Abstract
The developmental trajectories of theta band (4–7 Hz) event-related oscillations (EROs), a key neurophysiological constituent of the P3 response, were assessed in 2170 adolescents and young adults ages 12 to 25. The theta EROs occurring in the P3 response, important indicators of neurocognitive function, were elicited during the evaluation of task-relevant target stimuli in visual and auditory oddball tasks. Associations between the theta EROs and genotypic variants of 4 KCNJ6 single nucleotide polymorphisms (SNPs) were found to vary with age, sex, scalp location, and task modality. Three of the four KCNJ6 SNPs studied here were found to be significantly associated with the same theta EROs in adults in a previous family genome wide association study. Since measures of the P3 response have been found to be a useful endophenotypes for the study of a number of clinical and behavioral disorders, studies of genetic effects on its development in adolescents and young adults may illuminate neurophysiological factors contributing to the onset of these conditions.
Keywords: ERO, P3, Adolescent, Development, KCNJ6, Theta
1. Introduction
Studies of the age-variation of the effects of genotypic variants on the development of brain function will provide increasing insight into the processes of brain maturation and in particular the influences of neurophysiological factors during the transition from childhood through adolescence to adulthood. One important indicator of neurocognitive function is the P3 (or P300) response, evidenced by the production of a large positive waveform with a peak between 300 ms. and 700 ms. after the presentation of a target stimulus. The P3 response is elicited by infrequently presented target stimuli in a stream of more frequently occurring non-target stimuli in auditory or visual target detection (oddball) tasks, which call for the subject to respond to only the target stimulus. The P3 response has been proposed to index attentional and working memory resources (Polich, 2007). It has been associated with several anatomical loci including the supra-marginal gyrus, hippocampus, locus coeruleus, anterior cingulate cortex (ACC), insula, and the right-lateralized frontal and temporoparietal regions of the ventral attention network which may be part of a distributed circuit (Menon et al., 1997; Brázdil et al., 1999; Ardekani et al., 2002; Polich and Criado, 2006; Mantini et al., 2009; Sara and Bouret, 2012; Walz et al., 2014). Studies of visual and auditory target detection tasks using functional magnetic resonance imaging (fMRI) suggest that common, supramodal functional systems are involved as well as modality-specific systems (Walz et al., 2013; Linden et al., 1999). Frequency domain analysis suggests that the theta band event related oscillation (ERO) is a major constituent of the P3 response (Karakas et al., 2000a,b; Yordanova et al., 2003; Jones et al., 2006a,b; Rangaswamy et al., 2007). Theta EROs are important for processes underlying frontal inhibitory control, conscious awareness, recognition memory and episodic retrieval, as shown in a number of experimental contexts (Gevins et al., 1998; Jacobs et al., 2006; Klimesch et al., 1994, 2001, 2008; Vertes, 2005; Babiloni et al., 2009; Crowley et al., 2014).
There are many changes related to brain development during adolescence that may affect theta ERO power. On the neuronal level, there is a decrease in gray matter density and cortical thickness in adolescence, as well as increases in white matter, reflecting synaptic pruning and myelination (Sowell et al., 2004; Toga et al., 2006). On the structural/anatomical level, trajectories of brain volumes of different regions and tissue types, as well as other features of cortical anatomy, exhibit curvilinear properties which vary between regions (Lenroot et al., 2007; Shaw et al., 2008; Giedd et al., 2010; Raznahan et al., 2011a; Sullivan et al., 2011) and between sexes (Lenroot and Giedd, 2010; Lenroot et al., 2007; Peper et al., 2011; Koolschijn and Crone, 2013), as determined by magnetic resonance imaging (MRI) of subjects between the ages of 8 and 20. Sex differences are also present in functional MRI studies of the development of task-related brain activity in adolescents and young adults in a number of different tasks (Rubia et al., 2006; Christakou et al., 2009; Rubia et al., 2010, 2013; Rubia, 2013). Brain networks develop from a pattern of local connectivity to more global patterns of connectivity (Fair et al., 2008, 2009; Power et al., 2010; Supekar et al., 2009; Uddin et al., 2011; Vogel et al., 2010; Zielinski et al., 2010; Menon, 2013; Wu et al., 2013). Systematic changes of the electrophysiology of brain activity occur with age, both in the resting state and in a variety of task related conditions (Segalowitz et al., 2010; Sturman and Moghaddam, 2011). Among the most prominent are decrease in power in oscillatory activity in both resting state and task related activity (Yordanova and Kolev, 1997; Whitford et al., 2007). Sex differences in development have also been observed in task related activity (Nanova et al., 2008, 2011). Previous studies have examined patterns of the development of visual and auditory P3 peak amplitude in adolescents (Katsanis et al., 1996; Hill et al., 1999a; Stige et al., 2007; Nanova et al., 2008; van Beijsterveldt et al., 1998, 2001; Carlson and Iacono, 2006; Sumich et al., 2012). A previous study from our laboratory of adolescents and young adults (ages 12 to 25) described the developmental trajectories of theta band EROs measured in both auditory and visual tasks at three scalp locations (Chorlian et al., 2015). That study found that the developmental trajectories of the theta EROs are characterized by a general decrease in power with age, with large differences in temporal pattern between males and females, and relatively small differences between task modality and scalp location within each sex.
It is an important scientific objective to characterize the genetic basis of the large neurophysiological and neuroanatomical changes occurring in adolescence, which influence the cognitive and affective processes underlying behavioral changes. Twin studies suggest that there are genetic effects on P3 electrophysiological measures obtained in target detection tasks during adolescent development (Katsanis et al., 1997; van Beijsterveldt et al., 1998, 2001; Carlson and Iacono, 2006). A considerable number of studies have found high degrees of heritability for structural brain features measured in adolescents (see Douet et al. (2014) for a review) which exhibit a variety of developmental patterns as described above. Gene expression studies suggest large variations in gene expression during adolescence, both in humans and in rodents (Colantuoni et al., 2011; Kang et al., 2011; Naumova et al., 2013; Stead et al., 2006). Thus it might be expected that there would be considerable age variation in association between SNPs and neurophysiological measures during adolescence. To this end, genetic variants which have been shown to have a considerable association with neurophysiological measures in adults and in which the genetic factors are known to be related to cognitive function are attractive candidates for analysis.
The Collaborative Study on the Genetics of Alcoholism (COGA) previously reported an association with genome wide significance between the power of theta band (4–7 Hz) EROs in a visual oddball task and KCNJ6 single nucleotide polymorphisms (SNPs) in a predominantly adult sample (Kang et al., 2012). The protein encoded by KCNJ6 is known as GIRK2, which is widely distributed in the brain and is an important functional element in dopaminergic, cholinergic, GABAergic and glutamatergic synapses and hence regulation of neuronal excitability (Saenz del Burgo et al., 2008). To examine the age variation in the association between theta EROs and KCNJ6 SNPs during adolescence and young adulthood, the current study followed the methodology of a recent study of developmental trajectories of theta EROs (Chorlian et al., 2015), using the same sample of adolescents and young adults. Consideration of the results in that study (Chorlian et al., 2015), as described in section 3.1, informed the analysis employed in the present study. The developmental trajectories (time-series) of the association between KCNJ6 gene variants and theta band EROs during the P3 response to targets in adolescents and young adults are measured by the effect size of the genetic variants in a non-parametric regression model of the theta band EROs. These associations are described as a function of age, sex, task modality, and scalp location. The current study is not designed as a replication of the adult study; 19% of the subjects included in this study were subjects in Kang et al. (2012). This is the first developmental study of neurophysiological function that characterizes trajectories of the genetic association of task-related electrophysiological activity with specific SNPs.
2. Methods and Materials
2.1. Subjects
The sample comprised 2170 adolescents and young adults (1060 males and 1110 females) from the Prospective Study of the Collaborative Study on the Genetics of Alcoholism (COGA), examined within the age range of 12 to 25 years. COGA is a multisite collaboration designed to study the genetics of alcoholism (Begleiter et al., 1995) that has investigated members from multiplex alcoholic families (recruited through a proband in treatment) and a set of community (comparison) families, ascertained to be representative of the general population. These families were recruited during the years 1990 to 2000. The Prospective Study began in 2004 as a continuing study of adolescents and young adults from pedigrees ascertained in previous phases of COGA, and contained members from both the multiplex alcoholic families and the community (comparison) families. These families were recruited during the years 1990 to 2000. Over 80% of the subjects are from families originally recruited through an alcoholic proband, but fewer than 25% of the sample are first degree relatives of the probands, and many of the subjects from alcoholic families are only distantly related to the probands.
Participants in the study were reassessed at approximately two year intervals. Subjects were excluded from neurophysiological assessment if they had any of the following: (1) recent substance or alcohol use (i.e., positive breath-analyzer test and/or urine screen), (2) hepatic encephalopathy/cirrhosis of the liver, (3) history of head injury, seizures or neurosurgery, (4) uncorrected sensory deficits, (5) use of medication known to influence brain functioning, (6) history/symptoms of psychoses, (7) positive test for human immunodeficiency virus, (8) other acute/chronic medical illnesses that affects brain function and (9) and a score of less than 25 on the Mini Mental State Examination. This sample comprised subjects with one or more neurophysiological assessments: 475 had 1 assessment, 583 had 2, 576 had 3, 494 had 4, and 42 had 5 assessments. Data from six collection sites have been included in this study: SUNY Downstate Medical Center; University of Connecticut Health Science Center; Washington University School of Medicine in St. Louis; University of California at San Diego; University of Iowa, and Indiana University School of Medicine. Recruitment and assessment procedures have been described elsewhere (Reich, 1996; Begleiter et al., 1998; Edenberg et al., 2005), and are also available at this website: https://zork5.wustl.edu/niaaa/coga_instruments/resources.html. The experimental protocols were approved by each site’s institutional review board, and informed consent (for those over eighteen years of age) or assent (for those under eighteen years of age) was obtained from all participants.
2.2. Genotyping
Genotyping was performed at Washington University School of Medicine in St. Louis on an OpenArray platform, and at Indiana University School of Medicine in Indianapolis on the Sequenom MassArray system on a larger group of COGA subjects of which the sample described here is a subset. OpenArray genotyping is a multiplex TaqMan assay platform. The OpenArray Genotyping Plate Configurator was used to design assays. Arrays were scanned on the OpenArray NT imager and genotypes were called using the OpenArray SNP Genotyping analysis software. Sequenom Assays (iPLEX Gold) were designed using MassArray Assay Design Software (Sequenom, San Diego, CA). Hardy-Weinberg equilibrium (HWE) was computed separately in European Americans and African Americans, and cluster data was re-evaluated if HWE p < 0.05. All SNPs were cleaned for Mendelian inheritance using PEDCHECK (O’Connell and Weeks, 1998). SNP allele frequencies and heterozygosities were computed in PLINK (http://pngu.mgh.harvard.edu/purcell/plink; Purcell et al. (2007)) using data on founders only included in the larger group. Ethnic stratification was assessed with SNPrelate (Zheng et al., 2012) using 64 ancestry-informative SNPs, as part of a larger 96 SNP panel developed at the Rutgers University DNA and Cell Repository (RUID ™). The genotypic platform for this sample included six KCNJ6 SNPs. Initially five SNPs were selected that were significant in Kang et al. (2012) for theta band EROs: namely rs702859 (synonymous SNP in exon 4), rs858008, rs1787422, rs857975 and rs2835850 (located in the intronic region between exons 3 and 4 (according to HapMap Release 3.27); however, as rs857975 and rs2835850 were in strong LD with rs702859, they were not included in the study. An additional KCNJ6 SNP, rs2070995 (synonymous SNP in exon 3), was also available in this sample and included to provide the widest coverage of KCNJ6. Thus four of the available KCNJ6 SNPs were selected for this study: namely, rs858008, rs702859, rs1787422 and rs2070995; they were in modest LD with each other (0.04 < r2 < 0.53, 0.45 < D′ < 0.83; see table 1).
Table 1.
LD Matrix for KCNJ6 SNPs. Values of r2 below the diagonal; Values of D′ above the diagonal.
| rs702859 | rs858008 | rs1787422 | rs2070995 | |
|---|---|---|---|---|
|
| ||||
| rs702859 | 0.772 | 0.769 | 0.306 | |
| rs858008 | 0.325 | 0.823 | 0.305 | |
| rs1787422 | 0.435 | 0.502 | 0.347 | |
| rs2070995 | 0.017 | 0.023 | 0.031 | |
Minor allele frequencies for the KCNJ6 SNPs in the sample are provided in table 2. Allelic distribution for all four SNPs as a function of age was calculated using kernel density methods, and no genotype deviated from its mean frequency of occurrence by more than 4% at any age, and that deviations greater than 2.5% occurred at less than 4% of the ages.
