Abstract
Fungal-bacterial interactions are ubiquitous, yet their molecular basis is only poorly understood. In this study, a novel beneficial interaction between a strain of Pseudomonas putida and the fungus Saccharomyces cerevisiae was identified. When the bacteria were incubated alone in grape juice or in synthetic medium containing various concentrations of glucose, they lost viability rapidly during stationary phase. However, when the bacteria were incubated in these media in the presence of the fungus, their stationary phase survival improved dramatically. On agar plates containing glucose, the beneficial effects of the fungus were manifested in robust bacterial growth and exopolysaccharide production that led to visible mucoidy. In contrast, bacteria grew poorly and were nonmucoid in such media in the absence of the fungus. By using the available S. cerevisiae deletion library, yeast mutants that were unable to mediate this beneficial interaction were identified. These mutants revealed that the beneficial effect on bacterial physiology and survival was mediated by the ability of the fungus to metabolize the available glucose and consequent effects on the medium's pH. In natural environments where the concentration of glucose is high, it is likely that the presence of fungi has had profound beneficial effects on the physiology and survival of certain P. putida strains throughout their natural history.
Fungi and bacteria coexist in a myriad of different environments. One such environment is the rhizosphere, which contains different species of bacteria and fungi in close proximity. These microbes influence each other's physiology and metabolism as well as the health of the plants they might colonize. While many studies have investigated interactions between soil fungi and bacteria, most have focused on antagonistic interactions that prevent the establishment of plant pathogens in the rhizosphere (reviewed in reference 27). In studying the molecular basis of fungal-bacterial interactions under conditions of coexistence, we have discovered a beneficial interaction between Saccharomyces cerevisiae and Pseudomonas putida.
The bacterial side of fungal-bacterial interactions has been well studied, and much is known regarding bacterial effects on the growth and fitness of fungi, especially relating to antimicrobial production. Many studies have focused on the plant-associated pseudomonads, since Pseudomonas isolates are readily cultured from the soil, are easy to grow in the laboratory, have diverse metabolic functions in the environment, and produce many different antimicrobial compounds. Antimicrobials produced by plant-associated pseudomonads with activity towards fungi include 2,4-diacetylphloroglucinol, pyoluteorin, phenazine, and hydrogen cyanide (reviewed in references 1, 8, and 18). In addition to antimicrobial production, the production of siderophores, such as pyoverdine and pyochelin, by pseudomonads has been shown to protect plants from fungal pathogens (reviewed in reference 27). Interactions between Pseudomonas and fungi may also be nonantagonistic. For instance, it has long been known that certain Pseudomonas strains promote mushroom formation in Agaricus bispora (19). The exact mechanism of this growth promotion is not known, but it has been suggested that the Pseudomonas strains remove self-inhibitory compounds produced by the fungi that prevent mushroom formation (19).
Less is known about the fungal side of fungal-bacterial interactions, especially at the mechanistic level. The presence of nonpathogenic Fusarium species has been shown to increase the efficacy of some P. putida strains in controlling Fusarium wilt, a plant disease caused by some strains of Fusarium oxysporum (16). The molecular mechanism is again unknown, but it has been suggested that the presence of nonpathogenic Fusarium species stimulates the plant to release additional carbon sources that, in turn, enhance the production of siderophores by the pseudomonads (16). There is also evidence that there may be chemical signaling between fungi and bacteria in the environment. Medium pretreated with the phytopathogenic fungus Pythium ultimum decreased the expression of genes in Pseudomonas fluorescens that played a role in the ecological fitness of the P. fluorescens, suggesting that a soluble fungal product may decrease the fitness of a bacterium in the environment (5, 22).
As part of our ongoing studies on fungal-bacterial interactions, we discovered that in certain environments, a strain of P. putida rapidly lost viability during stationary phase when incubated alone, but this loss of viability was not observed when the bacteria were cocultured with S. cerevisiae. Here, we present this beneficial interaction and our genetic and biochemical analyses that revealed its molecular basis.
MATERIALS AND METHODS
Bacterial and yeast strains.
P. putida strain ZK3093 is a natural soil isolate from Colombia. Its isolation and characterization involved resuspending soil in sterile distilled water and plating on cetrimide agar (Oxoid Ltd., Basingstoke, Hampshire, England). Colonies that grew were streaked repeatedly (four times) for single-colony isolation. Species identification was carried out by amplifying the 16S rRNA by using PCR and sequencing the amplified rRNA genes with universal primers (7). The 16S rRNA sequence of ZK3093 was a 100% match to the 16S rRNA of several P. putida strains. The other Pseudomonas spp. tested (103 other strains in total) were isolated from soils from Massachusetts and Colombia in a similar fashion or by growth on King's B agar (10) containing 150 μg of ampicillin/ml, 15 μg of chloramphenicol/ml, and 100 μg of cycloheximide/ml.
The S. cerevisiae strain used in this study was BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Invitrogen Corp., Huntsville, Ala.).
Growth media.
Yeast extract-peptone (YP) medium contained 1% yeast extract, 2% peptone, and 1.5% agar. Semisynthetic acetate and YP media were prepared as described previously (9, 17). Buffered YP medium-2% glucose contained 150 mM MES [2-(N-morpholino)ethanesulfonic acid] and 50 mM MOPS [3-(N-morpholino)propanesulfonic acid] calibrated with sodium hydroxide to either pH 6 or pH 7. Synthetic dextrose minimal medium contained 0.67% Bacto yeast nitrogen base without amino acids, 2% glucose, and 2% agar (20). Grape juice was prepared by crushing organic grapes and filtering the juice with a 0.2-μm-pore-size filter.
Viability assay.
P. putida ZK3093 was incubated either alone or in coculture with S. cerevisiae BY4741 (inoculated 1:1) in grape juice and either YP medium-2% glucose or YP medium-0.5% glucose at 30°C on a roller. Samples were removed every 24 h, diluted in either YP medium-2% glucose or Luria-Bertani (LB) medium, and plated on YP medium-2% glucose with 10 mg of cycloheximide/liter or LB medium (to determine counts of viable bacteria). To determine counts of viable fungi, the dilutions were plated on synthetic dextrose minimal medium plus 20 mg of uracil/liter, 20 mg of histidine/liter, 20 mg of methionine/liter, and 100 mg of leucine/liter.
Cross-streak assay for bacterial growth and mucoidy in the presence of yeast.
P. putida ZK3093 was patched onto YP medium-2% glucose either alone or perpendicular to a patch of S. cerevisiae. The plates were incubated for 1 to 5 days at 30°C. The microbial growth was then analyzed visually.
