Chromosome Research. 2013;338(6107):679–82. http://doi.org/10.1126/science.1226028. [Google Scholar]
- Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, … Schneider R. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152(4):859–872. doi: 10.1016/j.cell.2013.01.032. http://doi.org/10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Uchida IA, Lin CC. Fluorescent staining ofhuman chromosomes: Identification ofsome common aberrations. CMA Journal. 1971;105(6):479–482. [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P. Mitotic chromosome condensation in vertebrates. Experimental Cell Research. 2012;318(12):1435–41. doi: 10.1016/j.yexcr.2012.03.017. http://doi.org/10.1016/j.yexcr.2012.03.017. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nature Genetics. 2001;27(3):304–308. doi: 10.1038/85871. http://doi.org/10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Varier Ra, Outchkourov NS, de Graaf P, van Schaik FMa, Ensing HJL, Wang F, … Timmers HTM. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. The EMBO Journal. 2010;29(23):3967–78. doi: 10.1038/emboj.2010.261. http://doi.org/10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle. 2006;5(February 2015):2555–2556. doi: 10.4161/cc.5.22.3463. http://doi.org/10.4161/cc.5.22.3463. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, … Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(Track II):10660–10665. doi: 10.1073/pnas.0600447103. http://doi.org/10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, … Timmers HTM. Selective Anchoring of TFIID to Nucleosomes by Trimethylation of Histone H3 Lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. http://doi.org/10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley Ja, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. The EMBO Journal. 2007;26(8):2041–2051. doi: 10.1038/sj.emboj.7601654. http://doi.org/10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke a, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. http://doi.org/10.1016/S0092-8674(00)00132-X. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, … Higgins JMG. Histone H3 Thr-3 Phosphorylation at Centromeres in Mitosis. Science (New York, NY) 2010;330(October):231–236. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Higgins JMG. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends in Cell Biology. 2013;23(4):175–84. doi: 10.1016/j.tcb.2012.11.005. http://doi.org/10.1016/j.tcb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Wang F, Ulyanova NP, Daum JR, Patnaik D, Kateneva AV, Gorbsky GJ, Higgins JMG. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. The Journal of Cell Biology. 2012;199(2):251–68. doi: 10.1083/jcb.201205106. http://doi.org/10.1083/jcb.201205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ulyanova NP, van der Waal MS, Patnaik D, Lens SMa, Higgins JMG. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Current Biology: CB. 2011;21(12):1061–9. doi: 10.1016/j.cub.2011.05.016. http://doi.org/10.1016/j.cub.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su J, Beliveau BJ, Bintu B, Moffitt JR, Wu C, Zhuang X. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353(6299):598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Demidov D, Lermontova I, Beeckman T, Van Damme D. Aurora Kinases Throughout Plant Development. Trends in Plant Science. 2016;21(1):69–79. doi: 10.1016/j.tplants.2015.10.001. http://doi.org/10.1016/j.tplants.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. http://doi.org/10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Krijger PHL, Geeven G, Anink-groenen LCM, Verschure PJ, De Laat W, … DeLaat W. Cause and Consequence of Tethering a SubTAD to Different Nuclear Compartments. Molecular Cell. 2016:1–13. doi: 10.1016/j.molcel.2016.01.001. http://doi.org/10.1016/j.molcel.2016.01.001. [DOI] [PMC free article] [PubMed]
- Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, … Neumann H. A cascade of histone modifications induces chromatin condensation in mitosis. Science (New York, NY) 2014;343(January):77–80. doi: 10.1126/science.1244508. http://doi.org/10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- De Wit E, Vos ESM, Holwerda SJB, Valdes-quezada C, Verstegen MJAM, Teunissen H, … Laat WDe. CTCF Binding Polarity Determines Chromatin Looping. Molecular Cell. 2015;60(4):1–9. doi: 10.1016/j.molcel.2015.09.023. http://doi.org/10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Xeros N. Deoxyriboside control and synchronization of mitosis. Nature. 1962;194:682–683. doi: 10.1038/194682a0. http://doi.org/10.1038/194682a0. [DOI] [PubMed] [Google Scholar]
- Xie J, Wooten M, Tran V, Simbolon C, Betzig E, Chen X, … Betzig E. Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Article Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell. 2015;163(4):1–14. doi: 10.1016/j.cell.2015.10.002. http://doi.org/10.1016/j.cell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of Histone H3-H4 tetramers during DNA replication dependent chromatin assembly. Science. 2010;328(April):1–5. doi: 10.1126/science.1178994. http://doi.org/10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science (New York, NY) 2010;330(6001):239–43. doi: 10.1126/science.1194498. http://doi.org/10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, … Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3189–3194. doi: 10.1073/pnas.0611419104. http://doi.org/10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Genome reactivation after the silence in mitosis: Recapitulating mechanisms of development? Developmental Cell. 2014;29(2):132–134. doi: 10.1016/j.devcel.2014.04.019. http://doi.org/10.1016/j.devcel.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Isbaner S, Heermann D. Mechanics of Sister Chromatids studied with a Polymer Model. Frontiers in Physics. 2013;1(October):1–11. http://doi.org/10.3389/fphy.2013.00016. [Google Scholar]
- Zhang Y, Xie S, Zhou Y, Xie Y, Liu P, Sun M, … Chen S. H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Reports. 2014;8(6):1989–2002. doi: 10.1016/j.celrep.2014.08.031. http://doi.org/10.1016/j.celrep.2014.08.031. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ding J, Li Y, Ren K, Sha J, Zhu M, Gao X. HnRNP U mediates the long-range regulation of Shh expression during limb development. Human Molecular Genetics. 2009;18(16):3090–3097. doi: 10.1093/hmg/ddp250. http://doi.org/10.1093/hmg/ddp250. [DOI] [PubMed] [Google Scholar]
- Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature Cell Biology. 2011;13(11):1295–304. doi: 10.1038/ncb2341. http://doi.org/10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MIJa, Ye Z, Kolovos P, Brouwer RWW, … Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(3):996–1001. doi: 10.1073/pnas.1317788111. http://doi.org/10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]