Table 2.
Minor allele frequencies of KCNJ6 SNPs in the sample. Population frequencies from HapMap CEU except rs702859 from ExAC.
| SNP | Sample | Population |
|---|---|---|
|
| ||
| rs702859 | 0.28 | 0.29 |
| rs858008 | 0.42 | 0.46 |
| rs1787422 | 0.16 | 0.20 |
| rs2070995 | 0.44 | 0.40 |
2.3. Electrophysiology
Two oddball tasks, one visual, the other auditory were used for this study (Cohen et al., 1994; Alexander et al., 1994). In each task, subjects were verbally instructed to suppress their eye blinks and to sit as still as possible. They were asked to respond to the target with a button press as quickly as possible, but not at the expense of accuracy, and not to respond to other stimuli.
2.3.1. Visual oddball task
A three-stimulus visual oddball task was employed with 280 visual stimuli of three different types: 35 targets (rarely occurring letter ‘X’) to which the subjects responded quickly and accurately with a button press, 210 non-targets (frequently occurring white squares) and 35 novels (rarely occurring random colored geometric figures) (probabilities of occurrence of 0.125, 0.750 and 0.125 respectively). Stimuli subtended a visual angle of 2.5 degrees with stimulus durations of 60 ms. and inter-stimulus intervals of 1625 ms. The stimuli were presented pseudo-randomly with the only constraint that a target or novel stimulus never preceded a target or novel stimulus.
2.3.2. Auditory oddball task
Subjects were presented binaurally with two tones of different frequencies. One stimulus was a low tone (600 Hz) and the other a high tone (1600 Hz) produced by a tone generator. Each stimulus had a 60 ms. duration (10 ms. rise and fall times and 40 ms. plateau) and an intensity level of 60 dB. A computer initiated the stimulus. The probabilities of the rare and the frequent tones were 0.125 and 0.875 respectively. The use of the low or high tone as the rare (target) tone was alternated across the subjects. The auditory stimuli were presented through headphones (model ER-3A Tubephone Insert Earphones, 50 Ω impedance; Etymotic Research, Elk Grove Village, IL); the earpiece and a short length of the Tubephone were fitted under the electrode cap, and the individual left and right transducer cases were situated on either side of the neck. Subjects received a maximum of 400 trials with a uniform interstimulus interval of 1500 ms.
2.3.3. Event-related potential recording
All six collaborating sites used identical experimental procedures and EEG acquisition hardware and software. Subjects were seated comfortably 1 m from a monitor in a dimly lit sound-attenuated RF-shielded booth (Industrial Acoustics Company, Bronx, NY, USA), and wore an electrode cap (Electro-Cap International, Inc., Eaton, OH, USA) as specified by the International 10–20 System for Electrode Placement (Fig. S2). The nose served as reference and the forehead served as ground. Electrode impedences were maintained below 5kΩ. Electrical activity was amplified 10,000 times using Neuroscan amplifiers and was recorded continuously over a bandwidth of 0.02–100.0 Hz on a Neuroscan system (Versions 4.1–4.5; Neurosoft, Inc., El Paso, TX, USA) at sampling rates of 256, 500 and 512 Hz, depending on the Neuroscan version, and stored for further analysis. During analysis all signals were re-sampled to 256 Hz and bandpass filtered between 0.05 and 55.0 Hz. Artifact rejection threshold was set at 100 μV. A minimum of 20 trials of 100 ms. pre-stimulus to 750 ms. post-stimulus artifact-free data for each stimulus was required for analysis.
2.3.4. ERO energy estimation
Estimates of localized power of non-stationary evoked potential time series were obtained using the S-transform, a time-frequency representation method developed by Stockwell (Stockwell et al., 1996). This method has been previously described and implemented in our laboratory to evaluate event-related signals in the time-frequency domain (Jones et al., 2006a,b; Kamarajan et al., 2006; Rangaswamy et al., 2007).
Event-related electrophysiological data for the target stimulus from the visual odd-ball task and the auditory oddball task were analyzed. The amplitude envelope of the S-transform time-frequency region was averaged across single trials, per individual, to obtain estimates of event-related total power. Mean power was calculated for each electrode within time-frequency regions of interest that were defined by frequency band range and time intervals. The measures used in this analysis were the total power in the theta band (3.0–7.0 Hz) oscillation at the frontal (Fz), central (Cz), and parietal (Pz) midline electrodes extracted from the 300–700 ms. time window, which corresponds to the window of the P3 component in the event-related waveforms. The time window and frequency band was chosen to capture relevant data from both tasks at the cost of being overly wide in both frequency and time ranges for each task individually, and used for reporting previous theta ERO results from our laboratory (Kang et al., 2012). The electrodes at the three scalp locations were chosen because of previous use in studies from our own laboratory (Jones et al., 2006a,b) and other developmental studies (Stige et al., 2007; Nanova et al., 2008). Correlations between measures were estimated using a method described in section 2.4.1.
2.4. Statistical Methodology
2.4.1. Developmental Models
In a previous study (Chorlian et al., 2015), the trajectories of the means of the theta band EROs were determined using a varying coefficient local linear regression model (loess). An identical model with the addition of genotypic covariates was applied to the same dataset, and the time-series of parameter estimates (β values) for the genotypic covariates was obtained in addition to the phenotypic trajectories. These time-series of parameter estimates, when scaled by the standard deviation of the data, are the association trajectories.
Phenotypic trajectory modeling
The value of a neurophysiological index observed at a specific age represents the cumulative effect of developmental changes up to the age at which the observation was made. The time course of this cumulative effect is the trajectory of the developmental process. Since there is no explicit biological model for the developmental trajectory, no parametric form is attributed to it. In the varying coefficient model (Fan and Zhang, 2008) used in this study, the dependent variable, the development trajectory of the phenotype, is an implicit (non-parametric) function of age, and the effects of the independent variables (covariates) on the trajectory can vary with age. (In this study, the developmental trajectories are those of the natural logarithm of the theta ERO power values from the scalp locations mentioned above. The logarithmic transformation of the data was used to eliminate the skewed distribution found at every age. In the paper all uses of the word “log” as short for “logarithm” mean natural logarithm, including all text on figures.) The developmental trajectory s(t) is the mean of the phenotypic values at age of interest estimated by a local linear regression calculation (loess) (Cleveland, 1979; Cleveland and Devlin, 1988; Wu and Zhang, 2002; Wang, 2003; Hoover et al., 1998). The local linear regression calculation is a sequence of regression calculations each centered at a specific age in which the intercept of the regression line is the estimate of the trajectory and the slope of the regression line the time derivative of the trajectory at the age at which the regression is centered. That is, a separate regression calculation is carried out to determine the mean and slope of the theta ERO power values for each central age, which ranges from ages 12 to 25 in one-tenth year intervals, using, in this case, age-invariant covariates. Each calculation contains data extending before and after the central age (except for the very first and last in the sequence); models centered one year apart share at least 86% of their data and models two years apart share at least 75%, so that there is considerable overlap in the data used for calculation in nearby ages. The regression is weighted by a function of distance of the observation from the central age, so that observations near the central age count more in determining the result than observations further away from the central age. Each of the six dependent variables, the theta ERO power values at each of the three scalp locations (electrodes) for each modality, is modeled separately in order to simplify the interpretation of the regression analysis. This methodology is adapted from the work of Cleveland (1979); Cleveland and Devlin (1988); Gasser et al. (1984a,b); Ramsay and Silverman (2002); Gasser et al. (2004); Molinari and Gasser (2004).
The model for each central age tk, k = 1 … 131, is represented by the following equation, in which yi, j is the dependent variable for subject i at observation j at age ti, j, Wh(t) the value of weighting function for the local linear regression model at age distance t from the central age, a function of the bandwidth (fraction of data included) h, β0,n,k the parameter to be estimated for covariate n for its effect on the intercept and β1,n,k the parameter to be estimated for covariate n for its effect on the slope, and Xi, j,m the value of covariate m of subject i at observation j and ei, j the error term. Different sets of covariates may be used for the estimation of the slope and the intercept. The weight Wh(ti, j − tk) will be non-zero only for an interval near tk and diminishes rapidly as the distance between ti, j and tk increases. Note that the beta values are indexed to emphasize that each is the product of a separate estimation for each central age. (In the equation, intercept terms appear on the first line and slope terms on the second line. The summations are over the repeated indices in the covariates and betas.)
The above is the model which represents the following weighted least squares minimization problem to be solved for the βs :
In the statistical model as used for this dataset the covariates for the intercept included sex, family type (Alcoholic or Community; see section 2.1), genotype (using an additive model) and a genotype × sex interaction, consistent with other studies of human development (Widén et al., 2010; Cousminer et al., 2013), and the first two principal components from the stratification analysis of the genetic dataset, which correspond to genetic ancestry (Astle and Balding, 2009); the only covariate for the slope is sex. Family type was used as a covariate in the regression analysis to address the possible differential effects, either genetic or environmental, of family history with regard to alcohol use disorders. Weights for individuals were adjusted to account for multiple observations on single individuals and co-presence of sibs in each of the local linear regression calculations. Weights for multiple observations from the same individual in any single regression calculation were adjusted by dividing each weight by the sum of all the weights used in the calculation for that individual if the sum was greater than 1. Weights for sibs were adjusted to account for commonality of SNPs; i.e. since on average pairs of sibs share 50% of their genotype, the weights for pairs of non-identical sibs was multiplied by .75, and correspondingly for larger numbers of sibs. The bandwidth for the kernel (Epanechnikov) was taken as 0.6 to minimize the mean squared error, although the variation in the size of the error was less than 2% over the range of bandwidths from 0.4 to 0.8. All calculations were carried out in Matlab2014a (The MathWorks Inc. Natick, MA, USA) using the function glmfit().
As there are 5555 observations spread over a 13 year age-range, there is sufficient data to provide estimates of the means of the variables at one-tenth year intervals and to provide estimates of the mean rates of change of the variables as well. Since 90% of the successive observations of individuals have an interval of greater than 1.75 years, no longitudinal modeling at the shorter time scale (6 months) at which significant changes in the measured variables can occur is possible. (The estimate for the time scale is derived from Sullivan et al. (2011)). Results are not independent between models for different ages since the data in each of the 131 regression models has considerable overlap with the data used in the models for nearby ages. Significance levels and effect sizes were obtained from the regression calculations and corroborated using a non-parametric bootstrap method with 1000 resamplings. The bootstrap also enabled the comparison of parameters estimated for different ages, since there was no simple direct method for calculating the covariance of the parameter estimates obtained from regressions with widely separated central ages.
The bootstrapping procedure is designed for independent observations and needed to be adapted to the presence of sibs and of multiple observations for each individual. It was decided to resample individuals, not observations. Individuals were selected with replacement from the dataset and each individual in the dataset was assigned the number of times she was picked in the selection procedure. In the interests of simplicity, the bootstrapping calculation repeated the actual data calculation with the full sample, including the adjustments for sibship and multiple observations for the same subject, but at the point when the regression calculations were carried out (i.e., the call to the Matlab procedure glmfit()), the weights assigned to each observation based on their distance from the central age were modified by being multiplied by the number of times the subject was included in the selection procedure. This was intended to preserve the overall character of the sibship alteration of weights, since if an individual was included in the sample multiple times, it was not considered desirable to count her as her own sib. Similarly, the multiplicity of observations from the same subject was preserved, since it was not considered desirable to diminish the effect of a subject being picked multiple times in the bootstrapping selection procedure.
Phenotypic correlation modeling
Age-centered and weighted correlations between theta ERO measures both within (intramodal) and between (intermodal) visual and auditory modalities for all pairs of locations were calculated using a method similar to that employed in the regression analysis, using the same age intervals and same weights as used in the regression analysis. These are fully described in our previous study (Chorlian et al., 2015).
2.4.2. Age variation of genotypic effects
Just as the regression calculations provide an estimate of the mean value of the dependent variable, the phenotype, at each central age, they correspondingly provide a parameter estimate (β) for the each of the covariates in the regression model. The parameter estimates are transformed into effect sizes by rescaling them in terms of the standard deviation of the dependent variable values. The use of effect sizes (and their corresponding p-values) from the regression calculations provides a sequence of association estimates spaced at 1/10th year intervals over ages 12 to 25. In this method, association is a function of age specified by a sequence of values, not by a functional form. Compared to using separate non-overlapping age ranges, the use of the local linear regression method provides greater power from the use of more observations for each parameter estimate and greater temporal resolution while resulting in the smoothing of estimates and a more complex interpretation of the results. Since there are 4 SNPs and 6 phenotypes (3 scalp locations: Fz, Cz, Pz × 2 modalities: auditory and visual) there are 24 association trajectories for each sex, each with 131 points. The calculation of the association trajectories provides a new set of data which is then subject to statistical analysis to determine its own pattern of age variation and sex specificity across the range of phenotypes.