A cellophane assay was used to determine if direct contact between S. cerevisiae and P. putida ZK3093 was necessary for the beneficial interaction. Sterile cellophane was placed on top of a YP medium-2% glucose plate. S. cerevisiae strain BY4741 was patched onto one half of the cellophane, and the plate was incubated at 30°C for 24 to 48 h. The cellophane and BY4741 were removed, and 400 μl of 4×-concentrated YP medium-2% glucose was added to the plate. The plate was allowed to dry, and P. putida strain ZK3093 was patched across both halves of the agar plate and incubated at 30°C for 24 to 48 h.
Yeast deletion screen.
The yeast deletion collection (29) was grown in 96-well microtiter plates containing 100 μl of YP medium-2% glucose/well at 30°C for 2 days. Next, the yeast was transferred to YP medium-2% glucose plates by using a prong and grown at 30°C for 2 days. Meanwhile, a 24-h culture of P. putida ZK3093 was diluted 1:100 into YP-2% glucose medium and grown in 96-well microtiter plates at 30°C for 24 h. ZK3093 was transferred from the microtiter plate by using a prong and was applied directly to the yeast colonies on the YP medium-2% glucose plate. The yeast and bacteria were coincubated at 30°C overnight. The plates were scored for the presence of mucoid and nonmucoid ZK3093. Yeast strains that yielded nonmucoid ZK3093 colonies were then retested in a cross-streak assay.
Regeneration of wild-type alleles.
The reconstructed strains of the yeast deletion mutants Δfum1, Δmdh1, Δsdh1, Δsdh4, Δtcm62, and Δemi5 were generated by replacing the deletion cassette in the genome with the wild-type copy of the corresponding gene. To generate the reconstructed strains, the wild-type copy of the deleted gene was amplified by PCR from the genome of BY4741. The deletion strains were transformed with their corresponding PCR products. Transformants were selected for growth on semisynthetic acetate medium since only strains with a functioning tricarboxylic acid (TCA) cycle will grow on the nonfermentable carbon source acetate. The replacement of the deletion cassette was confirmed by PCR analysis. Ten independent transformants were tested for the regaining of the wild-type phenotype by cross-streak assays with P. putida ZK3093.
Exopolysaccharide analysis.
Cells were scraped from plates and resuspended in 0.9% NaCl (2.5 to 5 ml per plate). Cells were removed by centrifugation (13,800 × g for 30 min at 4°C). Exopolysaccharide was precipitated from the supernatants with three volumes of ice-cold 95% ethanol at −70°C for 20 h. The precipitate was recovered by centrifugation (13,700 × g for 15 to 20 min at 4°C). The pellet was washed twice with 95% ethanol, washed once with 100% ethanol, resuspended in water, and dialyzed (Spectra/Por 1; molecular weight cutoff, 6,000 to 8,000; American Scientific Products, Inc., McGraw Park, Ill.) for 24 h against five changes of 4 liters of water. The sample was dried under a vacuum and analyzed for glycosyl composition at the Complex Carbohydrate Research Center, University of Georgia.
Glycosyl composition analysis was performed by combined gas chromatography-mass spectrometry (GC-MS) of the per-O-trimethylsilyl derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis. Methyl glycosides were first prepared from a dry sample by methanolysis in 1 M HCl in methanol at 80°C (18 to 22 h), followed by re-N-acetylation with pyridine and acetic anhydride in methanol (for detection of amino sugars). The samples were then per-O-trimethylsilylated by treatment with Tri-Sil (Pierce Biotechnology, Inc., Rockford, Ill.) at 80°C (0.5 h). These procedures were carried out as previously described (15, 31). GC-MS analysis of the per-O-trimethylsilyl methyl glycosides was performed on an Hewlett-Packard 5890 gas chromatograph interfaced to a Hewlett-Packard 5970 MSD, by using an All Tech EC-1 fused silica capillary column (30 m by 0.25 mm internal diameter).
Enzyme assays.
Yeast strains were grown in YP-dextrose medium, and P. putida strain ZK3093 was grown in either YP medium-2% glucose or YP medium-0.5% glucose at 30°C with shaking. Samples were taken at the time points indicated in Fig. 5 and 6. Cells were removed by centrifugation at 15,000 × g for 1 min. The concentrations of glucose, ethanol, succinate, acetic acid, and gluconic acid in the supernatant were measured by using enzyme assays, which were performed according to the instructions provided with the assay kits (r-Biopharm, Inc., Marshall, Mich.).
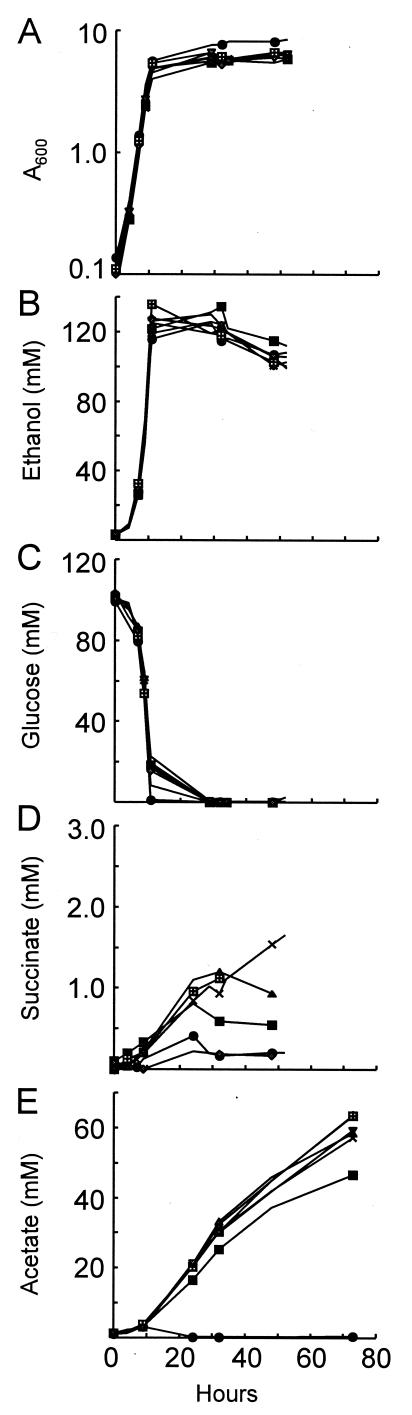
FIG. 5.
Phenotypes of S. cerevisiae mutants. S. cerevisiae strains were grown in YP medium-2% glucose and samples were taken at different time points. (A) Growth curve. The cells were pelleted and the concentration (mM) of (B) ethanol, (C) glucose, (D) succinate, and (E) acetic acid was measured in the supernatant by using enzyme assays as described in Materials and Methods. •, wild-type (BY4741); ▪, Δfum1; ♦, Δmdh1; X, Δsdh1; □, Δsdh4; ▾, Δtcm62; and ▴, Δemi5.