Identification of Significant SNPs
One of the principal objectives of this study is to describe the pattern of age specific variation of genotypic effects and to compare them across scalp location, modality, and sex within each SNP and between SNPs. Our interest is in the timing of significant genotypic effects on a single SNP level as a basis for these comparisons.
The evaluation of the significance of the association of the individual SNPs with theta EROs is determined by the selection of individual p-values based on control of the false discovery rate. Given the fact that this is a candidate gene study, in which at least a small proportion of the tested variables are expected to be significant, the control of the false discovery rate by the characteristics of data is appropriate. The method used to determine the false discovery rate for any particular level of p-value must be capable of dealing with correlated data, as p-values are correlated as a result of both the non-parametric regression method and the phenotypic correlations described in Chorlian et al. (2015) and summarized in section 3.1.2. The method used is derived from that in Storey and Tibshirani (2003), which uses the distribution of the calculated p-values to determine the prevalence of results which reflect the absence of association, and correspondingly, the prevalence of results which reflect the presence of association. (The part of the original algorithm which estimates the proportion of the non-associated results was modified to deal more robustly with truncated p-value distributions when necessary, and provides a somewhat more conservative estimate than does the original).
Specifically, the distribution of p-values from non-associated SNP-phenotype pairs should be uniform on the interval from 0 to 1, while the the distribution of p-values from associated SNP-phenotype pairs should be concentrated near 0. From the estimation of the occurrence of non-associated SNP-phenotype pairs, it is possible to estimate the occurrence of associated SNP-phenotype pairs, and determine the false discovery rate, sometimes called a q-value, as a function of thresholding at different p-values. As with any thresholding technique, choice of a threshold is arbitrary, and the correspondence between p-values and q-values for levels q < .05 and q < .01 are provided. This procedure is appropriate for the evaluation of all obtained p-values collectively, as the algorithm deals with correlated values appropriately. This provides information on a fine-grained time scale. However, given the large number of central ages at which the regression calculations were performed, in the interest of condensation of information and ease of interpretation results are also provided for 7 overlapping 3 year age ranges extending from age 12 to 24. (Ranges: 12–15, 13.5–16.5, 15–18, 16.5–19.5, 18–21, 19.5–22.5, 21–24.) Medians of p-values across the age-ranges were calculated to provide coarse-grained results. The concentration of measure of the range of p-values introduced by use of the median was considerable, particularly in the range 0 < p < 10−2 and introduced a corresponding bias in the direct application of the Storey-Tibshirani algorithm. As a consequence, the false discovery proportion threshold derived from the calculation on the raw p-values was used to determine the appropriate threshold for the control of the false discovery rate in the median p-values.
Relation of genotypic values to phenotypic correlations
Given that the phenotypic variables show high degrees of correlation, as shown in Figure 2 and described in section 3.1.2, it would be expected that regression models of correlated phenotypic variables using the same model structure and using the same independent variables would exhibit corresponding values for the parameter estimates, including the estimates for the effects of genotype. On the other hand, it also would be expected that some SNPs would be associated with biological factors acting in a spatially localized manner, while others would be associated with factors acting more globally. Because of the small number of SNPs available, a direct test of the similarity of phenotypic intramodal and intermodal correlations and the correspondences of association trajectories between different scalp locations (intramodal) and different modalities (intermodal) across SNPs was not possible.
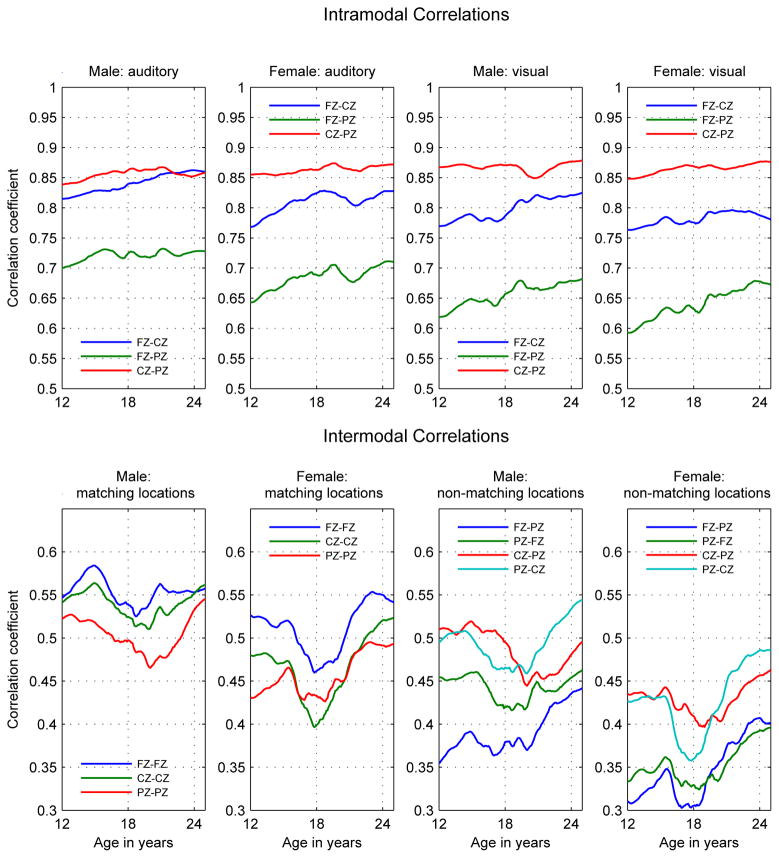
Figure 2.
Intramodal and Intermodal Phenotypic Correlations. Intramodal correlations in top panel: Left Columns: Auditory correlations; Right Columns: Visual Correlations. Intermodal correlations in bottom panel: Left Columns: Matching locations; Right Columns: Non-matching locations. (For non-matching locations the first electrode is the auditory the second the visual.) Note that the scales are different between in the top and bottom panels.
Noting the degree of heterogeneity between SNPs apparent in the association trajectories, and the apparent correspondences in significance of the association trajectories within the SNPs (see figure 3), a test of the null hypothesis that degree of within SNP correspondence could be the effect of a random occurrences of the significant SNP-phenotype association instances was undertaken. To be more specific, the within-SNP correspondences are between different scalp locations in the same modality, and between the different modalities at the same scalp location, where correspondence is based on simultaneous significance or non-significance of the coarse-grained p-values based on the threshold determined by controlling the false discovery rate at 5% for the median p-values. The measure for correspondence between scalp locations is the sum of number of significant values for the three scalp locations for each age-SNP-modality combination and for the correspondence between modalities is the sum of number of significant values for the two modalities for each age-SNP-scalp location combination. Permutation tests are applied to the null hypothesis that random permutation of SNP-phenotype association instances, preserving either the scalp locations for the intramodal correspondences and the modality for the intermodal correspondences, as well as the age relation, could be responsible for the observed pattern of correspondences. The tests permute the SNPs in each scalp location for the intramodal correspondences and in each modality for the intermodal correspondences while keeping all other relations constant. That is, it preserves the correlations which are the result of the regression method, as well as the phenotypes in the correspondences. A chi-squared test is used to compare the distribution of the correspondence measures between the observed data and the permuted data calculated for 1000 permutations of the SNPs.
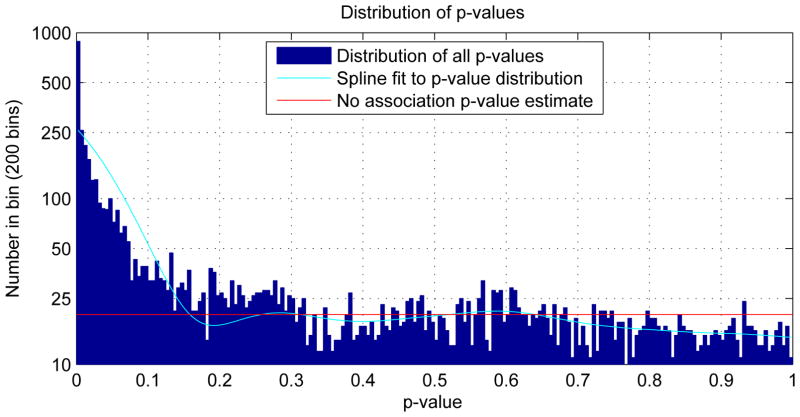
Figure 3.
Distribution of observed p-values and a conservative estimate of the distribution of the p-values of the cases in which there were no genotypic associations in the data. There were 6288 observed p-values of which 4000 were estimated to be instances with no genotypic association. The bins are .005 wide. This plot corresponds to figure 1 in Storey and Tibshirani (2003).
3. Results
3.1. Phenotypic (ERO) results
Results from a previous study of developmental trajectories of the phenotypes from the dataset used in this study (Chorlian et al., 2015) are summarized to provide a context for the results regarding the developmental trajectories of the genetic associations. Figures 1 and 2 are identical to Figures 1 and 2 in Chorlian et al. (2015).
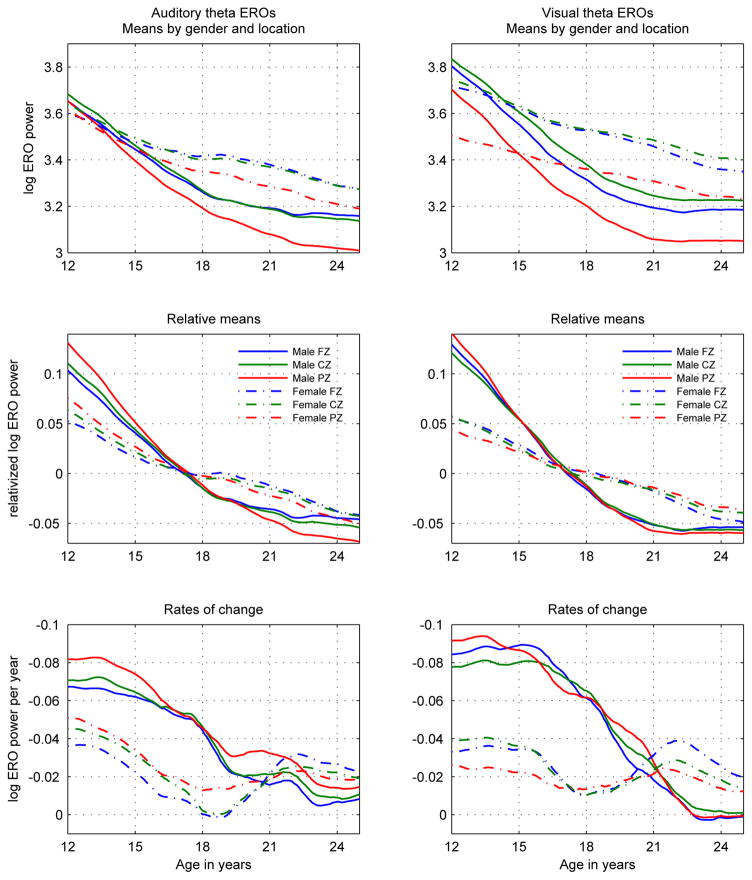
Figure 1.
Log-transformed theta ERO total power trajectory means (s(t)) in auditory (left column) and visual (right column) modalities presented in three views: Top row: Development curves: s(t). Middle row: Relative development curves s̃(t) = (s(t) − mean(s(t)))/mean(s(t)). Values are shown as relative to the mean value of the trajectory. Plots of relative values emphasize similarities and differences in overall shape, rather than in values. Bottom row: Rates of change of development curves: ds(t)/dt = (ds(t)/dt)/s(t). Each line in this graph represents the slope of the corresponding line in the graph in the top row at the corresponding age. The y-axis is inverted in order to illustrate the decrease in absolute value of the slopes with time. All line styles and colors of the graphs follow the legends in the middle row.
3.1.1. ERO power values
ERO power values had the following characteristics:
Male and female developmental trajectories of power were significantly different in their temporal characteristics, with more rapid decreases with age in theta ERO power in males than in females, and a less uniform rate of decrease with age in females than in males.
Male and female developmental trajectories of power were not significantly different in their regional characteristics. Relations between power at different locations were similar between males and females and the shapes of trajectories did not depend on location.
To illustrate the difference between male and female development in the trajectory plots, three different visual representations are employed in Figure 1: in the top row the values are plotted to compare the trajectories themselves; in the middle row the relative values are plotted to compare the shapes of the trajectories without regard to levels of value; in the bottom row the rates of change of the trajectories are plotted to explicitly represent the size of the changes in the trajectories. This figure is explained in more detail in Section 3.1 of Chorlian et al. (2015).
3.1.2. ERO power correlations
ERO power correlations had the following characteristics, as can be seen in Figure 2:
Both males and females exhibited increasing intramodal (between location) power correlations, with correlations between more distant locations showing the greatest proportional increase during the developmental period. Intramodal correlations were always larger than intermodal correlations.
Both males and females exhibited a decrease followed by a sharp increase in intermodal power correlations in a span centered at age 18 and ranging from ages 16 to 21, with the increase persisting to age 25. This, combined with the nearly monotonic increase within the intramodal power correlations indicates a divergence followed by a reconvergence in the developmental characteristics of the auditory and visual systems during the period from ages 16 to 21.
3.2. Genetic effects and gene × sex interactions
3.2.1. Trajectories of association
The trajectories of association were examined by the methods described in section 2.4.2. Most importantly, controlling the false discovery rate at 5% showed that about 24% of regression results were significant at a threshold of p < .009. Bootstrapping confirmed the statistical characteristics provided by the Matlab glmfit() function. This is not surprising given the approximate normality in the phenotypic data and the absence of extremely small minor allele frequencies in the genotypic data. Permutation tests showed that significant results were non-randomly distributed across the range of SNP-age-phenotype combinations. Mean phenotypic differences between genotypes as measured by effect sizes in each sex were smaller than differences between sexes. The small differences between estimates of effect size at successive ages in each sex makes it difficult to obtain accurate values for rates of change on the genotypic level, so that comparisons between genotypes are made in terms of the mean values of the trajectory (cumulative effect). This fact must be kept in mind in interpreting the results. These results are described in detail in the paragraphs below.
Identification of significant SNPs
Using the method described in Section 2.4.2, an algorithm adapted from Storey and Tibshirani (2003) was used to determine the false discovery rate based on an estimate of the probability of a true null hypothesis. This estimate is based on the distribution of p-values of the entire set of regression calculations. This procedure, when applied to the p-values of the genotypic parameters from the 6288 regression calculations provided a conservative estimate of the probability of a true null hypothesis at .63. This is illustrated in figure 3. To control the false discovery rate at 5%, the p-value threshold was set at p < 0.009, which includes 24% of the values tested. Correspondingly, as described in Section 2.4.2, using the proportion threshold to deal with median p-values for the condensed age ranges, 24% of the median p-values are less than 0.0136. To control the false discovery rate at 1%, the p-values threshold was set at p < 6.2 × 10−5, which includes 8% of the genotypic parameters. Correspondingly, 8% of the median p-values are less than .0014.
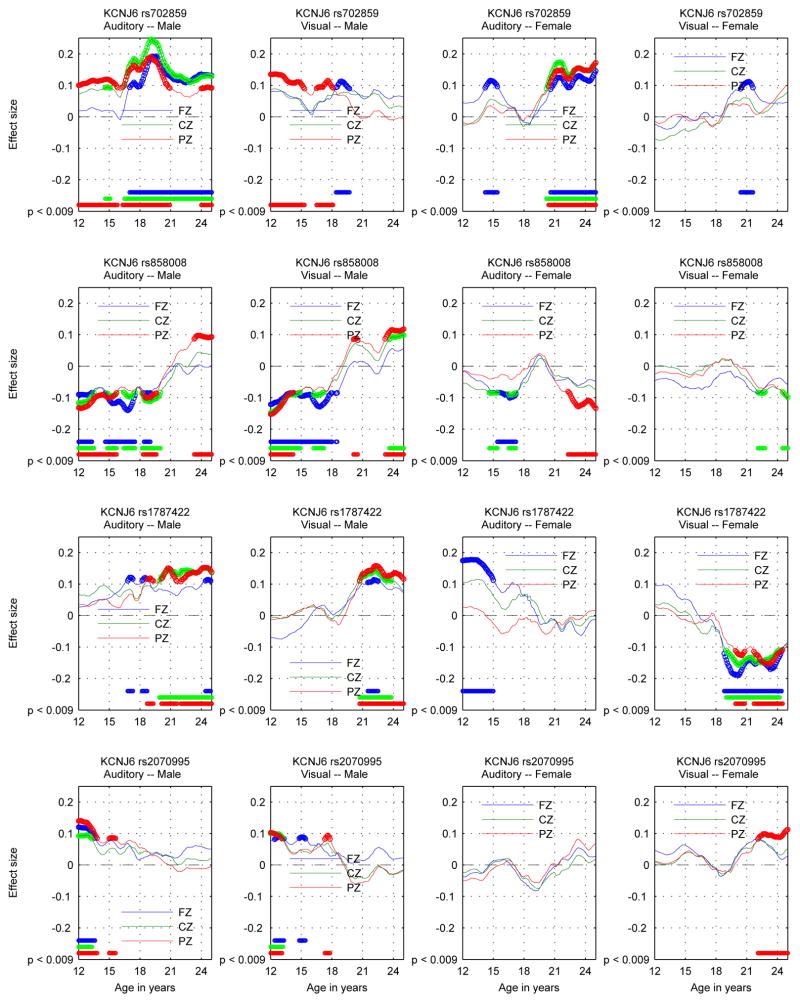
Figures 4 and 5 show the results of applying the criteria in the preceding paragraph to the data on both a coarse-grained and fine-grained time scale. The coarse-grained time scale provides an uncluttered view of the selection of significant SNPs by age ranges and suppresses inconsequential short-term variations in p-value. The fine-grained time scale provides plots of the effect sizes in a virtually continuous form, as well as indication of significance. The fine-grained time scale plot contains more information than the coarse-grained time scale plot but requires greater care in interpretation. For instance, many of the wiggles in the plot do not represent statistically significant variation of the effect sizes. Remarks on aspects of the results will refer to one or the other of the figures as appropriate.
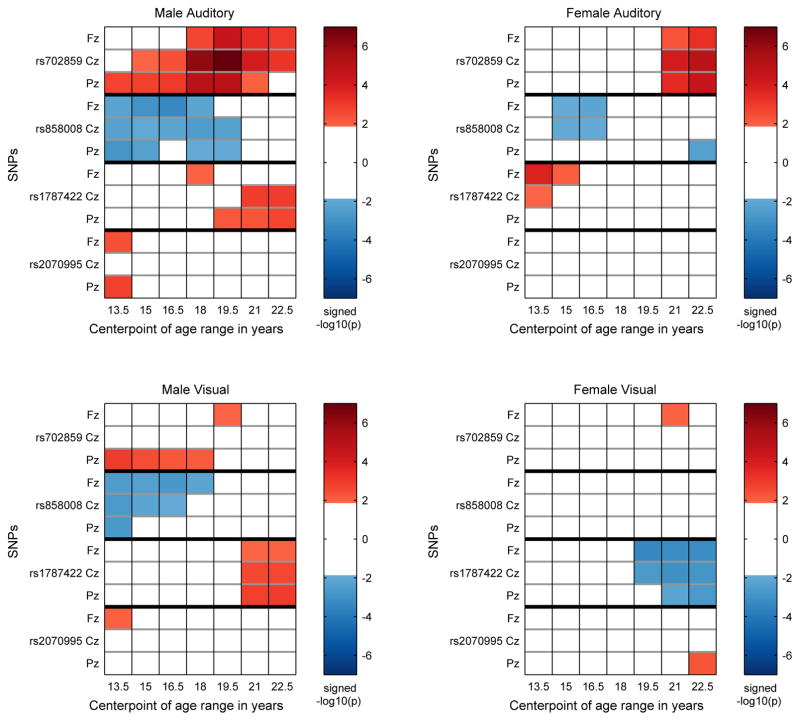
Figure 4.
KCNJ6 SNPs: Identification of significant associations between KCNJ6 SNPs and theta band ERO trajectories for a coarse age scale (seven overlapping 3 year age ranges) for each sex-modality combination. Each panel contains the information for one sex-modality combination for all 4 SNPs. The sex-modality combination is labeled at the top of each panel. For each SNP the rows are ordered Fz, Cz, Pz from top to bottom and ages increase from left to right. The median p-value for each sex-SNP-phenotype-age combination is −log10 transformed and multiplied by the sign of the effect size of the minor allele for each combination and color coded in the plot. Thresholding at p < .0136 controls the false discovery rate at 5% for the representation of age-specific p values by median p values in each age range. Markers on the color bars on the right of each plot indicate the signed median p-value on a −log10 scale. (−log10(.0136) = 1.87.)
Figure 5.
KCNJ6 SNPs: Phenotype-Genotype association trajectories shown by estimates of effect sizes for the minor allele in the additive genetic model in auditory and visual modalities for each SNP. One SNP per row. Males: two left columns; Females: two right columns. The auditory modality is to the left of the visual modality for each sex. Electrodes are color coded: FZ blue, CZ green, PZ red. Thick markers in plotted trajectories and at the bottom edge indicate ages where p < 0.009 for genotypic effects.
On the coarse-grained time scale, the identification of significant associations between KCNJ6 SNPs and theta band ERO trajectories for the seven overlapping three year age ranges extending from 12 to 24 (ranges: 12–15, 13.5–16.5, 15–18, 16.5–19.5, 18–21, 19.5–22.5, 21–24) by thresholding the median p-values at 0.0136 is shown in Figure 4. Each of the four panels illustrates values for a different sex-modality combination. Thresholding is at a level that controls the false discovery rate at 5%. Color coding is used to indicate the sign of the effect size of the minor allele. Table 3 provides a summary of the number of significant associations as a function of SNP, sex, and modality.
Table 3.
Distribution of the sum of number of significant SNP-phenotype combinations over all age ranges.
| SNP | Male | Female | ||
|---|---|---|---|---|
|
| ||||
| Auditory | Visual | Auditory | Visual | |
|
| ||||
| rs702859 | 16 | 5 | 6 | 1 |
| rs858008 | 13 | 8 | 5 | 0 |
| rs1787422 | 6 | 6 | 3 | 8 |
| rs2070995 | 2 | 1 | 0 | 1 |
On the finer-grained time scale, sex-modality specific trajectories of SNP-phenotype association as represented by the effect sizes determined in the regression model using genotype and genotype × sex interaction as covariates are shown in Figure 5 for each SNP-phenotype-sex association. The p < 0.009 criterion controls the false discovery rate at 5%. These spans of significance are indicated for each location at the bottom of each of the panels in Figure 5.
Effect sizes of genotype range from 0.1 to greater than 0.2 in absolute value when statistically significant at a p < 0.009 level, the level established when controlling the false discovery rate at 5%. The larger values are comparable to the effect size noted for significant KCNJ6 SNPs in Kang et al. (2012). The relation between effect size and variance explained is r2 = d2/(d2 + 4) where d is the effect size used here and r2 is the variance explained (Durlak, 2009). This implies that the values of r2 for significant results range from about .25% to 1%.
The significance of the genotypic results must be understood in terms of biological processes involved as well as effect size and number of samples. Sampling variation and and short term fluctuations in the measurement of electrophysiological variables mean that only durations of significant results of greater than 1 year should be given credence. Effect sizes of less than 0.1 when not part of a continuous significant interval greater than 2 years in duration should be discounted; this in particular applies to interpretation of the fine time scale figure. The use of age ranges as illustrated in the coarse time scale figure is an attempt to suppress the most likely false positive results.
Since the graphs show cumulative effects, the indications of significant difference at the bottom of each graph do not record the onset of the occurrence of the genotypic effect; the onset occurs earlier, perhaps at the time when the value of the effect size curve is near zero and the slope of the effect size curve increases in absolute value. This is difficult to quantify exactly, as the onset of genotypic effect is subject to the same uncertainty as the estimation of the ranges of significant effect. Similarly, the cessation of genotypic effect probably occurs by the time the effect size curve reaches an extremum.
3.2.2. Correspondence between association trajectories
Visual assessment of correspondences between association trajectories can be made by examination of the horizontal colored bars at the base of each panel for males (left two columns) and females (right two columns) in Figure 5. Intramodal correspondence can be examined by comparing the bars within each panel; Intermodal correspondence can be examined by comparing the similarly colored bars in adjacent panels of the male and female results in each of the four rows of panels, one for each SNP. Between SNP correspondence can be examined by comparing the similarly colored bars in the four columns of panels, one for each sex-modality combinations, on a pairwise basis.
Permutation tests, as described in section 2.4.2 were carried out to determine whether the intramodal and intermodal correspondences between association trajectories were SNP-specific, with the null hypotheses that they were not. These tests were applied to the data of the coarse-grained analysis. It should be noted that these tests depend on the heterogeneity of the timing of effects between SNPs, and would be more accurate if there were more SNPs.