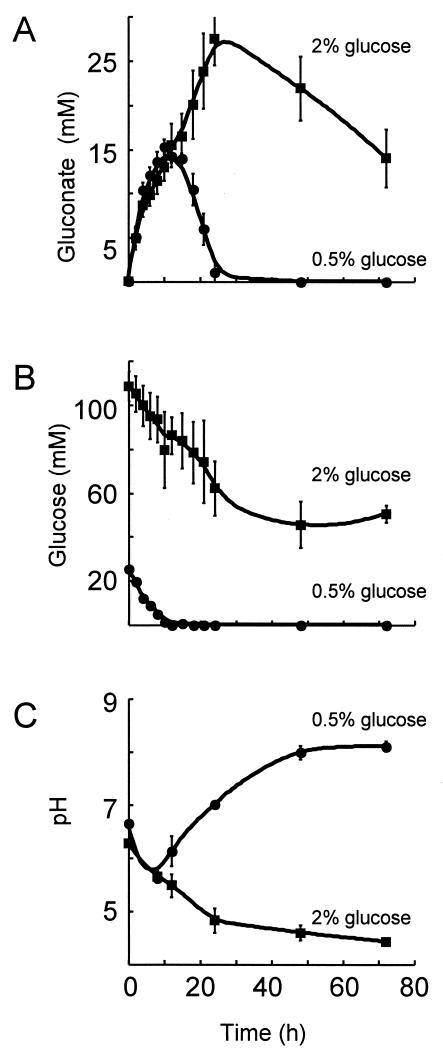
FIG. 6.
P. putida ZK3093 excretes more gluconate when grown in the presence of 2% glucose versus 0.5% glucose. ZK3093 was grown in either YP medium-2% glucose or YP medium-0.5% glucose, and samples were taken at different times. The cells were pelleted, and the concentrations of gluconate (A) and glucose (B) were measured in the supernatant enzymatically as described in Materials and Methods. The pH of the medium was measured at different times during growth (C). Samples were centrifuged and the pH of the spent medium was measured by using a pH meter. Mean values from three independent experiments ± SD are shown. •, 0.5% glucose; ▪, 2% glucose.
Determination of pH.
There were two methods used to determine the pH of agar medium. In one method, a pH indicator, bromcresol purple (40 μg/ml), was added to the medium. This indicator is yellow below pH 5.2 and purple above pH 6.8. For the second method, bacteria were grown on indicator plates for 24 h. The bacteria were then removed by scraping, and the agar was cut out, wrapped in Parafilm, and frozen at −20°C for 24 h. The agar was allowed to thaw, and the liquid removed was transferred to a fresh tube. The pH of the liquid was measured by using a pH meter.
RESULTS
Viability and morphology of P. putida ZK3093 in the presence and absence of S. cerevisiae BY4741.
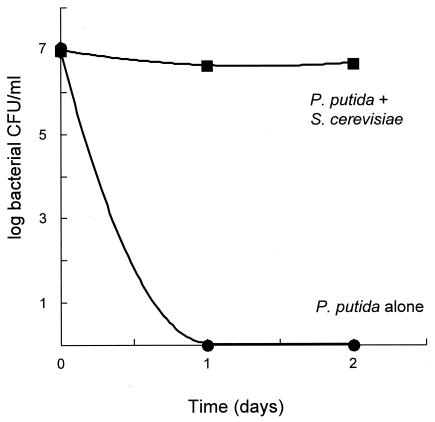
Our laboratory has been interested in bacterial survival during stationary phase and in fungal-bacterial interactions for some years. We wanted to initiate studies of fungal-bacterial interactions utilizing the model fungus S. cerevisiae because we reasoned that the power of yeast genetics would allow us to better characterize the eukaryotic side of any interesting interaction. We noticed a strikingly beneficial effect of the presence of the fungus on bacterial stationary phase survival under certain incubation conditions. One condition we tested was incubation in grape juice, because we reasoned that P. putida and S. cerevisiae might conceivably coexist on grapes and grape juice in natural settings. As seen in Fig. 1, when P. putida ZK3093 was incubated alone in sterile filtered grape juice, its viability (as judged by colony-forming ability) was completely lost in 1 day. In stark contrast, when the bacteria were coincubated with S. cerevisiae BY4741, bacterial colony counts remained constant for at least 2 days.
FIG. 1.
Bacterial stationary phase survival in grape juice. ZK3093 was grown either alone or in coculture with S. cerevisiae BY4741 in grape juice. Samples were taken every 24 h and plated onto LB plates to measure the numbers of bacterial CFU. Mean values from three independent cultures ± standard deviations (SD) were determined and graphed. Note that the SD marks are smaller than the symbols used and thus are not visible. •, ZK3093 alone; ▪, ZK3093 in coculture with BY4741.
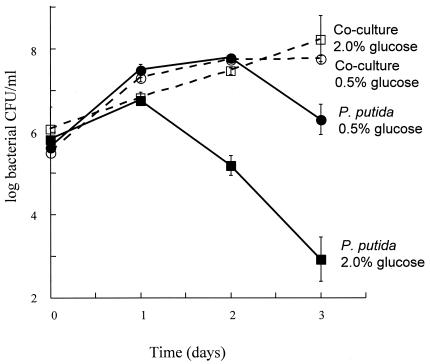
This beneficial effect of the fungus on bacterial stationary phase was also observed when we incubated the microbes in more traditional microbiological media. In particular, when liquid media based on YP medium plus various concentrations of glucose were used, the presence of S. cerevisiae BY4741 had a dramatic effect on bacterial colony counts in stationary phase (see Fig. 2). The viability of ZK3093 grown alone in YP medium-2.0% glucose decreased sharply between days 1 and 3. When the bacteria were grown alone in YP medium-0.5% glucose, there still was a significant drop in viable counts between days 2 and 3. However, under both conditions no drop in viability was observed during 3 days of incubation when the cultures contained S. cerevisiae BY4741. In both cocultures, the fungal cell counts reached 107 CFU/ml after day 2 and remained constant. Therefore, cocultivation under these conditions is beneficial for the bacteria without affecting fungal viability.
FIG. 2.
ZK3093 was grown either alone or in coculture with S. cerevisiae BY4741 in either YP medium-2% glucose or YP medium-0.5% glucose. Samples were taken every 24 h and plated onto YP medium-2% glucose plates with 10 mg of cycloheximide/liter to measure the numbers of CFU (bacteria only). Mean values from three independent cultures ± SD are shown except for the ZK3093 in 0.5% glucose culture, where the mean value is from six independent cultures ± SD. ▪, ZK3093 in 2% glucose; •, ZK3093 in 0.5% glucose; □, ZK3093 and BY4741 in 2% glucose; and ○, ZK3093 and BY4741 in 0.5% glucose.