Test of intramodal correspondences
There were 112 correspondence measures for the intramodal data, as defined in section 2.4.2, (7 age ranges × 4 SNPs × 2 modalities × 2 sexes), each of which was assigned a value equal to the sum of the number of p-values in the three different scalp locations less than the threshold, in this case .0136. As observed, there were 13 3’s, 14 2’s, 15 1’s, and 70 0’s. A permutation test of the null hypothesis that the observed measure came from a non SNP-specific combination of each of the same age range-modality-sex combinations showed that the null hypothesis could be rejected at the p < 4.15 × 10−4 level. This is the result of the fact that there were a much larger number of sets of scalp locations in which all three values were the same in the observed data than there were in the randomized data produced by the permutation process.
Test of intermodal correspondences
There were 162 correspondence measures for the intermodal data, each of which was assigned a value equal to the sum of the number of p-values in the two modalities less than the threshold, in this case .0136. Since there was only one occurrence of simultaneous significance for females, analysis was performed only on data from males. As observed, there were 18 2’s, 44 1’s, and 22 0’s. A permutation test of the null hypothesis that the duplets came from a non SNP-specific combination of each of the same age range-scalp location-sex combinations showed that the null hypothesis could be rejected at the p < .01 level. Again this is the result that there are more pairs in which both values were the same than could be expected from a random, non SNP-specific pairing.
3.2.3. Genotypic effects on phenotypic trajectories
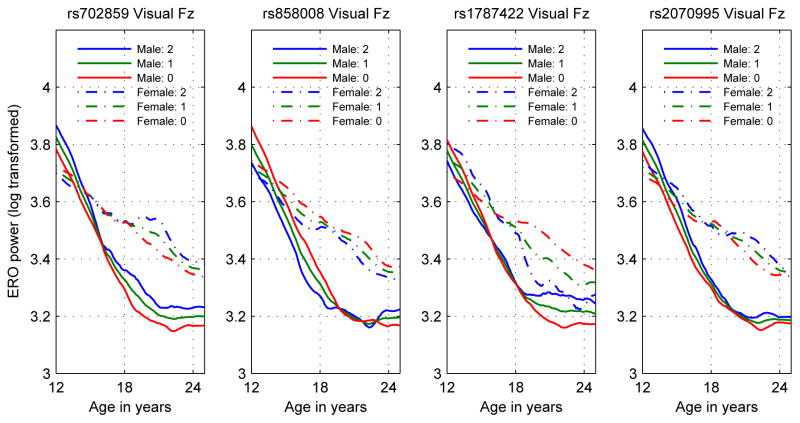
As a consequence of the sex-specific genotypic associations, trajectories of phenotypes when effects of sex and genotype are modeled show strong age-specific sex and genotype effects, as illustrated in the plots of the visual Fz phenotype of the four KCNJ6 SNPs studied in this analysis (Figure 6). Among the features, also observable in the previous plots, are opposite genotypic effects in males and females for rs1787422 and age reversal of genotypic effects in males for rs858008. The effect of family type (alcoholic or community family) was not significant using the same statistical methods as used for the genotypic variables, that is, controlling the false discovery rate to be less than 5%. This indicates that family history with regard to alcohol use disorders had little effect upon the phenotypic trajectories presented in the current study.
Figure 6.
Estimate of mean log transformed visual ERO values at the Fz electrode by genotype and sex for each of the KCNJ6 SNPs analyzed. Plot legend indicates values by sex and number of minor alleles.
4. Discussion
4.1. Introduction
4.1.1. Phenotypic development
The features of development found in the previous study (Chorlian et al., 2015), a general decrease in power with age, striking differences in temporal pattern between males and females, and increases in cross-regional and cross-modal phenotypic correlation, are consistent with important features of brain development. The general decrease in power corresponds to the neuronal level decrease in gray matter density and cortical thickness in adolescence, probably reflecting synaptic pruning and myelination, and an increase in white matter (Sowell et al., 2004; Toga et al., 2006). The sex-specific phenotypic divergence corresponds on the structural and functional levels to the considerable sex differences in the trajectories of the development of features of brain anatomy (Lenroot and Giedd, 2010; Lenroot et al., 2007; Peper et al., 2011; Koolschijn and Crone, 2013) and in task-related brain activity (Rubia et al., 2006; Christakou et al., 2009; Rubia et al., 2010; Sumich et al., 2012; Rubia et al., 2013; Rubia, 2013). The increases in cross-regional and cross-modal phenotypic correlation correspond to the development of brain networks from a pattern of local connectivity to more global patterns of connectivity (Fair et al., 2008, 2009; Power et al., 2010; Supekar et al., 2009; Uddin et al., 2011; Vogel et al., 2010; Zielinski et al., 2010; Menon, 2013; Wu et al., 2013).
Previous studies of P3 related electrophysiological measures have established its heritability in adolescents, and association with specific SNPs in adults (Section 4.2.1). Thus it would be expected that at some time in the process of developmental leading to adulthood, theta EROs would be associated with some SNPs found to be significantly associated with theta EROs in adults.
4.1.2. Genotypic development
SNP-phenotype associations were significant at a p < 0.009 level for some time span and location in each modality for at least one of the sexes in each of the four SNPs examined. SNP-phenotype-sex associations varied for the four SNPs in one or more of the following ways, as can be observed in Figures 4 and 5. The patterns displayed in the phenotype-genotype association trajectories described in this study were not predictable from previous studies of P3 related electrophysiology, but some are found in a number of other studies of development, which are discussed in the following sections.
-
Sex Differences:
Significant associations were opposite between sexes in the auditory modality for rs858008 (second row) and in the visual modality for rs1787422 (fourth row) and rs858008 (figure 5). All of these oppositions were stronger in the later stages of development. The are no opposite effects between modalities at the same scalp location or between scalp locations for the same modality in either males or female. The somewhat greater prevalence of effects in males as opposed to females as shown in Table 3 may only be a result of the use of the particular four SNPs genotyped from KCNJ6 (figure 4). Given the large difference between male and female development, divergent effect patterns in sex should not be surprising. Sex specific effects have been found in genes related to brain development and human height, which are described in Sections 4.2.2 and 4.2.3.
-
Age Variation:
No associations were present across the entire age range. There was a reversal of effect between different age ranges for both modalities in males for rs858008, although the effect in the later part of the age range is for a short interval (figure 5). There are a number of cases in which the effect size trajectories reverse with age without being significant on one or both sides of the reversal. There are too few SNPs to determine whether there is some meaningful pattern of the onset or cessation of significant associations. The considerable age variation in the effect sizes of the genotypic associations might correspond to the decidedly non-linear phenotypic developmental trajectories. Reversals with age in the sign of the effect sizes represent timing differences in development, in which variation in genotype is manifested in the variation in the initiation of some aspect of the developmental process. Similar age variation in effects have been found in genes related to brain development and human height, which are described in Sections 4.2.2 and 4.2.3.
-
Scalp location and Modality Effects:
The relatively large degree of SNP specific intramodal correspondence of phenotype-genotype association (p < 4.15×10−4) reflects both the high degree of phenotypic correlation and also the widespread distribution of the GIRK2 protein in the brain, similar to that found in developing and adult rodents (Fernánadez-Alacid et al., 2011; Saenz del Burgo et al., 2008). The lower degree of SNP specific intermodal correspondence in males (p < .01) reflects the lower degree of phenotypic intermodal correlation, but suggests that the genotypic effects of the KCNJ6 SNPs studied here are substantially independent of modality in males.
Effect of Subject Selection
Given the known association between low P3 amplitude and theta ERO values with risk for alcoholism (Euser et al., 2012; Rangaswamy and Porjesz, 2008; Jones et al., 2006a; Porjesz et al., 2005), we attempted to control the effect of selecting 80% of the subjects from families initially recruited through an alcoholic family by including a family type indicator (alcoholic or community family) in the regression model. The effect of family type was not found to be significant in the statistical analysis. Additionally, no significant association at the GWAS level between alcohol use disorder phenotypes and any of the KCNJ6 SNPs used in the study in adults was found using the same sample and methodology as that used in Kang et al. (2012). We know of no other study which shows association between these KCNJ6 SNPs and alcoholism case-control status.
Genotypic Aspects
Although none of the KCNJ6 SNPs studied here are coding SNPs, recent studies have shown that it is not only the coding SNPs that change the protein that influence a trait, but non-coding intronic and synonymous exonic SNPs can affect gene function. While synonymous SNPs (sSNPs), although in the coding region, do not change the protein sequence encoded by the gene, they can contribute to variation in effects in at least two ways: sSNPs could affect mRNA stability and thus protein expression and enzymatic activity and sSNPs can affect protein conformation which have functional and clinical consequences (Hunt et al., 2009; Sauna and Kimchi-Sarfaty, 2011). Intronic SNPs have effects on splicing and gene expression, both of which have functional and clinical consequences (Cooper, 2010; Harrison, 2015; Jo and Choi, 2015).
4.2. Genotypic effects in related studies
4.2.1. Comparison with other P3 studies
Twin studies suggest that there are genetic effects on P3 electrophysiological measures obtained in target detection tasks during adolescent development (Katsanis et al., 1997; van Beijsterveldt et al., 1998, 2001; Carlson and Iacono, 2006). As mentioned above, effects of SNPs from CHRM2 (Jones et al., 2006b), GRM8 (Chen et al., 2009), CRHR1 (Chen et al., 2010), HTR7 (Zlojutro et al., 2011), and KCNJ6 (Kang et al., 2012) on P3 related electrophysiological measures in adults have been documented. One study found an effect of CHRM2 SNPs on P3 electrophysiological measures in children and young adolescents (Hill et al., 2013).
Five earlier papers on genetic effects on the P3 amplitude in adolescents, a quantity closely related to theta ERO power (Jones et al., 2006a; Andrew and Fein, 2010), used non-genotypic ACE models applied to twins (Katsanis et al., 1997; van Beijsterveldt et al., 1998; Anokhin et al., 2001; van Beijsterveldt et al., 2001; Carlson and Iacono, 2006). All found some degree of genetic effect on P3 amplitude in some or all of the sample groups studied, with heritabilities from the ACE models ranging from 33% to 79%. The studies of Katsanis et al. (1997) and Carlson and Iacono (2006) included only male twins. The studies of van Beijsterveldt et al. (1998, 2001) and Anokhin et al. (2001) included both male and female twins; genetic effects were confirmed only for males. The study by Carlson and Iacono (2006), using a slope-intercept latent growth model for phenotypic data, found both slopes and intercepts to be heritable in the age range from 17 to 23. This result is consistent with those presented here. A genotypic effect beginning at age 17 and continuing past age 23 would produce a difference in slope attributable to a difference in genotype. An example of this is observable in panels 2 and 4 of Figure 6, where fitting a slope-intercept model to the separate genotypes in males over the age range of 17 to 23 would produce lines with different intercepts and different slopes. This type of genotypic effect would also be consistent with the first sentence of the conclusion, “... it appears that the genes influencing slope are largely, if not entirely, the same ones influencing intercept …” (Carlson and Iacono, 2006, p. 478). However, the heritability of slopes might be the result of age invariant genetic effects, and does not have as a consequence the age variation of genetic effects.
A recently published paper by Hill et al. (2013) which examined the relation of P3 amplitude to CHRM2 SNPs with a model using genotype × sex interactions, reported associations only in 8–12 year olds.
All of the four SNPs used in this study were analyzed in the family GWAS study by Kang et al. (2012). Three were found to be significantly associated with visual theta ERO power: two were directly measured (rs1787422, p < 2 × 10−7, rs858008, p < 5 × 10−6) and one was imputed (rs702859, p < 2 × 10−8); the other, rs2070995, was not significant at a p < 10−5 level. The results on KCNJ6 SNPs reported here are not directly comparable to those results because of the difference in the characteristics of the sample as well as the statistical methodology. The sample in Kang et al. (2012) was comprised of members of families of primarily European descent with mean age 31.2 years and an age range from 7 to 74 years. For each phenotype, only one value was used from each subject. The sample in this study is of heterogeneous descent between ages 12 and 25 and additionally multiple observations from the same individual were used. The regression method used here does not exploit family relationship data to establish genetic association so it is inherently less powerful than the generalized disequilibrium test used by Kang et al. (2012). For roughly the same level of effect size in the two studies, the effect sizes in Kang et al. (2012) meet a more stringent criteria (p < 10−5) than found in this study (p < .0009).
4.2.2. Comparison with other brain development studies
The genotypic variants of the COMT Valine158Methionine polymorphism (COMT Val(158)Met) have age and sex specific effects on brain structure and function in children and adolescents. An age reversal of genotypic effect of COMT Val(158)Met on attenuation in cortical thickness in bilateral PFC (medial and lateral aspects), temporal, and superior parietal regions was found in healthy controls from 9 to 22 years of age (Raznahan et al., 2011b). From ages 6 to 20 there is an age reversal in the genotypic effects on performance in a working memory task and a corresponding variation in the activation level of of the right inferior frontal gyrus and intraparietal sulcus (Dumontheil et al., 2011). These variants have effects on auditory evoked potentials in adults (Heitland et al., 2013; Saville et al., 2014). In 9 year olds, effects on IQ and executive function are found only in males, and are stronger in in pubertal than in prepubertal males (Barnett et al., 2007).
Results on studies of the temporal and spatial variation of gene expression in human brain tissue (Colantuoni et al., 2011; Kang et al., 2011; Naumova et al., 2013) suggest that genetic effects on neurophysiological systems are likely to show both age-related and regional variability. These studies indicate that changes in gene expression trajectories tend to peak in mid to late adolescence. It was also shown that different functional groups of genes have opposite directions of age-related changes of their expression levels. A study of the developmental trajectories of gene expression in the brains of rodents found systematic regional and functional variation of expression extending over the entire developmental period (Stead et al., 2006).
4.2.3. Comparison with other developmental studies
Age variation and sex differences in genotypic effects on development have been found in studies of the development of height in humans. A recent study by Cousminer et al. (2013) of the development of height in several samples ranging in age from pre-teen to adult revealed five loci (near MAPK3, PXMP3, VGLL3, ADCY3-POMC, and LIN28B) whose effect on height related phenotypes (measured by standard deviation from age/sex mean) varied with age and sex as calculated for six ages between 8 and 20. At all five loci, sex effects were significantly different between males and females at least one age. Opposite effects in males and females were found for the loci near VGLL3 and LIN28B. Since that study included results from samples considerbly larger than the sample studied here, it was able to detect significant effect sizes at a p < 10−5 level which were smaller by a factor of two than the effect sizes presented in this study. A previous study of LIN28B (Widén et al., 2010, p. 778) showed opposite effects for sexes in two SNPs from about age 8 to early adolescence. A study by Sovio et al. (2009) found age (infancy compared to puberty) but no sex specific genotypic effects for a number of SNPs (Table 2 in Sovio et al. (2009)) for height velocity. More peripheral instances of age variation in genotypic effect were shown in Chorlian et al. (2013) where some CHRM2 SNPs were found to affect risk for the onset of alcohol dependence in adolescents and young adults only in those who became alcohol dependent under the age of 16 and in Hill et al. (2013), mentioned above.
4.3. Summary
Given the large number of observations available, it became possible to estimate mean rates of change of development with considerable accuracy. This enabled the identification of striking sex differences in age-specific developmental patterns and the effects of genotype on development with a similar range of effect sizes as found in comparable GWA studies. Preliminary studies of other P3 related phenotypes derived from the same subjects with the same methodology show similar patterns of sex differences in mean rates of change but manifest different patterns of genotypic effect. This suggests that relatively large neurophysiological systems governed by multiple genetic factors may experience coordinated developmental patterns regulated by sex specific effects. A more comprehensive multivariate description of the genetic architecture of the P3 response can be obtained using EROs from different frequency bands and experimental conditions and using a number of different measures of the response, together with a number of different genetic factors known to affect neurophysiological and neuroanatomical features. Since measures of the P3 response have been found to be useful endophenotypes for the study of substance abuse disorders (Euser et al., 2012; Iacono and Malone, 2011; Rangaswamy and Porjesz, 2008), externalizing psychopathology (Gilmore et al., 2012), schizophrenia (Decoster et al., 2012; Johannesen et al., 2013), and ADHD (Szuromi et al., 2011), and is highly heritable (van Beijsterveldt and van Baal, 2002), studies of genetic effects on P3 development in adolescents and young adults may illuminate neurophysiological and neuroanatomical factors contributing to the onset of these conditions.
Highlights.
There is considerable age variation in the association of KCNJ6 SNPs with theta EROs, with different trajectories for different SNPs.
The association is considerably greater in males than in females.
Locational and and modality differences are small compared to sex differences.
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); University of Texas Health Science Center at San Antonio (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, J. McClintick, S. O’Connor, L. Wetherill, X. Xuei, Y. Liu (Indiana University); G. Chan (University of Iowa; University of Connecticut); D. Chorlian, N. Manz, C. Kamarajan, A. Pandey (SUNY Downstate); J.-C. Wang, M. Kapoor (Icahn School of Medicine at Mount Sinai) and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Additionally, we would like to thank the two anonymous reviewers who contributed their time and expertise to provide critiques and suggestions which greatly enhanced the paper in both substance and style.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
David B. Chorlian, Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry, SUNY Downstate Medical Center, Brooklyn, NY, USA
Madhavi Rangaswamy, Department of Psychology, Christ University, Bangalore, India.
Niklas Manz, Department of Physics, College of Wooster, Wooster, OH, USA.
Jacquelyn L. Meyers, Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry, SUNY Downstate Medical Center, Brooklyn, NY, USA
Sun J. Kang, Stratton VA Medical Center, Albany, NY, USA
Chella Kamarajan, Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry, SUNY Downstate Medical Center, Brooklyn, NY, USA.
Ashwini K. Pandey, Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry, SUNY Downstate Medical Center, Brooklyn, NY, USA
Jen-Chyong Wang, Mount Sinai School of Medicine, New York, NY, USA.
Leah Wetherill, Indiana University School of Medicine, Indianapolis, IN, USA.
Howard Edenberg, Indiana University School of Medicine, Indianapolis, IN, USA.
Bernice Porjesz, Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry, SUNY Downstate Medical Center, Brooklyn, NY, USA.
References
- Alexander JE, Polich J, Bloom FE, Bauer LO, Kuperman S, Rohrbaugh J, Morzorati S, O’Connor SJ, Porjesz B, Begleiter H. P300 from an auditory oddball task: inter-laboratory consistency. Int J Psychophysiol. 1994;17:35–46. doi: 10.1016/0167-8760(94)90053-1. [DOI] [PubMed] [Google Scholar]
- Andrew C, Fein G. Event-related oscillations versus event-related potentials in a P300 task as biomarkers for alcoholism. Alcohol Clin Exp Res. 2010;34(4):669–80. doi: 10.1111/j.1530-0277.2009.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokhin AP, van Baal GC, van Beijsterveldt CE, de Geus EJ, Grant J, Boomsma DI. Genetic correlation between the P300 event-related brain potential and the EEG power spectrum. Behav Genet. 2001;31(6):545–54. doi: 10.1023/a:1013341310865. [DOI] [PubMed] [Google Scholar]
- Ardekani BA, Choi SJ, Hossein-Zadeh GA, Porjesz B, Tanabe JL, Lim KO, Bilder R, Helpern JA, Begleiter H. Functional magnetic resonance imaging of brain activity in the visual oddball task. Brain Res Cogn Brain Res. 2002;14(3):347–56. doi: 10.1016/s0926-6410(02)00137-4. [DOI] [PubMed] [Google Scholar]
- Astle W, Balding DJ. Population Structure and Cryptic Relatedness in Genetic Association Studies. 2009 http://arxiv.org/pdf/1010.4681.
- Babiloni C, Vecchio F, Mirabella G, Buttiglione M, Sebastiano F, Picardi A, Di Gennaro G, Quarato PP, Grammaldo LG, Buffo P, Esposito V, Manfredi M, Cantore G, Eusebi F. Hippocampal, amygdala, and neocortical synchronization of theta rhythms is related to an immediate recall during Rey auditory verbal learning test. Hum Brain Mapp. 2009;30(7):2077–89. doi: 10.1002/hbm.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JH, Heron J, Ring SM, Golding J, Goldman D, Xu K, Jones PB. Gender-specific effects of the catechol-O-methyltransferase Val108/158Met polymorphism on cognitive function in children. Am J Psychiatry. 2007;164(1):142–9. doi: 10.1176/ajp.2007.164.1.142. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock VM, Porjesz B, Li TK, Schuckit MA, et al. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, et al. Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroencephalogr Clin Neurophysiol. 1998;108(3):244–250. doi: 10.1016/s0168-5597(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Brázdil M, Rektor I, Dufek M, Daniel P, Jurák P, Kuba R. The role of frontal and temporal lobes in visual discrimination task–depth ERP studies. Neurophysiol Clin. 1999;29(4):339–50. doi: 10.1016/s0987-7053(99)90047-3. [DOI] [PubMed] [Google Scholar]
- Carlson SR, Iacono WG. Heritability of P300 amplitude development from adolescence to adulthood. Psychophysiology. 2006;43(5):470–80. doi: 10.1111/j.1469-8986.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J, Jr, Kuperman S, O’Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):359–68. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Manz N, Tang Y, Rangaswamy M, Almasy L, Kuperman S, Nurnberger J, Jr, O’Connor SJ, Edenberg HJ, Schuckit MA, Tischfield J, Foroud T, Bierut LJ, Rohrbaugh J, Rice JP, Goate A, Hesselbrock V, Porjesz B. Single-nucleotide polymorphisms in corticotropin releasing hormone receptor 1 gene (CRHR1) are associated with quantitative trait of event-related potential and alcohol dependence. Alcohol Clin Exp Res. 2010;34(6):988–96. doi: 10.1111/j.1530-0277.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlian DB, Rangaswamy M, Manz N, Wang JC, Dick D, Almasy L, Bauer L, Bucholz K, Foroud T, Hesselbrock V, Kang SJ, Kramer J, Kuperman S, Nurnberger J, Jr, Rice J, Schuckit M, Tischfield J, Edenberg HJ, Goate A, Bierut L, Porjesz B. Genetic and neurophysiological correlates of the age of onset of alcohol use disorders in adolescents and young adults. Behav Genet. 2013;43(5):386–401. doi: 10.1007/s10519-013-9604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlian DB, Rangaswamy M, Manz N, Kamarajan C, Pandey AK, Edenberg H, Kuperman S, Porjesz B. Gender modulates the development of Theta Event Related Oscillations in Adolescents and Young Adults. Behav Brain Res. 2015 doi: 10.1016/j.bbr.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christakou A, Halari R, Smith AB, Ifkovits E, Brammer M, Rubia K. Sex-dependent age modulation of frontostriatal and temporo-parietal activation during cognitive control. Neuroimage. 2009;48(1):223–36. doi: 10.1016/j.neuroimage.2009.06.070. [DOI] [PubMed] [Google Scholar]
- Cleveland WS. Robust Locally Weighted Regression and Smoothing Scatterplots. JASA. 1979;74(368):829–836. [Google Scholar]
- Cleveland WS, Devlin SJ. Locally-Weighted Regression: An Approach to Regression Analysis by Local Fitting. JASA. 1988;83(403):596–610. [Google Scholar]