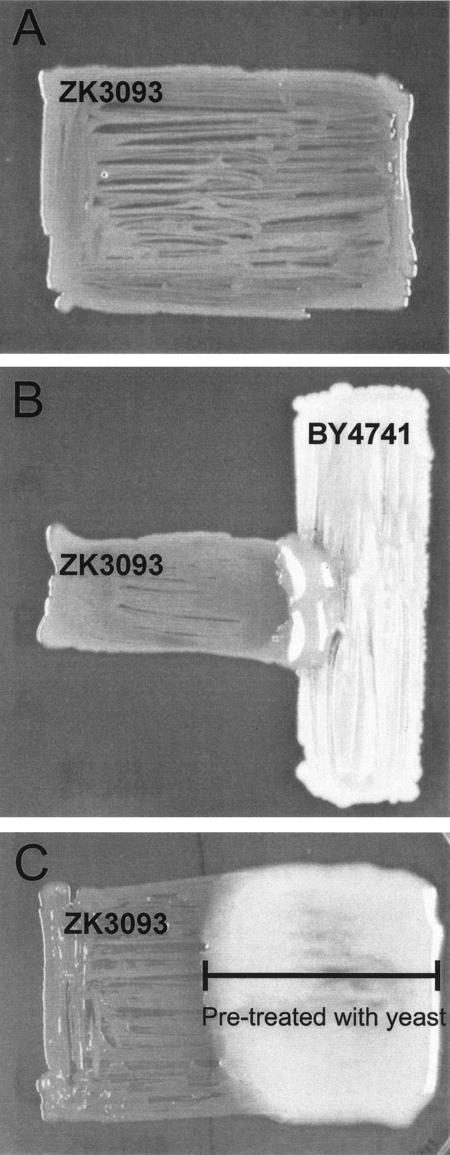
The beneficial effect on P. putida ZK3093 resulting from the presence of the fungus had an interesting manifestation when microbial growth was observed on solid medium (see Fig. 3). On agar plates containing YP medium-2% glucose, ZK3093 grows poorly by itself (Fig. 3A); after 3 days at 30°C, a bacterial patch is somewhat sparse. In contrast, when S. cerevisiae BY4741 is cross-streaked in the same plate (Fig. 3B), there is robust mucoid bacterial growth in the area of the cross-streak where both species are capable of close interaction. The beneficial effect of the presence of fungi on the growth of ZK3093 on solid YP medium-2% glucose was also observed when other fungi, namely Candida albicans, Penicillium chrysogenum, and Cladosporium sp., were used (results not shown).
FIG. 3.
S. cerevisiae permits robust mucoid growth of P. putida ZK3093. (A) P. putida strain ZK3093 was patched onto a YP medium-2% glucose plate and incubated at 30°C for 48 h. (B) S. cerevisiae strain BY4741 (vertical) was patched onto a YP medium-2% glucose plate perpendicular to the P. putida strain ZK3093 (horizontal) and incubated at 30°C for 48 h. (C) S. cerevisiae BY4741 was patched onto the right half of a cellophane-covered YP medium-2% glucose plate (bar) and incubated at 30°C for 24 h. The fungi and cellophane were removed, nutrients were added back, and P. putida ZK3093 was patched onto the plate and incubated at 30°C for 24 h.
Is direct contact between the fungi and bacteria necessary for the beneficial effect of fungi on ZK3093? To answer this question, we used the cellophane assay described in Materials and Methods. Briefly, a YP agar-2% glucose plate was pretreated with S. cerevisiae BY4741 by growing the fungus separated from the surface of the plate by a piece of sterile cellophane. The fungi were removed along with the cellophane, nutrients were added back, and ZK3093 was patched onto the plate. ZK3093 grew robustly and was mucoid only on the portion of the plate that was pretreated with S. cerevisiae BY4741 and not on the untreated section (Fig. 3C). Clearly, direct physical contact between the two microbes is not necessary to obtain the beneficial effect.
The readily observable effect of the fungus on bacterial growth and mucoidy on plates provided a convenient assay to characterize the interaction. First, we wanted to determine if this was a widespread phenomenon. We therefore monitored the mucoidy of several Pseudomonas spp. soil isolates on YP medium-2% glucose in the presence or absence of S. cerevisiae. Of 104 Pseudomonas spp. soil isolates we tested, 55 were mucoid while 49 were nonmucoid. Of those 49, 12 became mucoid in the presence of S. cerevisiae, similar to the results observed with ZK3093. Therefore, while the observed interaction between ZK3093 and S. cerevisiae BY4741 is not universal, it is certainly common.
The poor growth of ZK3093 in YP medium is a function of the glucose and overall sugar concentration.
In an effort to characterize the environmental influences leading to the poor growth in YP medium-2% glucose, we plated P. putida ZK3093 on various concentrations of glucose and in the presence of other sugars. When only glucose was added to YP medium, there was a dramatic change in physiology between 1 and 1.5% glucose. The strain grew well and displayed mucoidy at glucose concentrations below 1.5%, and grew poorly and was nonmucoid on plates containing 1.5% or higher glucose concentrations. This inhibitory effect appears specific for glucose, since growth was robust and mucoidy was apparent when the bacterial strain was grown on media containing 2% fructose, mannose, xylose, or galactose. In fact, ZK3093 was mucoid in the presence of mannose even up to a concentration of 5%, suggesting that the inhibition of mucoidy by glucose concentrations above 1% was not due to an osmotic effect. Quite interestingly, when other sugars were present, mucoidy was not observed in even lower concentrations of glucose, e.g., 0.5%.
ZK3093 mucoidy results from exopolysaccharide production.
Before proceeding any further in our characterization of the observed negative effects of glucose and the capacity of S. cerevisiae to suppress those effects, we wanted to determine whether the molecule responsible for the observed mucoidy was the same both in medium with sugars other than glucose and in the presence of fungi. Bacterial mucoidy is most often the result of exopolysaccharide production, so we proceeded to purify and characterize exopolysaccharides produced by ZK3093. We scraped off and resuspended in 0.9% NaCl all bacterial growth from fungus-pretreated or untreated YP medium-2% glucose plates and from untreated YP medium-2% mannose plates. The cells were removed by centrifugation, and exopolysaccharides present in the supernatant were precipitated with ethanol. No visible precipitate was recovered when ZK3093 was grown on YP medium-2% glucose plates in the absence of fungi. However, precipitates were recovered from both ZK3093 grown in YP medium-2% glucose plates that had been pretreated with fungi and from the YP medium-2% mannose plates. The precipitates were analyzed for carbohydrate composition as described in Materials and Methods. In both cases, the exopolysaccharide had nearly identical sugar composition, containing primarily glucose with small amounts of galactose, rhamnose, and mannose (Table 1). These results indicate that ZK3093 produces the same glucose-rich exopolysaccharide on both YP medium-2% mannose and in the presence of fungi on YP medium-2% glucose. Being mostly glucose, this exopolysaccharide differs in composition from marginalan, an acidic galactoglucan which was shown to be produced by several mushroom-associated P. putida strains (6).
TABLE 1.
Glycosyl composition of exopolysaccharide from ZK3093a
| Glycosyl residue | Mol% for pretreatment with:
|
|
|---|---|---|
| YP medium-2% glucose | YP medium-2% mannose | |
| Arabinose | ND | ND |
| Rhamnose | 3.7 | 3.7 |
| Fucose | ND | ND |
| Xylose | ND | ND |
| Glucuronic acid | ND | ND |
| Galaturonic acid | ND | ND |
| Mannose | 2.2 | 2.0 |
| Galactose | 8.2 | 7.9 |
| Glucose | 85.9 | 86.4 |
| N-Acetylglucosamine | ND | ND |
| 3-Deoxyl-d-manno-2-octulosonic acid | ND | ND |
Values are expressed as moles percent of total carbohydrate. ND, none detected.
Identification of fungal mutants deficient in the beneficial interaction.
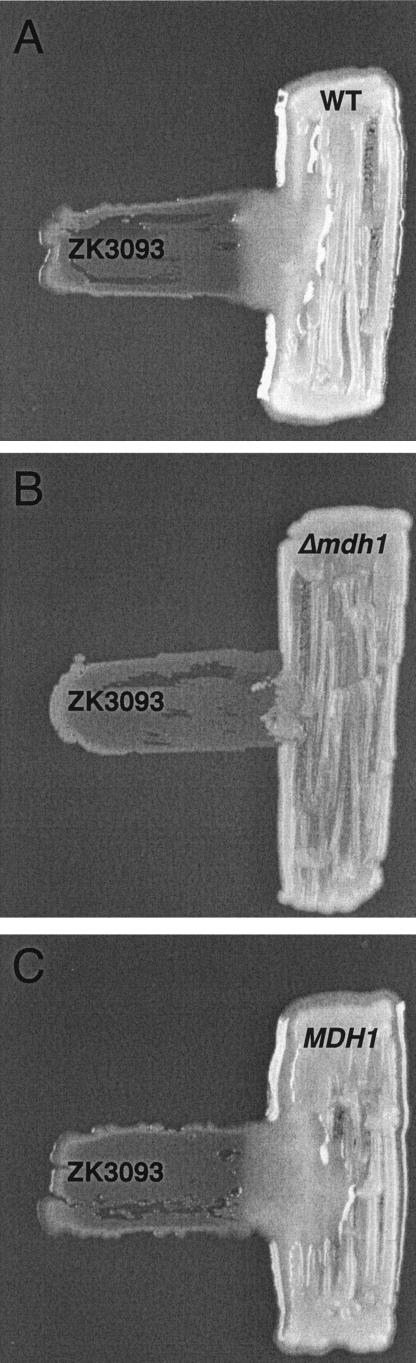
We next wanted to identify fungal mutants that were impaired in their ability to reverse the negative effects of glucose on P. putida ZK3093 physiology. To this end, we used the observed mucoidy as our assay to screen for S. cerevisiae mutants no longer able to carry out the beneficial interaction. We used the existing S. cerevisiae open reading frame deletion collection, which consists of precise deletions of every nonredundant open reading frame (greater than 100 bp) identified for S. cerevisiae (29). To identify the desired mutants, the deletion strains and ZK3093 were grown together on YP medium-2% glucose plates, as described in Materials and Methods, and the plates were visually inspected for mucoidy. Under these conditions, ZK3093 was only mucoid in the presence of the fungus. Mutants that were somehow compromised in the interaction, resulting in nonmucoid bacterial growth adjacent to them, were then rechecked by using a cross-streak assay. An example of one mutant with the desired phenotype is shown in Fig. 4. While ZK3093 was mucoid in the region of its growth adjacent to the wild-type yeast (BY4741) (Fig. 4A), ZK3093 was nonmucoid adjacent to Δmdh1, even after prolonged coincubations (up to 10 days) (Fig. 4B).
FIG. 4.
P. putida ZK3093 (horizontal) was patched onto a YP medium-2% glucose plate perpendicular to the S. cerevisiae strains BY4741 (A), Δmdh1::KanMX (B), and MDH1 (where the Δmdh1::KanMX allele on the chromosome was replaced with the wild-type copy of the MDH1 gene) (C). The plates were incubated at 30°C for 48 h. WT, wild type.
ZK3093 did not develop mucoidy when grown next to six S. cerevisiae mutants (Table 2). Four of these yeast mutants contained deletions of enzymes involved in the fungal TCA cycle: fumarase encoded by the FUM1 gene (30), malate dehydrogenase encoded by the MDH1 gene (14), and two subunits of the succinate dehydrogenase encoded by SDH1 and SDH4 (2, 3, 20). Fumarase, malate dehydrogenase, and succinate dehydrogenase catalyze consecutive reactions in the TCA cycle. The TCM62 and EMI5 genes were deleted from the remaining two mutants (Table 2). TCM62 encodes a chaperone that is essential for the assembly of the succinate dehydrogenase (4), which links TCM62 function to the TCA cycle. EMI5 encodes a gene of unknown function. One phenotype of the Δemi5 mutant, however, is that it cannot grow on nonfermentable carbon sources (23), a phenotype that it shares with mutants of the TCA cycle.
TABLE 2.
Genes identified in S. cerevisiae deletion mutant screening
| Gene name | Gene function | Comment |
|---|---|---|
| FUM1 | Fumarate hydratase | Enzyme in TCA cycle |
| MDH1 | Malate dehydrogenase | Enzyme in TCA cycle |
| SDH1 | Succinate dehydrogenase flavoprotein subunit | Enzyme in TCA cycle |
| SDH4 | Succinate dehydrogenase membrane anchor subunit | Enzyme in TCA cycle |
| TCM62 | Chaperone | Essential for succinate dehydrogenase biogenesis |
| EMI5 | Unknown |
To confirm that the observed phenotypes were indeed caused by the deletion mutations, each deletion cassette present in the genome was replaced with a wild-type copy of the gene. The interaction between S. cerevisiae and ZK3093 that yields mucoid bacterial growth was restored in the Δfum1, Δmdh1, Δsdh1, Δsdh4, Δtcm62, and Δemi5 strains by the reintroduction of the wild-type copy of the deleted genes. A representative example, where the reintroduction of the MDH1 gene into the Δmdh1 strain resulted in robust mucoid bacterial growth adjacent to the fungus, is shown in Fig. 4C.