- Cohen HL, Wang W, Porjesz B, Bauer L, Kuperman S, O’Connor SJ, Rohrbaugh J, Begleiter H. Visual P300: an interlaboratory consistency study. Alcohol. 1994;11:583–587. doi: 10.1016/0741-8329(94)90087-6. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–23. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum Genomics. 2010;4(5):284–8. doi: 10.1186/1479-7364-4-5-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousminer DL, Berry DJ, Timpson NJ, Ang W, Thiering E, Byrne EM, Taal HR, Huikari V, Bradfield JP, Kerkhof M, Groen-Blokhuis MM, Kreiner M, Kreiner-Mller E, Marinelli M, Holst C, Leinonen JT, Perry JR, Surakka I, Pietiläinen O, Kettunen J, Anttila V, Kaakinen M, Sovio U, Pouta A, Das S, Lagou V, Power C, Prokopenko I, Evans DM, Kemp JP, St Pourcain B, Ring S, Palotie A, Kajantie E, Osmond C, Lehtimäki T, Viikari JS, Kähönen M, Warrington NM, Lye SJ, Palmer LJ, Tiesler CM, Flexeder C, Montgomery GW, Medland SE, Hofman A, Hakonarson H, Guxens M, Bartels M, Salomaa V, Murabito JM, Kaprio J, Srensen TI, Ballester F, Bisgaard H, Boomsma DI, Koppelman GH, Grant SF, Jaddoe VW, Martin NG, Heinrich J, Pennell CE, Raitakari OT, Eriksson JG, Smith GD, Hyppönen E, Järvelin MR, McCarthy MI, Ripatti S, Widén E Early Growth Genetics (EGG) Consortium. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum Mol Genet. 2013;22(13):2735–47. doi: 10.1093/hmg/ddt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley MJ, van Noordt SJ, Wu J, Hommer RE, South M, Fearon RM, Mayes LC. Reward feedback processing in children and adolescents: medial frontal theta oscillations. Brain Cogn. 2014;89:79–89. doi: 10.1016/j.bandc.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoster J, De Hert M, Viechtbauer W, Nagels G, Myin-Germeys I, Peuskens J, van Os J, van Winkel R. Genetic association study of the P300 endophenotype in schizophrenia. Schizophr Res. 2012;141(1):54–9. doi: 10.1016/j.schres.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Douet V, Chang L, Cloak C, Ernst T. Genetic influences on brain developmental trajectories on neuroimaging studies: from infancy to young adulthood. Brain Imaging Behav. 2014;8(2):234–50. doi: 10.1007/s11682-013-9260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumontheil I, Roggeman C, Ziermans T, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T. Influence of the COMT genotype on working memory and brain activity changes during development. Biol Psychiatry. 2011;70(3):222–9. doi: 10.1016/j.biopsych.2011.02.027. [DOI] [PubMed] [Google Scholar]
- Durlak J. How to Select, Calculate, and Interpret Effect Sizes. J Pediatr Psychol. 2009;34(9):917–928. doi: 10.1093/jpepsy/jsp004. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Bierut LJ, Boyce P, Cao M, Cawley S, Chiles R, Doheny KF, Hansen M, Hinrichs T, Jones K, Kelleher M, Kennedy GC, Liu G, Marcus G, McBride C, Murray SS, Oliphant A, Pettengill J, Porjesz B, Pugh EW, Rice JP, Rubano T, Shannon S, Steeke R, Tischfield JA, Tsai YY, Zhang C, Begleiter H Repro-Gen Consortium. Description of the data from the Collaborative Study on the Genetics of Alcoholism (COGA) and single-nucleotide polymorphism genotyping for Genetic Analysis Workshop 14. BMC Genet. 2005;6(Suppl 1):S2. doi: 10.1186/1471-2156-6-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euser AS, Arends LR, Evans BE, Greaves-Lord K, Huizink AC, Franken IH. The P300 event-related brain potential as a neurobiological endophenotype for substance use disorders: a meta-analytic investigation. Neurosci Biobehav Rev. 2012;36(1):572–603. doi: 10.1016/j.neubiorev.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. The maturing architecture of the brain’s default network. Proc Natl Acad Sci U S A. 2008;105(10):4028–32. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, Schlaggar BL, Petersen SE. Functional brain networks develop from a ”local to distributed” organization. PLoS Comput Biol. 2009;5(5):e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zhang W. Statistical methods with varying coefficient models. Statistics and its Interface. 2008;1:179–195. doi: 10.4310/sii.2008.v1.n1.a15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernánadez-Alacid L, Watanabe M, Molnár E, Wickman K, Luján R. Developmental regulation of G protein-gated inwardly-rectifying K+ (GIRK/Kir3) channel subunits in the brain. Eur J Neurosci. 2011;34(11):1724–36. doi: 10.1111/j.1460-9568.2011.07886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T, Mller MHG, Köhler W, Molinari L, Prader A. Nonparametric regression analysis of growth curves. Ann Stat. 1984a;12(1):210–229. [Google Scholar]
- Gasser T, Köhler W, Mller MHG, Kneip A, Largo R, Molinari L, Prader A. Velocity and acceleration of height growth using kernel estimation. Ann Hum Biol. 1984b Sep-Oct;(5):397–411. doi: 10.1080/03014468400007311. [DOI] [PubMed] [Google Scholar]
- Gasser T, Gervini D, Molinari L. Kernel estimation, shape-invariant modeling and structural analysis. In: Hauspie R, Cameron N, Molinari L, editors. Methods in Human Growth Research. Cambridge University Press; 2004. pp. 179–204. [Google Scholar]
- Gevins A, Smith ME, Leong H, McEvoy L, Whitfield S, Du R, Rush G. Monitoring working memory load during computer-based tasks with EEG pattern recognition methods. Hum Factors. 1998;40(1):79–91. doi: 10.1518/001872098779480578. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Stockman M, Weddle C, Liverpool M, Alexander-Bloch A, Wallace GL, Lee NR, Lalonde F, Lenroot RK. Anatomic magnetic resonance imaging of the developing child and adolescent brain and effects of genetic variation. Neuropsychol Rev. 2010;20(4):349–61. doi: 10.1007/s11065-010-9151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Iacono WG. Brain electrophysiological endophenotypes for externalizing psychopathology: a multivariate approach. Behav Genet. 2010;40(2):186–200. doi: 10.1007/s10519-010-9343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ. Recent genetic findings in schizophrenia and their therapeutic relevance. J Psychopharmacol. 2015;29(2):85–96. doi: 10.1177/0269881114553647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitland I, Kenemans JL, Oosting RS, Baas JM, Böcker KB. Auditory event-related potentials (P3a, P3b) and genetic variants within the dopamine and serotonin system in healthy females. Behav Brain Res. 2013;249:55–64. doi: 10.1016/j.bbr.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Hill SY, Shen S, Locke J, Steinhauer SR, Konicky C, Lowers L, Connolly J. Developmental delay in P300 production in children at high risk for developing alcohol-related disorders. Biol Psychiatry. 1999a;46(7):970–81. doi: 10.1016/s0006-3223(99)00032-3. [DOI] [PubMed] [Google Scholar]
- Hill SY, Jones BL, Holmes B, Steinhauer SR, Zezza N, Stiffler S. Cholinergic receptor gene (CHRM2) variation and familial loading for alcohol dependence predict childhood developmental trajectories of P300. Psychiatry Res. 2013;209(3):504–11. doi: 10.1016/j.psychres.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- Hoover DR, Rice JA, Wu CO, Yang LP. Nonparametric smoothing estimates of time-varying coefficient models with longitudinal data. Biometrika. 1998;85(4):809–822. [Google Scholar]
- Iacono WG, Malone SM. Developmental Endophenotypes: Indexing Genetic Risk for Substance Abuse with the P300 Brain Event-Related Potential. Child Dev Perspect. 2011;5(4):239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Hwang G, Curran T, Kahana MJ. EEG oscillations and recognition memory: theta correlates of memory retrieval and decision making. Neuroimage. 2006;32:978–987. doi: 10.1016/j.neuroimage.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Jo BS, Choi SS. Introns: The Functional Benefits of Introns in Genomes. Genomics Inform. 2015;13(4):112–118. doi: 10.5808/GI.2015.13.4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesen JK, O’Donnell BF, Shekhar A, McGrew JH, Hetrick WP. Diagnostic specificity of neurophysiological endophenotypes in schizophrenia and bipolar disorder. Schizophr Bull. 2013;39(6):1219–29. doi: 10.1093/schbul/sbs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Chorlian D, Rangaswamy M, Kamarajan C, Padmanabhapillai A, Stimus A, Begleiter H. S-transform time-frequency analysis of P300 reveals deficits in individuals diagnosed with alcoholism. Clin Neurophysiol. 2006a;117(10):2128–43. doi: 10.1016/j.clinph.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Almasy L, Bierut L, Dick D, Goate A, Hinrichs A, Rice JP, Wang JC, Bauer LO, Crowe R, Foroud T, Hesselbrock V, Kuperman S, Nurnberger J, Jr, O’Connor SJ, Rohrbaugh J, Schuckit MA, Tischfield J, Edenberg HJ, Begleiter H. A cholinergic receptor gene (CHRM2) affects event-related oscillations. Behav Genet. 2006b;36(5):627–39. doi: 10.1007/s10519-006-9075-6. [DOI] [PubMed] [Google Scholar]
- Kamarajan C, Porjesz B, Jones K, Chorlian D, Padmanabhapillai A, Rangaswamy M, Stimus A, Begleiter H. Event-related oscillations in offspring of alcoholics: neurocognitive disinhibition as a risk for alcoholism. Biol Psychiatry. 2006;59(7):625–34. doi: 10.1016/j.biopsych.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Jr, Rice J, Schuckit M, Tischfield J, Bierut LJ, Edenberg HJ, Goate A, Foroud T, Porjesz B. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012;11(6):712–9. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas S, Erzengin OU, Basar E. A new strategy involving multiple cognitive paradigms demonstrates that ERP components are determined by the superposition of oscillatory responses. Clin Neurophysiol. 2000a;111(10):1719–32. doi: 10.1016/s1388-2457(00)00418-1. [DOI] [PubMed] [Google Scholar]
- Karakas S, Erzengin OU, Basar E. The genesis of human event-related responses explained through the theory of oscillatory neural assemblies. Neurosci Lett. 2000b;285(1):45–8. doi: 10.1016/s0304-3940(00)01022-3. [DOI] [PubMed] [Google Scholar]
- Katsanis J, Iacono WG, McGue MK. The association between P300 and age from preadolescence to early adulthood. Int J Psychophysiol. 1996;24(3):213–221. doi: 10.1016/s0167-8760(96)00063-3. [DOI] [PubMed] [Google Scholar]
- Katsanis J, Iacono WG, McGue MK, Carlson SR. P300 event-related potential heritability in monozygotic and dizygotic twins. Psychophysiology. 1997;34(1):47–58. doi: 10.1111/j.1469-8986.1997.tb02415.x. [DOI] [PubMed] [Google Scholar]
- Klimesch W, Schimke H, Schwaiger J. Episodic and semantic memory: an analysis in the EEG theta and alpha band. Electroencephalogr Clin Neurophysiol. 1994;91:428–441. doi: 10.1016/0013-4694(94)90164-3. [DOI] [PubMed] [Google Scholar]
- Klimesch W, Doppelmayr M, Yonelinas A, Kroll NE, Lazzara M, Rohm D, Gruber W. Theta synchronization during episodic retrieval: neural correlates of conscious awareness. Brain Res Cogn Brain Res. 2001;12:33–38. doi: 10.1016/s0926-6410(01)00024-6. [DOI] [PubMed] [Google Scholar]
- Klimesch W, Freunberger R, Sauseng P, Gruber W. A short review of slow phase synchronization and memory: evidence for control processes in different memory systems? Brain Res. 2008;1235:31–44. doi: 10.1016/j.brainres.2008.06.049. [DOI] [PubMed] [Google Scholar]
- Koolschijn PC, Crone EA. Sex differences and structural brain maturation from childhood to early adulthood. Dev Cogn Neurosci. 2013;5:106–18. doi: 10.1016/j.dcn.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Giedd JN. Sex differences in the adolescent brain. Brain Cogn. 2010;72(1):46–55. doi: 10.1016/j.bandc.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Gogtay N, Greenstein DK, Wells EM, Wallace GL, Clasen LS, Blumenthal JD, Lerch J, Zijdenbos AP, Evans AC, Thompson PM, Giedd JN. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. Neuroimage. 2007;36(4):1065–73. doi: 10.1016/j.neuroimage.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden DE, Prvulovic D, Formisano E, Vallinger M, Zanella FE, Goebel R, Dierks T. The functional neuroanatomy of target detection: an fMRI study of visual and auditory oddball tasks. Cereb Cortex. 1999;9(8):815–23. doi: 10.1093/cercor/9.8.815. [DOI] [PubMed] [Google Scholar]
- Lötsch J, Prüss H, Veh RW, Doehring A. A KCNJ6 (Kir3.2, GIRK2) gene polymorphism modulates opioid effects on analgesia and addiction but not on pupil size. Pharmacogenet Genomics. 2010;20(5):291–7. doi: 10.1097/FPC.0b013e3283386bda. [DOI] [PubMed] [Google Scholar]
- Mantini D, Corbetta M, Perrucci MG, Romani GL, Del Gratta C. Large-scale brain networks account for sustained and transient activity during target detection. Neuroimage. 2009;44(1):265–74. doi: 10.1016/j.neuroimage.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V, Ford JM, Lim KO, Glover GH, Pfefferbaum A. Combined event related fMRI and EEG evidence for temporal-parietal cortex activation during target detection. Neuroreport. 1997;8:3029–3037. doi: 10.1097/00001756-199709290-00007. [DOI] [PubMed] [Google Scholar]
- Menon V. Developmental pathways to functional brain networks: emerging principles. Trends Cogn Sci. 2013;17(12):627–40. doi: 10.1016/j.tics.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Molinari L, Gasser T. The human growth curve: distance, velocity and acceleration. In: Hauspie R, Cameron N, Molinari L, editors. Methods in Human Growth Research. Cambridge University Press; 2004. pp. 27–54. [Google Scholar]