Analysis of fungal mutants.
To determine how the fungal mutants may affect the fungal-bacterial interaction, we analyzed several factors that may be expected to be altered in mutants compromised in the TCA cycle. We compared the growth rates, ethanol production, glucose utilization, succinate production, and acetic acid production of the wild-type and mutant fungi. There was no significant difference between the growth rates of wild-type BY4741 and each of the mutant strains tested; their doubling times ranged between 1.84 and 1.9 h in YP medium-2% glucose (Fig. 5A). There was also no significant difference in either their ethanol production or their glucose utilization abilities (Fig. 5B and C). Some, but not all, of the mutants excreted more succinate than the wild type, but the increase was at most a twofold effect (Fig. 5D). In contrast to the relative normalcy of the other metabolites, all of the mutants excreted very large amounts of acetic acid (Fig. 5E). These results suggested that the large quantities of acetic acid excreted by the mutant yeast may play a role in preventing ZK3093 mucoidy.
Mucoidy of P. putida ZK3093 is influenced by the pH of the medium.
One possible way for acetic acid, excreted by the S. cerevisiae mutants, to interfere with ZK3093 mucoidy is through the acidification of the medium. In fact, the fungal mutants acidified the medium more dramatically than wild type, as determined by using pH indicator plates. Thus, we hypothesized that acidification of the medium could result in a compromised bacterial physiology manifested as an inability to produce exopolysaccharide.
To determine if the pH of the medium could directly influence bacterial mucoidy, ZK3093 was streaked onto YP medium-2% glucose plates that were buffered to either pH 6 or pH 7. ZK3093 was mucoid on plates buffered to pH 7 and nonmucoid on plates buffered to pH 6. To quantitate the effects of pH, we isolated exopolysaccharide, as described in Materials and Methods, and normalized the weight of the exopolysaccharide to the amount of cells. The normalized amount of exopolysaccharide produced by ZK3093 was 2.5 ± 0.3 mg/A600 when grown on medium buffered to pH 7 and 0.21 ± 0.06 mg/A600 when grown on medium buffered to pH 6.
Relationship between glucose concentration, pH of the medium, and ZK3093 mucoidy.
As mentioned before, the amount of glucose in the medium influenced ZK3093 mucoidy, with glucose concentrations at or above 1.5% leading to a nonmucoid phenotype and those below 1.5% resulting in a mucoid phenotype (Table 3). Since mucoidy was also affected by the pH of the medium, we tested whether there was a correlation between the final pH of the cultures and the mucoid phenotype. ZK3093 was streaked onto YP plates containing different concentrations of glucose and grown at 30°C for 24 h. ZK3093 was nonmucoid when grown on glucose concentrations of 1.5 or 2%. Under these conditions, the pH of the medium became more acidic, dropping to around 4.5 (Table 3). When grown on lower glucose concentrations, either 1 or 0.5%, ZK3093 was mucoid. Interestingly, the pH of the medium did not become more acidic, instead remaining around 7 (Table 3). Growth on 2% mannose or 2% galactose did not acidify the medium, and the cells were mucoid. Interestingly, when there was 1.5% mannose present, even 0.5% glucose acidified the medium and the bacteria were nonmucoid. These results indicate that there is a correspondence between mucoidy and medium pH; bacteria appear to shut down exopolysaccharide production (perhaps due to an overall compromised physiology) at acidic pHs.
TABLE 3.
Influence of carbon source on ZK3093 mucoidy and pHa
| Medium | Bacterial phenotype | Mean pH |
|---|---|---|
| YP-2% glucose | Nonmucoid | 4.4 ± 0.1 |
| YP-1.5% glucose | Nonmucoid | 4.6 ± 0.1 |
| YP-1.0% glucose | Mucoidb | 7.1 ± 0.2 |
| YP-0.5% glucose | Mucoid | 7.7 ± 0.1 |
| YP-2% mannose | Mucoid | 7.5 ± 0.1 |
| YP-1.5% mannose-0.5% glucose | Nonmucoid | 4.9 ± 0.2 |
| YP-2% galactose | Mucoid | 8.0 ± 0.03 |
Plates were visually inspected for mucoidy. Mean pH is the average ± standard deviation for three independent experiments.
The periphery of the patch was nonmucoid with a corresponding pH of 5.3.
ZK3093 excretes more gluconate at higher glucose concentrations.
Since there was a relationship between the nonmucoid phenotype of ZK3093 grown in glucose concentrations above 1% and the acidification of the medium, we hypothesized that the metabolism of glucose under these conditions led to medium acidification. There are two pathways for glucose catabolism in Pseudomonas. In one pathway, glucose is oxidized to gluconate in the periplasm by glucose dehydrogenase (13, 21, 24). The second pathway involves the active transport of glucose into the cell, where it is phosphorylated and subsequently metabolized (24). To determine if the effects of glucose concentrations above 1% on pH were due to the excretion of gluconate, we measured the amount of gluconate excreted by ZK3093 grown in two different glucose concentrations (2 and 0.5%) (Fig. 6).
To compare the amount of gluconate produced, ZK3093 was grown in either 2 or 0.5% glucose at 30°C and samples were taken at the indicated time points (Fig. 6). The cells were removed by centrifugation, and the concentrations of gluconate and glucose present in the medium were measured enzymatically. The amount of gluconate produced by ZK3093 grown in 0.5% glucose peaked at around 12 h of growth and then began to decrease, reaching negligible levels by 24 h (Fig. 6A). ZK3093 metabolized all of the glucose in the 0.5% culture by the 12-h time point, which corresponded to the peak in gluconate production (Fig. 6B). In contrast, ZK3093 grown in 2% glucose continued to accumulate gluconate in the medium until 24 h of growth (Fig. 6A). After 24 h, the gluconate concentration began to decrease, but even after 72 h it was still about 15 mM (Fig. 6A). The glucose concentration of the 2% culture decreased steadily in the first 24 h, but unlike that of the 0.5% glucose culture, it stabilized (Fig. 6B). The inability to completely metabolize the available glucose was a strong indication of a compromised physiology.
The pH of the culture medium reflected the concentration of gluconate. The pH of the 0.5% glucose culture reached its lowest point at 12 h, when the gluconate concentration reached its peak (Fig. 6A and C). As the gluconate was metabolized, the pH of the medium increased and reached 7 by 24 h (Fig. 6C). In the 2% glucose culture, however, the pH of the medium continued to decrease throughout the time course as gluconate production continued (Fig. 6A and C). Therefore, the pH decrease of the medium coincided with the excretion of gluconate into the culture medium. These results suggest that the nonmucoid phenotype of ZK3093 grown on 2% glucose was due to the acidification of the medium, which resulted from the conversion of glucose to gluconate. Thus, bacteria seem unable to control the activity of their glucose dehydrogenase and are therefore sensitive to high glucose concentrations. However, the presence of wild-type fungi in the near vicinity prevents the deleterious effects on the bacteria of too much glucose, presumably because of the ability of the fungi to quickly metabolize some of the glucose.
DISCUSSION
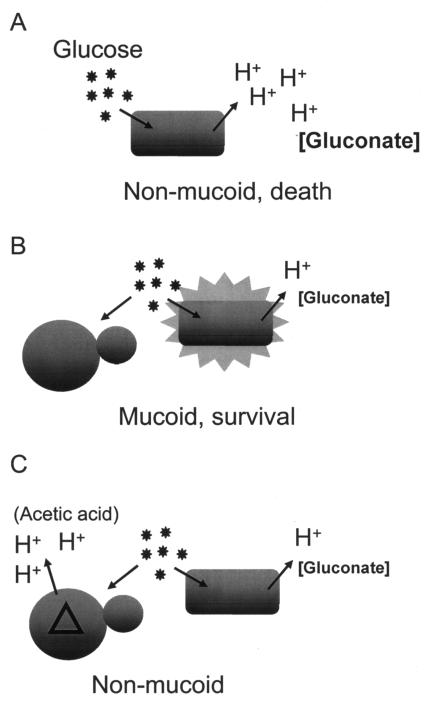
A model describing the influence of S. cerevisiae on P. putida ZK3093 on YP medium-2% glucose is presented in Fig. 7. On this high-glucose medium, the bacteria produce substantial amounts of gluconate. The resulting acidity leads to widespread deleterious physiological changes, which manifest in a reduction in exopolysaccharide production and eventual cell death (Fig. 7A). Other media do not cause acidity; hence, the bacteria are physiologically competent to produce exopolysaccharide and also maintain viability in stationary phase (see, for example, Table 3). When wild-type fungi are present, however, the fungi can rapidly metabolize the glucose to ethanol and low concentrations of acidic products. This metabolism probably leads to a lower glucose concentration available for the bacteria and, hence, less gluconate production (Fig. 7B), a situation similar to the growth of bacteria alone in lower glucose concentrations. The bacteria are nonmucoid in the presence of S. cerevisiae mutants that accumulate acetic acid (Fig. 7C), suggesting that the effector leading to poor physiology and nonmucoidy is acidity rather than gluconate itself. Fungal mutants blocked in glucose metabolism, rather than the acetic acid excreters that we found, might have the same effect. However, such mutants do not grow on YP medium-2% glucose and are absent from the deletion collection.
FIG. 7.
Model describing the influence of S. cerevisiae on ZK3093 mucoidy. (A) ZK3093 is nonmucoid and loses viability when grown in high concentrations of glucose. (B) ZK3093 is mucoid and shows increased survival in the presence of S. cerevisiae. (C) ZK3093 is nonmucoid in the presence of the acetic acid-excreting mutants isolated in the screen.
The way that Pseudomonas metabolizes glucose has interesting possibilities in the environment. It has been suggested that Pseudomonas may use the extracellular pathway of glucose metabolism when carbon sources are abundant as a way to sequester glucose in a form (gluconate) that cannot be used by other microbes (28), including S. cerevisiae. However, the consequent decrease in pH has a general toxic effect leading to a dramatic loss of bacterial viability. In other words, the bacteria seem to be committing a form of suicide by the overproduction of gluconate. So, in that regard, the presence of fungi is highly beneficial to the bacteria in environments containing large amounts of glucose or glucose in combination with other sugars. This may, in fact, be what is going on in the observed fungal rescue of bacterial viability in grape juice (Fig. 1).
In some fruits, glucose is a predominant sugar. In grapes, glucose and fructose accumulate in roughly equal amounts during berry ripening to concentrations of over 10% (11), well above the 2% that we have used. The high levels of glucose in grapes and the presence of S. cerevisiae on grapes suggest that in the environment, S. cerevisiae may influence the viability and physiology of P. putida strains in ways similar to those we have described here. This type of interaction may, in fact, be happening throughout the rhizosphere. In tomato root exudates, glucose and xylose are major sugar constituents (12). The glucose concentration in the rhizosphere was estimated based on the amounts measured in root exudates to be approximately 20 μM (12). This estimate, however, is a rough global estimate for the rhizosphere and does not take into account local concentrations. For instance, the concentrations of sugars may vary based on proximity to the root; microbes present near the root may experience higher sugar concentrations than microbes found farther from the root.
In the environment, multiple carbon sources are also available. As stated earlier, glucose and xylose are the major sugar components of tomato root exudates but additional sugars, such as fructose, maltose, ribose, and sucrose, are also present (12). Root exudates of barley, wheat, and cucumber contain galactose, fructose, arabinose, xylose, and maltose in addition to glucose (25, 26). In the presence of other carbon sources, the amount of glucose needed to negatively influence ZK3093 physiology, as measured by inhibition of exopolysaccharide production, is lower than when glucose is present alone (Table 3). This lack of mucoidy may also coincide with decreased bacterial viability in these environments. Thus, P. putida strains, such as ZK3093, might not survive in the rhizosphere unless they are in close proximity to fungal cells.
The pH of the environment is influenced by the metabolism of the microbes present and by the carbon sources available. P. putida strains may acidify the environment, depending on the types and concentrations of sugars available. This acidification would lead to the loss of viability if the bacteria were growing alone. However, the ubiquitous presence of fungi may greatly affect P. putida survival by changing the ratio of carbon sources available to the bacteria, changing the metabolism of the bacteria, and potentially influencing the pH of the environment.
Acknowledgments
We thank Dan Fraenkel for his invaluable insight into both yeast and Pseudomonas metabolism, Fred Winston for advice and access to the yeast deletion collection, and members of the Kolter laboratory for valuable discussions.
This work was supported by grants from the NIH (GM58213), the Ellison Medical Foundation (ID-SS-0248-02), and the DOE (DE-FG02-02ER63445). J.D.R. was the recipient of a postdoctoral fellowship from the PHS (5-F32-GM20871). The carbohydrate analysis was done by P. Azadi and coworkers at the Complex Carbohydrate Research Center, University of Georgia, who are supported in part by the Department of Energy-funded (DE-FG09-93ER-20097) Center for Plant and Microbial Complex Carbohydrates.