- Nanova P, Lyamova L, Hadjigeorgieva M, Kolev V, Yordanova J. Gender-specific development of auditory information processing in children: an ERP study. Clin Neurophysiol. 2008;119(9):1992–2003. doi: 10.1016/j.clinph.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Nanova P, Kolev V, Yordanova J. Developmental gender differences in the synchronization of auditory event-related oscillations. Clin Neurophysiol. 2011;122(5):907–15. doi: 10.1016/j.clinph.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Naumova OY, Lee M, Rychkov SY, Vlasova NV, Grigorenko EL. Gene expression in the human brain: the current state of the study of specificity and spatiotemporal dynamics. Child Dev. 2013;84(1):76–88. doi: 10.1111/cdev.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peper JS, Hulshoff Pol HE, Crone EA, van Honk J. Sex steroids and brain structure in pubertal boys and girls: a mini-review of neuroimaging studies. Neuroscience. 2011;2191:28–37. doi: 10.1016/j.neuroscience.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Polich J, Criado JR. Neuropsychology and neuropharmacology of P3a and P3b. Int J Psychophysiol. 2006;60(2):172–85. doi: 10.1016/j.ijpsycho.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Polich J. Updating P300: an integrative theory of P3a and P3b. Clin Neurophysiol. 2007;118(10):2128–48. doi: 10.1016/j.clinph.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Rangaswamy M, Kamarajan C, Jones KA, Padmanabhapillai A, Begleiter H. The utility of neurophysiological markers in the study of alcoholism. Clin Neurophysiol. 2005;116(5):993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Power JD, Fair DA, Schlaggar BL, Petersen SE. The development of human functional brain networks. Neuron. 2010;67(5):735–48. doi: 10.1016/j.neuron.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JO, Silverman BW. Applied Functional Data Analysis. Springer-Verlag; New York: 2002. [Google Scholar]
- Rangaswamy M, Jones KA, Porjesz B, Chorlian DB, Padmanabhapillai A, Kamarajan C, Kuperman S, Rohrbaugh J, O’Connor SJ, Bauer LO, Schuckit MA, Begleiter H. Delta and theta oscillations as risk markers in adolescent offspring of alcoholics. Int J Psychophysiol. 2007;63(1):3–15. doi: 10.1016/j.ijpsycho.2006.10.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B. Uncovering genes for cognitive(dys)function and predisposition for alcoholism spectrum disorders: A review of human brain oscillations as effective endophenotypes. Brain Res. 2008;1235:153–71. doi: 10.1016/j.brainres.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Shaw P, Lalonde F, Stockman M, Wallace GL, Greenstein D, Clasen L, Gogtay N, Giedd JN. How does your cortex grow? J Neurosci. 2011;31(19):7174–7. doi: 10.1523/JNEUROSCI.0054-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Greenstein D, Lee Y, Long R, Clasen L, Gochman P, Addington A, Giedd JN, Rapoport JL, Gogtay N. Catechol-o-methyl transferase (COMT) val158met polymorphism and adolescent cortical development in patients with childhood-onset schizophrenia, their non-psychotic siblings, and healthy controls. Neuroimage. 2011;57(4):1517–23. doi: 10.1016/j.neuroimage.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T. A genomic survey of alcohol dependence and related phenotypes: results from the Collaborative Study on the Genetics of Alcoholism (COGA) Alcohol Clin Exp Res. 1996;20:A133–137. doi: 10.1111/j.1530-0277.1996.tb01763.x. [DOI] [PubMed] [Google Scholar]
- Rubia K, Smith AB, Woolley J, Nosarti C, Heyman I, Taylor E, Brammer M. Progressive increase of frontostriatal brain activation from childhood to adulthood during event-related tasks of cognitive control. Hum Brain Mapp. 2006;27(12):973–93. doi: 10.1002/hbm.20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubia K, Hyde Z, Halari R, Giampietro V, Smith A. Effects of age and sex on developmental neural networks of visual-spatial attention allocation. Neuroimage. 2010;51(2):817–27. doi: 10.1016/j.neuroimage.2010.02.058. [DOI] [PubMed] [Google Scholar]
- Rubia K. Functional brain imaging across development. Eur Child Adolesc Psychiatry. 2013;22(12):719–31. doi: 10.1007/s00787-012-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubia K, Lim L, Ecker C, Halari R, Giampietro V, Simmons A, Brammer M, Smith A. Effects of age and gender on neural networks of motor response inhibition: From adolescence to mid-adulthood. Neuroimage. 2013;83C:690–703. doi: 10.1016/j.neuroimage.2013.06.078. [DOI] [PubMed] [Google Scholar]
- Saenz del Burgo L, Cortes R, Mengod G, Zarate J, Echevarria E, Salles J. Distribution and neurochemical characterization of neurons expressing GIRK channels in the rat brain. J Comp Neurol. 2008;510:581–606. doi: 10.1002/cne.21810. [DOI] [PubMed] [Google Scholar]
- Sara SJ, Bouret S. Orienting and reorienting: the locus coeruleus mediates cognition through arousal. Neuron. 2012;76(1):130–41. doi: 10.1016/j.neuron.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–91. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Saville CW, Lancaster TM, Stefanou ME, Salunkhe G, Lourmpa I, Nadkarni A, Boehm SG, Bender S, Smyrnis N, Ettinger U, Feige B, Biscaldi M, Mantripragada KK, Linden DE, Klein C. COMT Val158Met genotype is associated with fluctuations in working memory performance: converging evidence from behavioural and single-trial P3b measures. Neuroimage. 2014;100:489–97. doi: 10.1016/j.neuroimage.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Segalowitz SJ, Santesso DL, Jetha MK. Electrophysiological changes during adolescence: a review. Brain Cogn. 2010;72(1):86–100. doi: 10.1016/j.bandc.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, Greenstein D, Clasen L, Evans A, Rapoport JL, Giedd JN, Wise SP. Neurodevelopmental trajectories of the human cerebral cortex. J Neurosci. 2008;28(14):3586–94. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovio U, Bennett AJ, Millwood IY, Molitor J, O’Reilly PF, Timpson NJ, Kaakinen M, Laitinen J, Haukka J, Pillas D, Tzoulaki I, Molitor J, Hoggart C, Coin LJ, Whittaker J, Pouta A, Hartikainen AL, Freimer NB, Widen E, Peltonen L, Elliott P, McCarthy MI, Jarvelin MR. Genetic determinants of height growth assessed longitudinally from infancy to adulthood in the northern Finland birth cohort 1966. PLoS Genet. 2009;5(3):e1000409. doi: 10.1371/journal.pgen.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Toga AW. Mapping changes in the human cortex throughout the span of life. Neuroscientist. 2004;10:372–392. doi: 10.1177/1073858404263960. [DOI] [PubMed] [Google Scholar]
- Stead JD, Neal C, Meng F, Wang Y, Evans S, Vazquez DM, Watson SJ, Akil H. Transcriptional pro-filing of the developing rat brain reveals that the most dramatic regional differentiation in gene expression occurs postpartum. J Neurosci. 2006;26(1):345–53. doi: 10.1523/JNEUROSCI.2755-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stige S, Fjell AM, Smith L, Lindgren M, Walhovd KB. The development of visual P3a and P3b. Developmental Neuropsychology. 2007;32(1):563–584. doi: 10.1080/87565640701361096. [DOI] [PubMed] [Google Scholar]
- Stockwell RG, Mansinha L, Lowe RP. Localization of the complex spectrum: The S transform. IEEE Trans Signal Process. 1996;44:998–1001. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman DA, Moghaddam B. The neurobiology of adolescence: changes in brain architecture, functional dynamics, and behavioral tendencies. Neurosci Biobehav Rev. 2011;35(8):1704–12. doi: 10.1016/j.neubiorev.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Rohlfing T, Baker FC, Padilla ML, Colrain IM. Developmental change in regional brain structure over 7 months in early adolescence: comparison of approaches for longitudinal atlas-based parcellation. Neuroimage. 2011;57(1):214–24. doi: 10.1016/j.neuroimage.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumich AL, Sarkar S, Hermens DF, Ibrahimovic A, Kelesidi K, Wilson D, Rubia K. Sex differences in brain maturation as measured using event-related potentials. Dev Neuropsychol. 2012;37(5):415–33. doi: 10.1080/87565641.2011.653461. [DOI] [PubMed] [Google Scholar]
- Supekar K, Musen M, Menon V. Development of large-scale functional brain networks in children. PLoS Biol. 2009;7(7):e1000157. doi: 10.1371/journal.pbio.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuromi B, Czobor P, Komlósi S, Bitter I. P300 deficits in adults with attention deficit hyperactivity disorder: a meta-analysis. Psychol Med. 2011;41(7):1529–38. doi: 10.1017/S0033291710001996. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Sowell ER. Mapping brain maturation. Trends Neurosci. 2006;29:148–159. doi: 10.1016/j.tins.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar KS, Ryali S, Menon V. Dynamic reconfiguration of structural and functional connectivity across core neurocognitive brain networks with development. J Neurosci. 2011;31(50):18578–89. doi: 10.1523/JNEUROSCI.4465-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Individual differences in P300 amplitude: a genetic study in adolescent twins. Biol Psychol. 1998;47(2):97–120. doi: 10.1016/s0301-0511(97)00025-2. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CE, van Baal GC, Molenaar PC, Boomsma DI, de Geus EJ. Stability of genetic and environmental influences on P300 amplitude: a longitudinal study in adolescent twins. Behav Genet. 2001;31(6):533–43. doi: 10.1023/a:1013389226795. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CE, van Baal GC. Twin and family studies of the human electroencephalogram: a review and a meta-analysis. Biol Psychol. 2002;61(1–2):111–38. doi: 10.1016/s0301-0511(02)00055-8. [DOI] [PubMed] [Google Scholar]
- Vertes RP. Hippocampal theta rhythm: a tag for short-term memory. Hippocampus. 2005;15:923–935. doi: 10.1002/hipo.20118. [DOI] [PubMed] [Google Scholar]
- Vogel AC, Power JD, Petersen SE, Schlaggar BL. Development of the brain’s functional network architecture. Neuropsychol Rev. 2010;20(4):362–75. doi: 10.1007/s11065-010-9145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz JM, Goldman RI, Carapezza M, Muraskin J, Brown TR, Sajda P. Simultaneous EEG-fMRI Reveals Temporal Evolution of Coupling between Supramodal Cortical Attention Networks and the Brainstem. J Neurosci. 2013;33(49):19212–22. doi: 10.1523/JNEUROSCI.2649-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz JM, Goldman RI, Carapezza M, Muraskin J, Brown TR, Sajda P. Simultaneous EEG-fMRI reveals a temporal cascade of task-related and default-mode activations during a simple target detection task. Neuroimage. 2014;102(1):229–39. doi: 10.1016/j.neuroimage.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL. Nonparametric Regression Analysis of Longitudinal Data. 2003 http://anson.ucdavis.edu/~wang/paper/EOB3.pdf.
- Whitford TJ, Rennie CJ, Grieve SM, Clark CR, Gordon E, Williams LM. Brain maturation in adolescence: concurrent changes in neuroanatomy and neurophysiology. Hum Brain Mapp. 2007;28(3):228–37. doi: 10.1002/hbm.20273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widén E, Ripatti S, Cousminer DL, Surakka I, Lappalainen T, Järvelin MR, Eriksson JG, Raitakari O, Salomaa V, Sovio U, Hartikainen AL, Pouta A, McCarthy MI, Osmond C, Kajantie E, Lehtimäki T, Viikari J, Kähönen M, Tyler-Smith C, Freimer N, Hirschhorn JN, Peltonen L, Palotie A. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am J Hum Genet. 2010;86(5):773–82. doi: 10.1016/j.ajhg.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HL, Zhang JT. Local polynomial mixed-effects models for longitudinal data. JASA. 2002;97(459):883–897. [Google Scholar]
- Wu K, Taki Y, Sato K, Hashizume H, Sassa Y, Takeuchi H, Thyreau B, He Y, Evans AC, Li X, Kawashima R, Fukuda H. Topological organization of functional brain networks in healthy children: differences in relation to age, sex, and intelligence. PLoS One. 2013;8(2):e55347. doi: 10.1371/journal.pone.0055347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanova J, Kolev V. Developmental changes in the event-related EEG theta response and P300. Electroencephalogr Clin Neurophysiol. 1996;104(5):418–30. doi: 10.1016/s0168-5597(97)00054-3. [DOI] [PubMed] [Google Scholar]
- Yordanova J, Rosso OA, Kolev V. A transient dominance of theta event-related brain potential component characterizes stimulus processing in an auditory oddball task. 2003 doi: 10.1016/s1388-2457(02)00415-7. [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–8. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski BA, Gennatas ED, Zhou J, Seeley WW. Network-level structural covariance in the developing brain. Proc Natl Acad Sci U S A. 2010;107(42):18191–6. doi: 10.1073/pnas.1003109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Jr, Rice JP, Schuckit MA, Foroud T, Edenberg HJ, Porjesz B, Almasy L. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(1):44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]