REFERENCES
- 1.Bender, C., V. Rangaswamy, and J. Loper. 1999. Polyketide production by plant-associated pseudomonads. Annu. Rev. Phytopathol. 37:175-196. [DOI] [PubMed] [Google Scholar]
- 2.Bullis, B. L., and B. D. Lemire. 1994. Isolation and characterization of the Saccharomyces cerevisiae SDH4 gene encoding a membrane anchor subunit of succinate dehydrogenase. J. Biol. Chem. 269:6543-6549. [PubMed] [Google Scholar]
- 3.Chapman, K. B., S. D. Solomon, and J. D. Boeke. 1992. SDH1, the gene encoding the succinate dehydrogenase flavoprotein subunit from Saccharomyces cerevisiae. Gene 118:131-136. [DOI] [PubMed] [Google Scholar]
- 4.Dibrov, E., S. Fu, and B. D. Lemire. 1998. The Saccharomyces cerevisiae TCM62 gene encodes a chaperone necessary for the assembly of the mitochondrial succinate dehydrogenase (complex II). J. Biol. Chem. 273:32042-32048. [DOI] [PubMed] [Google Scholar]
- 5.Fedi, S., E. Tola, Y. Moënne-Loccoz, D. N. Dowling, L. M. Smith, and F. O'Gara. 1997. Evidence for signaling between the phytopathogenic fungus Pythium ultimum and Pseudomonas fluorescens F113: P. ultimum represses the expression of genes in P. fluorescens F113, resulting in altered ecological fitness. Appl. Environ. Microbiol. 63:4261-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fett, W. F., J. M. Wells, P. Cescutti, and C. Wijey. 1995. Identification of exopolysaccharides produced by fluorescent pseudomonads associated with commercial mushroom (Agaricus bisporus) production. Appl. Environ. Microbiol. 61:513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, J. G., L. L. Yan, F. E. Dewhirst, B. J. Paster, B. Shames, J. C. Murphy, A. Hayward, J. C. Belcher, and E. N. Mendes. 1995. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J. Clin. Microbiol. 33:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, D., C. Blumer, and C. Keel. 2000. Biocontrol ability of fluorescent pseudomonads genetically dissected: importance of positive feedback regulation. Curr. Opin. Biotechnol. 11:290-297. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstrations of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 11.Kliewer, W. M. 1965. Changes in concentration of glucose, fructose, and total soluble solids in flowers and berries of Vitis vinifera. Am. J. Enol. Vitic. 16:101-110. [Google Scholar]
- 12.Lugtenberg, B. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 13.Matsushita, K., M. Yamada, E. Shinagawa, O. Adachi, and M. Ameyama. 1980. Membrane-bound respiratory chain of Pseudomonas aeruginosa grown aerobically. J. Bacteriol. 141:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAlister-Henn, L., and L. M. Thompson. 1987. Isolation and expression of the gene encoding yeast mitochondrial malate dehydrogenase. J. Bacteriol. 169:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkle, R. K., and I. Poppe. 1994. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry, p. 1-15. In W. J. Lennarz and G. W. Hart (ed.), Methods in enzymology, vol. 230. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed]
- 16.Park, C.-S., T. Paulitz, and R. Baker. 1988. Biocontrol of Fusarium wilt of cucumber resulting from interactions between Pseudomonas putida and nonpathogenic isolates of Fusarium oxysporum. Phytopathology 78:190-194. [Google Scholar]
- 17.Przybyla-Zawislak, B., D. M. Gadde, K. Ducharme, and M. T. McCammon. 1999. Genetic and biochemical interactions involving tricarboxylic acid cycle (TCA) function using a collection of mutants defective in all TCA cycle genes. Genetics 152:153-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raaijmakers, J. M., M. Vlami, and J. T. de Souza. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Leeuwenhoek 81:537-547. [DOI] [PubMed] [Google Scholar]
- 19.Rainey, P. B. 1991. Effect of Pseudomonas putida on hyphal growth of Agaricus bisporus. Mycol. Res. 95:699-704. [Google Scholar]
- 20.Robinson, K. M., and B. D. Lemire. 1992. Isolation and nucleotide sequence of the Saccharomyces cerevisiae gene for the succinate dehydrogenase flavoprotein subunit. J. Biol. Chem. 267:10101-10107. [PubMed] [Google Scholar]
- 21.Schleissner, C., A. Reglero, and J. M. Luengo. 1997. Catabolism of d-glucose by Pseudomonas putida U occurs via extracellular transformation into d-gluconic acid and induction of a specific gluconate transport system. Microbiology 143:1595-1603. [DOI] [PubMed] [Google Scholar]
- 22.Smith, L. M., E. Tola, P. deBoer, and F. O'Gara. 1999. Signalling by the fungus Pythium ultimum represses expression of two ribosomal RNA operons with key roles in the rhizosphere ecology of Pseudomonas fluorescens F113. Environ. Microbiol. 1:495-502. [DOI] [PubMed] [Google Scholar]
- 23.Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman, T. Jones, A. M. Chu, G. Giaever, H. Prokisch, P. J. Oefner, and R. W. Davis. 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31:400-404. [DOI] [PubMed] [Google Scholar]
- 24.Temple, L. M., A. E. Sage, H. P. Schweizer, and P. V. Phibbs. 1998. Carbohydrate catabolism in Pseudomonas aeruginosa. In T. C. Montie (ed.), Pseudomonas. Biotechnology handbooks, vol. 10. Plenum Press, New York, N.Y.
- 25.Vancura, V. 1964. Root exudates of plants. I. Analysis of root exudates of barley and wheat in their initial phases of growth. Plant Soil 21:231-248. [Google Scholar]
- 26.Vancura, V., and A. Hovadik. 1965. Root exudates of plants. II. Composition of root exudates of some vegetables. Plant Soil 22:21-32. [Google Scholar]
- 27.Whipps, J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487-511. [DOI] [PubMed] [Google Scholar]
- 28.Whiting, P. H., M. Midgley, and E. A. Dawes. 1976. The role of glucose limitation in the regulation of the transport of glucose, gluconate and 2-oxogluconate, and of glucose metabolism in Pseudomonas aeruginosa. J. Gen. Microbiol. 92:304-310. [DOI] [PubMed] [Google Scholar]
- 29.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 30.Wu, M., and A. Tzagoloff. 1987. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1. J. Biol. Chem. 262:12275-12282. [PubMed] [Google Scholar]
- 31.York, W. S., A. G. Darvill, M. McNeil, T. T. Stevenson, and P. Albersheim. 1986. Isolation and characterization of plant cell walls and cell wall components, p. 3-40. In A. Weissbach and H. Weissbach (ed.), Methods in enzymology, vol. 118. Academic Press, Inc., Orlando, Fla.